Abstract
Averaged estimates of RNA polymerase II (RNAPII) elongation rates in mammalian cells have been shown to range between 1.3 and 4.3 kb min−1. In this work, nascent RNAs from an integrated human immunodeficiency virus type 1-derived vector were detectable at the single living cell level by fluorescent RNA tagging. At steady state, a constant number of RNAs was measured corresponding to a minimal density of polymerases with negligible fluctuations over time. Recovery of fluorescence after photobleaching was complete within seconds, indicating a high rate of RNA biogenesis. The calculated transcription rate above 50 kb min−1 points towards a wide dynamic range of RNAPII velocities in living cells.
Keywords: transcription, RNA polymerase II, live imaging; HIV, FRAP
Introduction
Eukaryotic transcription elongation by RNA polymerase II (RNAPII) has been measured in various ways as summarized in Ardehali & Lis (2009). Early experiments calculated heterogeneous nuclear RNA transcription in HeLa cells to occur at a rate of 3–6 kilobases (kb) per minute exploiting radioisotope pulse labelling of RNA (Sehgal et al, 1976). Work performed on the Drosophila Ubx and heat-shock genes (Shermoen & O'Farrell, 1991; O'Brien & Lis, 1993) or the giant dystrophin gene (Tennyson et al, 1995) involved averaged estimates on cell populations that yielded apparent elongation rates ranging from 1.1 to 2.5 kb min−1. Single-molecule experiments could be conducted only for prokaryotic and yeast RNA polymerases. In these cases, the average rates of elongation could be calculated in the range of 0.6–1.2 kb min−1 with variable pauses (Schafer et al, 1991; Yin et al, 1995; Wang et al, 1998; Galburt et al, 2007). More recently, bulk analysis of the first wave of transcription was obtained on several endogenous genes by quantitative reverse transcription–polymerase chain reaction (RT–PCR; 3.8 kb min−1; Singh & Padgett, 2009) or the use of tiling arrays (3.1 kb min−1; Wada et al, 2009). These studies provide the most reliable estimate of the average transcription rate on typical mammalian genes.
Pioneering studies by Singer and collaborators used quantitative RNA in situ hybridization at the serum-inducible rat β-actin gene in single cells to measure an average transcription rate of 1.1–1.4 kb min−1 (Femino et al, 1998). Then an RNA reporter system that uses the coat protein of phage MS2 fused to the green fluorescent protein was developed (Bertrand et al, 1998). By insertion of several MS2-binding sites (MS2 BS) in an inducible reporter gene, it has been possible to monitor dynamics of messenger RNAs (mRNAs) in a living cell (Forrest & Gavis, 2003; Golding & Cox, 2004; Shav-Tal et al, 2004; Le et al, 2005). However, to measure transcription elongation in mammalian cells, it has been necessary to artificially insert tandem arrays of an inducible transcription unit at a single chromatin location to gain sensitivity (Janicki et al, 2004). In this system, a maximum elongation rate of 4.3 kb min−1 was estimated (Darzacq et al, 2007). With a similar approach, on a construct based on the human immunodeficiency virus type 1 (HIV1), an elongation rate of 1.8 kb min−1 was obtained (Boireau et al, 2007). However, both studies relied on the insertion of tandem arrays of the constructs, between 75 (Boireau et al, 2007) and 200 copies (Darzacq et al, 2007), an artificial situation that has also been shown to induce heterochromatin at the site of repeats (Wang et al, 2006). Such pioneering studies could only open the way for further analysis to be carried out on single genes in single mammalian cells (Raj et al, 2006; Yunger et al, 2010; Lionnet et al, 2011).
Here we provide evidence of highly efficient transcription rates from an integrated lentiviral vector, as well as a detailed mathematical analysis for the interpretation of the new data. We also show that transcription from a single integration site exceeds >20-fold that measured from tandem arrays of the same transcription unit. The results highlight the problems of using large arrays in interpreting kinetic data.
Results And Discussion
Among retroviruses, which naturally integrate their genome in the host cell, HIV1 maintains a robust transcription in the presence of the viral Tat transactivator (Marcello et al, 2001). Therefore, cell lines harbouring a single integrated Tat-inducible lentiviral vector (supplementary Fig S1B online) represent an ideal tool to study RNA biogenesis from single transcription units in single living cells (Molle et al, 2007; De Marco et al, 2008; Maiuri et al, 2011). The HOS_A4 osteosarcoma cell line used throughout this study carries a single integrated inducible provirus with BSs for the MS2 protein for RNA tracking in living cells (see supplementary information online and De Marco et al (2008) for details). MS2 fused to the autofluorescent protein enhanced yellow fluorescent protein (EYFP–MS2nls) was used to monitor chimeric RNAs at the transcription site in HOS_A4 cells. We measured the brightness of the transcription spot and found that it was two- to threefold brighter, depending on the cell, than the surrounding nucleoplasm (Figs 1A and 2B). This indicates that 50–75% of the signal measured at this site is specific to transcription, providing a direct reflection of the amount of MS2 BS that are synthesized in the process. As a second test of whether this bright spot reflects transcription, we also treated cells with actinomycin D (Act-D) and camptothecin. This led to a reversible loss of the bright transcription sites in most nuclei (supplementary Fig S1A online). The remaining nuclei still showed a spot of comparable brightness to control cells, suggesting an all-or-none effect of the inhibitors.
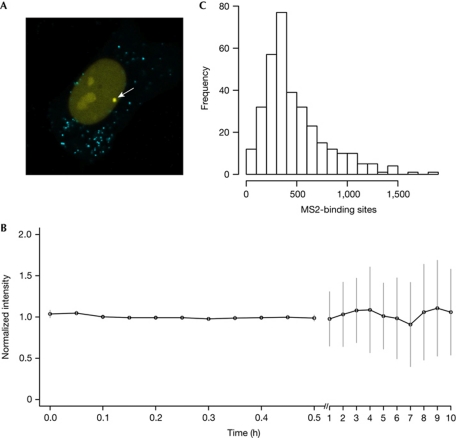
Figure 1.
Quantitative analysis of transcription from a single retroviral transcription unit integrated in a single living cell. (A) The HOS_A4 cell line. When the HOS_A4 clone was activated with Tat, a bright signal (arrow) was clearly visible in the nucleoplasm corresponding to nascent transcripts. The reporter ECFPskl localized to cytoplasmic peroxisomes. (B) Continuous transcription in single cells. The intensity of enhanced yellow fluorescent protein (EYFP) fluorescence at the transcription site was monitored in HOS_A4-expressing MS2–EYFPnls and activated by Tat for 30 min at 5-min intervals. Each acquisition was normalized to the intensity at time zero. Therefore, the figure represents the variability of intensity at steady state independently of the number of polymerases/nascent RNAs. The analysis was then extended for 10 h (N=15; error bars show s.d.). (C) Distribution and quantification of transcripts. Distribution of the number of MS2-binding sites at the transcription site at the steady state was measured in HOS_A4 cells activated by Tat by quantitative in situ hybridization (Boireau et al, 2007; N=337). Data were collected on different transcription sites at the same time point and therefore depend on the number of polymerases/nascent RNAs.
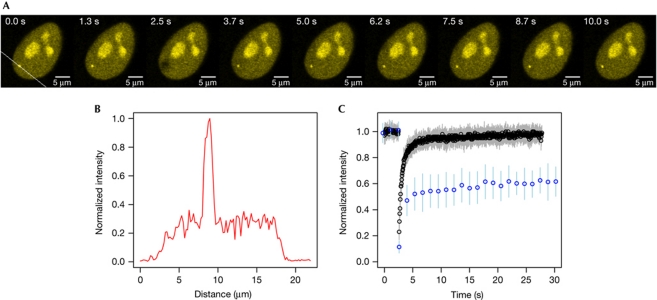
Figure 2.
FRAP analysis of the transcription site. (A) Series of frames from a typical fluorescence recovery after photobleaching (FRAP) experiment on HOS_A4 cells. Selected frames from supplementary Video S1 online taken at the indicated time points before and after the bleach are shown. (B) Fluorescence intensity profile of the transcription site. The intensity profile was measured along the white line across the transcription spot of HOS_A4 shown in the first frame of A. (C) Comparative analysis of FRAP recovery curves from single integrations and tandem arrays. Cells carrying a single integration (black circles, N=30; ±s.d., grey bars) or tandem arrays of the same construct (blue circles, N=36; ±s.d., blue bars) were bleached and recovery was monitored up to 30 s. Full recovery of tandem arrays is shown in supplementary Fig S4A online.
Given the evidence that this signal derives from transcription, we then continuously measured the intensity of the EYFP–MS2nls signal at this site in Tat-activated living HOS_A4 (Fig 1A). As shown in Fig 1B, little variation was observed in the intensity associated with the transcription sites for up to 10 h. The bursts or pulses observed in other systems were not detected (Golding et al, 2005; Chubb et al, 2006). These results indicate that, when the viral transactivator is not limiting, HIV1 transcription is continuously active and might be a mechanism to ensure high yields of virus production.
The minimum concentration of polymerases at the transcription site was estimated by a quantitative in situ hybridization on nascent RNAs (RNA ISH; Fusco et al, 2003; Boireau et al, 2007). At steady state, 476±17 MS2 BS, corresponding to 20±1 nascent RNAs, were counted on the transcription site assuming 100% probe-binding efficiency (Fig 1C). Considering that the length of the gene, from the MS2 repeats to the poly(A), is 5,406 bp, the density of equally spaced polymerases would be one for every 270 bp, which is consistent with previous observations on highly transcribed genes (O'Brien & Lis, 1993; Femino et al, 1998) and approaching ribosomal genes (Dundr et al, 2002).
Next, the kinetic behaviour of HIV1 RNA biogenesis from a single transcription unit was explored. To this end, the recovery of EYFP–MS2nls fluorescence bound to HIV RNA was measured after photobleaching. The signal from transcribed RNA was clearly visible above the background (Fig 2A,B) and recovered rapidly, regaining pre-bleach equilibrium after 10 s (supplementary Video S1 online; Fig 2A,C). This finding was unexpected as previous observations made on tandem arrays showed complete recovery only after more than 200 s (Boireau et al, 2007; Darzacq et al, 2007). To compare directly the same construct in the two configurations, we used a stable cell line carrying 35 tandem arrays of the same viral transcription unit (see supplementary information online and Kula et al (2011)). The number of nascent RNAs at the transcription site was also calculated by RNA quantitative ISH to be 54±16. When the same construct was compared directly either as a single integrated provirus or in tandem arrays, the former recovered 10 times more quickly than the latter (supplementary Video S2 online; Fig 2C; supplementary Fig S4A,B online). Furthermore, the fast recovery of the single integration was not an intrinsic property of the HOS_A4 cell clone used, as other clones integrated at different positions showed similar recovery curves (Fig 3A).
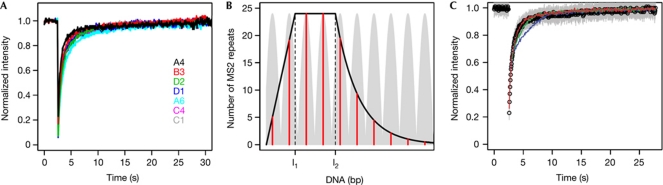
Figure 3.
An estimate of RNAPII transcription rates. (A) Fluorescence recovery after photobleaching (FRAP) of nascent transcripts from single integrations. Seven HOS clones obtained as described in De Marco et al (2008) were subjected to FRAP analysis after Tat induction. Each of them carries single integrations of the construct mapping in different sites of the genome. The line is the average over 10–40 cells for each clone. (B) The TranWave model. The diagram shows a representation of the TranWave model: a movie of the same is available as supplementary Video S3 online. Light grey indicates the positive travelling wave, black indicates the amplitude and red indicates the position of the sliding maxima. Each red bar represents an RNA polymerase II (RNAPII). L1 and L2 mark the boundaries of the transcribed region after the MS2-binding sites (MS2 BS; L1) and the SA7 (L2). (C) Monte Carlo coupled to TranWave simulations for single integrations. Simulations of different transcription rates at 25 kb min−1 (blue line), 50 kb min−1 (green line) and 100 kb min−1 (red line) with a fixed total number of MS2 BS and a fixed RNA-processing time determined experimentally as shown in Fig 1C and in supplementary Fig S4C online.
A lower-bound estimate of the transcription rate could be calculated by dividing the length of the gene (5,406 bp) for the fluorescence recovery after photobleaching (FRAP) recovery time (time to 95% recovery: 4 s for the single integration, 320 s for the tandem array). The full length of the gene was considered because the MS2-tagged transcript is not released until RNAPII reaches the end of the gene. This crude estimate of ∼80 kb min−1 for the single integration and 1 kb min−1 for several integrations clearly showed a huge difference of the transcription rate for the same transcription unit in the two configurations.
The preceding calculation ignores many important parameters such as processing of the RNA and diffusion of fluorescent proteins into and through the area of the bleach. To address these factors, we developed a new computational model (TranWave) to describe the equilibrium between production and processing of RNA at the transcription site (see supplementary information online for details). TranWave predicts the number of MS2 BS that are made available by RNAPII and measures the transcription rate of pre-mRNA synthesis. Assuming that the initiation rate and the transcription rate of RNAPII are constant, each polymerase sliding along the gene could be considered as the peak of a harmonic positive travelling wave (Fig 3B; supplementary Video S3 online). The amplitude of the wave increases linearly during the synthesis of the MS2 BS, is constant until the RNA is processed, and then decreases exponentially. Hence, the transcription rate of RNAPII could be considered as the propagation speed of the wave, which is the maximal velocity. When the transcription site is photobleached, the newly synthesized MS2 BS are instantaneously and irreversibly bound by the available free EYFP–MS2nls (see supplementary information online; supplementary Fig S2B,C online: Johansson et al, 1998; Boireau et al, 2007; Darzacq et al, 2007). In addition, the diffusive behaviour of EYFP–MS2nls in the nucleoplasm was studied using the implemented diffusion-binding model (Sprague et al, 2004; Mueller et al, 2008; supplementary Fig S3A online), and a two-dimensional Monte Carlo (MC) simulation was developed to describe the EYFP–MS2nls protein availability at the transcription site after the photobleaching (supplementary Fig S3B online). TranWave was initially tested for the cells carrying tandem arrays, and the transcription rate measured was 1.6±0.1 kb min−1, which is consistent with previous findings (supplementary Fig S4A,B online; Boireau et al, 2007). Finally, it was possible to analyse the transcriptional kinetics of the single integration. Knowing the number of MS2 BS at steady state at the transcription site (Fig 1C) and the mean processing time of the repeats measured by inverse FRAP (iFRAP) (0.73±0.04 s; supplementary Fig S4C online, consistent with the quick loss of transcript at the transcription site shown in supplementary Fig S1C online), it became possible to run the MC/TranWave simulations for three different transcription rates, 25, 50 and 100 kb min−1 (Fig 3C; supplementary Fig S6A,B online). The FRAP data were consistent with elongation rates above 50 kb min−1 and approximating 100 kb min−1. Possible imprecision on the polymerase counts, due to suboptimal probe binding in RNA quantitative ISH, or variations in the mean RNA processing time at the site of transcription, did not affect the model significantly (supplementary Fig S5 online). Thus, our estimates from a variety of FRAP analyses indicate an elongation rate that exceeds current estimates >20-fold (Ardehali & Lis, 2009), approaching values calculated only for certain DNA helicases and translocases (Bianco et al, 2001; Saleh et al, 2004).
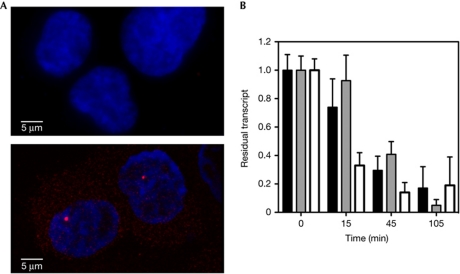
To confirm these findings, we performed an alternative test to estimate the elongation rate at this single-copy gene. We measured a total number of 1,113±353 mRNA particles for each cell at steady state by in situ hybridization on the whole cell volume (Fig 4A). We also measured the rate of RNA degradation in these cells by treating them with Act-D and then performing quantitative RT–PCR over time (Fig 4B). We found a degradation rate with a half time of 30 min. To maintain a steady-state concentration of 1,113±353 RNAs per cell, the transcription process must produce new RNAs every 1.2–2.4 s, approximately. This production time can be further subdivided into actual elongation and processing. The processing time was calculated by iFRAP to be equal to 0.73±0.04 s (supplementary Fig S4C online). Therefore, the 270 bp between two processing polymerases are transcribed in 0.4–1.6 s, resulting in elongation rates between 10 and 35 kb min−1. Furthermore, our calculation does not account for the fact that after the 1-h Act-D treatment some nuclei still show transcriptionally active spots, thus contributing to the residual RNA signal (supplementary Fig S1A online), and therefore result in an underestimation of the transcription rate.
Figure 4.
(A) RNA in situ hybridization of transcripts. The lentiviral transcript was detected by RNA in situ hybridization in HOS_A4 cells activated by Tat. Two cells are shown with nuclei stained with 4,6-diamidino-2-phenylindole (blue) and messenger RNA (mRNA) stained by Cy3 (red). The transcription site within the nucleus is clearly visible together with granules of mRNA in the nucleus and in the cytoplasm that could be quantified. Cells that where not activated are shown above as background control. (B) Decay rate of mRNA. HOS_A4 cells activated by Tat were treated with actinomycin D (Act-D; 10 μg ml−1) to inhibit transcription. Data are presented as residual transcripts at the indicated time points after Act-D treatment (±s.d.). Black bars in the histogram represent values for the primers mapping in the 5′-end of the transcript, grey bars for those detecting the ECFP gene. The unspliced pre-mRNAs (white bars) are highly unstable on transcription inhibition as expected.
In conclusion, we have shown here that RNA synthesis at a single-copy gene is significantly faster than at a tandem array. The tandem arrays have been widely used to study transcription processes in living cells; thus, our results indicate that behaviour at single-copy genes might be considerably different. Estimating kinetic parameters from the data of live cells is a complex process that is still under development. Thus, the elongation rates measured in this study, or in any of the preceding studies, might be imprecise. However, we also provided an independent elongation estimate for this single-copy gene that was in qualitative agreement with the kinetic calculations. This suggests that single-copy genes might show much faster elongation than previously expected. The high rate of transcription might be peculiar to the Tat/HIV1 axis, allowing efficient productive infection after induction of the latent provirus (Marcello, 2006). Chromatin conformation at the site of insertion (Lusic et al, 2003), as well as subnuclear positioning at the periphery of the nucleus (Dieudonné et al, 2009), might account for increased elongation and processing, providing a favourable environment at the site of transcription (Marcello et al, 2010).
Only recently a report measured transcription rates from a single engineered Flp recombinase target genomic locus (Yunger et al, 2010). The calculated transcription rate of 0.3–0.8 kb min−1 was in the lower range of previous measurements. The observation that RNAPII transcription rates can vary extensively adds a level of RNAPII plasticity for the control of transcription genome-wide. Specific gene loci might require increased transcription rates in response to various stimuli. In optimized conditions, these rates can approximate those evolved by a selfish virus such as HIV1. However, direct measurements on several endogenous alleles will be required for a full grasp of the dynamic range of RNAPII elongation in living cells.
Methods
Cell lines were described previously (De Marco et al, 2008; Kula et al, 2011). For live-cell experiments, cells were plated on glass-bottom plates and analysed at 37°C in a 5% CO2 humidified atmosphere in a non-fluorescent complete medium. For the time-series acquisitions, a stack of 21 images, 0.5 μm apart, was collected using a wide-field Leica DMRI inverted microscope ( × 63 objective, NA 1.3) controlled by Metamorph (Universal Imaging) and equipped with a CoolSnap K CCD camera (Roper scientific). The stacks were deconvolved with the ImageJ (http://rsb.info.nih.gov/ij/) plug-in ‘Iterative Deconvolve 3D’ (Dougherty, 2005). The transcription spot was tracked in two-dimensions (2D) with the ‘SpotTracker’ plug-in (Sage et al, 2005). FRAP experiments were conducted with a Zeiss confocal LSM510 Meta microscope ( × 63 objective, NA 1.4; Maiuri et al, 2011). Images of 128 × 128 pixels (24.4 × 24.4 μm) and optical thickness of 1.5 μm were acquired using <1% of the laser power. A circle with a diameter of 2.28 μm was bleached at 514 nm for 10 passages in <100 ms using a full-power laser. The bleached spots were tracked in 2D as above. For the data analysis, background was subtracted and fluorescence intensity was normalized as described in Phair & Misteli (2000). RNA ISH and RT–PCR were performed essentially as previously described (Boireau et al, 2007; Kula et al, 2011). Stack images were acquired with the wide-field microscope and deconvolved as described above. The transcription spots were identified and measured in 3D with the ImageJ plug-in ‘3D Object Counter’ (Bolte & Cordelieres, 2006). For the MC simulation, the random number generator of the C++ GSL library was used and a C++ script was written for the simulation. In addition, to compute the TranWave model, a C++ program was written. All the graphs have been made in R.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
This work was supported in part by an Human Frontier Science Program (HFSP) Young Investigators Grant, by the European Community FP6 STREP no. 012182, by the AIDS Program of the Istituto Superiore di Sanità of Italy and by Beneficentia Stiftung.
Author contributions: P.M. performed experiments with the help of A.K., A.DeM., A.K. and A.M. P.M. developed the TranWave with contributions from D.M. and J.G.M. P.M. and A.M. designed the experiments and analysed the data. A.M. conceived the study and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ardehali MB, Lis JT (2009) Tracking rates of transcription and splicing in vivo. Nat Struct Mol Biol 16: 1123–1124 [DOI] [PubMed] [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM (1998) Localization of ASH1 mRNA particles in living yeast. Mol Cell 2: 437–445 [DOI] [PubMed] [Google Scholar]
- Bianco PR, Brewer LR, Corzett M, Balhorn R, Yeh Y, Kowalczykowski SC, Baskin RJ (2001) Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature 409: 374–378 [DOI] [PubMed] [Google Scholar]
- Boireau S, Maiuri P, Basyuk E, de la Mata M, Knezevich A, Pradet-Balade B, Bäcker V, Kornblihtt A, Marcello A, Bertrand E (2007) The transcriptional cycle of HIV-1 in real-time and live cells. J Cell Biol 179: 291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, Cordelieres FP (2006) A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224: 213–232 [DOI] [PubMed] [Google Scholar]
- Chubb JR, Trcek T, Shenoy SM, Singer RH (2006) Transcriptional pulsing of a developmental gene. Curr Biol 16: 1018–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH (2007) In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol 14: 796–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marco A, Biancotto C, Knezevich A, Maiuri P, Vardabasso C, Marcello A (2008) Intragenic transcriptional cis-activation of the human immunodeficiency virus 1 does not result in allele-specific inhibition of the endogenous gene. Retrovirology 5: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieudonné M, Maiuri P, Biancotto C, Knezevich A, Kula A, Lusic M, Marcello A (2009) Transcriptional competence of the integrated HIV-1 provirus at the nuclear periphery. EMBO J 28: 2231–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty RP (2005) Extensions of DAMAS and benefits and limitations of deconvolution in beamforming. AIAA Paper 2005–2961 [Google Scholar]
- Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, Phair RD, Misteli T (2002) A kinetic framework for a mammalian RNA polymerase in vivo. Science 298: 1623–1626 [DOI] [PubMed] [Google Scholar]
- Femino AM, Fay FS, Fogarty K, Singer RH (1998) Visualization of single RNA transcripts in situ. Science 280: 585–590 [DOI] [PubMed] [Google Scholar]
- Forrest KM, Gavis ER (2003) Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol 13: 1159–1168 [DOI] [PubMed] [Google Scholar]
- Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E (2003) Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol 13: 161–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galburt EA, Grill SW, Wiedmann A, Lubkowska L, Choy J, Nogales E, Kashlev M, Bustamante C (2007) Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature 446: 820–823 [DOI] [PubMed] [Google Scholar]
- Golding I, Cox EC (2004) RNA dynamics in live Escherichia coli cells. Proc Natl Acad Sci USA 101: 11310–11315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding I, Paulsson J, Zawilski SM, Cox EC (2005) Real-time kinetics of gene activity in individual bacteria. Cell 123: 1025–1036 [DOI] [PubMed] [Google Scholar]
- Janicki SM et al. (2004) From silencing to gene expression: real-time analysis in single cells. Cell 116: 683–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson HE, Dertinger D, LeCuyer KA, Behlen LS, Greef CH, Uhlenbeck OC (1998) A thermodynamic analysis of the sequence-specific binding of RNA by bacteriophage MS2 coat protein. Proc Natl Acad Sci USA 95: 9244–9249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kula A, Guerra J, Knezevich A, Kleva D, Myers MP, Marcello A (2011) Characterization of the HIV-1 RNA associated proteome identifies Matrin 3 as a nuclear cofactor of Rev function. Retrovirology 8: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TT, Harlepp S, Guet CC, Dittmar K, Emonet T, Pan T, Cluzel P (2005) Real-time RNA profiling within a single bacterium. Proc Natl Acad Sci USA 102: 9160–9164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionnet T, Czaplinski K, Darzacq X, Shav-Tal Y, Wells AL, Chao JA, Park HY, de Turris V, Lopez-Jones M, Singer RH (2011) A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat Methods 8: 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusic M, Marcello A, Cereseto A, Giacca M (2003) Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter. EMBO J 22: 6550–6561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri P, Knezevich A, Bertrand E, Marcello A (2011) Real-time imaging of the HIV-1 transcription cycle in single living cells. Methods 53: 62–67 [DOI] [PubMed] [Google Scholar]
- Marcello A (2006) Latency: the hidden HIV-1 challenge. Retrovirology 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcello A, Dhir S, Dieudonné M (2010) Nuclear positional control of HIV transcription in 4D. Nucleus 1: 8–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcello A, Zoppé M, Giacca M (2001) Multiple modes of transcriptional regulation by the HIV-1 Tat transactivator. IUBMB Life 51: 175–181 [DOI] [PubMed] [Google Scholar]
- Molle D, Maiuri P, Boireau S, Bertrand E, Knezevich A, Marcello A, Basyuk E (2007) A real-time view of the TAR:Tat:P-TEFb complex at HIV-1 transcription sites. Retrovirology 4: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller F, Wach P, McNally JG (2008) Evidence for a common mode of transcription factor interaction with chromatin as revealed by improved quantitative fluorescence recovery after photobleaching. Biophys J 94: 3323–3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien T, Lis JT (1993) Rapid changes in Drosophila transcription after an instantaneous heat shock. Mol Cell Biol 13: 3456–3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair RD, Misteli T (2000) High mobility of proteins in the mammalian cell nucleus. Nature 404: 604–609 [DOI] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S (2006) Stochastic mRNA synthesis in mammalian cells. PLoS Biol 4: e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage D, Neumann FR, Hediger F, Gasser SM, Unser M (2005) Automatic tracking of individual fluorescence particles: application to the study of chromosome dynamics. IEEE Trans Image Process 14: 1372–1383 [DOI] [PubMed] [Google Scholar]
- Saleh OA, Pérals C, Barre FX, Allemand JF (2004) Fast, DNA-sequence independent translocation by FtsK in a single-molecule experiment. EMBO J 23: 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DA, Gelles J, Sheetz MP, Landick R (1991) Transcription by single molecules of RNA polymerase observed by light microscopy. Nature 352: 444–448 [DOI] [PubMed] [Google Scholar]
- Sehgal PB, Derman E, Molloy GR, Tamm I, Darnell JE (1976) 5,6-Dichloro-1-β-D-ribofuranosylbenzimidazole inhibits initiation of nuclear heterogeneous RNA chains in HeLa cells. Science 194: 431–433 [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, Spector DL, Singer RH (2004) Dynamics of single mRNPs in nuclei of living cells. Science 304: 1797–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shermoen AW, O'Farrell PH (1991) Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell 67: 303–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Padgett RA (2009) Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol 16: 1128–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague BL, Pego RL, Stavreva DA, McNally JG (2004) Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys J 86: 3473–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennyson CN, Klamut HJ, Worton RG (1995) The human dystrophin gene requires 16 h to be transcribed and is cotranscriptionally spliced. Nat Genet 9: 184–190 [DOI] [PubMed] [Google Scholar]
- Wada Y et al. (2009) A wave of nascent transcription on activated human genes. Proc Natl Acad Sci USA 106: 18357–18361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Koyama N, Nishida H, Haraguchi T, Reith W, Tsukamoto T (2006) The assembly and maintenance of heterochromatin initiated by transgene repeats are independent of the RNA interference pathway in mammalian cells. Mol Cell Biol 26: 4028–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J, Block SM (1998) Force and velocity measured for single molecules of RNA polymerase. Science 282: 902–907 [DOI] [PubMed] [Google Scholar]
- Yin H, Wang MD, Svoboda K, Landick R, Block SM, Gelles J (1995) Transcription against an applied force. Science 270: 1653–1657 [DOI] [PubMed] [Google Scholar]
- Yunger S, Rosenfeld L, Garini Y, Shav-Tal Y (2010) Single-allele analysis of transcription kinetics in living mammalian cells. Nat Methods 7: 631–633 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.