Abstract
Amelogenin is the major secretory product of ameloblasts and is critical for proper tooth enamel formation. Amelogenin isoforms and their cleavage products comprise over 80% of total secretory stage enamel protein. We have isolated and characterized four secreted amelogenin isoforms from developing porcine enamel : P190 (27-kDa), P173 (25-kDa), P132 (18-kDa) and P56 (6.5-kDa ; leucine rich amelogenin polypeptide or LRAP). P190 and P132 are low abundance amelogenins that contain a novel exon 4-encoded segment of lack the exon 3-encoded segment, respectively. P173 is the most abundant (major) amelogenin isoform. Cleavage of P173 by matrix metalloproteinase 20 (Mmp20) occurs at specific sites that generates a set of N-terminal cleavage products : P162 (23-kDa), P148 (20-kDa), P62/P63 (11-kDa), and Trp45 (6-kDa, tyrosine rich amelogenin polypeptide or TRAP). P148 is the most abundant protein in developing enamel and influences the conversion of amorphous calcium phosphate into hydroxyapatite in vitro. Mmp20 cleaves LRAP, the second abundant amelogenin isoform after Pro45 and Pro40. Processing by Mmp20 allows amelogenin cleavage products to serve separate functions. Over time, Mmp20 catalyzes additional cleavages that facilitate the progressive replacement of amelogenin by mineral, so enamel crystals thicken and widen with depth. Besides proteolytic processing, amelogenin protein-protein interactions are critical for function. Far-Western analyses demonstrate that the larger amelogenins (P173, P162, and P148) are only able to interact with larger amelogenins. No amelogenin-amelogenin interactions are observed for the smaller amelogenin cleavage products, TRAP or LRAP Amelogenin doesn’t interact with the 32-kDa glycosylated enamelin cleavage product, unless it it partially deglycosylated.
Keywords: tooth, enamel, amelogenin, amelogenesis imperfecta, protein-protein interaction
Introduction
Amelogenins are the most abundant and accumulate throughout the secretory stage enamel1,2). The amelogenin gene of 13 different mammalian species appears to residue either exclusively on the X-chromosome or jointly on the X- and Y-chromosomes3), and alternative splicing leads to the production of different amelogenin isoforms from a single primary RNA transcript4). Humans and pigs have two amelogenin genes ; mice have one. Approximately 90% of amelogenin mRNA expressed by human males is from the X chromosome5). AMELX mutations cause X-linked amelogenesis imperfecta6) and AmelX knock-out mice show enamel defects7), indicating that amelogenin is critical for proper dental enamel formation. Porcine amelogenin is expressed from both the X-and Y- chromosomes, although those genes encode secreted amelogenins with identical amino acid sequences8,9).
Many strategies have been used for the extraction, fractionation and isolation of amelogenins from developing teeth10,11). In the 1970’s, the non-dissociative extraction procedure has been developed at Tsurumi University12,13). This method was developed using the pig animal model to isolate and characterize amelogenins14,15) and later optimized for the efficient extraction of all enamel proteins and proteases16). Purification of porcine enamel proteins in quantity has fostered a better understanding of amelogenins and their roles in enamel biomineralization. Here we review our path to understanding the structure and proteolytic processing of porcine amelogenins, amelogenin-associated protein-protein interactions, and the possible function roles of amelogenin in enamel mineral formation.
Porcine Amelogenin Gene
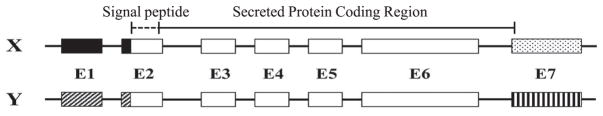
The cloning of porcine amelogenin cDNAs identified six isoforms (P190, P173, P157, P56, P41 and P40)17). The porcine amelogenin cDNA sequences showed nucleotide and amino acid variations within the signal peptide region encoded by exon 2, but not in the code for exons 3, 5, 6 and 7, which are included in the transcripts of the major amelogenin isoforms. The amelogenin cDNAs had two homologous but distinct exons 1 (1a and 1b), 2 (2a and 2b) and 7 (7a and 7b), which could be explained by the existence of two genes, or a single gene with alternative promoters and exons. Characterization of the porcine amelogenin genes showed that this pattern was due to there being X- and Y-chromosomal copies of the amelogenin gene9). The unusually high sequence homology between the two genes suggests that, in pig, the X and Y copies of the Amel locus have undergone gene conversion events that homogenized their sequences18).
Protein and PCR analyses show that a novel exon 4 is sometimes included in amelogenin mRNA transcripts and translated into protein (Fig. 1)19).
Fig. 1. Structure of porcine amelogenin genes.
Boxes indicate the 7 amelogenin exons. Unfilled boxes correspond to the amelogenin-coding region ; patterned boxes correspond to exonic non-coding regions. The DNA sequences in the X and Y copies of the porcine amelogenin genes are identical in the coding regions for the secreted protein (solid line), but differ in the coding regions for the signal peptides (dashed line).
Porcine Amelogenin Isoforms and Processing Pathway
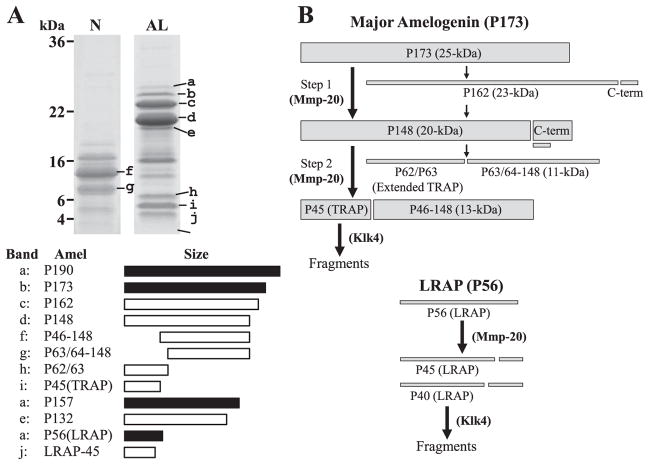
Four porcine amelogenin isoforms have been characterized at the protein level (Fig. 2A). The major porcine amelogenin (P173) has 173 amino acids and an apparent molecular weight on SDS-PAGE of 25-kDa. The second most abundant amelogenin isoform is the leucine-rich amelogenin polypeptide (LRAP)20) having 56 amino acids and an apparent molecular weight of 6.5-kDa. P190, the amelogenin isoform containing the novel exon 4-encoded segment has 190 amino acids and an apparent molecular weight of 27-kDa. The P157 amelogenin isoform lacks the exon 3-encoded segment. Its cleavage product lacking the C-terminal region has apparent molecular weight of 18-kDa (P132). These amelogenin isoforms undergo characteristic patterns of digestion by two extracellular proteases : Mmp2021) and kallikrein 4 (Klk4)22).
Fig. 2. Porcine amelogenin isoforms and cleavage products, and proteolytic processing pathway.
Based on the dissociative extraction method, ten amelogenin isoforms and their proteolytic cleavage products are observed on SDS-PAGE gel (N : extracts by Sorensen buffer (pH 7.4), AL : extracts by carbonate buffer (pH 10.8)). Of these amelogenins, P173 amelogenin isoform (b : P173) is a major amelogenin and its cleavage products (c : P162, d : P148, f : P46–148, g : P63/64–148, h : P62/63, i : P45 (TRAP)) are abundant during the secretory stage of enamel. The intact LRAP is found in the most outer layer enamel, but its cleavage product (j : LRAP-45) is usually detected in the mixture of outer and inner layer of enamel sample. P157 amelogenin isoform lacking exon 3 has not been identified, but its cleavage product (e : P132) has been characterized. P190 amelogenin isoform containing exon 4 encoded segment (a : P190) is the least abundant amelogenin. The major secreted amelogenin (P173) is usually processed in two steps (Fig. 2B). The initial cleavage is after Ser148 and generates P148, the most abundant amelogenin cleavage product in the matrix. The second cleavage is after Trp45 and generates tyrosine-rich amelogenin polypeptide (TRAP or P45) having an apparent molecular weight of 6-kDa and the 13-kDa amelogenins (P46–148). A minor processing pathway involves an initial cleavage after Pro162, generating the 23-kDa amelogenin (P162), which is cleaved again to generate P148. P148 is also cleaved in a less-often-used alternative pathway that generates extended TRAP having an apparent molecular weight of 7-kDa (P62/P63) and the 11-kDa amelogenin (P63/64–148). All these processing are caused by Mmp20 and Mmp20 showed little to no activity against TRAP LRAP (P56) is cleaved after Pro45 or Pro40 by Mmp20 to generate P45 (LRAP-45) and P40 (LRAP-40), and Mmp20 can not degrade any sites of LRAP other than them. The generated TRAP LRAP45 and LRAP-40 are fragmentated by Klk4 during the transition and maturation stage.
We recently tested the ability of Mmp20 and Klk4 to catalyze the cleavages that generate the P173 and LRAP cleavage products that accumulate in secretory stage23) and proved the processing pathway of porcine amelogenins (Fig. 2B). Mmp20 cleaves P173 after Trp45 and Ser148. In vivo, these cleavages generate the most abundant amelogenin cleavage products in porcine secretory-stage enamel : P148, TRAP, and the P46–148 amelogenins. Klk4 is not able to catalyze either of these cleavages. Mmp20 shows little or no activity against TRAP, whereas Klk4 degrades TRAP Mmp20 is also able to catalyze all of the cleavages that generate the less-abundant amelogenin cleavage products in secretory-stage enamel. That is, Mmp20 cleaves P173 after Pro162 and also after His62 and Ala63, that in vivo yields P162, "extended TRAP" (P62 and P63) and the 11-kDa (P63-148 and P64-148) amelogenins. Klk4 is not able to catalyze the cleavage of amelogenin sequences after Pro162 or Ala63, but catalyzes the cleavage after His62. However, cleavage after both His62 and Ala63 is used to generate the 11-kDa amelogenin. Mmp20 also cleaves LRAP after Pro45 (LRAP-45) and Pro40 (LRAP-40), whereas Klk4 does not cleave at these sites, but degrades LRAP by cleaving at other sites.
In situ hybridization studies have demonstrated that Mmp20 is expressed early in enamel formation, while Klk4 is not expressed until the transition stage2. Therefore, in developing pig teeth, Mmp20 is the only significant proteolytic activity in the enamel extracellular matrix during the secretory stage. It processes amelogenins into a group of cleavage products that accumulate and are only slowly degraded further by Mmp20. Both TRAP and LRAP accumulate because Mmp20 has little activity to degrade them. As Klk4 is not expressed during the secretory stage, it does not contribute to the processing of amelogenin. Klk4, however, is dedicated to its degradation during the transition and maturation stages.
Protein-Protein Interaction of Porcine Amelogenin
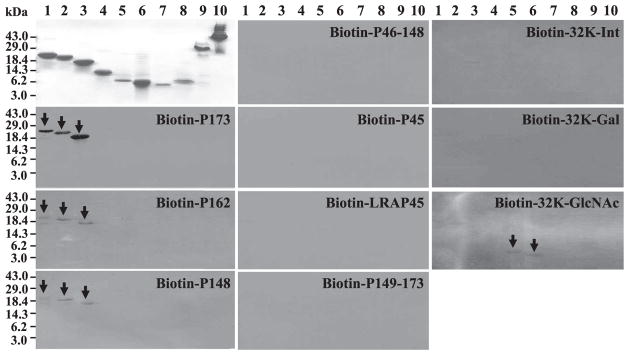
Besides proteolytic processing, amelogenin protein-protein interactions are also critical for structure and function. Amelogenin polymerizes into spherical structures named as “nanospheres” and it has believed to play a role in separating and spacing enamel crystallites25). Another model for amelogenin structure proposes that amelogenin "micelles" may form assemblies through ionic interactions involving their C-termini, which contain positively and negatively charged amino acids26,27). In the recent TEM study using porcine amelogenins, their self-assembly has showed the different state between the recombinant full-length porcine amelogenin (rP172) and the native porcine amelogenin cleavage product (P148) lacking the hydrophilic C-terminus of parent molecules28). The rP172 formed elongated assemblies of chain-like structures, while P148 exhibited no elongated structures, only loosely associated spherical particles. Thus, the hydrophilic C-terminus of amelogenin plays a necessary role in super-assembly. We assayed amelogenin protein-protein interaction using a Far-Western method. Intact porcine amelogenin (P173) and its cleavage products (P162, P148, P46–148, TRAP, P148–173), as well as LRAP-45 were biotiny-lated and purified. These biotin-labeled amelogenins were used to probe amelogenin protein-protein interactions by Far-Western blotting. The Far-Western result showed that only the larger biotin-labeled amelogenins (i. e. P173, P162 and P148) were able to interact with large amelogenins (Fig. 3). No amelogenin-amelogenin interactions were observed for the smaller amelogenin cleavage products or LRAP-45.
Fig. 3. Amelogenin-amelogenin and amelogenin—enamelin protein-protein interaction by Far-Western blots.
Coomassie Blue stained polyacrylamide gels are shown at the top of column 1. Replicas of these gels were transblotted to nitrocellulose and affinity stained using biotin-labed amelogenins or enamelins probes, the identity of which is provided at the top of each blot. Positive signals are indicated by arrows. Key to lanes : 1) P173, 2) P162, 3) P148, 4) P46–148, 5) P63, 6) P45 (TRAP), 7) LRAP-45, 8) P148–173, 9) intact 32-kDa enamelin, and 10) pig albumin.
Amelogenin is not glycosylated, but it has a domain called a tyrosine-rich N-terminal domain, which can bind to glycan chains having N-acetylglucosamine as its terminal sugar group29). We used porcine 32-kDa enamelin to study the amelogenin/32-kDa enamelin interactions. Enamelin is a glycoprotein that is secreted, along with amelogenins, into the enamel matrix. Its 32-kDa enamelin cleavage product, which is the most abundant accumulated enamelin in porcine secretory stage enamel, has three N-glycan chains that terminate with either sialic acid/galactose/N-acetylglucosamine or galactose/N-acetylglucosamine30,31). We purified 32-kDa enamelin and generated three enamelin molecules that varied only in the terminal group on their glycosidic chains. Intact 32-kDa enamelin(32-Int) has both sialic acid and β-galactose on its surface. Sialic acid was removed by neu-raminidase to give a product having only β-galactose on its surface (32-Gal). This product was further digested with β-galactosidase to give a product exposing N-acetylglucosamine on the terminal sugar of its glycosidic chains (32-GlcNAc). These three 32-kDa enamelins were conjugated to biotin and used for a Far-Western Blot analysis. Intact 32-kDa enamelin and β-galactose exposed 32-kDa enamelin did not interact with any of samples (Fig. 3)32). We conclude that amelogenins do not interact with the 32-kDa enamelin, and that the 32-kDa enamelin does not interact with itself. The N-acetylglucosamine exposed 32-kDa enamelin only interacted with smaller amelogenin cleavage products (P45 and P63). The results of our Far-Western analysis suggest that amelogenin assemblies are disrupted by the cleavages that generate P45 and P63, and that amelogenins released from these assemblies have increased affinity for specific glycoproteins.
Possible Function of Porcine Amelogenin
Amelogenin is the most abundant protein in secretory stage enamel and forms nanospheres that occupy the space between the mineral ribbons. The enamel ribbons slowly grow in width and thickness as amelogenins are degraded by Mmp20, so amelogenin functions in part, to maintain the appropriate space between crystals for crystal maturation27).
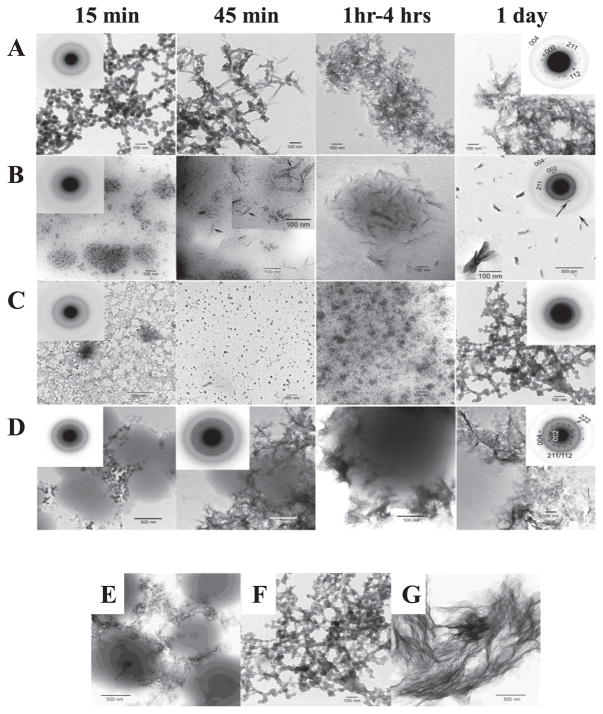
Knowing the primary structure of amelogenin and its processing by Mmp20 may enable amelogenin cleavage products to serve separate functions. Although enamel crystal ribbons elongate at a mineralization front where enamel proteins are secreted, the mineral particles in both outer and inner enamel layers are needle-shaped and assembled into parallel arrays, which is the typical organization of enamel mineral. Despite such similar appearance of the outer and inner enamel, the diffraction patterns of outer layer was similar to amorphous calcium phosphate (ACP), while those of inner layer was similar to low crystallinity carbonated hydroxyapatite (LCHA) suggesting that newly formed enamel mineral is ACP and it converts to LCHA33). To gain information how amelogenins affect mineral formation, our collaboration group identified mineral phase by TEM in selected area electron diffraction (SAED) mode using the recombinant full-length porcine amelogenin (rP172) and the recombinant and the native porcine amelogenin cleavage product (rP147 and P148)34). The result showed that monodispersed particles and/ or network of amorphous calcium phosphate (ACP) were observed in all samples at 15 min. After 1 day, the mineral phase in control, rP172, and rP147 solutions exhibited randomly arranged plates or bundles of aligned needles corresponding to the hydroxyapatite (HA) phase. In contrast, the mineral phase in P148 solution had still ACP phase after 1 day, suggesting P148 amelogenin can inhibit spontaneous calcium phosphate formation in vitro and stabilize ACP (Fig. 4A—D). Additionally, the mineral phase showed different patterns in the P148 and rP147 solutions. The difference between these two amelogenins is that P148 has the first methionine and is phosphorylation at Ser16. Therefore, the next objective was to test if phosphorylation of amelogenin affects ACP stability. Following the dephosphorylation from P148, the mineral phase in dephosphorylated P148 solution indicated bundles of plates corresponding to hydroxyapatite phase after 1 day (Fig. 4E—G). This finding supports that the phosphorylated N-terminal region of amelogenin binds to forming calcium phosphate nuclei and ACP and inhibits their transformation to HA.
Fig. 4. TEM micrographs of calcium phosphate mineral products formed in the presence of amelogenins examined at selected times (15 min, 45 min, 1 hour-4 hours, and 1 day), and the comparison with or without amelogenin phosphorylation examined after 1 day. (Courtesy Dr. H Margolis.).
As shown in the control (A), rP172 (B) and P148 (C) at 15 min, ACP was initially formed in the control and in the presence of rP172 and P148, based on the observed (insets) SAED patterns. In the presence of rP147 (D), both spherical particles and small plate-like crystals were observed, consistent with a somewhat diffuse SAED pattern. Subsequent changes in mineral particle shape and organization with time are described in the text. As described, in the presence of rP172 (B) ACP particles were observed (inset) to align and form needle-like particles at 45 min. After 1 day, based on SAED analyses, randomly arranged plate-like apatitic crystals were found in the control (A, inset, showing circular distribution of apatitic reflections), whereas aligned bundles of needle-like apatitic crystals were formed in the presence of rP172 (B, insets, showing a higher magnification of a bundle of aligned crystals and an arc-like diffraction pattern in the direction of the 002 reflection). In contrast, ACP was observed in the presence of P148 (C, inset), even after 1 day. In the presence of rP147 (D), crystal formation took place over time outside of observed protein masses. Randomly arranged plate-like HA crystals were observed at 1 day that were similar in these respects to those seen in the control, consistent with the SAED patterns obtained (D, 1 day, inset, showing circular distribution of apatitic reflections). Plate-like apatitic crystals that were slightly but significantly smaller than those found in the control were observed when mineralization was carried out in the presence of the non-phosphorylated rP147 (E), whereas bundles of significantly larger plate-like crystals were observed when mineralization was carried out using the partially dephosphorylated P148, dephoso-P148 (G). In sharp contrast, assemblies of spherical ACP particles were observed in the presence of P148 (F).
The anionic components of hydroxyapatite consist of PO43− and OH−. In aqueous solution, PO43− is in equilibrium with HPO42− and H2PO4−, while OH− is in equilibrium with H2O. During the formation of hydroxyapatite, deprotonation of H2PO4−, HPO42− and H2O occurs to replace PO43− and OH- that enter the crystal lattice. At pH = 7.4, an average of 11 protons are generated for each unit cell of hydroxyapatite that forms. Studies using recombinant mouse amelogenin (rM179) demonstrated that the histidine residues in amelogenin are able to absorb protons35). The result of proton binding showed that rM179 can absorb between 11 and 15 protons as the pH drops from 8 to 5 through 14 histidines in a single amelogenin molecule. The P148, which is most abundant amelogenin at the secretory stage of amelogenin, also has 14 histidine residues in one molecule. Therefore, it may also be able to absorb protons that are generated by secretory stage mineral formation, and may buffer enamel fluid in vivo.
Conclusion Remarks
By focusing on the purifying ample quantities of recombinant amelogenin or native amelogenin from developing teeth, characterizing its structure, its processing by proteases, its proton binding, its protein-protein interactions, and interactions with mineral, we have gained valuable information that allow us better understand its functional roles in enamel formation.
Acknowledgments
I thank the National Institute of Craniofacial and Dental Research of the National Institutes of Health (NIDCR/NIH) and The Japanese Ministry of Education, Culture, Sports, Science and Technology, Grant-in-Aid for Young Scientists. I also thank Dr. Kazuhiko Kawasaki at Pennsylvania State University for helpful comments and many principle investigators who have allowed us to collaborate with them and supported our research with their grant funding, including Drs. Margolis, Bartlett, Hu and Simmer.
References
- 1.Uchida T, Tanabe T, Fukae M, Shimizu M, Yamada M, Miake K, Kobayashi S. Immunochemical and immunohistochemical studies, using antisera against porcine 25 kDa amelogenin, 89 kDa enamelin and the 13–17 kDa nonamelogenins, on immature enamel of the pig and rat. Histochemistry. 1991;96 :129–138. doi: 10.1007/BF00315983. [DOI] [PubMed] [Google Scholar]
- 2.Tanabe T, Fukae M, Uchida T, Shimizu M. The localization and characterization of proteinases for the initial cleavage of porcine amelogenin. Calcif Tissue Int. 1992;51 :213–217. doi: 10.1007/BF00334549. [DOI] [PubMed] [Google Scholar]
- 3.Nakahori Y, Takenaka O, Nakagome Y. A human X-Y homologous region encodes amelogenin. Genomics. 1991;9 :264–269. doi: 10.1016/0888-7543(91)90251-9. [DOI] [PubMed] [Google Scholar]
- 4.Gibson CW, Golub E, Ding WD, Shimokawa H, Young M, Termine J, Rosenbloom J. Identification of the leucine-rich amelogenin peptide (LRAP) as the translation product of an alternatively spliced transcript. Biochem Biophys Res Commun. 1991;174 :1306–1312. doi: 10.1016/0006-291x(91)91564-s. [DOI] [PubMed] [Google Scholar]
- 5.Salido EC, Yen PH, Koprivnikar K, Yu LC, Shapiro LJ. The human enamel protein gene amelogenin is expressed from both the X and the Y chromosomes. Am J Hum Genet. 1992;50 :303–316. [PMC free article] [PubMed] [Google Scholar]
- 6.Lagerström M, Dahl N, Nakahori Y, Nakagome Y, Backman B, Landegren U, Pettersson U. A deletion in the amelogenin gene (AMG) causes X-linked amelogenesis imperfecta (AIH1) Genomics. 1991;10 :971–975. doi: 10.1016/0888-7543(91)90187-j. [DOI] [PubMed] [Google Scholar]
- 7.Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen E, Thyagarajan T, Sreenath T, Wright JT, Decker S, Piddington R, Harrison G, Kulkarni AB. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276 :31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- 8.Hu CC, Fukae M, Uchida T, Qian Q, Zhang CH, Ryu OH, Tanabe T, Yamakoshi Y, Murakami C, Dohi N, Shimizu M, Simmer JP. Cloning and characterization of porcine enamelin mRNAs. J Dent Res. 1997;76 :1720–1729. doi: 10.1177/00220345970760110201. [DOI] [PubMed] [Google Scholar]
- 9.Ikawa T, Kakegawa A, Nagano T, Ando H, Yamakoshi Y, Tanabe T, Simmer JP, Hu CC, Fukae M, Oida S. Porcine amelogenin is expressed from the X and Y chromosomes. J Dent Res. 2005;84 :144–148. doi: 10.1177/154405910508400207. [DOI] [PubMed] [Google Scholar]
- 10.Termine JD, Belcourt AB, Christner PJ, Conn KM, Nylen MU. Properties of dissociatively extracted fetal tooth matrix proteins. J Biol Chem. 1980;20 :9760–9768. [PubMed] [Google Scholar]
- 11.Fincham AG, Moradian-Oldak J, Simmer JP. The structural biology of the developing dental enamel matrix. J Struct Biol. 1999;126 :270–299. doi: 10.1006/jsbi.1999.4130. [DOI] [PubMed] [Google Scholar]
- 12.Fukae M, Ijiri H, Tanabe T, Shimizu M. Partial amino acid sequences of two proteins in developing porcine enamel. J Dent Res. 1979;58 :1000–1001. doi: 10.1177/002203457905800201011. [DOI] [PubMed] [Google Scholar]
- 13.Shimizu M, Tanabe T, Fukae M. Proteolytic enzyme in porcine immature enamel. J Dent Res. 1979;58 :782–789. doi: 10.1177/00220345790580023001. [DOI] [PubMed] [Google Scholar]
- 14.Fukae M, Shimizu M. Amino acid sequence of the main component of porcine enamel proteins. Jpn J Oral Biol. 1983;25 :29. [Google Scholar]
- 15.Yamakoshi Y, Tanabe T, Fukae M, Shimizu M. In: Amino acid sequence of porcine 25 kDa amelogenin. Tooth Enamel V, Fearnhead RW, editors. Florence Publishers; Tokyo: 1989. pp. 314–321. [Google Scholar]
- 16.Yamakoshi Y, Hu JC-C, Ryu OH, Tanabe T, Oida S, Fukae M, Simmer JP. A comprehensive strategy for purifying pig enamel proteins, in Biomineralization : formation, diversity, evolution and application. In: Kobayashi I, Ozawa H, editors. Proceedings of the 8th International Symposium on Biomineralization; Niigata, Jpn. Sept 25—28, 2001; Hadano, Jpn: Tokai University Press; 2003. pp. 326–332. [Google Scholar]
- 17.Hu CC, Bartlett JD, Zhang CH, Qian Q, Ryu OH, Simmer JP. Cloning, cDNA sequence, and alternative splicing of porcine amelogenin mRNAs. J Dent Res. 1996;75 :1735–1741. doi: 10.1177/00220345960750100501. [DOI] [PubMed] [Google Scholar]
- 18.Marais G, Galtier N. Sex chromosomes : how X-Y recombination stops. Curr Biol. 2003;13 :R641–643. doi: 10.1016/s0960-9822(03)00570-0. [DOI] [PubMed] [Google Scholar]
- 19.Hu CC, Ryu OH, Yamakoshi Y, Zhang CH, Cao X, Qian Q, Simmer JP. Pig amelogenin gene expresses a unique exon 4. Connect Tissue Res. 2002;43 :435–440. doi: 10.1080/03008200290001140. [DOI] [PubMed] [Google Scholar]
- 20.Fincham AG, Belcourt AB, Termine JD, Butler WT, Cothran WC. Dental enamel matrix : sequences of two amelogenin polypeptides. Biosci Rep. 1981;1 :771–778. doi: 10.1007/BF01114799. [DOI] [PubMed] [Google Scholar]
- 21.Bartlett JD, Simmer JP, Xue J, Margolis HC, Moreno EC. Molecular cloning and mRNA tissue distribution of a novel matrix metalloproteinase isolated from porcine enamel organ. Gene. 1996;183 :123–128. doi: 10.1016/s0378-1119(96)00525-2. [DOI] [PubMed] [Google Scholar]
- 22.Simmer JP, Fukae M, Tanabe T, Yamakoshi Y, Uchida T, Xue J, Margolis HC, Shimizu M, DeHart BC, Hu CC, Bartlett JD. Purification, characterization, and cloning of enamel matrix serine proteinase 1. J Dent Res. 1998;77 :377–386. doi: 10.1177/00220345980770020601. [DOI] [PubMed] [Google Scholar]
- 23.Nagano T, Kakegawa A, Yamakoshi Y, Tsuchiya S, Hu JC, Gomi K, Arai T, Bartlett JD, Simmer JP. Mmp-20 and Klk4 cleavage site preferences for amelogenin sequences. J Dent Res. 2009;88 :823–828. doi: 10.1177/0022034509342694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu JC, Sun X, Zhang C, Liu S, Bartlett JD, Simmer JP. Enamelysin and kallikrein-4 mRNA expression in developing mouse molars. Eur J Oral Sci. 2002;110 :307–315. doi: 10.1034/j.1600-0722.2002.21301.x. [DOI] [PubMed] [Google Scholar]
- 25.Fincham AG, Moradian-Oldak J, Simmer JP, Sarte P, Lau EC, Diekwisch T, Slavkin HC. Self-assembly of a recombinant amelogenin protein generates supramolecular structures. J Struct Biol. 1994;112 :103–109. doi: 10.1006/jsbi.1994.1011. [DOI] [PubMed] [Google Scholar]
- 26.Fukae M, Yamamoto R, Karakida T, Shimoda S, Tanabe T. Micelle structure of amelogenin in porcine secretory enamel. J Dent Res. 2007;86 :758–763. doi: 10.1177/154405910708600814. [DOI] [PubMed] [Google Scholar]
- 27.Fukae M. Enamel formation biochemical aspects. J Oral Biosci. 2009;51 :46–60. [Google Scholar]
- 28.Wiedemann-Bidlack FB, Beniash E, Yamakoshi Y, Simmer JP, Margolis HC. pH triggered self-assembly of native and recombinant amelogenins under physiological pH and temperature in vitro. J Struct Biol. 2007;160:57–69. doi: 10.1016/j.jsb.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ravindranath R, Moradian-Oldak J, Fincham A. Tyrosyl motif in amelogenins binds N-acetyl-D-glucosamine. J Biol Chem. 1999;274 :2464–2471. doi: 10.1074/jbc.274.4.2464. [DOI] [PubMed] [Google Scholar]
- 30.Yamakoshi Y. Carbohydrate moieties of porcine 32 kDa enamelin. Calcif Tissue Int. 1995;56 :323–330. doi: 10.1007/BF00318054. [DOI] [PubMed] [Google Scholar]
- 31.Yamakoshi Y, Pinheiro FH, Tanabe T, Fukae M, Shimizu M. Sites of asparagine-linked oligosaccharides in porcine 32 kDa enamelin. Connect Tissue Res. 1998;39 :39–46. doi: 10.3109/03008209809023910. [DOI] [PubMed] [Google Scholar]
- 32.Yamakoshi Y, Hu J-C, Fukae M, Tanabe T, Oida S, Simmer J. Amelogenin and 32 kDa enamelin protein-protein interactions. In: Kobayashi I, Ozawa H, editors. Biomineralization (Biom 2001) formation, diversity, evolution and application. Tokai University Press; Kanagawa, Jpn: 2004. pp. 338–342. [Google Scholar]
- 33.Beniash E, Metzler RA, Lam RS, Gilbert PU. Transient amorphous calcium phosphate in forming enamel. J Struct Biol. 2009;166 :133–143. doi: 10.1016/j.jsb.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwak SY, Wiedemann-Bidlack FB, Beniash E, Yamakoshi Y, Simmer JP, Litman A, Margolis HC. Role of 20-kDa amelogenin (P148) phosphorylation in calcium phosphate formation in vitro. J Biol Chem. 2009;284 :18972–18979. doi: 10.1074/jbc.M109.020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryu OH, Hu CC, Simmer JP. Biochemical characterization of recombinant mouse amelogenins : protein quantitation, proton absorption, and relative affinity for enamel crystals. Connect Tissue Res. 1998;38 :207–214. doi: 10.3109/03008209809017038. [DOI] [PubMed] [Google Scholar]