Abstract
This paper investigates the problem of atlas registration of brain images with gliomas. Multi-parametric imaging modalities (T1, T1-CE, T2, and FLAIR) are first utilized for segmentations of different tissues, and to compute the posterior probability map (PBM) of membership to each tissue class, using supervised learning. Similar maps are generated in the initially normal atlas, by modeling the tumor growth, using reaction-diffusion equation. Deformable registration using a demons-like algorithm is used to register the patient images with the tumor bearing atlas. Joint estimation of the simulated tumor parameters (e.g. location, mass effect and degree of infiltration), and the spatial transformation is achieved by maximization of the log-likelihood of observation. An Expectation-Maximization algorithm is used in registration process to estimate the spatial transformation and other parameters related to tumor simulation are optimized through Asynchronous Parallel Pattern Search (APPSPACK). The proposed method has been evaluated on five simulated data sets created by Statistically Simulated Deformations (SSD), and fifteen real multichannel glioma data sets. The performance has been evaluated both quantitatively and qualitatively, and the results have been compared to ORBIT, an alternative method solving a similar problem. The results show that our method outperforms ORBIT, and the warped templates have better similarity to patient images.
Index Terms: Statistical atlas, deformable registration, brain tumor, EM algorithm, tumor growth modeling, reaction-diffusion equation
I. Introduction
Glioblastoma multiforme (GBM), a primary malignant brain tumor, is the most common form of the glioma tumors, which in spite of multi-modality treatments, remains as an incurable and rapidly fatal disease. The anatomic location of a glioma influences prognosis and treatment options. A few studies aim at discovery of the distribution of gliomas in different anatomic areas of the brain. For instance, Larjaavara et al. [1], demonstrate that such distribution of location of gliomas is an uneven function within the brain, with the densest occurrence in the frontal lobe. Duffao et al. [2] find that low grade gliomas are often observed in secondary functional areas of the brain. While in these works, manual localization of glioma (in standard space) has been utilized, a deformable registration framework can be very useful for objective numerical evaluations of such clinical findings since various patient images can be mapped to a common space or atlas. Such an statistical atlas derived from glioma images can be a powerful tool for summarizing the population data, examining the spatial relationships between pathology and other anatomical locations, knowledge discovery and learning of glioma behavior.
Although a plethora of methods for normal-to-normal brain registration exist [3]–[17], the problem of registering images of tumor patients to standardized templates has been relatively unexplored, and proven to be extremely challenging. Due to large deformations and lack of clear definition of anatomical detail in patients images, direct application of the available registration methods to images of tumor patients can lead to poor registration around the tumor region. In most of glioma bearing MR images, the confounding effects of edema and tumor inltration, which cause changes in the image intensities, render the task of finding correspondences difficult. In fact, the fundamental assumption of existence of correspondences between the atlas and the patients images, which is ubiquitous in the most of the available deformable registration methods, is violated due to the anatomical changes caused by tumor emergence and tissue death. Another difficulty, is the presence of the mass-effect in the patient image. Mass-effect causes deformations of the adjacent structures and ventricles by excessive pushing, rendering it difficult to apply standard image warping algorithms.
This paper presents a framework to circumvent these difficulties by building upon our work in [18], [19], i.e., creating a topologically equivalent atlas by simulating tumor in the atlas space and use it for registration to patient images. The parameters of this tumor simulation are estimated as part of the registration process. Apart from methodological details, one feature that is different in our method compared to [18], [19], is the tumor type studied here. The method in [18], [19] used a simple pressure model to simulate the tumor mass-effect on the atlas and did not consider tumor infiltration and presence of edema, the most important issues in glioma patients, which are the focus herein. The glioma images usually indicate severe complexities around the tumor and may include edemas that render the registration task extremely challenging. To the best of our knowledge, this work represents the first report of such registration task in the literature.
As in [18], [19], we capture the total deformation (between atlas and patients image) using two different components: the mass effect, and the deformation due to the inter-subject differences. Explicit mass-effect simulation by biophysical models [20]–[22] is performed in the atlas space prior to warping to target patient image, thereby allowing more realistic warps to be achieved via image warping. Furthermore, other information such as estimated tumor density can be directly used to compute similarities to the patient images.
A. Related Work
A few publications have proposed the spatial normalization of pathological brain images. Masking of pathologies is utilized in Brett et al. [23] and Stefanescu et al. [24] where the warping close to tumors is only driven by the information of the neighboring structures. Nowinski et al. [25] use feature points (Talairach transformation), followed by a radial mass-effect model to warp atlas onto the patient image. In order to create topologically equivalent pair of images, Kyriacou et al. [26] remove the tumor from the patient image, other methods embed a tumor in the atlas (atlas seeding). Atlas seeding has also the advantage that allows more realistic warping, in the form of mass-effect, to be captured. In these methods, a deformation field is first created by mass-effect models, e.g. simplified radial growth models [25], [27], and later refined by a non-rigid deformation based on optical flow [28]–[30]. Incorporation of more advanced biomechanical models of the tumor growth to simulate tissue loss and compute displacements, was introduced in a series of previous publications by our group. For instance Mohamed et al. [31], [32], developed a PCA based statistical method to learn the tumor-growth deformations across different subjects, or within the same subject [18]. The statistical approach was chosen to reduce the high computational cost of the finite element based biomechanical models for tumor growth simulation leaving the burden of simulations to off-line training. Statistical models, however, are not very accurate and also are limited by the parameters used during training.
Recently, Zacharaki et al. proposed ORBIT [18], [19]. Similar to our framework, ORBIT simulates the tumor growth and mass-effect on the atlas prior to warping to the patient image, and it also estimates the best parameter set for this simulation. As ORBIT is originally designed to work with tumors with minimal edema and infiltration, tumor simulation is achieved using the pressure model described in [22], a framework which models tissue necrosis and its replacement by the tumor and also computes the mass-effect. As a result ORBIT, only estimates seed location and the tumor growth factor as the required parameter set.
In contrast to ORBIT, the primary objective of this work is the registration of brain images with GBM, a particular kind of primary tumors which is known to be infiltrative. The tumor may take on a variety of appearances, depending on the amount of hemorrhage, necrosis, or its age and might indicate no clear edges. Mass effect from the tumor and edema may compress the ventricles. Therefore, images with GBM usually have complex and inhomogeneous imaging patterns, which can not be appropriately simulated using the simple pressure tumor model as done in ORBIT.
This paper, proposes a new framework to handle some of these complexities through the following elements: 1) As illustrated in [33], using multi-modality MR imaging and supervised SVM based classification, the original intensity data is transfered into the space of the probability maps (PBMs) of various tissue types, which can be viewed as the memberships to normal and abnormal tissue classes. 2) A more realistic biomechanical tumor model [20] appropriate for GBM, is integrated to our registration framework, and the relevant parameters for the tumor growth modeling (such as seed location, diffusion coefficients etc.) are estimated. Given an atlas and its corresponding set of healthy tissue probability maps, the estimated tumor density map and the mass-effect is used for explicit computation of the PBMs in the atlas space, allowing the similarities between the corresponding PBMs of the patient and atlas to be measured. 3) We anticipate that the edema can not be estimated through the diffusion-reaction based modeling of tumor growth in [20]. Presence of vasogenic cerebral edema depends on the intracranial pressure gradient and involves complex mechanisms where the bulk flow and not diffusion should be considered as the main cause for the spread of edema through the white matter [34]. The proposed method is capable of handling the lack of edema and such unrealistic molding issues, through the segmentation of the patient space into three different regions roughly as: edema, non-edema and outliers (w.r.t. registration model).
Because no a priori information about the tumor parameters and the registration is given, our problem of joint estimation of the warping, tumor growth parameters and the segmentation labels, is an estimation problem from incomplete data. The classical approach toward such problems in the literature, is to utilize the Expectation-Maximization algorithm as it has been followed in this work. In short, EM is utilized for three purposes: First, to estimate the outliers region, where no correspondence can be identified between the patient and atlas spaces, Second, to estimate the plausible regions where a partial or full correspondence between the actual patient and estimated PBMs in the atlas can be achieved, Third, to provide a framework to estimate the deformation field. In this respect, we regard the deformation field as a high-dimensional parameter set that should be estimated through EM.
This paper is organized as follows. In section II, we briefly review our tumor generator model and introduce the relevant parameters that are estimated. In section III, the basis of the simulation of the atlas (moving) PBMS is illustrated. The details of our EM algorithm are given in section IV, where we derive the update equation for the deformation field. In section VI the efficacy of this framework on both synthetic and clinical cases is evaluated, and the sensitivity of registration to the tumor parameter estimation is presented. The paper concludes in section VII.
II. Biomechanical Tumor Modeling
The framework for modeling the glioma tumor growth and its mechanical impact on the surrounding tissue is the same as the outlined modeling by Hogea et al in [20], [35]. In this section a brief review is given, mainly to illustrate the set of parameters that are used to simulate the tumor growth, hereafter called q. The modeling framework consists of a reactive-advective-diffusive mass transport for tumor cells, coupled with elasticity for the brain [21], [36]. The system of PDEs governing the deformable model to simulate the glioma growth consists of:
| (1) |
where Cq, is the generated tumor density corresponding to the set of parameters q, and u, v are displacement field caused by the presence of the tumor and velocity field, respectively. The parameters include: D ∈ {DWM, DGM} the diffusion coefficients, the Young modules μ, λ and proliferation coefficient ρ. Other quantities include m = (λ, μ, D) and the f(Cq, p) is a function of the tumor density that controls the behavior of the mass-effect, defined as:
| (2) |
The parameter p2 regulates both the spatial location and the strength of the mechanical deformation caused by the tumor, while p1 is simply a scaling factor. Given any arbitrary boundary and initial conditions over Cq, u and v, the system of equations in (1) are solved using a fast Eulerian continuum approach on a unmeshed grids of nodes [20]. For the set of all experiments we let zero initial conditions for the displacement and the velocity field and assume the following for the tumor density:
| (3) |
Where we chose d to be the same as the image resolution to allow the initial distribution to be fairly localized at x0. Given this definition, the set of (tumor) parameters which are estimated within our algorithm consist of:
| (4) |
III. Method
In this section the basis of our algorithm is illustrated. We emphasize that throughout the paper we will refer to the fixed space as the patient space and the moving (warped) space as the atlas, in other words we warp the atlas to the patient space.
A. Computing Fixed Feature Images Using SVM
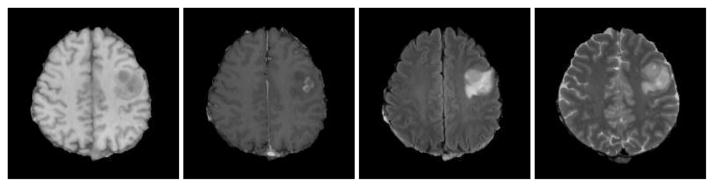
One of the basic challenges for intensity based tumorous image registration task, is that the relation between the observed image intensity and the density of cancerous cells, is either unknown or very complex. For example, as seen in Fig. 1, in the T1 weighted modality edema and non enhancing tumor areas have very close range of intensity values to gray matter, however the corresponding FLAIR image, reveals the edema in much better way by a hyper-intense signal around the tumor. Therefore, image intensity does not seem to be a reliable source of information for our registration task.
Fig. 1.
Imaging profile of a glioma; from left to right: T1, Contrast enhanced T1, FLAIR and T2 images.
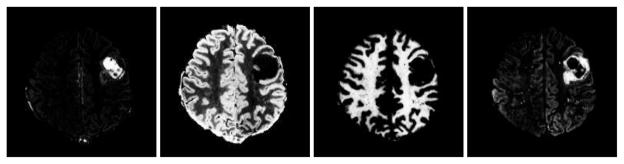
As discussed in [33] multi-modality imaging can help differentiating various tissue types in patients with brain tumors. Based on multi-parametric imaging techniques, in [33] supervised classification using SVM was performed to segment the brain images into six different classes, namely: white matter (WM), gray matter (GM) and CSF, enhancing tumor (ET), non-enhancing tumor (NET), edema or swelling around the tumor (ED). (Necrosis was manually segmented as tumor.) In this paper a similar SVM segmentation framework has been utilized for classification of the pathological images. More specifically, we use four modalities, namely T1/T2/FLAIR and T1-CE (constrast-enhanced perfusion), as the multichannel imaging profile of the patient, and train a SVM model to compute the posterior probability maps [38] of different tissue types in the patient space. Sample results of such computed PBMs are given in Fig. 2. In the next step, these PBMs are registered to their corresponding estimated pairs in the atlas. The estimated PBMs of diseased tissue types in the atlas (moving template) are constructed using the estimated tumor density map and mass-effect. This step has been explained in further detail in section III-B.
Fig. 2.
Sample PBMs computed from SVM; from left to right: F1 (tumor), F3 (gray matter), F4 (white matter) and F5 (edema).
For simplicity, we assume that the ET and NET classes can be integrated under a unique tumor label (TU). In addition, since the necrosis is in fact the death of the cancerous cells as a result of too much density and the lack of enough nutrition, we regard it as a part of TU with probability of one, and the computed PBMs from SVM are masked to take this effect. Therefore we can hypothesize that a set of L = 5 feature images Fj(x), corresponding to different tissue PBMs computed by SVM, is defined in a fixed space ΩF and x ∈ ΩF whereas 1 ≤ j ≤ L enumerates the set of class labels: {TU, CSF, GM, WM, ED}.
B. Constructing Moving Feature Images
As explained in the previous sections, diffusion-reaction equation is used for tumor growth modeling. However, edema is not modeled explicitly as this would entail complex poroelastic material [39]. Because of missing the edema in atlas, only four feature images corresponding to the probability maps of {TU, CSF, GM, WM}, are computed in the moving space ΩM. Since the tumor density is in fact the partial volume of the space that has been occupied by the tumor, deterministic relations between the the original PBMs of healthy moving template and those obtained after embedding the tumor can be established similar to [40]. Let a tumor density map 0 ≤ Cq(x) ≤ 1 be defined in the moving space ΩM. Such relations can be written as: 1
| (5) |
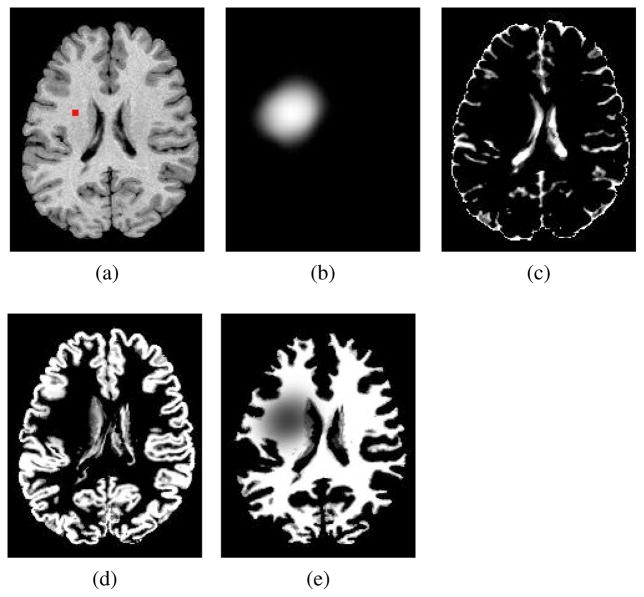
where u(x) is a mapping from ΩM to ΩM and explains the mass-effect of the tumor, and Pl(·), l ∈ {CSF, GM, WM} denotes the tissue PBMs that represent the atlas structure prior to tumor growth. Note that we have as they indicate modified probability maps. A sample output of equation (5) has been depicted in Fig. 3 that illustrates how moving side feature images, i.e., are produced.
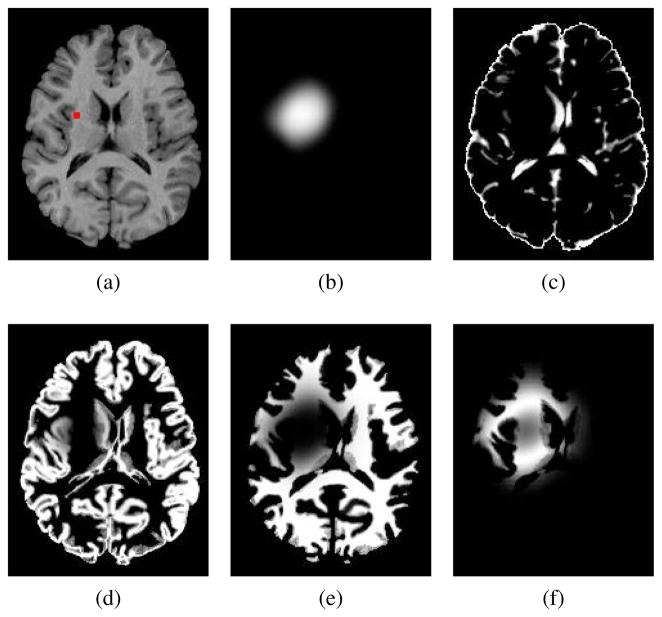
Fig. 3.
Constructing the set of moving feature images. (a) T1 healthy template image segmented by FAST [37] to make original template tissue probability maps (not indicated), the marker indicates a tentative seed location designated to create moving PBMs: (b) created tumor density map ; (c)–(e) and : tissue probability maps with mass-effect and soft-masking by tumor density map in (b), see the set of equations in (5).
IV. EM ALGORITHM WITHIN THE REGISTRATION FRAMEWORK
As stated in section I, our alternating framework for registration and estimation of the tumor parameters has been motivated by the EM algorithm. The cost function of our registration algorithm can be intuitively interpreted as a conditional l2 similarity measure between corresponding estimated moving (atlas) and real patient PBMs. However, the lack of edema information by tumor modeling in the atlas, prevents such a naive similarity to be a confident measure throughout the entire image domains. In fact, since edema is confined to white matter [40] and remains unobserved, the estimated white matter probability around the tumor is not reliable. The remedy taken in this paper, is to use Expectation-Maximization algorithm explained in section IV-A to segment ΩF in two different regions: a sub-domain D0 in ΩF within which a full vectorial l2 similarity measure between and Fj(x), 1 ≤ j ≤ 4 is plausible, and D1 in which the white matter information is excluded from driving the registration. This partitioning is guided by the patient edema’s PBM, and is statistically estimated through the E step as illustrated in the next section.
A. Problem Definition and The Cost Function
Let 1 ≤ i ≤ N represent the index of an arbitrary voxel xi in ΩF and h(x) define the transformation from space ΩF to ΩM. Let a set of N i.i.d observation vectors be defined as:
| (6) |
As explained in section IV, we consider partitioning of ΩF into D0 and D1. To that end we assume that the pdf of i th observation vector , can be modeled as a mixture of the two pdfs (f0, and f1) and estimate the weights of the mixture in the E step. We define f0, and f1 as:
| (7) |
where Φ = {h(xi), σ1, ···, σ4|xi ∈ ΩF, 1 ≤ i ≤ N} is the set of unknown parameters to be estimated. Note that the last channel information in f1 has been replaced by a uniform distribution 2, posing no similarity constraint on white matter.
In order to make a more compact notation in the next steps, we introduce variance matrices defined as:
| (8) |
Using these definitions equations in (7) can be written as:
| (9) |
Our problem of joint registration and estimation of the q (tumor parameters) can be defined as the optimum solution of the following problem:
| (10) |
and:
| (11) |
where πk(xi) is the value of the prior probability of class k at voxel xi ∈ ΩF. In the literature, this is achieved by applying the classical principle of the Expectation-Maximization algorithm, minimizing an upper bound of the right hand side of (10). This requires the derivatives of the cost function w.r.t the set of unknown parameters to be computed first. However, we have no analytical expression for the derivatives w.r.t to q and the numerical computation of those derivatives and following a line search algorithm to minimize the cost, can be extremely expensive in terms of CPU cycles. As a remedy we only estimate Φ within the classical EM algorithm and optimize q by APPSPACK, a derivative-free parallel pattern search algorithm [41]. In other words, in stead of solving (10), we decouple the minimization and solve the following problem:
| (12) |
Where inner minimization w.r.t Φ is achieved by EM, and outer minimization is done by APPSPACK. In practice this means that, the tumor parameters q are not updated through the consecutive iterations of E and M steps until convergence where the computed cost value is returned to APPSPACK as a new sample of the cost function. The computed cost value is then utilized by a search algorithm implemented in APPSPACK to inquire another sample of the cost function and identify a new minimum. Since this procedure requires several evaluations of the cost function, multiple instances of EM algorithms with different tumor parameters need to be executed. To that end, we use APPSPACK which allows us to gain a significant efficiency, thanks to its parallel execution. The procedure has been illustrated through the algorithm 1.
Algorithm 1.
Estimation of the optimum deformation field and tumor parameters: (Φo, qo)
| 1: | Emin ← +∞ |
| 2: | while APPSPACK queue is not empty do |
| 3: | Pop a q from queue. |
| 4: | Using EM Compute: Φt = argminΦ −logf(Yq|Φ). |
| 5: | Et ← −logf(Yq|Φt). |
| 6: | if Et < Emin then |
| 7: | Emin ← Et. |
| 8: | Φo ← Φt. |
| 9: | qo ← q. |
| 10: | end if |
| 11: | Return Et to APPSPACK. |
| 12: | end while |
The parameter estimation can be made robust by modeling the “outliers’ class. This is particularly important in our registration application, since it allows relaxation of the similarity constraint whenever no actual correspondence can be established between the patient and atlas images. To that end, the ML cost in (10) is modified by implicitly introducing a third rejection class. Similar to segmentation framework outlined in [42], at every iteration: m, given the current estimate of the unknown parameters Φ(m−1), the modified likelihood for parameter estimation is written as:
| (13) |
In which λ is a small constant. Increasing λ also increases the robustness on the parameter estimation (registration), but also adversely affects the sensitivity w.r.t segmentations. In this paper, we found λ ≃ 1e−3 to be a good compromise between robustness and sensitivity. stands for posterior probability at iteration m of class k at voxel i, computed in E step as 3:
| (14) |
We use the edema’s PBM measured by our SVM classifier in the previous section for setting: π0(·) = 1 − F5(·) and π1(·) = F5(·). As explained in [42], robust estimation of Φ can now be achieved by maximizing:
| (15) |
where:
| (16) |
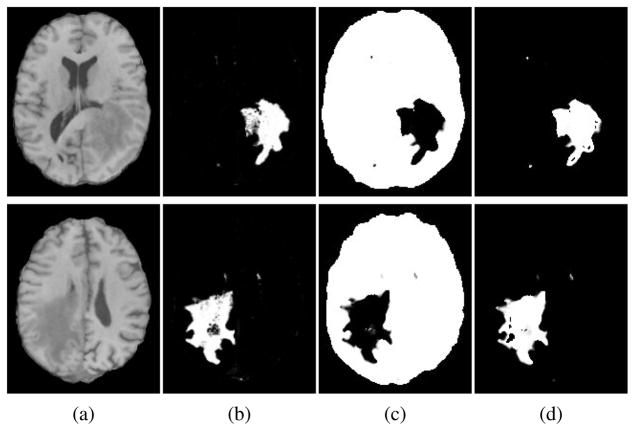
The computed weights , k = 0, 1 at every voxel i define the membership values of that voxel to each cluster k. Therefore they represent an statistical estimation for the partitioning of the ΩF to D0 and D1. A couple of sample partitioning has been indicated in Fig. 4, which also illustrates that how the extracted edema’s PBM from SVM, are utilized as spatial prior functions for such segmentations.
Fig. 4.
Automatic partitioning of ΩF through equation (16) in the E step: (a)Patient T1 image, (b)edema’s probability map (F5) measured by our SVM classifier is used as the prior map for D1 i.e. π1(·), (c)computed membership function for class one (w0), and (d)computed membership function for class zero (w1) in steady state.
B. Estimation of Φ
Because the number of unknown parameters to be estimated in Φ is very large (≃ N3), the estimation problem is highly ill-posed. Robust estimation could be achieved by smoothness constraint over deformation field i.e. his modeled through a Markov Random Field, however, such an approach makes the closed form solutions difficult. Here we choose to relax this constraint and derive an explicit form for his and then project the obtained deformation to the space of the acceptable deformations ( merely smooth or diffeomorphic transformations). Therefore, although the deformation field is explicitly estimated according to maximum-likelihood principle, it is smoothed by convolution with a Gaussian filter, similar to Thirions’ demons framework [43].
In M step, in addition to updating the variances, we derive the update equation for the deformation field using directional derivative principle. To that end, the variation of (15) w.r.t an infinitely small and arbitrary test function ψ, is assigned to zero, i.e:
| (17) |
where . Replacing (7) in (15) and keeping the second order terms, as the proof is given in the appendix, the update rule of the velocity field can be obtained by the solution to the following 1 ≤ i ≤ N independent linear systems:
| (18) |
where matrix Wi is defined as:
| (19) |
∇M is a 3 × 4 matrix defined by column-wise concatenating of the gradient vectors of the moving PBMs, i.e.:
| (20) |
is defined as:
| (21) |
Solution to (18), similar to [43], can be made more stable by adding a norm penalizing term for the update velocity vi at the left hand side:
| (22) |
where is proportional to the maximum norm of the update (||vi||). In this paper we define it as:
| (23) |
where denotes the jth element of the vector . The reason for such selection is empirical and can be explained as follows: because of the initial approximate alignment, the variances are in practice much smaller than one, i.e. . Therefore in the regions far separated from the tumor where , the registration is dominated by , whereas in regions where i.e. adjacent to tumor, takes large values and therefore the norm of the update vector ||vi|| is further penalized. This property is important and minimizes unrealistic warping in the vicinity of tumors.
Another distinction of our formulation from the original demons algorithm presented in [43], [44], is the mechanism for estimation and masking out the outliers by the engagement of the time varying weights in the update equation (22). In fact, it is easy to see that for any outliers making and , the update vector vi will be very small, and therefore its contribution in the subsequent smoothing step is minimized. Update equations for the variances, σj, 1 ≤ j ≤ 3, with similar principle to [42] can also be derived as:
| (24) |
and:
| (25) |
From these equations, are constructed in accordance with the definitions in (8). In order to obtain the updated deformation field, we solve (22) and set4:
| (26) |
C. Estimation of q
Simultaneous estimation of the warping and tumor parameters is a highly ill-posed problem. In order to make a better estimation of q, we propose to constraint the solutions in such a way that the estimated tumor densities have the same mass (expected probability) as the target real tumor in the patient image. In particular given a target tumor PBM in the fixed domain PTU (x), the following constraint is desired:
| (27) |
The approximation is because we are only able to carry out the forward tumor simulation, and no target tumor mass can be accurately specified before hand.5 The tumor growth is pursued until the total mass of the created tumor density exceeds the target mass minimally. The difference between target and created mass values on atlas, depends on the size of time step of the forward simulation process and in order to minimize it smaller time steps are desired. In practice we observed that choosing the time step of 5 days, is sufficiently small to keep this error within the five percent of the target mass.
V. Implementation Details
For every patient, our preprocessing pipeline starts with skull stripping and cerebellum removal of all modalities (T1/T2/FLAIR/T1-CE). Next, these images are co-registered using the FLIRT algorithm [45] to construct the set of the voxel-wise feature vectors required by SVM classifier. In order to train our SVM classifer, an expert radiologist in our center was requested to delineate some representative regions of the different tissue types for a couple of sample data sets as described in section III-A. Using these ROIs, a non-linear Gaussian kernel based SVM model is trained according to principles illustrated in [38]. For the subsequent test subjects, before application of the SVM, all modalities are histogram matched to their corresponding modalities in the training samples.
Prior to application of our deformable registration, the computed PBMs of each patient and the atlas should be linearly registered. Our atlas has the same image dimension of 256×256×128 and voxel size of 0.9375×0.9375×1.5 mm3 as utilized in [20]. In order to keep the consistency with this optimal lattice specifications, we linearly register the computed PBMS to our atlas using FLIRT. The affine transformed PBMs obtained in this way, are then utilized as the fixed reference feature images.
Our deformable registration approach, is implemented using a multi-scale framework. In order to minimize the risk of convergence to local optimums, registration starts with lower resolutions and the computed deformation field after interpolation is set as the initial field in the next level. A down sampling pattern of 4:2:1 is used to construct the three-level pyramids of both the estimated and patient PBMs.
In order to make faster tumor simulations, the set of equations in (1) is solved on a lattice of 65 × 65 × 65 nodes, down-sampled from the original atlas space [20]. The estimated tumor density is then up-sampled to create the diseased PBMs using the equations in (5). In order to find the best tumor simulation parameters, the APPSPACK optimization library launches several parallel MPI registration jobs assigned with different set of tumor parameters on a Linux cluster with Dual Intel Xeon 2.80 GHz CPUs. Each process returns the registration cost value to APPSPACK until there is no other point left in the queue as specified in Algorithm 1. Furthermore, in order to reduce the computation burden this procedure is only executed in the coarsest level of the pyramids and for a few (ten) iterations. After estimation of the best q, the registration is iterated with the finest resolution and more number of iterations.
The initial seed location was set as the center of the search span which is defined by the user. Affine registered patient image to atlas, was utilized to evaluate the extent of the tumor. We observed that the span of 4 × 4 × 4 cm3, was sufficiently large to cover the major parts of the tumorous bulks and to ensure that the estimated seed will be located inside of the search span. In addition, the following search span was utilized for the rest of other parameters whenever not mentioned: 0.01 ≤ ρ ≤ 0.1, 0.01 ≤ p1 ≤ 12, 0 ≤ p2 ≤ 0.02 and 1e−13 ≤ Dg, Dw ≤ 1e−7.
Registration module has been coded in C++ using Insight Toolkit library, and the template class was based on the contributed code of Vercauteren et al [44]. Inherited properties from this object oriented programming, allows us to enforce different constraints on the deformation field such as diffeomorphism (compositive update rule), simple smoothness (additive update rule), linear elasticity and viscosity. In this work we have been using diffeomorphic transformation model, and in each iteration the computed deformation is smoothed with a Gaussian kernel of σ = 2. This value is kept fixed during all of our experiments.
VI. Results
The registration accuracy is evaluated on both simulated and real PBMs of glioma. The simulated PBMs are produced by embedding a tumor on a synthetic brain image (shown in Fig. 5.a) made by deforming our standard atlas (shown in Fig. 3.a). The deformation field was exactly known and was generated by Statistically Simulated Deformations (SSD) introduced in [46]. The same standard atlas of Fig. 3.a was then registered to simulated patient PBMs, and the estimated deformation field was compared to the ground truth which was available to us using SSD.
Fig. 5.
Using statistically simulated deformations (SSD) to create reference feature images (PBMs). (a) Sample generated T1 image by SSD using Fig. 3.a as the base template, the marker indicates the seed location designated to create reference (fixed) PBMs: (b) created tumor density map F1; (c)–(e) F2; F3 and F4: tissue probability maps with mass-effect and soft-masking by the created tumor density map in (b), (f) simulated edema F5 using (28).
To assess the registration performance of our real glioma images, we use the same subject independent evaluation method utilized in [47]. In particular we measure Jaccard ratios to evaluate the overlap between the warped labels from atlas, and the reference labels obtained from our SVM classifier. This is because, in general we found the glioma images to be extremely complex (especially around the tumor) to allow finding exact correspondences between the patient image and the atlas. Therefore the evaluation method utilized in [18] was not reliable this study.
A. Simulating Reference PBMs For Validation Purpose
For the purpose of measuring the registration accuracy using SSD, reference (fixed) PBMs should be simulated. In a real patient case, these are supplied through our SVM classifier as shown in Fig. 2. The difficulty arisen in simulating these PBMs is because of the fact that we do not create edema through application of our biophysical tumor model, whereas in real images, edema is present. To fill out this gap we propose an ad hoc formulation for edema, we emphasize that this is only meant for data simulation for validation purposes and the registration algorithm itself is independent of this formulation.
Edema simulation is based on the observation that the probability of finding the edema is maximized somewhere in between the tumor bulk (i.e., Cq = 1.0) and the healthy tissue (i.e., Cq = 0.0). Therefore the simulated PBM of edema is proposed to be in the form of Cq(1 − Cq), which has the maximum on Cq = 0.5. We also assume that the edema should be confined into white matter. Given these intuitions, the proposed set of equations to simulate our reference PBMs for validation experiments is very similar to (5) but includes the edema’s PBM:
| (28) |
where Fsim4 stands for edema’s PBM, u :ΩF → ΩF is the mass-effect deformation estimated by our tumor generator model [20], q0 is the tumor parameter set used to create the tumor density map Cq0 and u, and hssd : ΩF → ΩM is the statistically simulated deformation learned from a bank of normal-to-normal warping sets [46]. Note that these PBMs sum up to one, as required for an actual set of tissue probability maps in a real patient. We use FAST [37], to segment a T1 template atlas image given in ΩM, into three different labels, and compute the posterior probability map for each tissue label, i.e. PCSF, PGM and PWM. The computed tissue maps are then plugged in (28) to hand in the simulated reference PBMs. As seen in (28), the data in atlas (PCSF, PGM and PWM) is linked to the reference feature images, by the compositive mapping of hssd ∘ u(x). Because both the mass effect u and hssd are computed, the ground truth for the total deformation field is known and can be used for validation purposes. A sample set of the constructed PBMs using (28) has been given in Fig. 5.
B. Evaluation Using Simulated PBMs by SSD
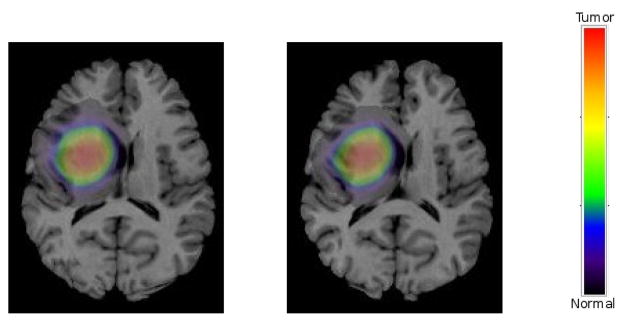
We simulated five sets of reference PBMs and evaluated both registration accuracy and estimated tumor parameters in accordance with principles illustrated in section VI-A. Different seed locations with various diffusion coefficients were utilized to created these reference PBMs. As a result, they were different in both tumor locations and diffusion patterns. It should be emphasized that the same template used by SSD to create the reference PBMs in (28), is utilized to create moving PBMs as illustrated in section III-B. Estimation of the best tumor parameter set and deformation field is achieved using the procedure in Algorithm 1 and the principles illustrated in section IV-B. A sample registration result is shown Fig. 6, in which the target T1 image is made by warping the template image in Fig. 3.a through hssd ∘ u(x). The result shows a good match between the target and warped images and tumor densities. The color bar aside indicates the region covered by red color is fully replaced by the tumor, while the rest of spectrum is used to indicate diffusion and infiltration.
Fig. 6.
Registration results using simulated patient PBMs; Left: The patient T1 image corresponding to the target PBMs shown in Fig. 5(b)–(f) overlayed by the target tumor density, Right: registered atlas overlayed by the warped tumor density.
The five sets of tumor parameters used to create the synthetic reference PBMs, have been summarized in Table I. Each row corresponds to one experiment specified by a unique q0 in (28). In these experiments, we chose p1 = 0 and ρ = 0.025 during both simulation of reference PBMs and registration. This setting corresponds to have u(x) = x. Therefore the registration accuracy can be directly computed by comparing the estimated deformation field to the SSD created deformation, i.e. hssd. Table II, summarizes the estimated tumor parameters. It shows that the tumor seeds in patient and the atlas are registered with high spatial accuracy, since the estimated seed locations in atlas are mapped very closely to the patient’s tumor seed locations. (To see this compare the xc, yc, zc in corresponding rows in Table I). However, the estimated diffusion coefficients are different from the values in Table I utilized to simulate the reference patient PBMs. This is because, the tumor growth PDE model in (1) has been solved in different domains (patient vs. atlas), which have different portions of GM, WM and CSF. Therefore it is not possible to estimate the same diffusion parameters. Registration accuracy in terms of root-mean-square error, has been summarized in Table III. As shown, the rms errors are evaluated on three zones. These regions are delineated by thresholding the reference tumor density map, i.e. Cq0, in two different levels, leaving the entire image domain into far, intermediate and near zones (w.r.t tumor location). For example, the third column corresponds to rms registration error on and in the vicinity of the tumor, where the tumor density is high. The last row is the column wise average of reported errors. As seen, the average rms registration error on the far zone (healthy areas) is comparable to the diagonal size of the image voxels (2.1 mm), and deteriorates on the intermediate and near zones (with no significant differences on the averages). The the maximum rms of (3.25 mm) can be found in the near zone, and falls within the diagonal size of the voxel size used for tumor simulation (4.8 mm). It should be pointed out that we do not compare our method to ORBIT using the images created by SSD [46]. The reason is because SSD uses HAMMER [12] generated deformation fields as training samples. Those deformation fields are easily captured by ORBIT, since ORBIT has been build on the basis of HAMMER (they are basically same registration procedures on healthy parts of brain). Therefore, the simulated images are highly biased in favor of ORBIT, making such comparison not to be fair.
Table I.
Coordinates of the tumor centers (in ΩF) and the diffusion coefficients used to simulate five sets of reference PBMs.
| Set no. | xc | yc | zc | dw | dg |
|---|---|---|---|---|---|
| 1 | 101.493 | 63.4347 | 79.3623 | 1e-8 | 0.5e-8 |
| 2 | 147.798 | 153.792 | 108.928 | 2e-8 | 0.5e-8 |
| 3 | 101.499 | 116.723 | 107.319 | 1e-8 | 0.5e-8 |
| 4 | 140.288 | 99.9364 | 122.25 | 2e-8 | 0.5e-8 |
| 5 | 105.94 | 150.00 | 101.81 | 1e-8 | 0.5e-8 |
Table II.
Estimated tumor centers (mapped to ΩF) and diffusion coefficients of the five simulated patients PBMs corresponding to table I
| Set no. | xc | yc | zc | dw | dg |
|---|---|---|---|---|---|
| 1 | 101.5 | 62.7969 | 79.0937 | 0.65e-8 | .5e-8 |
| 2 | 148 | 153.906 | 110.0 | 1.4e-8 | 1e-8 |
| 3 | 101.547 | 116.25 | 107.094 | 0.5e-8 | 0.5e-8 |
| 4 | 139.75 | 100.453 | 122.094 | 2.21e-08 | 0.505e-8 |
| 5 | 105.1 | 149.23 | 100.94 | 1.4e-08 | 0.505e-8 |
Table III.
Registration accuracy ( in mm) w.r.t ground truth deformation in three different zones of tumor density.
| C ≤ 1e − 5 | 1e − 5 ≤ C ≤ 0.5 | 0.5 ≤ C ≤ 1 | |
|---|---|---|---|
| 1 | 2.49 | 3.1 | 2.18 |
| 2 | 2.638 | 3.04 | 3.58 |
| 3 | 2.67 | 3.14 | 3.08 |
| 4 | 2.37 | 3.28 | 3.73 |
| 5 | 2.5 | 3.18 | 3.63 |
| Avg. | 2.53 | 3.14 | 3.25 |
C. Sensitivity of The Cost Function W.R.T q
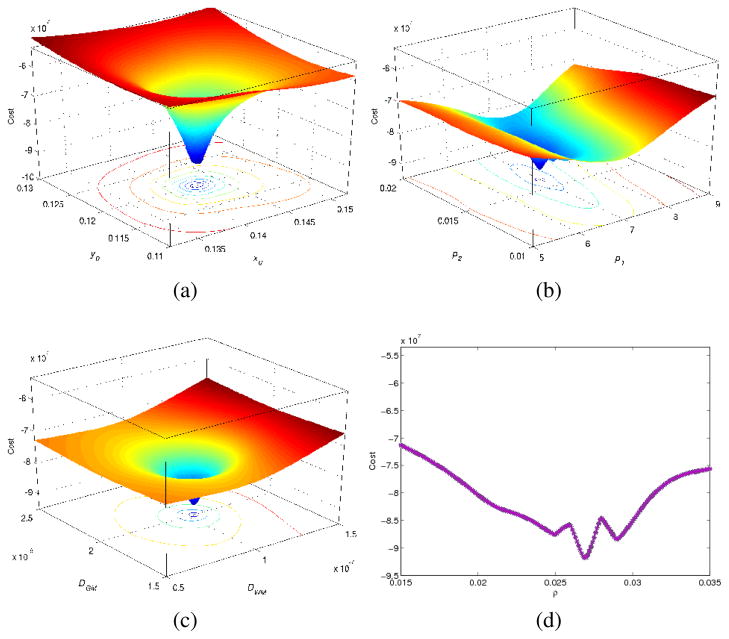
In order to test the potential of optimizing the tumor growth parameters using the proposed optimality criterion, we have plotted the total cost as a function of the error in estimating q. We should emphasize that for an inter-subject registration task, because the PDEs introduced in (1) should be solved in two different domains, it is not possible to estimate exactly the same tumor parameters. Therefore, the results in this section have been obtained using an intra-subject registration task. To that end, we only simulate a mass-effect on the template, i.e., we put hssd(x) = x in our estimated PBMs in (28), and try to estimate the original q0. The computed cost values are then interpolated within the 2D planes intersecting the estimated q, as shown in Fig. 7. As shown, the cost function in general remains locally convex with regard to the most of the parameters and they have been estimated reasonably around the target values (central values of the indicated ranges). As seen in panels (b) and (d), there are some fluctuations around the target values. We believe that this is because of lacking edema as a part of our estimated moving feature images, and also numerical errors arisen when the registration errors are very small. In addition, Fig. 7 indicates that the largest sensitivity is achieved versus the change in the tumor seed location, and this is followed by diffusion coefficients, masseffect parameters and proliferation coefficient.
Fig. 7.
2D profiles of the cost function versus estimated tumor parameters (q). Sensitivity across: (a)tumor seed coordinates, (b)mass-effect parameters, (c)diffusion coefficients, (d)proliferation coefficient. The estimated parameters are reasonably around the target values. The largest sensitivity is achieved versus change in the tumor seed location.
D. Robustness and Convergence Analysis
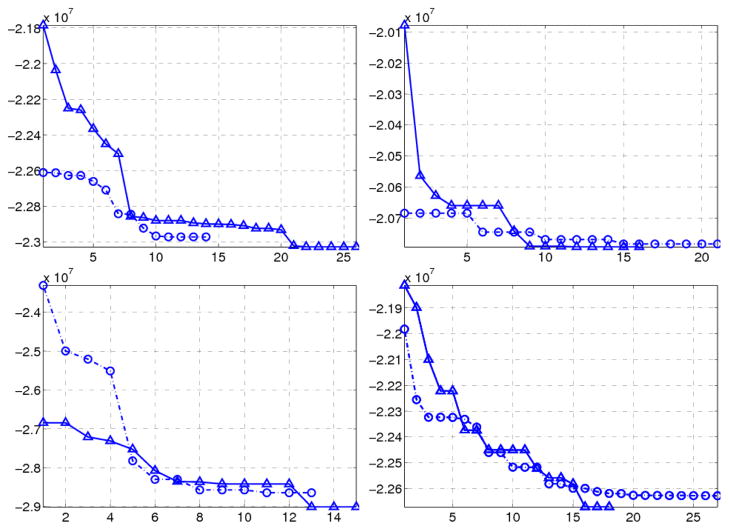
Because of the larger number of tumor parameters (compared to ORBIT), and the combination of two different optimization algorithms the overall robustness of the algorithm was of major interest. We evaluated the robustness and convergence behavior of the algorithm using four real glioma patients. For every individual a pair of registration experiments with different search spans of tumor parameters q was performed. To that end, the search span for q in the first experiment was enlarged by 50 % in every dimension to define a new search span for the second experiment. Of especial interest was to evaluate that how the converged values of q are different from each other. For an ideally robust estimation with a global minimum, those values should be same. However, our experiments showed that the cost function was not convex and therefore different tumor parameters were estimated. For every individual each panel in Fig. 8 shows that how the cost values drop to different local minimums. Note that the horizontal axis counts the number of “successful’ registrations in which APPSPACK have found a new minimizing value. For every patient the corresponding converged tumor parameters using different search spans of q, have been listed in a single row of Table IV (because of moderate sensitivity on p2, its value was fixed to zero in these experiments). The comparison of the converged tumor parameters for each patient shows that the estimated tumor center locations are minimally different compared to e.g. diffusion coefficients, and they can be estimated more robustly. For the third subject, the dimensionality of the search space was reduced by fixing the diffusion coefficients, therefore optimization was done w.r.t other tumor parameters. Consequently, overall a more robust estimation was achieved on the rest of parameters.
Fig. 8.
Convergence plots of four real sample subjects. Vertical and horizontal axes indicate the cost value and number of minimizing registrations. Each panel corresponds to a single subject and represents the minimization of the cost function with two different search span for tumor parameters.
Table IV.
Estimated values of the tumor parameters q corresponding to Fig. 8 using APPSPACK
| Subj. | xc | yc | zc | p1 | ρ | dw | dg |
|---|---|---|---|---|---|---|---|
|
| |||||||
| 1 | 13.2 | 14.0 | 9.0 | .05 | 6.2 | 6e-9 | 1.5e-9 |
| 13.2 | 14.5 | 9.0 | .03 | 7 | 8e-9 | 5.5e-9 | |
|
| |||||||
| 2 | 11.5 | 10.8 | 8.9 | .06 | 6 | 1.5e-10 | 5e-13 |
| 11.3 | 11.4 | 9.4 | .06 | 5 | 1.5e-11 | 1e-12 | |
|
| |||||||
| 3 | 11.0 | 11.0 | 12.4 | .01 | 5.0 | 1.5e-9 | 1e-10 |
| 11.0 | 11.0 | 12.7 | .01 | 7.5 | 1.5e-9 | 1e-10 | |
|
| |||||||
| 4 | 14.3 | 7.2 | 9.8 | .04 | 3.3 | 1e-7 | 1.5e-9 |
| 14.3 | 7.2 | 9.8 | .02 | 3.5 | 5e-8 | 1.0e-9 | |
In addition, the impact of convergence into different local minimums on the registration errors has been studied by measuring the Jaccard overlap ratios in Table V(refer to section VI-E1 for definition and further details). The table indicates that lower cost values (E in the second column) generally imply larger overlap ratios on various tissue labels and therefore a better registration quality.
Table V.
Jaccard ratios of various tissue labels corresponding to converged local minimums in Fig. 8
| Subj. | E(×107) | L-Vent | Tum | L-GM | L-CSF |
|---|---|---|---|---|---|
|
| |||||
| 1 | −2.29 | 54.5% | 38.2% | 57.19% | 20.0% |
| −2.30 | 55.1% | 42.2% | 56.13% | 24.0% | |
|
| |||||
| 2 | −2.05 | 65.5% | 48.6% | 60.0% | 20.9% |
| −2.08 | 65.7% | 47.6% | 60.0% | 25.2% | |
|
| |||||
| 3 | −2.86 | 60.0% | 38.4% | 49.0% | 17.4% |
| −2.90 | 61.2% | 40.4% | 49.7% | 19.4% | |
|
| |||||
| 4 | −2.262 | 56.5% | 68.9% | 67.0% | 18.5% |
| −2.267 | 58.6% | 72% | 68.9% | 18.9% | |
E. Registration of Real Glioma Images
Fifteen multi-channel data sets of real gliomas with different grades, and sizes were selected for registration with the normal template (atlas) and the comparison was made to ORBIT [19]. The preprocessing step and the implementation details are illustrated in section V.
As the reference input, ORBIT requires a label image which should be created by segmentation of the patient image. For that purpose the hard segmentations (tissue labels) obtained through our SVM was used for each subject. Therefore, the advantage of using multi-parametric images for segmentation has also been utilized in ORBIT. Moreover, since some of our glioma images contain large portions of edema, and the ORBIT does not handle this type of tissue label, we replace edema by WM. This is based on our hypothesis that edema only spreads into white matter. Therefore the performance of ORBIT was increased substantially. Other details and parameters were same as in [19]. In terms of computational cost, having the tumor parameters our registration method runs faster, however since we estimate 8 parameters, versus four parameters in ORBIT, the total computation cost stays similar to ORBIT and depending on the tumor size may vary between (6 ~ 14h).
1) Quantitative Assessment
As illustrated in section VI, the complexity of glioma images, especially around the tumor, makes the task of finding of corresponding landmarks between the patient image and the atlas difficult and unreliable. Therefore the evaluation method utilized in [18] was not a robust measure for this study. In this paper similar to [47], the rater independent Jaccard ratio, measuring the overlap between target (from SVM) and warped labels from atlas, has been utilized for quantitative evaluations. More specifically for each label k we measure:
| (29) |
where n represent the number of voxels in a set, Tk is the set of voxels that have been labeled as k in patient, and Wk is the set of voxels obtained by warping of voxels originally labels as k in atlas.
In order to be discriminative on performance for local (tumor vicinity) and global regions (rest of the healthy portions), the warped tumor density created in atlas space, was segmented by a threshold of 1e−5 to divide the patient image space into two separate regions denoted as local (L) and global (G). Next, the intersections of these regions with SVM segmented GM, CSF and TUM, along with manually segmented ventricles V ent were used to create different target labels. Note that the normal part of the WM (excluding edema) is not used in the quantitative evaluation. This is because we do not model edema in atlas, and WM in atlas is in fact a mixture of edema and white matter where we do not have a border to separate them.
The measured jaccard ratios for all of our fifteen test subjects are shown using bar and scatter plots in Fig. 9. In addition a paired t-test was used to identify statistically meaningful differences between measurements. Table VI summarizes the mean JR measures made by averaging though all test subjects both for ORBIT and our method. As seen our method outperforms the ORBIT in most of the labels, by minimum margin of 6.3% on tumors and maximum 21.15% on G-CSF.
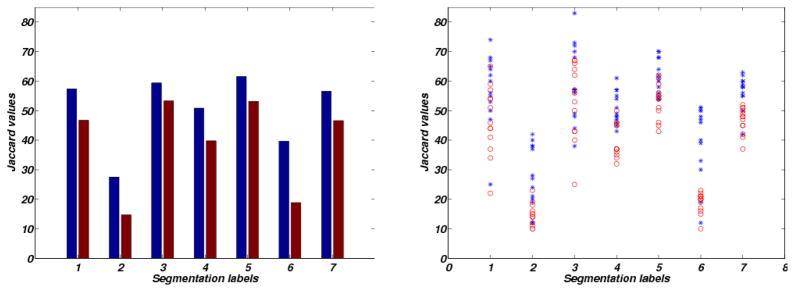
Fig. 9.
Plots of jaccard values for different segmentation labels enumerated as 1:L-Vent, 2:L-CSF, 3:TUM, 4:L-GM, 5:G-Vent, 6:G-CSF, 7:G-GM; Left: mean values, Right: scattered samples. The red and blue colors correspond to ORBIT and our method’s result.
Table VI.
Mean jaccard values of different segmentation labels. Significant differences are indicated in bold (p-value ≤ 0.05).
| Label | Our proposed method | ORBIT |
|---|---|---|
| L-Vent | 57.38 | 46.77 |
| L-CSF | 28.54 | 15.69 |
| TUM | 59.38 | 53.31 |
| L-GM | 50.85 | 39.77 |
| G-Vent | 62.54 | 53.15 |
| G-CSF | 40.0 | 18.85 |
| G-GM | 57.0 | 47.62 |
2) Qualitative Assessment
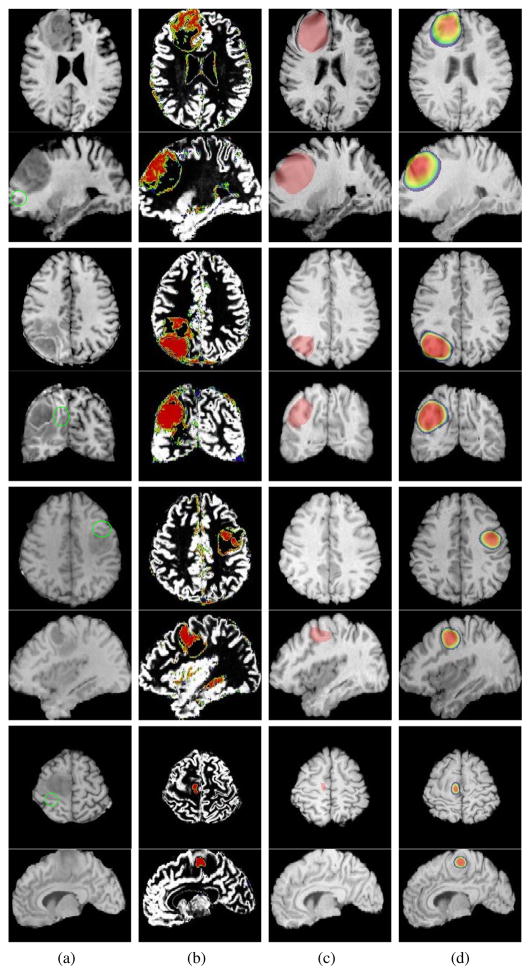
In addition to quantitative results based on the label overlaps, the registration performance of our method is also visually evaluated. The registration results of eight test subjects of different grades of glioma, with the normal atlas and its comparison to ORBIT are illustrated in Fig. 10 and Fig. 11. The normal atlas in these experiments was the same atlas in Fig. 3 used in our previous sections. From left to right, first column show the T1 images (other modalities are not shown), the second column show the PBMs (reference tissue probability maps) of tumor and gray matter, obtained through our SVM classifier. This is followed by the ORBIT and our proposed method’s results. Similar color bar in Fig. 6 has been utilized to overlay the warped tumor density on the warped atlas images in our method. It is important to note that the regions overlayed by red are fully replaced by the tumor. The presence of other structures in these areas, denote how they have been captured by the tumor, and should not be taken as their real existence. This complexity in rendering the results is indispensable, because we had no clues on the change of image intensity levels by presence of tumor. Also, since ORBIT does not generate diffused tumor densities, only the estimated tumor labels are presented. Green markers in the first column indicate some locations where our method has achieved a better similarity in comparison to ORBIT.
Fig. 10.
Registration results with the same atlas shown in Fig. 3.a. The blank spaces in between the rows separates the patients. Columns from left to rigth: (a) T1 fixed reference images, (b) Overlay of the extracted PBMs of GM, and TUM, (c) Warped template images to the reference using ORBIT (overlayed by the estimated tumor label), (d) Warped template using our algorithm (overlayed by the warped tumor density map originally generated in the atlas space). The markers in the first column around the tumor indicate some location where our method has had a better performance in comparison to ORBIT.
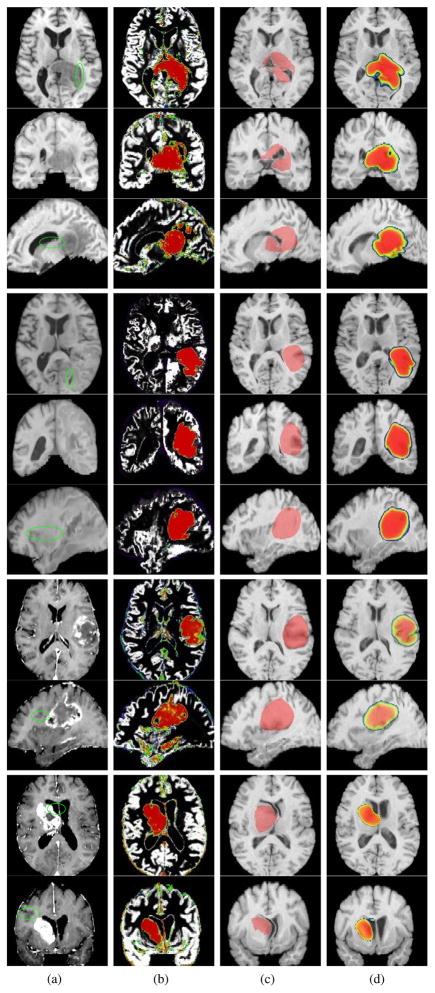
Fig. 11.
Continued from Fig. 10
In general, very good similarities between the warped atlas overlayed by the warped tumor densities, and the patient images can be observed for all cases using our method. Also, the warped tumor densities resemble the reference PBMs, and overall match the pathological areas.
The patients indicated in Fig. 10 have been diagnosed with various grades of gliomas and the tumors with infiltration. In the first sample ORBIT has captured the infiltrated tumor parts as a bulk mass, and some unrealistic mass-effect has been generated (top rows in panel c). Whereas the warped tumor density using our method clearly shows infiltration, and registered template resembles the patient image. The second example, indicates a case where the infiltration and the mass effect have been underestimated by ORBIT, whereas using our method the target tumor and the mass effect have been captured in a better way. The third and forth (separated by white spaces from top) samples in Fig. 10 indicate patients with lower grades of gliomas and smaller tumor bulks. The axial and sagital slices of the third and forth cases in panel (c), shows the ORBIT’s failure to capture some parts of the tumor (as no tumor label is observed in the corresponding slice), whereas the result of our method shows some tumor diffusion, matching corresponding reference tumor PBM and a better registration performance has been achieved using our method (sample locations have been marked in the first column).
The first three rows on the top of Fig. 11 show an example patient, in which the ventricles have been heavily involved. As seen, ORBIT fails to capture the tumor pattern correctly, as the ventricles are not squeezed by its estimated tumor, whereas our method has a better performance since the created tumor resembles the patients tumor and ventricles indicate the mass effect. In the last example Fig. 11 (bottom rows), tumor is less infiltrative and has a bulk specified by the enhancement in the borders. ORBIT has over estimated the mass-effect and excessively pushed the ventricles, whereas such a pattern is not observed in our result and the ventricles are more similar to those of the patient.
VII. Conclusion and Future Research Orientation
A deformable registration algorithm for glioma images with a normal atlas(template) is proposed. We utilized multi-parametric imaging techniques to compute the tissue probability maps of the patient with an expert trained Support Vector Machine. This step was critical to provide matching information on and around the tumor. Using fuzzy probabilistic maps is also especially important for gliomas, because those tumors are diffusive and therefore every voxel is expected to include some tumor portion. On the atlas, we used reaction-diffusion equation to simulate a tumor growth and estimate the mass-effect. Normal probability maps of the healthy atlas, were modified to reflect the tumor invasion, and registered to their corresponding probability maps in the patient image. In the atlas, since we do not model the edema, which is assumed to be confined into the white matter, the edema’s probability map (PBM) remains as an unobserved additive component with the simulated white matter PBM. Therefore in edema region, the information of the white matter PBM is ignored and the registration is achieved by matching of CSF, gray matter and tumor PBMs between the patient and the tumor embedded atlas. Therefore we believe that the registration quality is better than the case in which the whole edema region is masked out from driving the registration ( [23] and [24]). Such unrealistic modeling issues such as the lack of edema in the estimated PBMs on the atlas, is handled using EM algorithm, that estimates the spatial transformation and simultaneously segments the edema in the patient image.
The tumor parameters such as seed location, mass-effect coefficients, proliferation coefficient and diffusion parameters are estimated through APPSPACK [41]. The performance of the method was evaluated using both statistically simulated brain images, with known deformation field, and the real glioma images. Our experiments show that the proposed method has a very good registration performance, and the warped atlas resembles the target patient images.
To be accurate on registration errors, we used statistically simulated brain images and PBMs as target patient images. This allowed us to compare our computed deformation fields to the ground truth deformations, available from our simulations. Table III summarizes mean square error results, and indicates the average of registration accuracy is in the order of one voxel size.
Comparison was made to ORBIT [19], this comparison was chosen especially because, for tumor images ORBIT has shown to outperform other registration methods. Both of our qualitative and quantitative results indicate that our method has a better performance, especially for highly infiltrated tumors. Examples for this claim can be found in Fig. 10 and Fig. 11, where some tumors indicate significant infiltration.
For real glioma images, our quantitative comparison of registration efficiency was based on the tissue overlap ratios as utilized in [47], [48]. Jaccard ratio was especially preferred mainly because it allows rater independent evaluation of registration, and the complexity of the images did not allow reliable landmark localization. To be more specific, JRs were measured in both local (w.r.t tumor) and global (considered as healthy) regions as shown in Table VI. The table demonstrates that our proposed method has a better performance compared to ORBIT. It should be pointed out that the main objective of such table is to establish a basis for the comparison and the reported values can be further improved upon having a better quality on SVM classifier. In particular, poor overlap ratio on CSF, is partly because those structures are very tiny and therefore the overlap ratio is sensitive to small displacements. From our t-test scores, we notice that the improvement of registration in areas close to tumor was not equally significant as the rest of the brain. This is because, the current tumor growth models employed in both registration methods, have a limited ability to simulate anisotropic tumor patterns as they are observed in real glioma images. Such inadequately realistic tumor simulation causes poor matches and therefore although we have achieved an improvement (6%) over ORBIT, the difference is not statistically significant. In fact, the performance of the proposed method depends on the target tumor shape of the patient, and is the best for blob-like tumors. We expect upon integration of a better modeling, e.g. by fusion of DTI tensors for anisotropic tumor propagation along the white matter fiber tracts as done in [21], the quality of the registration to improve.
In addition, the current framework does not consider patients with multiple tumor bulks. In reality, metastasis might occur in glioma which results in several disjointed tumor areas. For such cases, several tumor seeds can be embedded in the atlas, however, this increases the number of parameters to be estimated and can result in a significantly longer processing time.
Although not covered in this paper, the sensitivity analysis of the registration quality w.r.t patient PBMs is an important issue that should be addressed. We acknowledge that having a robust and accurate classification of the patient images, increases the registration quality. However, for noisy segmentations a compromise between the robustness and sensitivity of the algorithm can be established by adjusting the λ. 6
One limitation in computing tissue probability maps is that, the current imaging modalities have intrinsic limitation to detect lower tumor cell densities [49], [50]. As a result, we acknowledge that with current modalities the computed PBMs on infiltrated areas might not be accurate. One interesting branch of future research is to enhance the accuracy of the measured PBMs by fusion of other information sources such as spectroscopies made around the tumor.
In the current form of our work, the edema is not directly simulated, mainly because it contributes to more complexity of our simulations, and increases the number of total required parameters, which would increase the computational cost in turn. Integration of edema modeling (such as one proposed by Nagashima et al [39]) can be considered in the next steps. Also in the current implementation, the classification of the subject multi-modality images is not updated during the registration process. Given an estimate of the registration of the atlas and tumor parameters, one can consider further refining the classification of the subject. The natural way for that purpose is to update the segmentation labels in the E step of our algorithm. This extension, however, was not initially considered since we wanted to favor the user’s knowledge to be more specific on making segmentations of tumor and edema. The developed registration package is available for download via the homepage of Section of Biomedical Image Analysis, University of Pennsylvania: https://sbia-svn.uphs.upenn.edu/
Acknowledgments
The authors would like to thank Dr. C. Hogea for developing the reaction-diffusion model, Dr. E.I. Zacharaki for helping us to run ORBIT and Dr. E. R. Melhem from the Department of Radiology, University of Pennsylvania, for providing us the patients data sets.
This work was supported by National Institute of Health Grant 5R01NS042645.
Nomenclature
velocity vector norm penalizer at voxel i and iteration m
- λ
a small constant employed for outlier detection
- ψ
arbitrary test function used to derive the update equation
deformation field vector at voxel i and iteration m
- q
the set of tumor parameters needed for tumor simulation
the residual vector used in linear system at voxel i
variance matrices of class 0 and 1
- u
the mass-effect deformation field
- vi
the velocity field vector at voxel i
the coefficients matrix of linear system at voxel i
observation vector at ith voxel in ΩF made by differencing target and warped PBMs
- Yq
the set of N i.i.d observation difference vectors
- ΩF
domain of fixed (patient) PBMs
- ΩM
domain of moving (atlas) PBMs
- Φ
set of unknown parameters to be estimated by EM
- π0,π1
a priori probability maps for class 0 and 1
- ρ
proliferation coefficient
variance of jth difference channel
- Cq
the simulated tumor density map
- D0,D1
subregions of ΩF
- DWM,DGM
diffusion coefficients of white and gray matter
- f0,f1
probability distribution functions
- Fj
jth fixed (patient) PBM
- Fsimj
jth simulated target PBM for the purpose of validation
ith moving PBM generated in atlas
- N
the number of voxels in ΩF, p1, p2, mass-effect parameters
robust posterior probability of voxel i belonging to class k at iteration m
- x,y, z
tumor seed coordinates
Appendix A Proof of update equations (18)
To compute the derivative according to (17), we first compute the differential w.r.t h(xi) and keep variances σi fixed at the current estimate, i.e . We only consider those terms in (15) which involve h(xi):
| (30) |
by re-arranging the terms:
| (31) |
Let:
using the definition of directive differential operator < ·, · > in (17) and its linear properties we have:
and using (20):
| (32) |
where ∇M has been defined in (20). Keeping terms with up to the second order of ||vi||, (32) can be summarized as:
| (33) |
Replacing (33) in (32), the right hand side of (17) can be written as:
| (34) |
where and , have the same definitions in (19, 21). Since for every test function ψ defined in ΩF the above equality should hold true we must have:
| (35) |
which is solved for the vi.
Footnotes
In a discrete analogy, equations in (5) can be explained using expectations in multivariate hypergeometric distribution. For the sake of brevity the final equations are mentioned here.
given the fact that Fj and are probability maps staying between 0 to 1, is a random variable between [−1,1], hence the corresponding uniform distribution is 0.5.
we will refer to the iteration number in parenthesized upper indexes throughout the rest of paper. In addition vectors and matrices are notified in bold fonts.
In fact, we follow [43], [44] and in stead of addition, a composition update rule is used i.e.: , to achieve a faster convergence.
Another alternative would be to introduce a “soft constraint’ as a prior information term to penalize the difference between the mass values. However, that would have required a weighting parameter to be selected by user, therefore it was avoided here.
To see this, note that regions with incorrect segmentation labels result in poor match to atlas and therefore are captured as outliers where and ||vi|| ≈ 0. In such locations the registration is driven by the neighborhood information. Since are inversely proportional to λ, increasing the λ makes the registration more robust but less sensitive (w.r.t patient PBMs).
Contributor Information
Ali Gooya, Email: ali.gooya@uphs.upenn.edu, Section of Biomedical Image Analysis, Department of Radiology, University of Pennsylvania, Suite 380, 3600 Market Street, 19104 PA, USA.
George Biros, Email: biros@gatech.edu, College of Engineering Biomedical Engineering, Georgia Institute of Technology, 1324 Klaus Advanced Computing Building, 266 Ferst Drive, Atlanta GA 30332-0765.
Christos Davatzikos, Email: christos.davatzikos@uphs.upenn.edu, Section of Biomedical Image Analysis, Department of Radiology, University of Pennsylvania, Suite 380, 3600 Market Street, 19104 PA, USA.
References
- 1.Larjavaara S, Mntyl R, Salminen T, Haapasalo H, Raitanen J, Jskelinen J, Auvinen A. Incidence of gliomas by anatomic location. Neuro Oncol. 2007;9:319–325. doi: 10.1215/15228517-2007-016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duffao H, Capalle L. Preferential brain locations of low grade gliomas. Cancer. 2004;15:2622–2626. doi: 10.1002/cncr.20297. [DOI] [PubMed] [Google Scholar]
- 3.Ashburner J, Friston KJ. Voxel-based morphometry: The methods. Neuro Image. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 4.Christensen GE, Johnson HJ. Consistent image registration. IEEE Trans Med Imag. 2001;20:568–582. doi: 10.1109/42.932742. [DOI] [PubMed] [Google Scholar]
- 5.Chui HL, Rangarajan A. A new point matching algorithm for non-rigid registration. Comput Vision and Image Understand. 2003;89:114–141. [Google Scholar]
- 6.Davatzikos C. Spatial transformation and registration of brain images using elastically deformable models. Comput Vision and Image Understand. 1997;66:207–222. doi: 10.1006/cviu.1997.0605. [DOI] [PubMed] [Google Scholar]
- 7.Ferrant M, Warfield S, Guttman C, Mulkern R, Jolesz F, Kikinis R. 3-d image matching using a finite element based elastic deformation model. Proc MICCAI. 1999:202–209. [Google Scholar]
- 8.Gee JC. On matching brain volumes. Pattern Recognition. 1999;32:99–111. [Google Scholar]
- 9.Johnson HJ, Christensen GE. Consistent landmark and intensity-based image registration. IEEE Trans Med Imag. 2002;21:450–461. doi: 10.1109/TMI.2002.1009381. [DOI] [PubMed] [Google Scholar]
- 10.Pizer S, Fritsch DS, Yushkevich PA, Johnson VE, Chaney EL. Segmentation, registration and measurement of shape variation via image object shape. IEEE Trans Med Imag. 1999;18:851–865. doi: 10.1109/42.811263. [DOI] [PubMed] [Google Scholar]
- 11.Rueckert D, Sonoda LI, Hayes C, Hill D, Leach M, DJH Non-rigid registration using free-form deformations: Application to breast MR images. IEEE Trans Med Imag. 1999;18:712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- 12.Shen D, Davatzikos C. HAMMER: Hierarchical attribute matching mechanism for elastic registration. IEEE Trans Med Imag. 2002;21:1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- 13.Studholme C, Cardenas V, Blumenfeld K, Schuff N, Rosen HJ, Miller B, Weiner M. Deformation tensor morphometry of sematic dementia with quantitive validation. Neuro Image. 2004;21:1387–1398. doi: 10.1016/j.neuroimage.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 14.Thompson P, Toga AW. A surface-based technique for warping three-dimensional images of the brain. IEEE Trans Med Imag. 1996;15:402–417. doi: 10.1109/42.511745. [DOI] [PubMed] [Google Scholar]
- 15.Vemuri BC, Ye J, Chen Y, Leonard CM. Image registration via level-set motion: Applications to atlas-based segmentation. Med Image Anal. 2003;7:1–20. doi: 10.1016/s1361-8415(02)00063-4. [DOI] [PubMed] [Google Scholar]
- 16.Pluim JP, Maintz JB, Viergever MA. Mutual-information based registration of medical images: A survey. IEEE Trans Med Imag. 2003;22:986–1004. doi: 10.1109/TMI.2003.815867. [DOI] [PubMed] [Google Scholar]
- 17.Lorenzen MP, Davis B, Gerig G, Bullitt E, Joshi S. Multimodal image set registration and atlas formation. Med Image Anal. 2006;10:440–451. doi: 10.1016/j.media.2005.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zacharaki EI, Shen D, Lee SK, Davatzikos C. ORBIT: A multiresolution framework for deformable registration of brain tumor images. IEEE Trans Med Imag. 2008;27:1003–1018. doi: 10.1109/TMI.2008.916954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zacharaki EI, Hogea CS, Shen D, Biros G, Davatzikos C. Non-diffeomorphic registration of brain tumor images by simulating tissue loss and tumor growth. Neuroimage. 2009;46:762–774. doi: 10.1016/j.neuroimage.2009.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hogea C, Davatzikos C, Biros G. An image-driven parameter estimation problem for a reaction-diffusion glioma growth model with mass effects. J Math Biol. 2008;56:793–825. doi: 10.1007/s00285-007-0139-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clatz O, Sermesant M, Bondiau PY, Delingette H, Warfield SK, Malandain G, Ayache N. Realistic simulation of the 3-d growth of brain tumors in MR images coupling diffusion with mass effect. IEEE Trans Med Imag. 2005;24:1334–1346. doi: 10.1109/TMI.2005.857217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hogea CS, Abraham F, Biros G, Davatzikos C. A framework for soft tissue simulations with applications to modeling brain tumor mass-effect in 3-d images. 3rd Canadian Conf. Comput. Rob. Vis. (CRV); 2006. pp. 24–33. [DOI] [PubMed] [Google Scholar]
- 23.Brett M, Leff AP, Rorden C, Ashburner J. Spatial normalization of brain images with focal lesions using cost function masking. Neuroimage. 2001;14:486–500. doi: 10.1006/nimg.2001.0845. [DOI] [PubMed] [Google Scholar]
- 24.Stefanescu R, Commowick O, Malandain G, Bondiau PY, Ayache N, Pennec X. Non-rigid atlas to subject registration with pathologies for conformal brain radiotherapy. Proc MICCAI. 2004:704–711. doi: 10.1007/11566489_114. [DOI] [PubMed] [Google Scholar]
- 25.Nowinski WL, Belov D. Toward atlas-assisted automatic interpretation of MRI morphological brain scans in the presence of tumor. Acad Radiol. 2005;12:1049–1057. doi: 10.1016/j.acra.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 26.Kyriacou SK, Davatzikos C, Zinreich SJ, Bryan RN. Nonlinear elastic registration of brain images with tumor pathology using a biomechanical model. IEEE Trans Med Imag. 1999;18:580–592. doi: 10.1109/42.790458. [DOI] [PubMed] [Google Scholar]
- 27.Ganser KA, Dickhausa H, Metznerb R, Wirtzb CR. A deformable digital brain atlas system according to talairach and tournoux. Med Image Anal. 2004;8:3–22. doi: 10.1016/j.media.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 28.Cuadra MB, Pollo C, Bardera A, Cuisenaire O, Villemure JG, Thiran JP. Atlas-based segmentation of pathological MR brain images using a model of lesion growth. IEEE Trans Med Imag. 2004;23:1301–1314. doi: 10.1109/TMI.2004.834618. [DOI] [PubMed] [Google Scholar]
- 29.Dawant BM, Hartmann SL, Gadamsetty S. Brain atlas deformation in the presence of large space-occupying tumours. Proc MICCAI. 1999;1679:589–596. doi: 10.1002/igs.10029. [DOI] [PubMed] [Google Scholar]
- 30.Cuadra MB, Craene M, Duay V, Macq B, Pollo C, Thiran JPh. Dense deformation field estimation for atlas-based segmentation of pathological MR brain images. Comput Methods Programs Biomed. 2006;84:66–75. doi: 10.1016/j.cmpb.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 31.Mohamed A, Zacharaki EI, Shen D, Davatzikos C. Deformable registration of brain tumor images via a statistical model of tumor-induced deformation. Med Image Anal. 2006;10:752–763. doi: 10.1016/j.media.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 32.Mohamed A, Davatzikos C. Finite element modeling of brain tumor mass-effect from 3-d medical images. Proc MICCAI. 2005:400–408. doi: 10.1007/11566465_50. [DOI] [PubMed] [Google Scholar]
- 33.Verma R, Zacharaki EI, Ou Y, Cai H, Chawla S, Lee SK, Melhem ER, Wolf R, Davatzikos C. Multiparametric tissue characterization of brain neoplasms and thier recurrence using pattern classification of MR images. Acad Radiol. 2008;15:566–977. doi: 10.1016/j.acra.2008.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stummer W. Mechanisms of tumor-related brain edema. Neurosurgical FOCUS. 2007:1–7. doi: 10.3171/foc.2007.22.5.9. [DOI] [PubMed] [Google Scholar]
- 35.Hogea C, Davatzikos C, Biros G. Brain tumor interaction biophysical models for medical image registration. SIAM J Sci Comput. 2008;30:3050–3072. [Google Scholar]
- 36.Swanson KR, E, Murray D. A quantitative model for differential motality of gliomas in grey and white matter. Cell Prolif. 2000;33:317–329. doi: 10.1046/j.1365-2184.2000.00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden markov random field model and the expectation maximization algorithm. IEEE Trans Med Imag. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 38.Chang C-C, Lin C-J. LIBSVM: a library for support vector machines. 2001 software available at http://www.csie.ntu.edu.tw/cjlin/libsvm.
- 39.Nagashima T, Tamaki N, Takada M, Tada Y. Formation and resolution of brain edema associated with brain tumors: A comprehensive theoretical model and clinical analysis. Acta Neurochir Suppl(Wien) 1994;60:165–167. doi: 10.1007/978-3-7091-9334-1_44. [DOI] [PubMed] [Google Scholar]
- 40.Prastawa M, Bullitt E, Moon N, Leemput KV, Gerig G. Automatic brain tumor segmentation by subject specific modification of atlas priors. Acad Radiol. 2003;10:1341–1348. doi: 10.1016/s1076-6332(03)00506-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffin JD, Kolda TG. Tech Rep. Sandia National Laboratories; Albuquerque, NM and Livermore, CA: Jul, 2006. Asynchronous parallel generating set search for linearly-constrained optimization. [Google Scholar]
- 42.Leemput KV, Maes F, Bello F, Vandermeulen D, Colchester A, Suetens P. Automated segmentation of multiple sclerosis lesions by model outlier detection. IEEE Trans Med Imag. 2001;20:677–688. doi: 10.1109/42.938237. [DOI] [PubMed] [Google Scholar]
- 43.Thirion JP. Image matching as diffusion process: An analogy with maxwell’s demons. Med Ima Anal. 1998;2:243–260. doi: 10.1016/s1361-8415(98)80022-4. [DOI] [PubMed] [Google Scholar]
- 44.Vercauteren T, Pennec X, Perchant A, Ayache N. Diffeomorphic demons: Efficient non-parametric image registration. Neuroimage. 2009;45:61–72. doi: 10.1016/j.neuroimage.2008.10.040. [DOI] [PubMed] [Google Scholar]
- 45.Jenkinson M, Bannister PR, Brady JM, Smith SM. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- 46.Zhong X, Dinggang S, Bilge K, Christos D. Statistical representation and simulation of high-dimensional deformations: Application to synthesizing brain deformations. Proc MICCAI. 2005:500–508. doi: 10.1007/11566489_62. [DOI] [PubMed] [Google Scholar]
- 47.Postelnicu G, Zollei L, Fischl B. Combined volumetric and surface registration. IEEE Trans Med Imag. 2008;28:508–522. doi: 10.1109/TMI.2008.2004426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crum WR, Rueckert D, Jenkinson M, Kennedy D, Smith SM. A framework for detailed objective comparison of non-rigid registration algorithms in neuroimaging. Proc MICCAI. 2004:679–686. [Google Scholar]
- 49.Konukoglu E, Clatz O, Bondiau PY, Delingette H, Ayache N. Extrapolating glioma invasion margin in brain magnetic resonance images: Suggesting new irradiation margins. Medical Image Analysis. 2010;14(2):111–125. doi: 10.1016/j.media.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 50.Tracqui P, et al. A mathematical model of glioma growth: The effect of chemotherapy on spatio-temporal growth. Cell Proliferation. 1995;28:17–31. doi: 10.1111/j.1365-2184.1995.tb00036.x. [DOI] [PubMed] [Google Scholar]