Abstract
Bimolecular fluorescence complementation was used to engineer CD8 molecules so that CD8αα and CD8αβ dimers can be independently visualized on the surface of a T cell during antigen recognition. Using this approach, we show that CD8αα is recruited to the immunological synapse almost as well as CD8αβ, but because the kinase Lck associates preferentially with CD8αβ in lipid rafts, CD8αα is the weaker co-receptor. During recognition of the strong CD8αα ligand H2-TL, CD8αα is preferentially recruited. Thus, recruitment of the two CD8 species correlates with their relative binding to the available ligands, rather than with the co-receptor functions of the CD8 species.
Keywords: bimolecular fluorescence complementation, co-receptor, T-cell activation
Introduction
CD8 is the co-receptor for major histocompatibility complex class I (MHC-I)-restricted recognition of antigen. It binds directly to non-polymorphic regions of the MHC-I heavy chain. Through its cytoplasmic tail, CD8 interacts with the Src-family protein tyrosine kinase Lck (Veillette et al, 1988; Barber et al, 1989). Antigen recognition by the T-cell receptor (TCR) brings CD8 and therefore Lck into close proximity with the TCR (Zamoyska, 1998; Yachi et al, 2005), allowing Lck to phosphorylate elements of the TCR–CD3 complex and to precipitate the signalling cascade. CD8 exists on the T-cell surface in two isoforms, CD8αα and CD8αβ. CD8αβ is expressed by mature T cells and developing thymocytes, whereas CD8αα is expressed in certain subsets of T cells, including TCRαβ-expressing gut intraepithelial cells, some γδ T cells and some natural killer and dendritic cells. Both CD8 dimers are expressed on CD8αβ+ intraepithelial cells, and there is evidence for transient upregulation of CD8αα on activated CD8αβ+ T cells (reviewed by Cheroutre & Lambolez, 2008).
CD8αβ is a much stronger co-receptor than CD8αα, the expression of CD8β enhancing TCR sensitivity about 100-fold over cells expressing only CD8α (Karaki et al, 1992; Renard et al, 1996; Yachi et al, 2005), and indeed CD8αα might be a negative regulator of T-cell activation (Cheroutre & Lambolez, 2008). CD8β is reported to mediate interactions between the CD8 and the TCR–CD3 complex (Doucey et al, 2003). Because the affinities of the two isoforms for MHC-I molecules are similar (Garcia et al, 1996; Kern et al, 1999; Wang et al, 2009), and because Lck interacts with the cytoplasmic domain of CD8α, the likely mechanism for this difference in activity is the location of CD8αβ in membrane rafts. The CD8β molecule, similar to the other T-cell co-receptor CD4, but unlike CD8α, can be palmitoylated, resulting in its localization in lipid rafts (Arcaro et al, 2000, 2001). Lck is also targeted to rafts through palmitoylation and myristoylation (Rodgers et al, 1994), so that the presence of Lck and CD8αβ in lipid rafts provides both a motif and an opportunity for their interaction. CD8α also mediates interaction with another lipid raft resident molecule, LAT (Bosselut et al, 1999).
At present there are few or no data on the differential roles of CD8αα and CD8αβ during antigen recognition. The findings noted above indicate that CD8αβ plus lipid rafts should be strongly recruited to the immunological synapse (IS). On the other hand, if CD8αα and CD8αβ bind equally well to MHC-I, there is no a priori reason why CD8αα should not be recruited to the IS as strongly as CD8αβ, even without lipid rafts. Moreover, there is some doubt about the relevance of lipid rafts in the formation of the IS (Bunnell et al, 2002). For this reason, we decided to test the differences, if any, that are present in the molecular dynamics of CD8αα and αβ on the surface of T cells before and during stimulation.
Bimolecular fluorescence complementation (BiFC) allows identification of molecules that interact with each other in live cells. In this technique, the two proteins of interest are expressed as chimaeras with either the N-terminal or C-terminal half of a green fluorescent protein-related molecule. If the two proteins interact, the two halves of the fluorescent protein (FP) refold to form a complete FP (Hu et al, 2002; Hu & Kerppola, 2003). By using halves from different FPs, different spectra are obtained, allowing multicolour experiments to determine how and where one protein interacts with two different partners (Hu & Kerppola, 2003; Shyu et al, 2006). We adapted this technique to CD8, such that CD8αα and CD8αβ would be separately visible on the same cell. We then used fluorescence deconvolution microscopy to follow CD8αα and αβ dimers to investigate whether there is differential recruitment of the two forms of CD8 to the IS during antigen recognition.
Results And Discussion
Expression of CD8αα-Cerulean and CD8αβ-Venus
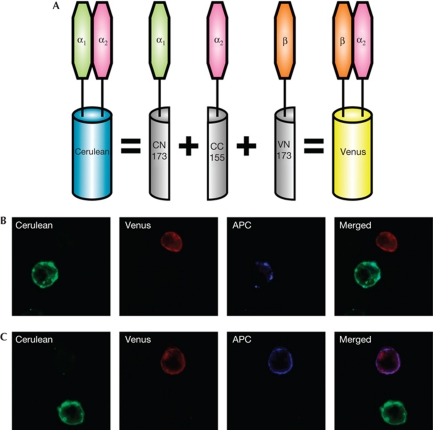
We prepared constructs for BiFC to allow expression of CD8αα as a chimaera with the cyan (C)FP Cerulean and CD8αβ as a chimaera with the yellow FP Venus (Shyu et al, 2006). The strategy for expressing CD8 as different coloured chimaeric proteins is shown in Fig 1A. The CD8α attached to the C terminus of CFP (which folds with both Cerulean and Venus N termini) was the CD8α2 allele recognized by anti-Ly2.2 monoclonal antibody, whereas that attached to the N terminus of Cerulean was CD8α1 (epitope recognized by anti-Ly2.1). When stably expressed in the OT-I T-hybridoma line (Yachi et al, 2005), the CD8αα and CD8αβ fluoresced as predicted (Fig 1B,C). This is a new use of BiFC, which was originally developed to show the formation of dimers between different molecules (Hu et al, 2002; Hu & Kerppola, 2003).
Figure 1.
Principle of the bimolecular fluorescence complementation (BiFC) assay and specific CD8α1 and CD8β detection. (A) CD8α2–CC155 forms CD8αα when expressed with CD8α1–CN173, or CD8αβ when expressed with CD8β–CN173. Note that non-fluorescent CD8αα or CD8αβ can also be formed from CD8α1 or CD8α2 homodimers, or from CD8α1–CD8β heterodimers. (B,C) Cells expressing only the fluorescent molecules CD8α2α1 (Cerulean: shown as green) or CD8α2β (Venus, shown as red) were mixed at a 1:1 ratio and stained with allophycocyanin (APC)–anti-CD8α1 (B) or APC–anti-CD8β (C). The antibody staining is shown in blue.
The relative efficiency of formation of the two CD8 complexes was compared by flow cytometry, with expression of comparable amounts of fluorescent CD8αβ and CD8αα proteins, as well as the ovalbumin (OVA)-specific TCR (supplementary Fig S1 online). This does not take into account the formation of non-fluorescent CD8αβ and CD8αα dimers, which also occurs. The surface concentration of TCR was 7–9% of that of the mature T cells, similar to the difference between CD4+8+ and CD4+8− or CD4−8+ thymocytes, or between CD4+8+ thymocytes and mature T cells (Crispe et al, 1987). Although the hybridomas stained ∼2- to 3-fold brighter than primary T cells for CD8α1 and CD8β, the surface concentration of CD8 was 2- to 3.5-fold lower than the concentration on the mature T cells (supplementary Fig S1 online). We investigated the partitioning of the different molecules in membrane fractions on the basis of detergent solubility. As expected, CD8αα was primarily in the detergent-soluble non-raft fraction and CD8αβ primarily in the detergent-resistant raft fraction (supplementary Fig S2 online).
Co-receptor-dependent TCR–MHC binding
We used MHC-peptide (MHCp) tetramers (Kb-OVA, recognized by OT-I TCR) to investigate the ability of CD8αα-Cerulean and CD8αβ-Venus to enhance TCR binding to MHCp. Cells expressing fluorescent CD8αα, CD8αβ or both were mixed and allowed to bind phycoerythrin-labelled Kb-OVA tetramers (supplementary Fig S3A online). Cells expressing OT-I TCR but not CD8 rapidly bound a small quantity of Kb-OVA. Cells coexpressing OT-I TCR and either CD8αβ-Cerulean or CD8αα-Venus bound labelled tetramer more effectively, whereas expression of both CD8αα-Cerulean and CD8αβ-Venus resulted in the highest rate of tetramer accumulation. CD8αα+αβ-expressing hybridoma cells grown for several weeks showed a gradual loss of expression of either co-receptor. Tetramer binding to cells that had lost one of the co-receptors showed tetramer binding rates similar to those of the single co-receptor-expressing cells (supplementary Fig S3B online). Cells expressing more co-receptor molecules showed stronger tetramer binding (supplementary Fig S3C online). Surprisingly, tetramer binding to TCR plus CD8αα—the weaker co-receptor—was stronger than that binding to TCR plus CD8αβ. When the cells were subgated for increasing ratio of one CD8 species to the other, tetramer binding increased as the proportion of CD8αα increased relative to CD8αβ. This can be explained because, similarly to CD8αα, unstimulated TCRs are not raft associated (Montixi et al, 1998). Earlier data showing increased binding of multimeric MHCp to CD8αβ compared with CD8αα used a T hybridoma in which CD8αβ was reported to be constitutively associated with TCR in lipid rafts (Arcaro et al, 2001; Doucey et al, 2003). This is not always the case (Yachi et al, 2005), which might explain the discrepancy in the results. Thus, initial binding to TCR, shown by the CD8-independent, small, rapid rise in binding seen with all of the cell populations, is followed by a stage in which the proximity and/or mobility of the CD8 molecules makes a difference to the ability to increase tetramer binding.
Separate capping of CD8αα-Cerulean and CD8αβ-Venus
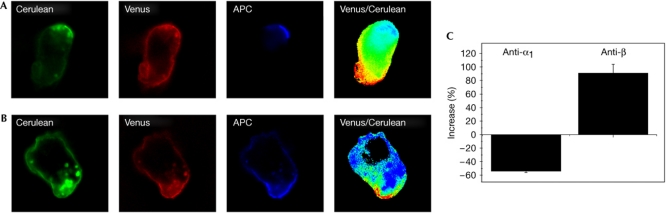
To induce patching and capping of the CD8αα or -αβ molecules, cells expressing both species were incubated at 4°C with monoclonal antibodies recognizing CD8α1 (anti-Ly2.1) or CD8β, followed by crosslinking reagents. After further incubation at 37°C, cells were fixed and analysed. Both CD8αα-Cerulean (Fig 2A) and CD8αβ-Venus (Fig 2B) were preferentially capped by their cognate antibody, although there was some co-capping of the other species. We calculated the percentage of increase (Fig 2C; see Methods) to show the preferential recruitment of one species over the other in a defined region of interest. A positive value represents preferential recruitment of CD8αβ, whereas a negative value represents preferential recruitment of CD8αα. The anti-CD8α1 preferentially capped CD8αα-Cerulean, whereas the anti-CD8β preferentially capped CD8αβ-Venus, indicating that these species were mostly physically independent of each other. Lck associated with both CD8αα and CD8αβ. Capping of CD8αα or CD8αβ with specific antibodies co-capped Lck. In cells expressing both CD8αα and CD8αβ, Lck was co-capped with either species (supplementary Fig S4 online).
Figure 2.
Separate capping of CD8αα and CD8αβ. (A,B) Representative images of co-capping with anti-CD8α1 (anti-Ly2.1) (A) or anti-CD8β (B) monoclonal antibodies on cells expressing all three bimolecular fluorescence complementation (BiFC) constructs. On panels labelled Venus/Cerulean, the intensity ratio of Venus/Cerulean is represented using a high/low scale in which blue indicates a lower ratio value (more CD8α2α1) and red a higher ratio value (more CD8α2β). (C) Shows quantification of the differential capping of CD8α2α1 and CD8α2β as percentage of increase. Negative values reflect preferential recruitment of CD8α2α1 and positive values the preferential recruitment of CD8α2β. Error bars show s.e.m., n=16 for anti-α1 and n=30 for anti-β. APC, allophycocyanin.
CD8αα and CD8αβ recruitment to the IS
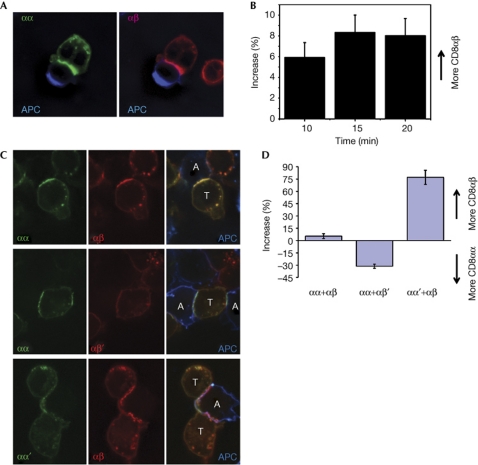
The interaction of a CD8+ T cell with an antigen-presenting cell (APC) results in recruitment of CD8αβ to the IS (Yachi et al, 2005, 2006; Gascoigne et al, 2010). Both CD8αα-Cerulean and CD8αβ-Venus were recruited to the IS during the interaction of the T cells with APC (Fig 3A,B), with only a slight bias towards recruitment of CD8αβ. The comparable recruitment of both species was surprising given that CD8αβ is a much better co-receptor. It suggests that the enhanced co-receptor function is not because of any preferential ability of CD8αβ to go to the IS, nor is it because of its stronger ability to bind to MHC-I. One potential reason could be that the strong recruitment of both molecules is due to some interaction between CD8 molecules. To test this directly, we made mutations in the MHC-I-binding sites of CD8α and CD8β, and expressed them with CD8α2–CC155 such that binding-competent CD8αα was expressed with mutated CD8αβ (CD8αβ′; β-chain class I-binding mutation S101A (Devine et al, 2006)), or binding-competent CD8αβ was expressed with mutated CD8αα (CD8αα′; α-chain class I-binding mutation N107A (Devine et al, 2002)). These binding mutants of CD8 were not recruited to the IS in response to antigen recognition, and did not interfere with the recruitment of the non-mutant species (Fig 3C,D). We next measured tetramer binding using the same analysis strategy as in supplementary Fig S3B (Fig S5 online). We gated samples of cells expressing CD8αα+CD8αβ′ or CD8αα′+CD8αβ, cells that had lost one of these species or non-fluorescent cells that had lost both. In both cell types, the addition of mutant CD8 did not substantially alter tetramer binding by the non-mutant species. Cells showing only the fluorescent signal characteristic of the mutant species bound significantly less tetramer. This binding was, however, higher than that of CD8– cells. This probably indicates the presence of non-fluorescent (non-mutant) CD8α2–CC155 homodimers, which were capable of binding to the tetramer.
Figure 3.
CD8αα or αβ recruitment to the IS depends on the ability to bind to MHC-I. (A) T cell expressing CD8αα-Cerulean and CD8αβ-Venus interacting with a Cy5-labelled antigen-presenting cell (APC). For clarity, Cerulean and Venus channels merged with the Cy5 channel are presented separately. (B) Time course of differential recruitment of CD8αα-Cerulean and CD8αβ-Venus in cells responding to RMA-S cells presenting Kb-OVA. Graph shows the percentage of increase ±s.e.m., n⩾20. (C) T cells expressing CD8αα+CD8αβ, CD8αα+CD8αβ′ or CD8αα′+CD8αβ fluorescent proteins during interaction with ovalbumin (OVA) peptide-loaded EL4 cells. Cerulean and Venus channels are presented separately (left and centre) and merged with Cy5 (APC) channel in right panels. T cells and APCs are labelled on the merged image (T and A, respectively). (D) Preferential recruitment of wild-type co-receptor species compared with non-major histocompatibility complex-I-binding mutants to the immunological synapse (IS). Negative values reflect preferential recruitment of CD8αα and positive values the preferential recruitment of CD8αβ. Graph shows the percentage of increase ±s.e.m. n=17, 18 and 15.
These results show that the recruitment of both CD8 species in response to antigenic stimulation is due to each species independently interacting with the MHC-I molecule, and not due to interactions between CD8 molecules. Thus, the co-receptor prowess of CD8αβ is not related to a stronger ability to bind to MHC-I, nor to a stronger ability to be recruited to the IS during antigen recognition, nor even to its association with lipid rafts per se, leaving only its ability to preferentially bind to Lck. This is in accord with a recent modelling study indicating that the most important function of co-receptors is to target Lck to the TCR-recognizing antigen (Artyomov et al, 2010).
Preferential CD8αα recruitment by H2-TL
The non-classical MHC-I molecule thymus leukaemia antigen (H2-TL) binds to CD8αα with much higher affinity than to CD8αβ (Leishman et al, 2001; Liu et al, 2003; Attinger et al, 2005). We therefore tested whether CD8αα or CD8αβ recruitment to the T-cell–APC (T–APC) interface would be altered in the presence of H2-TL, owing to the non-cognate interaction of CD8αα with H2-TL. We stimulated the OT-I hybridomas expressing the two fluorescent forms of CD8 with RMA-S cells or with RMA-S cells transfected with H2-TL (RMA-S–TL; Attinger et al, 2005). These RMA-S cells were first incubated at 32°C in the presence or absence of OVA peptide. This peptide binds to H2-Kb expressed on the cell surface at this temperature, thus stabilizing the H2-Kb molecule when the temperature is later raised to 37°C (Ljunggren et al, 1990; Yachi et al, 2005, 2006, 2007). Because H2-TL does not bind to peptides (Weber et al, 2002; Liu et al, 2003), it does not require Tap2 for its cell-surface expression. The RMA-S–TL cells therefore express H2-TL constitutively (Attinger et al, 2005).
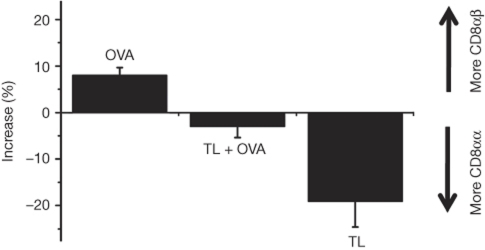
CD8αβ was recruited slightly more effectively than CD8αα when OVA was presented on Kb on RMA-S cells (Fig 4). Cells presenting both H2-TL and OVA-Kb recruited CD8αα and CD8αβ to the interface at the same rate. Finally, presentation of H2-TL alone caused preferential recruitment of CD8αα to the T–APC interface. Thus, recognition of H2-TL enhanced CD8αα recruitment, and recognition of H2-Kb enhanced CD8αβ recruitment. The presence of both H2-TL and OVA-Kb removed the difference in the preferential recruitment of the two CD8 species.
Figure 4.
Ligand dependence of the differential recruitment of CD8αα and CD8αβ. Data are presented for T cells expressing CD8αα-Cerulean and CD8αβ-Venus responding to RMA-S cells presenting Kb-OVA (OVA), RMA-S cells expressing thymus leukaemia antigen (H2-TL or TL) and also presenting Kb-OVA (TL+OVA) and RMA-S cells expressing TL alone (TL). Error bars represent s.d. n=25 for each group.
Concluding Remarks
BiFC was successfully used to engineer CD8αα and CD8αβ dimers to enable them to be independently visualized on the T-cell surface. Both αα and αβ dimers behaved independently when crosslinked by antibodies, but both were strongly recruited to the IS by recognition of antigen. The strong recruitment of CD8αα was surprising, as it is a much weaker co-receptor than CD8αβ. Synapse recruitment of either species was lost when its ability to bind to MHC-I was compromised, showing that this recruitment was a genuine effect of MHC recognition. Lck and CD8αβ were mainly present in lipid rafts, whereas CD8αα was excluded, although Lck was able to co-cap with both CD8 species. Thus, the ability of CD8αβ to function as a strong co-receptor is related to its ability to associate with Lck in the appropriate membrane domains, not to binding to MHC or to recruitment to the IS. However, we found that recognition of H2-TL separated CD8αα recruitment from that of CD8αβ, owing to the much stronger interaction of CD8αα with H2-TL. Thus, in this case, the recruitment of the two CD8 species correlated with the relative binding to the available ligands.
Methods
DNA constructs and gene expression. CD8α1 (methionine at residue 78) was generated from the CD8α2 (Val78) construct by single point mutation. BiFC fragments CC155, Cr173 and Vn173 (Hu et al, 2002; Hu & Kerppola, 2003) were fused to the C terminus of CD8α2, CD8α1 and CD8β, respectively, with spacers between the proteins to allow proper folding and flexibility (Yachi et al, 2005). Mutations in the MHC-I-binding sites of CD8 were as follows: CD8α1 N107A to make CD8αα′ . In the CD8αα structure, N107 on one α-subunit interacts with residues M228, E229 and L230 of Kb, and in the other α-subunit it interacts with Kb E222 (Devine et al, 2002). CD8β S101A was made for CD8αβ′ (Devine et al, 2006). S101 interacts with residues T225, M228 and L230 of Kb (Wang et al, 2009). Constructs cloned in pLN1 retroviral expression vector were transduced into hybridomas expressing the OT-I TCR but not CD8 (Yachi et al, 2005, 2006). Expressors were selected by FACS, sorted and cloned. The following antibodies were used: anti-Ly2.1 (that is, anti-CD8α1; clone 49-31.1, mouse IgG3), anti-Ly2.2 (that is, anti-CD8α2; clone AD4(15), mouse IgM, Cedarlane) and anti-CD8β (YTS156.7.7, rat IgG2b, BioLegend).
Imaging. Fluorescence imaging was performed on a Marianas system (Intelligent Imaging Innovations, Santa Monica, CA). To avoid crosstalk, images were acquired sequentially using the following filter sets: Cerulean, Ex: 430/25 nm, Em: 470/30 nm; Venus, Ex: 510/20 nm, Em: 550/50 nm, Cy5 or APC Ex: 622/36 nm, Em: 700/75 nm (Zal & Gascoigne, 2004; Yachi et al, 2005, 2006). Image analysis was performed using the Slidebook software. After background subtraction, the IS (or cap) and a non-IS (or non-cap) area were delimited manually on the image. Average fluorescence intensities for both areas were measured, and the area intensity ratio (AR)=Isynapse/Inon-synapse was calculated for each channel. The percentage of increase was then calculated as ((ARVenus/ARCerluean)−1) × 100. This parameter defines the preferential recruitment of one fluorophore compared with the other in the region of interest, independently from their actual concentration or fluorescence intensity. The −1 term was introduced so that preferential recruitment of Cerulean (CD8αα) or Venus (CD8αβ) is easily identified by a negative or a positive value, respectively.
Statistics. The mean difference hypothesis of Student's two-tailed t-test assuming different variances and a confidence level of 95% was performed using Microcal Origin.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
This study was supported by the National Institutes of Health (NIH) grants R01GM065230 and R01AI074074 to N.R.J.G., an Irvington Institute Fellowship of the Cancer Research Institute to J.-P.C., and an NIH Training grant T32HL07195 to P.P.Y. We thank T. Kerppola (University of Michigan) and C. Hu (Purdue University) for the BiFC system, H. Cheroutre (La Jolla Institute for Allergy and Immunology) for RMA-S-TL and R. Zamoyska (Univesity of Edinburgh, UK) for anti-CD8β serum. This is Manuscript #21149 from The Scripps Research Institute.
Author contributions: N.R.J.G. and J.-P.C. designed the project; J.-P.C. made and transfected the BiFC constructs; and J.-P.C. and V.R. conducted the experiments, with contributions from J.A. and P.P.Y.; N.R.J.G. J.-P.C. and V.R. wrote the manuscript. Figures were prepared by J.-P.C. and V.R.
Footnotes
The authors declare that they have no conflict of interest.
References
- Arcaro A et al. (2001) CD8β endows CD8 with efficient coreceptor function by coupling T cell receptor/CD3 to raft-associated CD8/p56lck complexes. J Exp Med 194: 1485–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcaro A, Gregoire C, Boucheron N, Stotz S, Palmer E, Malissen B, Luescher IF (2000) Essential role of CD8 palmitoylation in CD8 coreceptor function. J Immunol 165: 2068–2076 [DOI] [PubMed] [Google Scholar]
- Artyomov MN, Lis M, Devadas S, Davis MM, Chakraborty AK (2010) CD4 and CD8 binding to MHC molecules primarily acts to enhance Lck delivery. Proc Natl Acad Sci USA 107: 16916–16921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attinger A, Devine L, Wang-Zhu Y, Martin D, Wang JH, Reinherz EL, Kronenberg M, Cheroutre H, Kavathas P (2005) Molecular basis for the high affinity interaction between the thymic leukemia antigen and the CD8αα molecule. J Immunol 174: 3501–3507 [DOI] [PubMed] [Google Scholar]
- Barber EK, Dasgupta JD, Schlossman SF, Trevillyan JM, Rudd CE (1989) The CD4 and CD8 antigens are coupled to a protein-tyrosine kinase (p56lck) that phosphorylates the CD3 complex. Proc Natl Acad Sci USA 86: 3277–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosselut R, Zhang W, Ashe JM, Kopacz JL, Samelson LE, Singer A (1999) Association of the adaptor molecule LAT with CD4 and CD8 coreceptors identifies a new coreceptor function in T cell receptor signal transduction. J Exp Med 190: 1517–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunnell SC, Hong DI, Kardon JR, Yamazaki T, McGlade CJ, Barr VA, Samelson LE (2002) T cell receptor ligation induces the formation of dynamically regulated signaling assemblies. J Cell Biol 158: 1263–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheroutre H, Lambolez F (2008) Doubting the TCR coreceptor function of CD8αα. Immunity 28: 149–159 [DOI] [PubMed] [Google Scholar]
- Crispe IN, Shimonkevitz RP, Husmann LA, Kimura J, Allison JP (1987) Expression of T cell antigen receptor β-chains on subsets of mouse thymocytes. Analysis by three-color flow cytometry. J Immunol 139: 3585–3589 [PubMed] [Google Scholar]
- Devine L, Rogozinski L, Naidenko OV, Cheroutre H, Kavathas PB (2002) The complementarity-determining region-like loops of CD8α interact differently with β2-microglobulin of the class I molecules H-2 Kb and thymic leukemia antigen, while similarly with their α3 domains. J Immunol 168: 3881–3886 [DOI] [PubMed] [Google Scholar]
- Devine L, Thakral D, Nag S, Dobbins J, Hodsdon ME, Kavathas PB (2006) Mapping the binding site on CD8β for MHC class I reveals mutants with enhanced binding. J Immunol 177: 3930–3938 [DOI] [PubMed] [Google Scholar]
- Doucey MA, Goffin L, Naeher D, Michielin O, Baumgartner P, Guillaume P, Palmer E, Luescher IF (2003) CD3δ establishes a functional link between the T cell receptor and CD8. J Biol Chem 278: 3257–3264 [DOI] [PubMed] [Google Scholar]
- Garcia KC, Scott CA, Brunmark A, Carbone FR, Peterson PA, Wilson IA, Teyton L (1996) CD8 enhances formation of stable T-cell receptor/MHC class I molecule complexes. Nature 384: 577–581 [DOI] [PubMed] [Google Scholar]
- Gascoigne NRJ, Zal T, Yachi PP, Hoerter JAH (2010) Co-receptors and recognition of self at the immunological synapse. Curr Top Microbiol Immunol 340: 171–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- Hu CD, Kerppola TK (2003) Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat Biotechnol 21: 539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaki S, Tanabe M, Nakauchi H, Takiguchi M (1992) β-Chain broadens range of CD8 recognition for MHC class I molecule. J Immunol 149: 1613–1618 [PubMed] [Google Scholar]
- Kern P, Hussey RE, Spoerl R, Reinherz EL, Chang HC (1999) Expression, purification, and functional analysis of murine ectodomain fragments of CD8αα and CD8αβ dimers. J Biol Chem 274: 27237–27243 [DOI] [PubMed] [Google Scholar]
- Leishman AJ, Naidenko OV, Attinger A, Koning F, Lena CJ, Xiong Y, Chang HC, Reinherz E, Kronenberg M, Cheroutre H (2001) T cell responses modulated through interaction between CD8αα and the nonclassical MHC class I molecule, TL. Science 294: 1936–1939 [DOI] [PubMed] [Google Scholar]
- Liu Y, Xiong Y, Naidenko OV, Liu JH, Zhang R, Joachimiak A, Kronenberg M, Cheroutre H, Reinherz EL, Wang JH (2003) The crystal structure of a TL/CD8αα complex at 2.1 Å resolution: implications for modulation of T cell activation and memory. Immunity 18: 205–215 [DOI] [PubMed] [Google Scholar]
- Ljunggren HG et al. (1990) Empty MHC class I molecules come out in the cold. Nature 346: 476–480 [DOI] [PubMed] [Google Scholar]
- Montixi C, Langlet C, Bernard AM, Thimonier J, Dubois C, Wurbel MA, Chauvin JP, Pierres M, He HT (1998) Engagement of T cell receptor triggers its recruitment to low-density detergent-insoluble membrane domains. EMBO J 17: 5334–5348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renard V, Romero P, Vivier E, Malissen B, Luescher IF (1996) CD8β increases CD8 coreceptor function and participation in TCR-ligand binding. J Exp Med 184: 2439–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers W, Crise B, Rose JK (1994) Signals determining protein tyrosine kinase and glycosyl-phosphatidylinositol-anchored protein targeting to a glycolipid-enriched membrane fraction. Mol Cell Biol 14: 5384–5391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu YJ, Liu H, Deng X, Hu CD (2006) Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. Biotechniques 40: 61–66 [DOI] [PubMed] [Google Scholar]
- Veillette A, Bookman MA, Horak EM, Bolen JB (1988) The CD4 and CD8 T cell surface antigens are associated with the internal membrane tyrosine-protein kinase p56lck. Cell 55: 301–308 [DOI] [PubMed] [Google Scholar]
- Wang R, Natarajan K, Margulies DH (2009) Structural basis of the CD8αβ/MHC class I interaction: focused recognition orients CD8β to a T cell proximal position. J Immunol 183: 2554–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber DA, Attinger A, Kemball CC, Wigal JL, Pohl J, Xiong Y, Reinherz EL, Cheroutre H, Kronenberg M, Jensen PE (2002) Peptide-independent folding and CD8αα binding by the nonclassical class I molecule, thymic leukemia antigen. J Immunol 169: 5708–5714 [DOI] [PubMed] [Google Scholar]
- Yachi PP, Ampudia J, Gascoigne NRJ, Zal T (2005) Nonstimulatory peptides contribute to antigen-induced CD8-T cell receptor interaction at the immunological synapse. Nat Immunol 6: 785–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachi PP, Ampudia J, Zal T, Gascoigne NRJ (2006) Altered peptide ligands induce delayed and reduced CD8-TCR interaction—a role for CD8 in distinguishing antigen quality. Immunity 25: 203–211 [DOI] [PubMed] [Google Scholar]
- Yachi PP, Lotz C, Ampudia J, Gascoigne NRJ (2007) T cell activation enhancement by endogenous pMHC acts for both weak and strong agonists but varies with differentiation state. J Exp Med 204: 2747–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zal T, Gascoigne NRJ (2004) Photobleaching-corrected FRET efficiency imaging of live cells. Biophys J 86: 3923–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamoyska R (1998) CD4 and CD8: Modulators of T-cell receptor recognition of antigen and of immune responses? Curr Opin Immunol 10: 82–87 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.