Abstract
The epigenome has been hypothesized to provide the interface between the environment and the nuclear DNA (nDNA) genes. Key factors in the environment are the availability of calories and demands on the organism’s energetic capacity. Energy is funneled through glycolysis and mitochondrial oxidative phosphorylation (OXPHOS), the cellular bioenergetic systems. Since there are thousands of bioenergetic genes dispersed across the chromosomes and mitochondrial DNA (mtDNA), both cis and trans regulation of the nDNA genes is required. The bioenergetic systems convert environmental calories into ATP, acetyl-Coenzyme A (acetyl-CoA), S-adenosyl-methionine (SAM), and reduced NAD+. When calories are abundant, ATP and acetyl-CoA phosphorylate and acetylate chromatin, opening the nDNA for transcription and replication. When calories are limiting, chromatin phosphorylation and acetylation are lost and gene expression is suppressed. DNA methylaton via SAM can also be modulated by mitochondrial function. Phosphorylation and acetylation are also pivotal to regulating cellular signal transduction pathways. Therefore, bioenergetics provides the interface between the environment and the epigenome. Consistent with this conclusion, the clinical phenotypes of bioenergetic diseases are strikingly similar to those observed in epigenetic diseases (Angelman, Rett, Fragile X Syndromes, the laminopathies, cancer, etc.), and an increasing number of epigenetic diseases are being associated with mitochondrial dysfunction. This bioenergetic-epigenomic hypothesis has broad implications for the etiology, pathophysiology, and treatment of a wide range of common diseases.
1. INTRODUCTION
“The modern definition of epigenetics is information heritable during cell division other than the DNA sequence itself” (Feinberg, 2007). Hence, epigenetics is thought to provide a flexible interface between the organism and its environment. Until now, the DNA sequences of interest to biology and medicine have been in the chromosomal DNA, which is transmitted during meiotic and mitotic cell division according to the rules of Gregor Mendel. However, most common metabolic and degenerative diseases and multiple cancers are familial, but are not classically Mendelian in their transmission. Therefore, such “complex diseases” have been attributed to epigenetic changes in response to the environmental change (Feinberg, 2007, 2008).
However, deficiency in energy metabolism has also emerged as an alternative explanation for the etiology of complex diseases over the past 21 years. The primary limiting factor for growth and reproduction of all biological systems is energy and the first reports that mitochondrial DNA (mtDNA) mutations can cause disease (Goto et al., 1990; Holt et al., 1988; Holt et al., 1990; Shoffner et al., 1990; Wallace et al., 2007; Wallace et al., 1988a; Wallace et al., 1988b) have been followed by reports that a broad spectrum of metabolic and degenerative diseases can have a mitochondrial etiology (Wallace et al., 2007). Moreover, environmentally adaptive mtDNA variants have been associated with predisposition to virtually the entire range of common “complex” diseases (Wallace, 2008).
However, similar symptoms imply a common pathophysiology. Therefore, epigenetic and mitochondrial genetic diseases must be interrelated through their impinging on a common function.
To understand nuclear-mitochondrial interactions, we must consider the early stages in the endosymbiontic event that created the eukaryotic cell about 2 billion years ago (Lane, 2002, 2005; Wallace, 2007). In the beginning, the proto-nucleus-cytosol was limited by energy. This limitation was alleviated by its symbiosis with an oxidative α-protobacterion, the proto-mitochondrion. Therefore, growth and replication of the nucleus became limited by mitochondrial energy production and thus calorie availability. This necessitated the regulation of nuclear replication and gene expression by calorie availability mediated by mitochondrial energetics. This was achieved by coupling modulation of nDNA chromatin structure and function by modification via high energy intermediates: phosphorylation by ATP, acetylation by acetyl-Coenzyme A (Ac-CoA), deacetylation by nicotinamide adenine dinucleotide (NAD+), and methylation by S-adenosyl-methionine (SAM).
Conversely, the nucleus had to develop mechanisms for modulating mitochondrial growth and replication. This was additionally complicated by the successive transfer of genes from the proto-mitochondrial DNA to the nDNA, with the cytosolic translation products being directionally imported back into the mitochondrion (Wallace, 2007). This process proceeded over a billion years with the result that the nDNA-encoded genes of the mitochondrial genome are now dispersed throughout the chromosomes (Wallace, 2007). Therefore, new mechanisms had to evolve to permit the coordinate expression of the mitochondrial genes based on nuclear requirements for energy for growth and reproduction. As a result, this became one of the early driving forces for the evolution of interchromosomal coordinate transcriptional regulation.
Over the subsequent 1.2 billion years, the nucleus-cytosol became increasingly specialized in specifying structure while the mitochondrion became entirely dedicated to energy production. Ultimately, this subcellular specialization became sufficiently refined and efficient that it permitted the advent of multicellularity and thus plants and animals (Wallace, 2007). Still, all subsequent tissue development, species radiation, and environmental adaptation were rooted in this fundamental energetic-epigenetic cooperation.
2. MITOCHONDRIAL BIOENERGETICS
Complex structures can only be maintained by the continual flux of energy. Life, therefore, is the interaction between structure, energy, and information, with information required for both structure and energetics (Wallace, 2007).
2.1. Mitochondrial Energetics: OXPHOS
Energetics in animals is based on the availability of reducing equivalents, consumed as carbohydrates and fats. Glucose is cleaved into pyruvate via glycolysis, and the pyruvate enters the mitochondrion via pyruvate dehydrogenase (PDH) resulting in acetyl CoA, NADH + H+, and CO2. The acetyl CoA then enters the Tricarboxylic Acid (TCA) Cycle which strips the hydrogens from the hydrocarbons generating NADH + H+. Fatty acids are oxidized within the mitochondrion by β oxidation to generate acetyl CoA, NADH + H+, and FADH2, the latter contained in the electron transfer factor (ETF). Two electrons (reducing equivalents from hydrogen) are transferred from NADH + H+ to the OXPHOS complex NADH dehydrogenase (complex I) or from FADH2 containing enzymes such as the ETF dehydrogenase or succinate dehydrogenase (SDH, complex II) to reduce ubiquinone (coenzyme Q10, CoQ) to ubiquinol CoQH2. The electrons from CoQH2 are transferred successively to complex III (bc1 complex), cytochrome c, complex IV (cytochrome c oxidase, COX), and finally to oxygen (½ O2) to give H2O.
The energy that is released as the electrons flow down the ETC is used to pump protons out across the mitochondrial inner membrane through complexes I, III, and IV creating a proton electrochemical gradient (ΔP = ΔΨ + ΔμH+). The potential energy stored in ΔP is used for multiple purposes: to import proteins and Ca++ into the mitochondrion, to generate heat, and to synthesize ATP within the mitochondrial matrix. The energy to convert ADP + Pi to ATP comes from the flow of protons through the ATP synthetase (complex V) back into the matrix. Matrix ATP is then exchanged for cytosolic ADP by the inner membrane adenine nucleotide translocators (ANTs) (Wallace, 2007).
The efficiency by which dietary reducing equivalents are converted to ATP by OXPHOS is known as the coupling efficiency. This is determined by the efficiency by which protons are pumped out of the matrix by complexes I, III, and IV and the efficiency by which proton flux through complex V is converted to ATP. The uncoupler drug 2,4-dinitrophenol (DNP) and the nDNA-encoded uncoupler proteins 1, 2, and 3 (Ucp1, 2, and 3) render the mitochondrial inner membrane leaky for protons, by-passing complex V and dissipating the energy as heat (Wallace, 2007).
2.2. OXPHOS Complexes and the mtDNA
OXPHOS complexes are assembled from proteins encoded in both the nDNA and the mtDNA. Complex I is composed of 45 polypeptides, seven (ND1, 2, 3, 4, 4L, 5, and 6) encoded by the mtDNA; complex II from four nDNA polypeptides; complex III from 11 polypeptides, one (cytochrome b, cyt b) encoded by the mtDNA; complex IV form 13 polypeptides, three (COI, II, III) from the mtDNA; and complex V from about 16 polypeptides, two (ATP6 & 8) from the mtDNA. The mtDNA also encodes the 22 tRNAs and 12S and 16S rRNAs for translation of the 13 mtDNA polypeptides. Of the five complexes, only complexes I, III, IV, and V transport protons, and these are the same complexes for which key polypeptides are retained on the mtDNA. Therefore, it follows that the complex I, III, IV, and V electron and proton transfer proteins have been retained on the maternally-inherited and non-recombining mtDNA to avoid mixing OXPHOS complex proteins which could be incompatible and affect the coupling efficiency (Wallace, 2007).
Each mammalian cell contains hundreds of mitochondria and thousands of mtDNAs. When a mutation arises in a mtDNA, it creates a mixed population of normal and mutant mtDNAs, a state known as heteroplasmy. When a heteroplasmic cell divides, the two types of mtDNAs are randomly distributed into the daughter cells, resulting in genetic drift toward either pure mutant or wild type. Over time this replicative segregation results in segregation of the mutant mtDNAs into pure mutant or normal populations, termed homoplasmic cells (Wallace, 2007). As the percentage of mutant mtDNAs increases, mitochondrial energetic function decreases. When energy output is insufficient for normal tissue function, a threshold is crossed, symptoms appear, and apoptosis or necrosis may be initiated and clinical symptoms ensue (Wallace, 2005b; Wallace et al., 2007).
2.3. Reactive Oxygen Species (ROS)
As a by-product of OXPHOS, mitochondria generate much of the endogenous ROS of the cell. Under normal physiological conditions, ROS production is highly regulated, at least in part controlled by complex I (Evans et al., 2000; Hansen et al., 2006; Jones, 2006a; Kelley and Parsons, 2001; McCord, 2000). However, when the ETC becomes highly reduced, the excess electrons can be passed directly to O2 to generate superoxide anion (O2.−). The O2.− generated by complex I is released into the mitochondrial matrix where it is converted to hydrogen peroxide (H2O2) by the matrix manganese superoxide dismutase, MnSOD (Sod2 gene). Superoxide generated from complex III is released into the mitochondrial intermembrane space where it is converted to H2O2 by Cu/ZnSOD (Sod1) which is positioned in the mitochondrial intermembrane space and cytosolic. Mitochondrial H2O2 can diffuse into the nucleus-cytosol. If H2O2 encounters a reduced transition metal or is mixed with O2.−, the H2O2 can be further reduced to hydroxyl radical (OH.), the most potent oxidizing agent of the ROS (Figure 1). ROS can damage cellular proteins, lipids, and nucleic acids. Hence, excessive mitochondrial ROS production can exceed the antioxidant defenses of the cell, and the cumulative damage can ultimately destroy the cell.
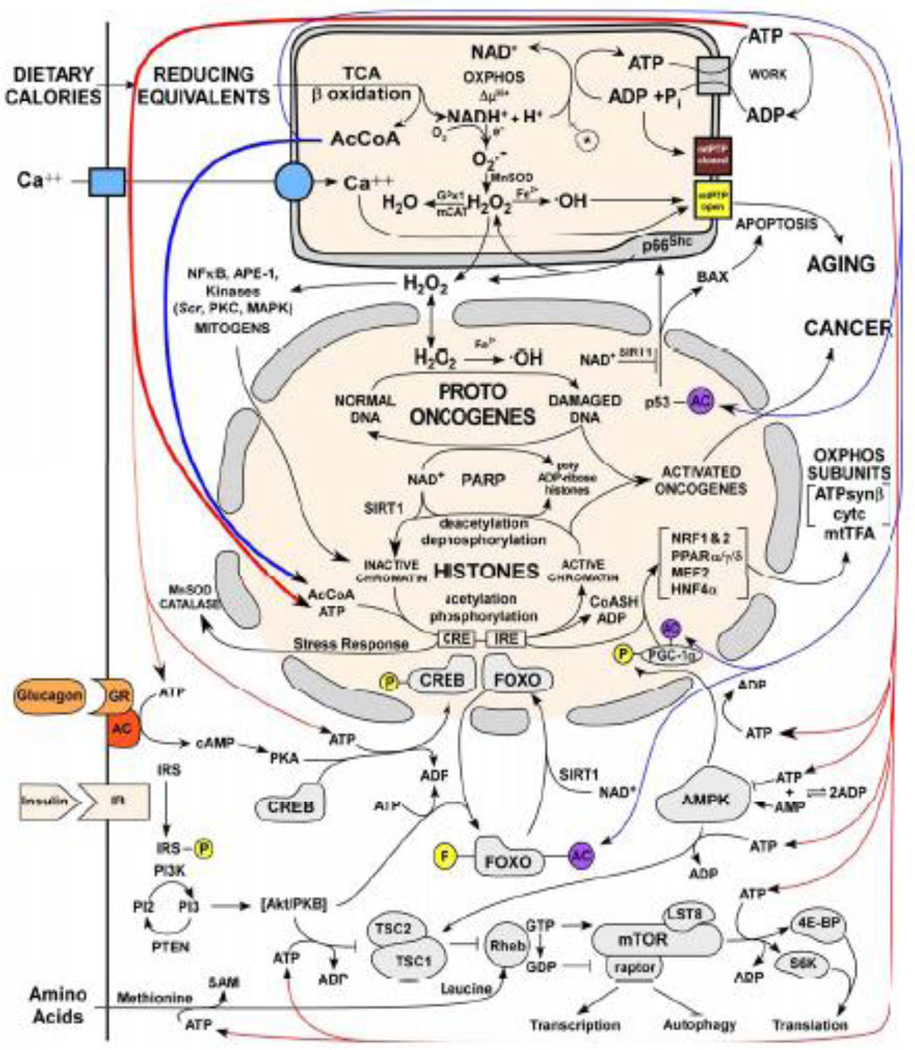
Figure 1. Energetic Regulation of the Epigenome.
Mitochondrial energetics links the epigenome to calorie availability through high energy intermediates and redox reactions. The mitochondrion is at the top of the figure, the nucleus in the middle, and the cytosol in at the bottom. Calories as reducing equivalents enter the cell and mitochondria at the upper left resulting in generation of acetyl-CoA, reduction of NAD+, and ATP. Energy then flows from top to bottom. ATP drives the phosphorylation of nuclear and cytosolic signal transduction proteins, acetyl-CoA acetylates chromatin and signal transduction proteins, and NAD+ acts through the Sirtuins to deacetylate proteins. Abbreviations: AcCoA = acetyl-CoA, NAD+ = nicotinamide adenine dinucleotide (oxidized) & NADH (reduced), mtPTP = mitochondrial permeability transition pore, GPx1 = glutathione peroxidase 1, mCAT = mitochondrially-targeted catalase, PARP = poly ADP ribose polymerase. Other abbreviations are described in the text.
2.4. The Mitochondrial Permeability Transition Pore (mtPTP)
The mitochondria evolved a self-destruct system, the mtPTP. The mtPTP is activated when the biochemical health of the mitochondria and cell decline, specifically when mitochondrial energy production declines, ROS generation increases, and excessive Ca++ is released into the cytosol and taken up by the mitochondrion. When the mtPTP is activated, it opens a channel in the mitochondrial inner membrane, short circuits ΔP, and initiates programmed cell death (apoptosis) (Wallace, 2005c).
2.5. Mitochondrial Dynamics
The mitochondria within a mammalian cell are in constant motion and undergoing repeated rounds of fission and fusion. Mitochondrial fusion and fission not only merges the mitochondrial inner and outer membranes but also mixes mitochondria matrices and redistributes the mtDNAs. The mammalian mitochondrial fusion machinery involves three major proteins: mitofusin 1 (Mfn1), 2 (Mfn2), and the Optic Atrophy-1 Protein (Opa1) (Chen et al., 2005; Chen et al., 2003a; Cipolat et al., 2004), while the mitochondrial fission machinery involves dynamin-related protein 1 (Drp1), Fis1, and Mff (Gandre-Babbe and van der Bliek, 2008; James et al., 2003; Smirnova et al., 2001; Yoon et al., 2003).
3. THE ENERGETIC EVOLUTION OF THE EPIGENOME
Prior to the advent of free oxygen in the biosphere, substrate-level phosphorylation was the primary mechanism for generating ATP in non-photosynthetic organisms. After the generation of free oxygen by photosystem II the redox range of biology was greatly expanded and OXPHOS became the most efficient system for generating ATP from the reducing equivalents present in organic molecules (Lane, 2002, 2005; Wallace, 2007).
3.1. The Energetic Evolution of the Epigenome
In the proto-nucleus-cytosol cell, ATP, from glycolysis, would have provided the high energy intermediate linking calories to cellular growth and replication. Consequently, phosphorylation-dephosphorylation must have been the first post-translational modification evoked for regulating DNA-protein interactions. When substrates were plentiful, ATP concentrations would increase, leading to increased phosphorylation of DNA-bound proteins. The resulting negative charge added to the protein by the phosphate group would then increase repulsion of the proteins from the DNA negatively charged sugar-phosphate backbone, opening chromatin for DNA transcription and replication. Phosphorylation also was used to modulate enzyme activities, signal transduction pathways, and the transcriptional apparatus in relation to energy availability.
As atmospheric oxygen became a dominant terminal electron acceptor, OXPHOS became the major source of ATP in non-photosynthetic eukaryotic cells. This cemented the symbiosis between the oxidative proto-mitochondrion and the proto-nucleus-cytosol. As a result acetyl-CoA became the metabolic intermediate that linked glycolytic pyruvate and fatty acid catabolism with the mitochondrial TCA cycle and OXPHOS and the concentration of acetyl-CoA became the cytosolic correlate for caloric availability.
Therefore, acetyl-CoA must have been the next energetic substrate to be used to couple cellular energetics with nuclear gene expression and cellular proliferation. Acetylation of lysines on DNA binding proteins neutralizes their positive charge, reducing protein affinity for DNA. As a result, when carbohydrates and or fats were abundant, protein acetylation increased; DNA binding proteins became acetylated and detached; and transcription, replication, and cell proliferation were stimulated. When carbohydrates and fats were limited (fasting-starvation), acetyl-CoA levels decreased; acetylation decreased; chromatin became condensed; and cellular gene expression, replication and proliferation were suppressed. In mammalian cells the carbon for acetyl-CoA destined for histone acetylation is derived from glucose through the mitochondrial TCA cycle via citrate. Glucose atoms transverse glycolysis to pyruvate and the pyruvate enters the mitochondrion where it is converted to acetyl-CoA via pyruvate dehydrogenase. The mitochondrial acetyl-CoA is condensed with oxaloacetate by citrate synthetase to generate citrate and the citrate is exported from the mitochondrion into the cytosol and nucleus by mitochondrion inner membrane citrate carrier. Cytosolic and nuclear citrate can then be converted to acetyl-CoA via ATP-citrate lyase, an enzyme found in both the nucleus and the cytosol. The nuclear acetyl-CoA is then preferentially used for histone acetylation. In the absence of glucose and the presence of supraphysiological levels of acetate, the mammalian cytosolic acetyl-CoA synthetase (AceCS1) can also generate acetyl-CoA. This can diffuse into the nucleus for use in histone acetylation (Rathmell and Newgard, 2009; Wellen et al., 2009). The facultative anaerobe, Saccaromyces cerevisiae, lacks ATP-citrate lyase. Therefore, it generates acetyl-CoA for histone acetylation via cytosolic and nuclear acetyl-CoA synthetases (Asc2p and Asc1p)(Takahashi et al., 2006), which might reflect its unique metabolism. The importance of acetylation as an energy surrogate is also extended to the regulation of transcription factors, enzymes, and signal transduction pathways. However, the kinetics of acetylation of histones versus other substrates differs (Rathmell and Newgard, 2009; Wellen et al., 2009).
Reduced energy supplies (fasting) would not only reduce acetyl-CoA levels but also reduce available reducing equivalents, resulting in the oxidation of NADH to NAD+. Since NAD+ but not NADH is a substrate for the class III histone deacetylases, the Sirtuins, starvation would activate the Sirtuins to deacetylate the DNA-binding proteins. This would increase the positive charge of the chromatin proteins, causing chromatin condensation, and suppressing transcription, replication, growth, and proliferation.
The third macromolecular modification system for modulating nuclear gene expression and replication in response to energy availability and nutrient supply is methylation by S-adenosyl-L-methionine (SAM). Methylation can increase macromolecular interactions through increased van der Waals forces. Therefore, methylation is more versatile than phosphorylation or acetylation for chromatin regulation.
SAM is produced in the cytosol by the reaction L-methionine + ATP to give SAM + Pi + PPi. Ths, SAM links energy production to the methylation of lysines and arginines in proteins and cytosines in DNA. The production of methionine from homocysteine is also regulated by mitochondrial metabolism. The single carbon metabolisms of the mitochondrion and cytosol are linked through the exchange of serine and glycine, which are interconvert within the two compartments through methylene-tetrahydrofolate via the mitochondrial and cytosolic serine hydroxymethyltransferases. The mitochondrial bifunctional enzyme converts methylene-tetrahydrofolate to formyl-tetrahydrofolate, thus diverting mitochondrial one carbon units from serine production to format, which is used in purine biosynthesis. Mitochondrial bifunctional enzyme is expressed in embryonic but not differentiated cells. When mitochondrial bifunctional enzyme is not expressed, mitochondrial serine production increases and the serine is exported to the cytosol. In the cytosol, the single carbon units of serine are transferred through methylene-tetrahydrofolate and methyl-tetrahydrofolate to convert homocystine to methionine, which can then be converted to SAM. The synthesis of serine within the mitochondrion requires NAD+. When OXPHOS is inhibited, the mitochondrial NADH/NAD+ ratio increases, thus inhibiting the mitochondrial production of methylene-tetrahydrofolate and thus serine. This reduces methionine and SAM production (Naviaux, 2008).
All of these macromolecular modification systems were in place prior to the advent of multicellularity 800 million years ago (Wallace, 1982, 2007). Multicellularity and terminal differentiation created two types of cell, the mortal somatic cells and the immortal germ and pluripotent stem cells. Since immortal cells predated mortal cells, it follows that immortal germ and stem cells should still utilize phosphorylation and acetylation to link energy availability with cell growth and reproduction and that stem cells should have the most open chromatin (Wen et al., 2009). However, to form tissues, the terminally differentiated cells must stop replicating. Hence, large blocks of chromatin must be shut down (Wen et al., 2009). Moreover, new genes were needed to block the cell cycle when activated by acetylation of the surrounding cellular chromatin. The p21 gene is one current example, being up-regulated when its promoter is acetylated and functioning to block replication (Glozak and Seto, 2007; Xiao et al., 2000). Cells which circumvented such replication blockades would then need to be destroyed before they could disrupt the tissue structure. This led to the development of mtPTP and pro-apoptotic proteins like Bax, which are also induced by histone acetylation at their promoters (Glozak and Seto, 2007). Additional layers of control were added once cellular proliferation became disconnected from energetics and increasingly complex structures were formed (Figure 2).
3.2. Energetic Regulation of the Epigenome
The nDNA of the modern eukaryotic cell is packaged in nucleosomes encompassing 146 to 147 base pairs of DNA wrapped around a complex of two copies each of histones H2A, H2B, H3, and H4. In yeast, these histones and the C-terminal domain of RNA Polymerase II (POLII) can be phosphorylated by kinases using ATP. Changes in histone serine/threonine phosphorylation in yeast are linked to changes in the extracellular environment. A change in carbon source results in the phosphorylation of serine 10 in H3 (H3-S10Pi), which is linked to acetylation of lysine 14 in H3 (H3-K14Ac). This stimulates transcription. ATP-driven phosphorylation of the POLII C-terminal domain also modulates transcription and phosphorylation of serine 10 in H2B (H2-S10Pi) is associated with stress and cell death (Fuchs et al., 2009).
3.2.1. Energetic Regulation at the Chromatin Level
Acetylation of histones is catalyzed by histone acetyltransferases (HATs) which catalyze the reaction of acetyl-CoA with the ε-amino group of lysines in histones. There are four classes of HATs known in eukaryotic cells, Gcn5/PCAF, MYST, p300/CBP, and Rtt109. All have structurally similar acetyl-CoA binding sites (Marmorstein and Trievel, 2009; Smith and Denu, 2009). The p300/CBP HAT complex was first characterized as an activator of cAMP responsive genes. Elevated cAMP activates protein kinase A to phosphorylate CREB at serine 133. Phospho-CREB recruits CBP to cAMP response elements (CREs) in the DNA, and CBP uses its intrinsic HAT activity to open the chromatin for transcription (Tini et al., 2002).
Histone deacetylases (HDACs) fall into four classes. Three classes have similar deacetylase mechanisms involving Zn2+ and include class I (HDAC1–3 & 8), class II (HDAC 4–7 & 9–10), and class IV (HDAC 11). The class III Sirtuin HDACs catalyze deacetylation by the reaction of acetyl-lysine + NAD+ → lysine + nicotinamide + 2’-O-acetyl-ADP ribose (OAADPr) (Smith and Denu, 2009).
Methylation of chromatin, with the methyl group donated by SAM, can occur on either lysines or arginines. Lysine methylation is catalyzed by histone lysine methyltransferases and can result in mono-, di-, or tri-methylation. There are two histone lysine methyltransferase classes: the SET domain family and the DOT1 family. The SET domain class methylates both nucleosomal and free histones while the DOT1 class only methylates nucleosomal histones (Smith and Denu, 2009). Histone demethylation involves two classes of reactions. The first class includes lysine-specific demethylase 1 (LSD1) and is specific for mono- and dimethylated histones. LSD1 employs FAD to oxidize the -N-CH3 bond, which is then cleaved by water to generate formaldehyde. The resulting FADH2 is reoxidized by O2 to generate H2O2. The second class is the Jumonj family of histone demethylases. These demethylate trimethyl-lysines in a reaction using O2 and 2-oxoglutarate as co-reactants generate succinate and CO2 and release formaldehyde (Marmorstein and Trievel, 2009; Smith and Denu, 2009).
Arginine methylation is catalyzed by protein arginine methyltransferases (PRMT) in which the methyl group for SAM is transferred to the arginine guanidinium side chain There are two classes of PRMTs: PRMT1 first generates monomethylarginine and then asymmetric N, N’ dimethylarginine while PRMT2 initially generates monomethylarginine but then generates symmetric N,N’ dimethylarginine. JMJD6, a homologue of he JHDM family of lysine demethylases, provides an arginine demethylase (Smith and Denu, 2009). A number of additional chromatin modifications are also known including ubiquitination; sumoylation; and protein arginine deiminase, which hydrolyzes the guanidinium side chain to generate citrulline and ammonia (Marmorstein and Trievel, 2009; Smith and Denu, 2009)
The distribution of histone modifications has been hypothesized to provide a “histone code” of gene expression (Fuchs et al., 2009; Strahl and Allis, 2000). In yeast, the 5’ coding regions of active genes have an increased density of H2A-K7Ac, H3-K9Ac, H3-K14Ac, H3-K18Ac, H4-K5Ac, and H4-K12Ac while the 3’ coding regions of active genes are enriched for H2B-K16Ac, H4-K8Ac, and H4-K16Ac (Wyrick and Parra, 2009). Furthermore, H3-K4me is associated with active transcription, H3-K4me3 is prevalent near promoters, and H3-K4me1 is more abundant in coding regions (Fuchs et al., 2009). In mammalian cells, active chromatin histones H3, H4, and H2A are acetylated including H3-K9Ac and histones H3 and H4 are methylated including H3-K4me, H3-K4me2 and H3-K4me3, the later found at the 5’ end of transcribed regions; H3-K36me3 found at the 3’ end of transcribed regions; plus H3-K9me, H3-K27me, and H4-K20me. By contrast, in inactive chromatin histones are hypoacetylated and H3 is methylated to generate H3-K9me3, H3-K27me3, and H3-K79me3. Methylated H3-K9 specifically interacts with Heterochromatin Protein 1 (HP1). H4-K16Ac and H4-K20me have also been noted in cancer cell chromatin (Glozak and Seto, 2007; Schneider and Grosschedl, 2007).
Beyond local histone modifications, the nuclear genome is organized into structural and functional chromosomal territories (Schneider and Grosschedl, 2007). In general, gene-rich chromosomes such as human chromosome 19 are located toward the interior of the nucleus while gene-poor chromosomes like 18 are located toward the periphery of the nucleus (Cremer et al., 2003). The functional genes within the nuclear interior are organized into “transcriptional factories” in which widely scattered genes can be colocalized into the same focus of active transcription. This permits positive and negative transcriptional regulatory factors bound to particular cis DNA elements to influence the expression of genes dispersed on the same of different chromosomes (Cai et al., 2006; Fraser and Bickmore, 2007; Gondor and Ohlsson, 2006; Schneider and Grosschedl, 2007; Zhao et al., 2006).
The chromatin in the central nucleus is generally gene-rich regions, more diffuse, and associated with increased gene expression (ridges). The chromatin toward the nuclear periphery is gene-poor, more compact, and has reduced gene expression (antiridges) (Goetze et al., 2007). Transcribed regions of the chromatin can form loops which can either interact with genes within the loop (Cai et al., 2006; Gondor and Ohlsson, 2006) or between chromatin loops on the same or different chromosomes (Schneider and Grosschedl, 2007; Zhao et al., 2006). In the case of the IL3, 4, 5 gene cluster on mouse chromosome 11, activation of T helper 2 cells results in the 200 kb region becoming folded into a series of small loops through the induction and binding of the special AT-rich sequence binding protein 1 (SATB1). SABT1 binds to the base of each loop and is accompanied by the acetylation of H3-K9 and H3-K14; the binding of the regulatory factors GATA3, STAT6, c-maf, and Brg1; and the recruitment of RNA polymerase II (Cai et al., 2006). Therefore, the regional clustering of related transcriptional units plus the development of transcriptional factories between chromosomes can provide the capacity for the transcription regulation of the widely dispersed nDNA genes of the mitochondrial genome.
Transcriptionally inactive regions of the genome can be bound in large organized chromatin K9 modifications (LOCKs) blocks. These regions can range in size up to 4.9 Mb and have been identified by the immunoprecipitation of chromatin enriched for H3-K9Me2 or H3-K27me3. They are commonly demarcated by CCCTC-binding factor (CTCF) insulator sites, and are envisioned to represent relatively inactive chromatin regions associated with the periphery of the nuclear envelope. In undifferentiated mouse ES cells only about 4% of the chromatin is in LOCKs, but in differentiated ES cells this increases to 31%. In brain 10% of the chromatin is in LOCKs while in liver 46% is in LOCKs. In the human chromosome 15q11-13 imprinted region associated with Angelman and Prader-Willi syndromes, placental chromatin contains two Mb LOCKs, one encompassing the SNRPN gene associated with Pader-Willi and the other encompassing the GABRB3 gene, the Angelman syndrome UBE3A gene lying between the two LOCKs (Wen et al., 2009). Regions of the chromatin ranging in size from 0.1 to 10 Mb have also been found to be preferentially associated with nuclear lamina, specifically lamin B1, located on the inside of the nuclear envelope. These lamina-associated domains (LADs) are generally low gene expression domains and are flanked by CTCF binding sites, CpG islands, and promoters oriented away from the LADs (Guelen et al., 2008). Approximately 82% of LOCKs correspond to LADs (Wen et al., 2009).
3.2.2. Energetic Regulation at the Signal Transduction Level
The post-translational modification of a wide spectrum of metabolic and regulatory proteins by phosphorylation and acetylation unifies the energetic regulation of chromatin with the physiological state of the cell. Phosphorylation modulates the activity of a significant portion of the cellular proteins. The great majority of receptors for environmental signals transmit these signals via tyrosine or serine-threonine phosphorylation. Most of the intermediate steps in signal transduction pathways are also mediated by kinases and phosphatases (Alberts et al., 2002).
3.2.2.1. Global Energetic Regulation of Metabolism
A more global regulation of metabolism by energy availability can be mediated by the conversion of ATP to cAMP by adenylylcyclase. cAMP is linked to cellular energetics throughout the kingdoms of life. Classic examples include modulation of the lactose operon in E. coli; providing the chemical gradient along which Dictyostelium discoidum amoebe migrate during starvation to form the fruiting body (Alberts et al., 2002); regulation of mitochondrial OXPHOS, ROS, and longevity in Drosophila (Tong et al., 2007); the modulation of peroxisome-proliferator-activated receptor γ-coactivator-1α (PGC-1α) transcription (Daitoku et al., 2003); and phosphorylation of complex I and IV proteins in mammalian cells (Bellomo et al., 2006; Chen et al., 2004; Lee et al., 2005; Papa et al., 2002; Technikova-Dobrova et al., 2001).
Acetylation-deacetylation of regulatory and metabolic proteins is also ubiquitous. The p53 protein can be acetylated by p300/CBP at six lysines and by PCAK at another lysine. Acetylation at lysine 382 by p300/CBP increases its activity, while the deacetylation of lysine by Sirt1 reduces its activity. Acetylation of transcriptional co-repressor PLZF by p300 at five lysines within the Zn-finger domain increases its DNA binding and enhances its suppression of growth promoting genes. Acetylation of GATA1 by p300/CBP within its Zn-fingers is important for chromatin binding and transcriptional activation resulting in hematopoietic terminal differentiation. NF-κB, which is a heterodimer of RelA/p65 and p50 or p52 subunits, is maintained in an inactive form in the cytoplasm complexed with IκB. Upon stimulation, IκB is phosphorylated by IκB kinase and degraded permitting NF-κB to be translocated to the nucleus. Acetylation of p65 by p300/CBP or PCAF at K218, K221, and K310 enhances DNA binding and transcriptional activity and inhibits IκB binding. Deacetylation by HDAC3 promotes IκB binding and nuclear egress. Deacetylation of NFκB by SirT1 at K310 decreases expression of NF-κB target genes (Glozak and Seto, 2007). Acetylation of PGC-1α by GCN5 attenuates its transcriptional co-activator activity suppressing mitochondrial biogenesis. Deacetylation by Sirt1 restores PGC-1α activity and increases mitochondrial biogenesis (Gerhart-Hines et al., 2007). Acetylation of FOXO by p300/CBP causes its removal from the nucleus, while deacetylation by Sirt1 facilitates its transfer back into the nucleus (Accili and Arden, 2004).
DNA methylation, DNA repair, and transcription come together through acetylation by the association of p300/CBP HAT, thymidine DNA glycosylase (TDG), and the bifunctional protein apurinic/apyrimidinic endonuclease /redox factor-1 (APE/Ref1Red/Ox). Methylation of cytosine in CpG dinucleotides is important in regulation of gene expression. However, deamination of cytosine and methyl-cytosine result in uracil and thymine, respectively, accounting for approximately 30% of all de novo nDNA mutations. Correction of the resulting G/T and G/U base pairs involves DNA glycosylases that recognize and excise the mispaired thymine or uracil. Mammals have two thymine DNA glycosylases, TDG and MBD4. MBD4 is concentrated in heterochromatin, but TDG is enriched in transcriptionally active euchromatin. When TDG binds to the G/T or G/U base mismatches, this leads to the additional binding of both p300/CBP and APE-1/Ref1Red/Ox. The p300/CBP acetylates the local histones to open the chromatin while the APE-1/Ref1Red/Ox cleaves the apyrimidinic DNA strand for repair (Tini et al., 2002). APE-1/Ref1Red/Ox is a bifunctional protein that not only participates in DNA repair, but also regulates a variety of transcription factors through thiol/disulfide conversions. Since thiol-disulfide chemistry is regulated by mitochondrial and cytosolic redox state (Hansen et al., 2006; Jones, 2006b, 2008; Kemp et al., 2008; Schafer and Buettner, 2001) the p300/CBP-TDG-APE-1/RefRed/Ox complex integrates all aspects of the energetic regulation of the epigenome (Figure 1).
The coordination between cellular metabolism and transcriptional regulation is classically illustrated by the response of mammalian metabolism to the availability of carbohydrate calorie resources. When carbohydrates were abundant to our ancestors, such as during the plant growing season, consumption of plant starch resulted in increased blood glucose levels. This stimulates the pancreatic β cells to release insulin. Insulin signals the availability of carbohydrates to the rest of the tissues in the body, shifting metabolism toward glycolysis for ATP generation and the storage of excess calories as fat. When carbohydrates are scarce, glucose becomes limiting to animals, serum glucose levels fall, insulin secretion declines, and the pancreatic α cells secrete glucagon. Glucagon then signals via cAMP to mobilize stored fat and up-regulate mitochondrial OXPHOS to burn fat and generate energy to survive periods of glucose deprivation (Wallace, 2007) (Figure 1).
When insulin is secreted due to increased serum glucose, it binds to insulin receptors of target tissue cells. This activates PI3 kinase (PI3K) which activates the Akt/PKB protein kinase. Akt then phosphorylates and inactivates the FOXO transcription factors. When unphosphorylated, FOXO transcription factors are active and bind to insulin response elements (IREs) in the promoters of nDNA genes. One FOXO target gene is the transcriptional co-activator, PGC-1α. PGC-1α interacts with a variety of tissue-specific transcription factors to induce key nDNA-encoded mitochondrial genes to up-regulate OXPHOS. Thus, when glucose is abundant, the FOXOs are phosphorylated and inactive, PGC-1α is down-regulated, mitochondrial OXPHOS is depressed, and metabolism shifts toward glycolysis. When blood glucose is low, insulin secretion decline, and the FOXOs are unphosphorylated and active. This induces PGC-1α and OXPHOS. Low blood glucose also stimulates glucagon secretion, which binds to the glucagon receptor of target calls and activates adenylylcyclase to produce cAMP. Elevated cAMP activates PKA to phosphorylate CREB which enters the nucleus and binds CRE cis elements, one of which is up-stream of PGC-1α. This up-regulates PGC-1α and OXPHOS expression. Therefore, when carbohydrates are limiting, the expression of PGC-1α is doubly induced by the dephosphorylation of the FOXOs and the phosphorylation of CREB, switching cellular metabolism toward OXPHOS to burn stored fats to survive periods of carbohydrate deprivation. Antioxidant defenses are also up-regulated in conjunction with OXPHOS to offset the effects of increased mitochondrial ROS production (Figure 1).
The switch from glycolysis to OXPHOS may also be mediated by the cytosolic NAD+/NADH levels through Sirt1. Catabolism of glucose by glycolysis reduces cytosolic NAD+ to NADH and produces pyruvate, which is further metabolized in the mitochondrion. By contrast, fatty acid oxidation occurs exclusively in the mitochondrion, leaving cytosolic NAD+ oxidized. Therefore, when glucose in abundant, the cytosolic NAD+ is reduced and the NADH can not be used by Sirt1 as a co-reactant to deacetylate the FOXOs and PGC-1α. Hence, the FOXOs and PCG-1α remain inactive down-regulating mitochodntrial biogenesis. When fats are oxidized, cytosolic NAD+ remains abundant, permitting the deacetylation of the FOXOs and PGC-1α and the up-regulation of OXPHOS.
3.2.2.2. Metabolic Regulation via Nuclear Receptors
PGC-1α is one of three members of a gene family of transcriptional co-activators (PGC-1α, PGC-1β, and PERC) that modulate mitochondrial function by regulating genes for energetics including mitochondrial biogenesis, thermogenesis, and fatty acid oxidation (Kelly and Scarpulla, 2004; Scarpulla, 2002). PGC-1α was cloned through its interaction with PPARγ (Yang et al., 2006). PPARγ is a member of the nuclear receptor (NR) superfamily, a large family of ligand-dependent transcriptional factors. This superfamily consists of 48 members in human and 49 in mouse which are central to the regulation of metabolism, development, reproduction, and immune response. All members of the nuclear receptor superfamily share a common architecture containing a N-terminal ligand independent activation domain (AF-1), a DNA binding domain (DBD) consisting of two highly conserved zinc-finger motifs which interact with the hormone response elements (HRE, 5’-AGGTCA-3’), a hinge region, and the C-terminal ligand binding domain (LBD). Various members of the NR superfamily can bind to DNA as monomers homodimers, or heterodimers. Based on their ligand binding, the NRs are divided into three major groups, “endocrine receptors” which encompass the steroid receptors and heterodimeric receptors, the “adopted orphan receptors” which include the lipid sensors and “enigmatic orphans,” and the “orphan receptors” whose ligands are unknown.
The steroid receptors of the endocrine class include the estrogen receptors (ER) α and β, androgen receptor, glucocorticoid receptor and others, all of which function as homodimers. The heterodimeric endocrine receptors include the thyroid hormone receptors α and β which dimerize with RXR.
The orphan receptors bind lipids and include PPARα, γ, and δ which also heterodimerize with RXR. PPARα is expressed in liver, heart, muscle and kidney and regulates fatty acid oxidation and apolipoprotein synthesis. PPARγ is most abundant in adipose tissue and regulates adipogenesis, but it is also expressed in liver and skeletal muscle. PPARδ is ubiquitously expressed and promotes mitochondrial fatty acid oxidation, energy expenditure, and thermogenesis. Mice over expressing PPARδ in skeletal muscle exhibit a shift in their muscle fiber distribution from glycolytic type II fibers to oxidative type I fibers with an associated remarkable increase in exercise endurance (Wang et al., 2004).
The enigmatic “adopted orphan receptors” include the estrogen-related receptors (ERR) α, β, and γ. The ERRs often act as monomers binding consensus half sites found in many genes involved in mitochondrial function. Under physiological conditions, the ERRs are not regulated by ligands, but instead by binding the inducible co-activators PGC-1α and PGC-1β. The interactions of ERRs with the PGC-1s then induce mitochondrial β-oxidation and oxidative metabolism. PGC-1α and β can interact with other NRs to up-regulate mitochondrial genes. While both can interact with PPARα and δ in muscle, heart, and white adipose tissue to induce mitochondrial fatty acid oxidation, PGC-1α can interact with FXR and HNF-4α in liver to induce gluconeogenesis during fasting, and PGC-1β can interact with SREBP-1c (steroid regulatory element binding protein-1c) and FOXA2 to promote lipogenesis. NR heterodimers involving RXR can interact with transcriptional co-repressors such as SMRT/NCOR plus HDACs in the absence of ligands to inhibit transcription, but in the presence of ligands can shift to binding transcriptional co-activators p300/CBP or SRC/p160 to activate transcription. On activation or induction of PGC-1, the ERR binds PGC-1 and this nucleates an active transcriptional complex that can include p300/CBP and SRC/p160 (Mangelsdorf and Evans, 1995; Shulman and Mangelsdorf, 2005; Sonoda et al., 2008).
While the interaction of PGC-1α and β with the NRs appear to be master regulators of oxidative metabolism, other steroid receptors also play an integral and often unexpected role in regulating mitochondrial function. It is now known that the estrogen receptors ERα and β are present not only in the nucleus-cytosol, but also in the mitochondrial matrix (Duckles et al., 2006; Pedram et al., 2006; Stirone et al., 2005). In cerebral vascular endothelial cells, estrogen induces mtDNA-encoded polypeptides such as COI and also the nDNA encoded mitochondrial polypeptides COX subunit IV (COX14), cytochrome c, and the nDNA gene mitochondrial nuclear transcription factor-1 (NRF1), as well as citrate synthetase and COX activities (Duckles et al., 2006; Stirone et al., 2005). Estrogen also decreases mitochondrial ROS production in association with an increase in MnSOD specific activity without increases in protein level in both vascular endothelial cells (Razmara et al., 2007; Razmara et al., 2008; Stirone et al., 2005) and also mammary tumor cells (Pedram et al., 2006). Therefore, the NR superfamily exerts broad control on many aspects of mitochondrial physiology in response to endogenous and exogenous environmental factors.
3.2.2.3. Metabolic Regulation via TOR Kinase
In addition to the transcriptional regulation of mitochondrial function, mitochondrial bioenergetics and biogenesis is modulated by post-translational modifications. One important mediator of cellular and mitochondrial energetics is AMP-activated protein kinase (AMPK). Under conditions where energy production is compromised (e.g. hypoxia, starvation, OXPHOS inhibitors) or when energy consumption accelerates (e.g. muscle contraction), the ATP/ADP ratio declines. This drives the adenylate kinase reaction (2 ADP → ATP + AMP). The resulting increased AMP/ATP ratio then activates AMPK. Activated AMPK phosphorylates PGC-1α; up-regulates PGC-1α expression and OXPHOS (Jager et al., 2007); induces glucose uptake and glycolysis; activates fatty acid oxidation, and suppresses fatty acid synthesis; stabilizes p53, p21, and p27 to inhibit cell growth and proliferation; and represses the target of rapamycin (TOR) kinase to inhibit protein synthesis (Hardie, 2007). When nutrients again become available, the AMP/ATP ratio declines, AMPK activity is suppressed, and TOR signaling is activated to resume protein synthesis and stimulate cell growth (Wullschleger et al., 2006) (Figure 1).
The TOR serine/threonine kinases link nutrient availability to regulation of anabolism or catabolism. Mammalian cells have two TOR (mTOR) complexes: mTORC1 and mTORC2. mTORC1 consists of mTOR, raptor, and mLST8 and it regulates protein synthesis, metabolism, ribosomal biogenesis, transcription, and autophagy (Figure 1). mTORC2 is composed of mTOR, rictor, and mLST8 and it regulates via Akt/PKB cell proliferation, survival, metabolism, transcription and actin organization and thus motility. Rapamycin, when complexed with FKBP12, inhibits mTORC1 but not mTORC2.
mTORC1 is regulated by the insulin signaling pathway, stress and hypoxia, cellular energy levels through AMPK, and nutrient levels. Insulin and insulin-like growth factors activate the insulin receptor which phosphorylates insulin receptor substrate (IRS), thus activating phosphatidyl-inositol 3 kinase (PI3K). This activates Akt/PKB to phosphorylate and inactivate the tuberous sclerosis protein complex (TSC). TSC is composed of the proteins hamartin (TSC1) and tuberin (TSC2) and TSC2 acts as a GTPase activating protein (GAP) for the small GTPase Rheb. Rheb binds to the kinase domain of mTOR. When bound to GTP Rheb activates mTOR and when bound to GDP it inhibits mTOR. Thus, when insulin and IGFs are present, TSC is inactive, and Rheb retains GTP and activates mTOR. When insulin and IGF are absent, TSC is active, Rheb GTP is converted to GDP, and mTOR is inhibited. Activated mTORC1 phosphorylates downstream proteins S6K1 and 4E-BP to increase protein synthesis, and other targets to stimulate growth related processes. The Ras/ERK pathway can also phosphorylate TSC, stimulating growth. When ATP is limiting and the AMP/ATP ratio rises, AMPK is activated and phosphorylates TSC, enhancing GAP activity, converting Rheb GTP to GDP, and thus inhibiting mTOR. Therefore, ATP deficiency inhibits protein synthesis and growth processes. Similarly, hypoxia inhibits mTORC1 signaling and limits protein synthesis and growth via activation of hypoxia inducible factors-1α and 2α (HIF-1α, HIF-2α). The HIF-1α-HIF-1β complex induces the expression of the REDD complex (REDD1 & 2) which releases TSC2 from its association with the inhibitory 14-3-3 protein permitting TSC1/2 to inhibit mTORC1 and inhibit growth (DeYoung et al., 2008). Increased amino acids stimulate growth through mTORC1. The elevated amino acids, in particular leucine, induce an influx of Ca++ into the cell, which activates calmodulin. The Ca++/calmodulin complex binds hVps34 which is attached to the mTORC1 complex. This activates hVps34 to generate PI(3)P which activates mTORC1 leading to phosphorylation of S6K1, 4E-BP, and other mTORC1 targets proteins and increased protein synthesis and growth (Gulati et al., 2008). Nutrient deprivation, by contrast, is associated with reduced cytosolic Ca++, inhibition of mTORC1, dephosphorylation of target proteins and inhibition of protein synthesis and growth. mTORC1 also phosphorylates and inhibits the ATG1 protein kinase which mediates the early steps in autophagy. Therefore, when energy and nutrients are plentiful, mTORC1 is active, ATG1 is phosphorylated and autophagy is inhibited. When nutrients and energy are limiting, mTORC1 is inactive, ATG1 becomes dephosphorylated, and autophagy recovers nutrients from cellular macromolecules (Wullschleger et al., 2006).
3.2.2.4. Metabolic and Circadian Regulation via Sirtuins & NAD+
Cellular energetics is also modulated by the cellular redox state through the NADH/NAD+ ratio. Since NAD+, but not NADH, is a substrate for many of the class III sirtuin deacetylases, sirtuin activity links protein deacetylation to the cellular NADH/NAD+ ratio. Mammals possess seven sirtuins: SIRT1, -2, -6, and -7 located in the nucleus; SIRT1 and -2 in the cytosol; and SIRT 3, -4, and -5 in the mitochondrion. SIRT1 is the best studied, deacetylating proteins using NAD+. Acetylation of FOXO transcription factors by p300/CBP reduces their activities, while deacetylation by SIRT1 increases activity. Thus, when calories are limited, NADH is oxidized to NAD+. This activates SIRT1 to deacetylate the FOXOs, up-regulating OXPHOS to begin oxidizing stored fat. PGC-1α can also be acetylated by GCN5 reducing its activity when calories are plentiful, and deacetylation by SIRT1 and NAD+ when reducing equivalents are scarce increasing PCG-1α activity in muscle and up-regulating mitochondrial fatty oxidation (Gerhart-Hines et al., 2007). SIRT1 also regulates mitophagy through deacetylation of members of the Atg family of proteins (Lee et al., 2008). SIRT3 deacetylation of various OXPHOS proteins also modulates mitochondrial metabolic rate (Ahn et al., 2008).
SIRT1 deacetylase activity is also linked to circadian rhythms (Asher et al., 2008; Nakahata et al., 2008). Circadian rhythms are generated by a transcriptional negative feedback loop in which two transcription factors BMAL1 and CLOCK form a heterodimeric complex that drives the expression of two negative regulators, PER and CRY. Expression of PER and CRY attenuates the activity of the heterodimeric BMAL1/CLOCK complex and turns down PER and CRY expression. The CLOCK protein has HAT activity which is enhanced by BMAL1 and is essential to maintain the circadian rhythm (Doi et al., 2006). CLOCK-driven acetylation of histones H3 and H4 and BMAL1 are all cyclical, as is PER2 acetylation (Asher et al., 2008; Doi et al., 2006). BMAL1 and PER2 are also more stable when acetylated (Asher et al., 2008; Nakahata et al., 2008). SIRT1 also binds and deacetylates CLOCK, BMAL1, and PER2. Therefore, deacetylation by SIRT1 plus NAD+ is also regulating circadian rhythm. SIRT1 expression level is circadian in mouse liver, being maximum at Zeitgeber time (ZT) 16 during the active dark period (ZT = 0 being the beginning of a 12 hour light cycle) and minimum at ZT4 during the quiescent light period. SIRT1 deacetylation is required for the oscillation of BMAL1 expression, since SIRT1 knockout cells have significantly reduced expression of the core circadian genes including Clock, Bmal1, Per1, and Cry1 (Asher et al., 2008; Nakahata et al., 2008). Consistent with the SIRT1 requirement for NAD+ for its deacetylase activity, oscillations of the circadian clock are coupled to oscillations of the cellular NAD+ levels. The synthesis of NAD+, in turn, is regulated by the expression of nicotinamide phosphoribosyltransferase (NAMPT), the rate-limiting step in mammalian NAD+ synthesis. The Nampt gene in mouse contains an E-box (CACGTG) cis element that binds and is regulated by the CLOCK-BMAL1-SIRT1 complex. This causes the expression of NAMPT and thus NAD+ levels to oscillate. Therefore, circadian rhythms are regulated both by the transcriptional CLOC-BMAL1 and CRY-PER feedback loop and also by the SIRT1-CLOCK-BMAL1 and NAMPT-NAD+ metabolic feedback loop (Nakahata et al., 2009; Ramsey et al., 2009; Wijnen, 2009). Since cellular energetics drives oxidation-reduction reactions and modulates the NAD+/NADH redox ratio, and the redox state of NAD+ is modulated by caloric availability, circadian rhythms, food intact, and activity must interact (Eckel-Mahan and Sassone-Corsi, 2009).
The circadian oscillations of the Clock, Bmal1, Per2, and Cry1 genes are also paralleled by oscillations in the expression of 28 of the 45 nuclear receptor genes (Yang et al., 2006). The mRNA levels of the PPAR family members showed tissue-specific oscillations that correlated with those of the Clock, Bmal1, Per2, and Cry1 genes in mouse white adipose tissue (WAT), brown adipose tissue (BAT), liver, and skeletal muscle (Yang et al., 2006). In BAT, all three PPAR genes, as well as PGC-1α and UCP-1, reach their maximal expression during ZT4, the light period when mice are at rest, and minimum at ZT16 during the dark period when mice are active. Thus, BAT thermogenesis is maximum during resting and minimum during periods of maximum activity, thus balancing heat generation by ATP hydrolysis during exercise with heat generation during rest by thermogenesis. In WAT, by contrast, PPARγ is maximum during the active dark period, ZT16, and minimal during the resting light period, ZT 4. This parallels the expression of the SREBP-1c, adiponectin, and leptin and is consistent with energy production being maximal during the dark period when mice are most active (Yang et al., 2006). Since the circadian regulation of the PPAR genes is at the transcription level, part of the regulation must be the result of changes at the chromatin level. Therefore, calorie intake and energy metabolism directly regulate epigenomic gene expression, and reciprocally epigenomic changes regulated food intake and energy metabolism.
3.2.2.5. PGC-1α and Sirtuin Mutant Mice
The spectrum of phenotypic and physiological parameters regulated by energy metabolism has been demonstrated by generating mice in which the genes of the PGC-1 and Sirtuin families have been inactivated. In mice lacking PGC-1α (−/−) only about half of the pups survive through early postnatal period into adulthood. These animals weigh 10–15% less at 2 months of age, and have abnormal BAT with large lipid droplets, but normal liver, heart, skeletal muscle, and pancreas (Lin et al., 2004). The PGC-1β knockout mice show grossly normal development, growth, and fertility (Sonoda et al., 2007).
Both the PGC-1α and PGC-1β knockout mice show reduced cold tolerance, decreased mitochondrial energy metabolism, and altered circadian rhythms indicating that the two transcription co-activators have overlapping functions. However, the two knockout mice differ in their tissue specificities demonstrating that PGC-1α and PGC-1β have different roles in regulating metabolism. PGC-1α knockout mice accumulate large lipid droplets in BAT and up-regulate gluconeogenesis in liver under normal conditions, while PGC-1β knockout mice accumulate large lipid droplets in their livers after being fed a high-fat diet. PGC-1α −/− mice are hyperactive in both light and dark cycles while PGC-1β-deficient mice have decreased activity during the dark cycle. Finally, PGC-1α –deficient but not PGC-1β-deficient mice develop neuronal degeneration (Lin et al., 2004; Sonoda et al., 2007).
The activation of PGC-1α by NAD+-linked deacetylation by Sirt1 directly ties OXPHOS induction to calorie restriction (Cheng et al., 2003; McBurney et al., 2003) (Boily et al., 2008; Wallace, 2005c). SIRT1 is the mammalian orthologue of the yeast Sir2, and Sir2 orthologues which regulate life span in association with calorie restriction in multiple species (Lin et al., 2000; Rogina and Helfand, 2004; Wallace, 2005c). Thus, longevity and mitochondrial energetics are linked.
The homozygous knockout of Sirt1 in the mouse on a 129/Sv background results in a 50% prenatal mortality, with the remaining animals being small and dying within a month. Sirt1 deficiency on a CD1 and 129/Sv mixed background results in animals which survive to adulthood, though both males and females are sterile (McBurney et al., 2003). On the 129/Sv background, the Sirt1 −/− mutation is associated with retinal and heart defects (Cheng et al., 2003). Microarray analysis of Sirt1-silenced animals suggests that Sirt1 acts more on mammalian signal transduction pathways and transcription factors than on chromatin structure (McBurney et al., 2003).
Sirt1 −/− mice remain small and are also less active, yet their daily food intake is normal. This is because they are hypermetabolic, Sirt1−/− liver mitochondria being more uncoupled and thus less efficient at generating ATP. Caloric restriction of Sirt1−/− mice does not extend lifespan, supporting the hypothesis that life span is extended by the up-regulation of mitochondrial OXPHOS through the Sirt1-mediated deacetylation of the FOXO and PGC-1α transcription factors (Boily et al., 2008).
Mice in which the mitochondrial Sirt3 and Sirt5 genes have been inactivated are viable (Haigis et al., 2006; Lombard et al., 2007). Sirt3 −/− mice have no histological abnormalities or marked changes in the expression of TFAM, MnSOD, COX IV, cyt c, and Ucp-1 in BAT even though mitochondrial proteins such as glutamate dehydrogenase (GDH) have markedly increased acetylated lysine levels (Lombard et al., 2007). However, the hyperacetylation of mitochondrial proteins seen in Sirt3 −/− mouse embryo fibroblasts and Sirt3 −/− tissues is associated with a 50% reduction in mitochondrial ATP production and complex I activity (Ahn et al., 2008). Thus, Sirt3 appears to be regulating mitochondrial basal metabolic rate. Since Sirtuins are activated by NAD+, an increased NAD+/NADH ratio resulting from a starvation state would deacetylate the OXPHOS proteins increasing mitochondrial basal metabolic rate and facilitating the oxidation of stored fats to generate ATP.
Sirt4 is an ADP-ribosylase. While Sirt4 −/− mice appear normal in development, growth, and fertility, they show increased circulating insulin levels. Exposure of Sirt4 −/− islets to glucose significantly increases insulin secretion. Injection of glutamine into Sirt4 −/− mice also increases plasma insulin and Sirt4 −/− islets exposed to glutamine or leucine have increased insulin secretion. These effects are mediated by the Sirt4-regulating ADP-ribosylation of pancreatic GDH which decreases its activity thus negatively regulating amino acid stimulated insulin secretion (Haigis et al., 2006).
Therefore, the acetylation of the FOXO transcription factors and the PGC-1α and β transcriptional coactivators via acetyl-CoA depress OXPHOS, shifting metabolism toward glycolysis in the fed state. However, during fasting the shift of the NADH/NAD+ ratio toward NAD+ results in the Sirt1-mediated deacetylation of these factors. Similarly, during fasting when NAD+ is prevalent, GDH is ADP-ribosylated and inactivated, suppressing amino acid stimulated insulin secretion. This inhibits the storage of calories as fat, activates the FOXOs and induces and activates PGC-1α, thus inducing mitochondrial OXPHOS to oxidize fatty acids and amino acids to provide energy during fasting. Therefore, regulation of energetic gene expression and metabolism is directly linked to the availability and nature of the calories for the animal, intimately linking mitochondrial energy metabolism and the epigenome.
4. MITOCHONDRIAL GENETIC DISEASE
The elucidation of a broad spectrum of mtDNA and nDNA mitochondrial gene mutation diseases has revealed that mitochondrial dysfunction can result in the entire range of clinical phenotypes associated with metabolic and degenerative diseases as well as cancer and aging (Wallace et al., 2007). Mitochondrial diseases have been shown to affect the highly oxidative tissues including the brain, heart, muscle, kidney, and endocrine systems; as well as the metabolic systems resulting in diabetes, obesity, and influence a range of age-related disorders. Thus, the symptoms of mitochondrial diseases correspond to those attributed to epigenomic changes.
4.1. Recent mtDNA Mutations and Diseases
Clinically relevant mtDNA variants fall into three classes: recent deleterious mutations resulting in maternally transmitted disease, ancient adaptive variants that predispose individuals to disease in different environments, and the age-related accumulation of somatic mtDNA mutations that erode function and provide the aging clock. The extraordinary clinical variability of mtDNA diseases results from different mutations in the same gene causing different phenotypes and from the same mutation at different levels of heteroplasmy causing different phenotypes (Wallace, 2005c).
Pathogenic mtDNA mutations include both rearrangement mutations and base substitution mutations. Rearrangement mutations can either be de novo deletion mutations or maternally transmitted insertion mutations which are unstable and generate deletion mutations in post-mitotic cells. Most deletion mutations remove at least one tRNA and thus affect protein synthesis (Wallace et al., 2001). MtDNA rearrangement syndromes are invariably heteroplasmic and have been associated with maternally-inherited diabetes and deafness, Chronic Progressive External Ophthalmoplegia (CPEO), the Kearns-Sayre Syndrome (KSS), and the Pearson marrow/pancreas syndrome. Differences in mtDNA rearrangement phenotypes appear to stem from differences between insertions and deletions, the diversity of tissues that contain the rearrangement, and the percentage of mtDNAs harboring the rearrangement in each tissue (Wallace et al., 2007).
Base substitution mutations can alter either polypeptide genes (polypeptide mutations) or rRNAs and tRNAs (protein synthesis mutations). Pathogenic polypeptide mutations encompass a broad spectrum of multisystem diseases including LHON (Wallace et al., 1988a), Leigh syndrome (Holt et al., 1990), and mitochondrial myopathy (Andreu et al., 1999a; Andreu et al., 1998; Andreu et al., 1999b). Mitochondrial tRNA and rRNA protein synthesis mutations can result in multisystem diseases including Myoclonic Epilepsy and Ragged Red Fiber Disease (MERRF) (Shoffner et al., 1990; Wallace et al., 1988b); Mitochondrial Encephalomyopathy, Lactic Acidosis, and Stroke-Like Episodes (MELAS) (Goto et al., 1990); encephalomyopathy; mitochondrial myopathy and exercise intolerance; CPEO and KSS; gastrointestinal syndrome; dystonia; diabetes; deafness; cardiomyopathy; renal failure, Alzheimer Disease (AD); Parkinson Disease (PD). (Wallace et al., 2007).
4.2. Ancient mtDNA Mutations and Disease
As modern humans migrated out of Africa, mtDNA mutations accumulated along radiating maternal lineages. When a mtDNA acquired a functional mutation beneficial in a particular environment, then that mtDNA became enriched by selection and radiated within that environment to give a cluster of related mtDNA haplotypes, known as a haplogroup. Hence, haplogroups correlate with the geographic origin of the indigenous population being studied and are designated by sequential alphabetical letters (Mishmar et al., 2003; Ruiz-Pesini et al., 2004; Ruiz-Pesini and Wallace, 2006; Wallace, 2005c; Wallace et al., 1999; Wallace et al., 2003).
These same ancient adaptive mtDNA mutations influence individual predisposition to a wide spectrum of common diseases today. Various mtDNA haplogroups have been associated with neurodegenerative diseases (Brown et al., 2002; Brown et al., 1997; Brown et al., 1995; Carrieri et al., 2001; Chagnon et al., 1999; Ghezzi et al., 2005; Jones et al., 2007; Khusnutdinova et al., 2008; Shoffner et al., 1993; Torroni et al., 1997; Udar et al., 2009; van der Walt et al., 2004; van der Walt et al., 2003), diabetes and metabolic syndrome (Crispim et al., 2006; Fuku et al., 2007; Mohlke et al., 2005; Nishigaki et al., 2007; Saxena et al., 2006), infectious disease including AIDS progression (Baudouin et al., 2005; Hendrickson et al., 2008; Hendrickson et al., 2009; Raby et al., 2007), longevity (De Benedictis et al., 1999; Ivanova et al., 1998; Niemi et al., 2003; Rose et al., 2001; Tanaka et al., 2000; Tanaka et al., 1998), and cancer (Bai et al., 2007; Booker et al., 2006; Brandon et al., 2006; Darvishi et al., 2007; Gottlieb and Tomlinson, 2005; Wallace, 2005a). From these reports, it is clear that mtDNA functional variation modulates predisposition to a wide range of metabolic and degenerative diseases as well as influenceing cancer risk and longevity.
4.3. Somatic mtDNA Mutations in Aging and Cancer
Mutations in the mtDNA have been observed to accumulate with age in a variety of post-mitotic tissues in a wide range of species, and in a spectrum of complex of age-related diseases (Wallace, 2005c). Increasing the mtDNA mutation rate in mice increases their aging rate (Kujoth et al., 2005; Trifunovic et al., 2004) while decreasing the somatic mtDNA mutation rate by introducing catalase into the mitochondrial matrix extends mouse life span (Schriner et al., 2005). Therefore, the accumulation of somatic mtDNA mutations provides an aging clock that helps define an animal’s life span and contributes to the delayed-onset and progressive course of complex diseases (Wallace, 2005c). Moreover, both somatic and germline mtDNA mutations have been associated with cancer (Brandon et al., 2006; Ishikawa et al., 2008; Petros et al., 2005; Wallace, 2005a).
4.4. Mitochondrial Diseases of nDNA-mtDNA Interaction
Mutations in nDNA-encoded OXPHOS genes have also been linked to a variety of multisystem disorders (Wallace et al., 2007). These include mutations that inactivate OXPHOS complex structural and assembly genes (Procaccio and Wallace, 2004; Zhu et al., 1998), mutations that destabilize the mtDNA (Carrozzo et al., 2007; Kaukonen et al., 2000; Mandel et al., 2001; Nishino et al., 1999; Palmieri et al., 2005; Saada et al., 2001; Spelbrink et al., 2001; Van Goethem et al., 2001), and mutations that perturb mitochondrial fusion and fission (Delettre et al., 2000; Zuchner et al., 2004).
5. MITOCHONDRIAL EXPLANATIONS FOR EPIGENETIC DISEASES
Therefore, mitochondrial gene defects can result in virtually all of the symptoms associated with the common “complex” diseases, confirming the importance of mitochondrial bioenergetics in health. Since mitochondrial metabolism also regulates the substrates for epigenomic regulation, it follows that changes in mitochondrial metabolism may also perturb the epigenomic state. Furthermore, in cells and animals in which the epigenomic elements NRs, PGC-1α and β, and the Sirtuins are inactivated, mitochondrial function is also perturbed. Similarly, compounds such as resveratrol and its derivatives, which activate SIRT1, can up-regulate mitochondrial energy production and have been found to ameliorate metabolic and age-related phenotypes (Baur et al., 2006; Baur and Sinclair, 2006; Feige et al., 2008; Figarella-Branger et al., 1992; Lagouge et al., 2006; Milne and Denu, 2008; Milne et al., 2007; Pearson et al., 2008; Wood et al., 2004). It therefore follows that alterations in the epigenome should also perturb mitochondrial function. Epigenetic alteration in mitochondrial function could occur at either the local intra-chromosomal domain level to suppress mitochondrial regulatory or maintenance genes or at the inter-chromosomal level to disrupt transcriptional factories resulting in the loss of coordinate regulation of energetic genes.
5.1. Altered Intrachromosomal Domains and Energetics
Evidence that mitochondrial dysfunction is associated with epigenomic changes can be found for a wide range of classical epigenomic diseases. In cancer, a genetic disease, changes in the epigenome such as “Loss of Imprinting” (LOI) and hypomethylation are common. LOI of the insulin-like growth factor II (IGF2) provides one example of how changes in the cancer epigenome might affect mitochondrial function. Half of Wilms tumors as well as lung cancer, breast cancer, ovarian cancer, and glioma show LOI of the IGF2 locus. IGF2 is an imprinted locus normally expressed only from the paternal allele. Hence, LOI increases IGF2 expression (Bjornsson et al., 2007; Feinberg, 2007). In the mouse, LOI of Igf2 is associated with enhanced Igf2 signaling due to augmentation of Akt/PKB signaling (Kaneda et al., 2007). Activation of the Akt/PKB pathway suppresses mitochondrial OXPHOS and drives the cell toward glycolysis (Figure 1). Glycolysis is also enhanced in liver cancer by reversing the hyermethylation of the promoter of hexokinase II. Following epigenomic activation, hexokinase II transcription is further regulated by glucose; hypoxia via HIF-1α, cAMP via CREB, insulin and glucagon, and mutated p53 (Goel et al., 2003; Wallace, 2005a). Hexokinase II binds glucose and ATP, converting them into glucose-6-phosphate, the commitment step of glycolysis. Akt/PKB also phosphorylates hexokinase II causing it to bind to the mitochondrial outer membrane pore protein porin (voltage dependent anion channel, VDAC) with high affinity. The binding of hexokinase II to VDAC also antagonizes the pro-apoptotic action of Bax and Bak. The expression profile of the ANT isoforms is also altered in transformed cells (Torroni et al., 1990) and the ANTs export the mitochondrial ATP from the matrix to the intermembrane space so that VDAC can export ATP into the cytosol. Thus the binding of hexokinase II to VDAC means that ATP leaving the mitochondria binds preferentially to hexokinase II. This co-opts mitochondrial ATP production to drive glycolysis. Therefore epigenetic changes in cancer specifically suppress mitochondrial function in favor of glycolysis.
The chromatin modifications of cancer cells can be directly modulated by mitochondrial OXPHOS. Cancer cells which have been cured of their mtDNA but either chemical or genetic treatments have been found to have altered methylation patterns in 2–9% of the CpG islands surveyed. This is comparable to the 2–5% alteration in methylation regions associated with epigenomic changes during differentiation. In three out of four cases, removal of the mtDNA leads to the hypomethylation commonly associated with LOI in cultured cancer cells. When mtDNAs were reintroduced to the mtDNA-deficient cancer cells, 30% of the hypomethylated sitres become remethylated. Thus, there is a direct cause and effect relationship between mitochondrial function and epigenomic methylation in cancer cells (Naviaux, 2008; Smiraglia et al., 2008).
The p53 protein also regulates OXPHOS through induction of the complex IV chaperone protein “Synthesis of Ctyochrome c Oxidase 2” (SCO2) which is responsible for inserting Cu into COX. By contrast, p53 negatively regulates phosphoglycerate mutase of glycolysis and Akt. Therefore, mutational inactivation of p53 in cancer cells suppresses OXPHOS and induces glycolysis (Matoba et al., 2006).
Epigenetic alteration in genomic metylation in association and in part because of mitochondrial OXPHOS deficiency act in concert with alterations in the expression of IGF2, hexokinase II, the ANTs, and the inactivation of p53, to shift cancer cell energy production of toward glycolysis. This helps to explain Otto Warburg’s observation over 70 years ago that cancer cells generate excess lactate indicating hyperactive glycolysis, in the presence of oxygen, “aerobic glycolysis” (Wallace, 2005a).
LOI in the IGF2 gene region on chromosome 11q15.5 can also result in the dymorphology condition, Beckwith-Wiedemann Syndrome (BWS). About 15% of BWS of patients show LOI at the IGF2 locus reactivating the maternal allele and resulting in the overproduction of IGF2,. IGF2 overproduction leads to prenatal overgrowth, midline abdominal wall defects, ear creases and pits, and neonatal hypoglycemia. BWS cases associated with IGF2 LOI are also prone to Wilm’s tumor (Bjornsson et al., 2007; Feinberg, 2007, 2008). Again, over-expression of IGF2, acting via the PI3K-Akt-FOXO-PGC-1α pathway, would suppress mitochondrial OXPHOS (Figure 1).
In the mouse, the BWS locus is located on chromosome 7. The expression of the paternally expressed Igf2 gene and the maternally expressed H19 are regulated by the surrounding chromatin loop structure. The Igf2 and H19 genes are encompassed in an approximately 100 kb imprinted region, separated by a CTCF insulator site with an enhancer down stream of H19. The paternal and maternal transcription of the Igf2 and H19 genes is regulated by the interaction of differentially methylated regions (DMRs), three in Igf2 and one at the CTCF insulator site. On the maternal chromosome, the DMR of H19 associates with DMR1 near the 5’ end of Igf2. This places H19 in the active chromatin domain with the enhancer and the Igf2 gene on the other side of the CTCF site in an inactive chromatin domain shutting off Igf2. On the paternal chromosome, the H19 DMR interacts with the 3’ Igf2 DMR2 domain. This creates a loop which brings the Igf2 gene adjacent to the H19 enhancer on the same side as the enhancer, thus permitting its transcription. The H19 gene on this chromosome is methylated and silenced (Murrell et al., 2004). Therefore, LOI in both cancer and BWS link epigenomic alterations to mitochondrial dysfunction.
Prader-Willi syndrome (PWS) and Angelman syndrome (AS) are classical examples of epigenomic diseases resulting from genetic errors in imprinted loci on chromosome 15q11-13. Preliminary evidence is implicating mitochondrial dysfunction in AS as well. PWS is associated with hypotonia, hyperphagia, and obesity while AS is associated with hyperactivity, thinness, and autism. PWS involves a paternal deficiency in 15q11-13 while AS involves a maternal deficiency. Current evidence suggests that the expression of the small nuclear ribonuclear polypeptide N (SNRPN) and the adjacent imprinting control region (ICR) and the expression of the ubiquitin-protein ligase E3A (UBE3A) are reciprocally imprinted on the maternal and paternal chromosomes. Therefore, genetic inactivation of the expressed UBE3A gene is responsible for AS and inactivation of the expressed SNRPN and ICR loci are associated with the PWS phenotype (Feinberg, 2007). In an analysis of mice mutated at the UBE3A locus, we have found that the brain mitochondria are small and dense and that the activity of the combined respiratory complex III is reduced (Su et al., 2009). Therefore, mitochondrial dysfunction may contribute to the etiology of AS. While the mechanism by which UBE3A deficiency causes mitochondrial dysfunction is unknown, UBE3A is an E3 ubiquitin ligase and ubiquination is important in maintaining the structural and functional integrity of the mitochondrion (Escobar-Henriques et al., 2006; Neutzner and Youle, 2005). The mitochondrial ubiquitin ligase MARCH-V is important in regulating the mitochondrial fusion and fission dynamics through Drp1 and Fis1 (Karbowski et al., 2007; Nakamura et al., 2006) and the human deubiquitinating enzyme ubiquitin-specific protease 30 (USP30) is embedded in the mitochondrial outer membrane and important in maintaining mitochondrial morphology (Nakamura and Hirose, 2008). Therefore, loss of the 15q11-13 loci could perturb mitochondrial function and explain some of the pathophysiology of these diseases.
Tuberous sclerosis complex (TSC) is another cell over-growth disorder, which affects virtually every organ of the body. Clinical features include cutaneous hypomelanotic macules, facial angiofibromas, etc.; central nervous system subependymal nodules, and cortical tubers, learning difficulties, and seizures; renal angiomyolipomas and cysts; cardiac rhabdomyomas; and retinal hamartoma. TSC is caused by mutations in either the hamartin TSC1 gene or the tuberin TSC2 gene. TSC1 + 2 act through Rheb to regulate TORC1, and TOR is implicated in regulation of mitochondrial OXPHOS (Chen et al., 2008; Shadel, 2008)(Figure 1).
About 25–50% of TSC subjects manifest autistic traits. Autism has been associated with alteration in neurite growth and synaptic plasticity and there is growing evidence that the mitochondria play a central role in these processes (Kang et al., 2008; Li et al., 2004; Mattson and Liu, 2002; Schuman and Chan, 2004). Therefore, TSC forms a connection between cell over-growth syndromes, neurological disorders such as autism, and mitochondrial energy metabolism.
The Fragile X Mental Retardation (FMR) Syndrome is also associated with mental retardation and autism. The FMR protein (FMRP) is an RNA binding protein that has recently been shown to be required for the expression of the mitochondrial and cytosolic Cu/Zn superoxide dismutase (SOD1) (Bechara et al., 2009). Since SOD1 functions to eliminate mitochondrial intermembrane space superoxide, its suppression could increase mitochondrial oxidative stress and thus cause symptoms through mitochondrial dysfunction (Wallace, 2005c).
Rett syndrome is an autism spectrum disorder caused by mutations in the X-linked MeCP2 gene (Amir et al., 1999; Loat et al., 2008). Rett patients have been reported to harbor mitochondrial structural aberrations and reductions in skeletal muscle OXPHOS complexes I, III, IV, though not II. Furthermore, mice in which MeCP2 is inactivated have increased complex III and core protein 1 gene (Ugcrc1) expression, together with reduced OXPHOS coupling (Eeg-Olofsson et al., 1989; Heilstedt et al., 2002; Kriaucionis et al., 2006). The MeCP2 protein binds to methyl CpG nDNA domains and modulates gene expression, either activation or repression, in part by recruiting histone-modifying enzymes such as deacetylases and methyltransferases (Chen et al., 2003b; Klein et al., 2007). MeCP2 can impart long-term silencing by binding to CpG rich domains and stimulating the deacetylation of histones H3 and H4 resulting in chromatin condensation. MeCP2 can also perform histone deacetylase-independent repression which is relevant to X chromosome inactivation and genomic imprinting (Matijevic et al., 2009). One of the targets of MeCP2 is promoter III of the neurotrophin gene, BDNF (Brain-Derived Neurotrophic Factor). It was first thought the MeCP2 repressed BDNF expression (Chen et al., 2003b), but MeCP2 was subsequently reported to enhance BDNF expression (Klein et al., 2007). Neurotrophic factors such as BDNF as well as cAMP stimulate the phosphorylation of CREB, which is important in neuronal plasticity and neurite outgrowth. Furthermore, the translation of BDNF mRNA and p250GAP mRNA are inhibited by the microRNA, miR132, and both BDNF and p250GAP regulate neuronal morphogenesis. The expression of miR132 is also induced by phospho-CREB demonstrating a feedback loop for regulating neuronal morphogenesis (Klein et al., 2007; Vo et al., 2005). Neurotrophic factors such as BDNF and NGF, bind and activate the tropomyosin-related kinase (Trk) receptor B (TrkB). Major down-stream pathways of the neurotrophic factors are the PI3K-Akt- and MAPK pathways and PKC. These in turn regulate AP1, NFκB, and the FOXOs to regulate energy production pathways, antioxidant enzymes, and anti-apoptotic proteins (Mattson, 2008).
In a rat brain preparation of mitochondria plus synaptosomal membranes, BDNF was found to act through the MAPK pathway to increase OXPHOS coupling efficiency, presumably increasing ATP production. This modulation occurs through complex I, not complex II, and occurs when glutamate and malate are used as the mitochondrial NADH generators, but not when pyruvate and malate were used. Thus, the BDNF regulation of mitochondrial respiration is specific for glutamate as a substrate, and glutamate along with γ-aminobutyric acid (GABA) are coupled through the TCA cycle to neuronal metabolism and transmission. BDNF can also modulate the stimulation of oxygen consumption by calcium, which is also consistent with increasing coupling efficiency (Markham et al., 2004). Surprisingly, the BDNF receptor, TrkB, has been localized to the mitochondrion, as well as to the plasma and endoplasmic reticulum membranes, in both muscle and nerve cells. The TrkB receptor has three isoforms, full length TrkB with tyrosine kinase activity, and two truncated forms (TrkB-T1 and TrkB-T2) without the tyrosine kinase domain. The truncated forms of TrkB appear to predominate in the mitochondrion, and preliminary studies suggest that they are localized to the mitochondrial inner membrane (Wiedemann et al., 2006). Since TrkB1 has been reported to mediate BDNF-evoked calcium signaling in glial cells, it is possible that TrkB-T1 is directly regulating mitochondrial ATP production and mitochondrial calcium homeostasis through modulating the mitochondrial membrane potential. Therefore, inactivation of MeCP2 could selectively repress the expression of the nDNA encoded mitochondrial genes, providing a coherent argument that at least part of the pathophysiology of Rett syndrome involves mitochondrial dysfunction.
MeCP2 also binds to the PWS imprinting control region (ICR) and down-regulates Ube3a and the adjacent Gaba receptor gene (Gabrb3). MeCP2 also binds to the H19 imprinted region altering regulation of the adjacent Igf2 expression (Samaco et al., 2005). This co-regulation of the dispersed genes within a chromosomal region suggests that MeCP2 might regulate chromatin transcriptional loop domains (Murrell et al., 2004) and perhaps disrupts a mitochondrial gene transcriptional factory. Therefore, Rett Syndrome provides a strong case for how perturbation of the epigenome could reduce mitochondrial function.
Timothy syndrome, which presents with autism and Long QT syndrome, provides further evidence for connecting neurodegenerative disease with Ca++ regulation and thus mitochondrial function. Timothy Syndrome has been linked to mutations in the T-type voltage-gated Cav1.2 channel encoded by the CACNA1C gene, most commonly involving the G406R allele (Gargus, 2009). Cav1.2 shows its highest expression in the hippocampus, amygdale, and putamen. A survey of the CACNA1H gene, a close paralog of the Timothy syndrome Ca++ channel, in 461 autism DNAs revealed six new mutations. Functional studies of two of these mutant proteins, R212C and R902W, revealed extended Ca++ influx into the cytosol (Splawski et al., 2006). Cytosolic Ca++ levels are regulated by mitochondrial Ca++ uptake, driven by the mitochondrial inner membrane potential (ΔP). Excessive Ca++ uptake would limit ATP production, increase oxidative stress, and activate the mtPTP causing cell death.
Evidence is also accumulating implicating mitochondrial dysfunction in some forms of autism spectrum disorder (ASD), in addition to AS, TSC, Rett syndrome, FMR and Timothy syndrome. A subset of ASD patients is associated with chromosomal gene copy number variants (CNV). Ten percent of sporadic autism patients have CNVs, as compared to 3% of multiplex families and 1% of controls (Sebat et al., 2007; Zhao et al., 2007). A subset of these variants may coincide with important nDNA mitochondrial gene loci (Smith et al., 2008). Alterations in mitochondrial structure and function have been repeatedly observed in autistic patients (Correia et al., 2006; Filipek, 2005; Gargus and Imtiaz, 2008; Haas et al., 1996; Holtzman, 2008; Lombard, 1998; Oliveira et al., 2007; Poling et al., 2006), and alterations in the mtDNA have been observed in some cases of ASD (Fillano et al., 2002; Graf et al., 2000; Pons et al., 2004). The activity of the mitochondrial inner membrane Ca-regulated aspartate/glutamate carrier (AGC) gene, SLC25A12, and/or its expression have been reported to be increased in autistic patients, at least in part due to elevated brain calcium levels (Lepagnol-Bestel et al., 2008; Palmieri et al., 2008; Ramoz et al., 2004). SLC25A12-deficient mice show impaired myelination, confirming that alterations in AGC activity can affect neurological function (Hong et al., 2007). Since AGC regulates the increased flux of reducing equivalents into the mitochondrion, increased AGC activity would initially increase ATP production, but ultimately overload the ETC with electrons increasing oxidative stress and causing the loss of neuronal spines and synapses characteristic of autism pathology (Lepagnol-Bestel et al., 2008; Palmieri et al., 2008; Ramoz et al., 2004). Therefore, mitochondrial dysfunction may account for some forms of ASD.
The importance of mitochondrial ROS production in cell function and stem cell biology has been demonstrated in studies of mice in which the Bmi1 member of the Polycomb family has been inactivated. The Polycomb family of transcription repressors mediates gene silencing by regulating chromatin structure. Bim1 is essential for the maintenance and self-renewal of both the hematopoietic and neuronal stem cells, and Bim1−/− mice have severe neurological abnormalities, alterations in hematopoietic cells, growth retardation, and shortened life span. Bone marrow cells from Bim1-deficient mice show an inhibition of mitochondrial ETC, reduced O2 consumption, reduced ATP production, and increase mitochondrial ROS production. The increased mitochondrial ROS is a major factor for the reduced stem cell function. The reduction of the Bim1−/− induced ROS using N-acetylcysteine antioxidant is associated with the recovery of the bone marrow derived cells and partial restoration of thymic atrophy. In part, the effects of the increased mitochondrial ROS are due to activation of the DNA damage response pathway. This was confirmed by inactivation of the Chk2 gene which partially restored the normal phenotype (Liu et al., 2009). Therefore, genes central to stem cell function and thus development can be directly regulated by mitochondrial ROS signaling.
The mitochondrion also plays an important role in calcium homeostasis. In mice, neuronal spine formation and maintenance can be modulated by the inactivation of the mitochondrial anti-apoptotic protein, Bcl-w, and the glutamate receptor δ (Grid2), an excitatory Ca++ channel expressed in Purkinje cells. Bcl-w and Grid2-deficient mice develop enormously elongated mitochondria in their Purkinje dendrites indicating blocked mitochondrial fission. This is associated with aberrant dendrites, spines, and synapses and results in severe ataxia. Therefore, mitochondrial calcium not only regulates mitochondrial metabolism but also mitochondrial fission, directly affecting synapse formation, learning, and memory (Liu and Shio, 2008).
5.2. Altered Transcriptional Factories and Energy Deficits
While the above examples suggest the inhibition of mitochondrial function through alterations in the transcriptional regulation of individual chromosomal domains, the maintenance of the mitochondria likely also requires the coordinate expression of many mitochondrial loci dispersed across multiple chromosomes. These dispersed mitochondrial genes might require inter-chromosomal factories for coordinate expression. Therefore, we might expect that epigenomic defects that disrupt transcriptional factories might also be associated with mitochondrial dysfunction. The laminopathies could represent this class of epigenomic defect.
The laminopathies are caused by mutations in the LMNA gene, the product of which is involved in the production of the type V intermediate filament lamina that line the inside of the nuclear envelope, but are also found in the nucleoplasm. The lamina are produced from three genes: lamins A and C produced by alternative splicing from the LMNA gene, lamin B1 produced by the LMNB1 gene, and lamins B2 + B3 produced from the LMNB2 gene. The B-type lamins are ancient and constitutive and remain associated with the nuclear envelope at interphase and during mitosis. The A-type lamins (A/C) arose along with the metazoans, are expressed in association with differentiation, dissociate from the nuclear envelope during mitosis (Liu and Zhou, 2008), and a substantial portion are found centrally within the nucleus (Galiova et al., 2008).
The lamin A protein consists of a globular N-terminus domain, followed by an α-helical rod-like domain containing four coiled-coil repeats (1A, 1B, 2A, 2B), a nuclear localization sequence, an Ig-like domain, and a C-terminal domain which includes the end of the mature lamin A followed by a C-terminal pre-peptide extension ending in the sequence Cys-Ser-Ile-Met. In newly synthesized normal prelamin A, the Cys in the Cys-Ser-Ile-Met peptide is farnesylated by farnesyltransferase (FT) resulting in binding to the endoplasmic reticulum (ER). The ER metalloprotease, ZMPSTE24, then cleaves off the terminal Ser-Ile-Met peptide, and the free Cys carboxyl group is methylated. ZMPSTE24 again cleaves prelamin A 15 amino acids N-terminal from the farnesylated Cys. This releases soluble lamin A to be imported into the nucleus where it is assembled with lamin B into the nuclear lamina as well as being located in the nucleoplasm. Lamin C encompasses the N-terminal 2/3 of the lamin A protein (Liu and Zhou, 2008). The lamin B proteins are also farnesylated, but not proteolytically processed. Hence, they remain membrane bound and diffuse along the ER into the nuclear envelope where they nucleate the assembly of the soluble lamin A and C polypeptides into the nuclear lamina.
Over 200 pathogenic mutations have been identified in the LMNA gene yielding a plethora of clinical phenotypes. These phenotypes can be recapitulated by the introduction of Lmna and Zmpste24 gene mutations into the mouse, thus proving that lamin A and C defects cause these diseases (Stewart et al., 2007).
The pathophysiology of these diseases remains a mystery. A common finding among the LMNA mutations is the presence of highly distorted nuclei and marked repositioning of heterochromatin within the nucleus (Liu and Zhou, 2008; Park et al., 2009; Worman and Bonne, 2007). The nuclear lamina binds the Sun 1 and 2 proteins, which extend through the inner nuclear membrane, interact with the Nesprin-2 Giant and Nesprin 3 proteins that extend through the outer nuclear membrane, and which bind to the cytosolic actin cytoskeleton. Emerin, which interacts with lamin A, also links the nuclear envelope to the microtubule network and anchors the centrosome (Hale et al., 2008; Lee et al., 2007). LMNA-induced alterations in the cytoskeleton should alter the distribution and movement of mitochondria which are tethered to the cytoskeleton. Since treatment of cells with microtubule inhibitors alters mitochondrial OXPHOS activity and increases ROS production (Wagner et al., 2008), LMNA mutations could contribute to reduced mitochondrial function and increased ROS production.
Lamin A deficiency also results in redistribution of chromatin within the nucleus. In immortalized mouse embryo fibroblasts, 50% of the nuclear periphery is occupied by heterochromatin, but in LMNA −/− cells, the level of peripheral heterochromatin is significantly reduced. Moreover, the number of chromocenters is reduced in the nuclear interior of LMNA −/− cells and the heterochromatin clusters in larger internal nuclear aggregates. LMNA −/− cells also have smaller chromosomal domain areas than LMNA +/+ cells. Among the HP1α, β, and γ proteins important in heterochromatization, the HP1β develops chain-like aggregates. Treatment of LMNA +/+ or −/− cells with the HDAC inhibitor, Trichostatin (TSA), causes the areas of the chromosomal domains of LMNA −/− cell to increase and approach the size and distribution as those of LMNA +/+ cells and also restores the distribution of the HP1β and SC-35 foci (Galiova et al., 2008). Therefore, LMNA mutations that disorganize the chromatin domain structure and could result in loss of coordinate transcriptional control of the dispersed energy gene.
Consistent with this conjecture, lamin A mutations have been associated with an altered distribution and function of key transcription factors (e.g., c-fos, β-catenin, etc.) (Liu and Zhou, 2008; Pekovic et al., 2007; Pekovic and Hutchison, 2008), activation of the MAPK stress pathway (Muchir et al., 2007), and the Notch pathway (Scaffidi and Misteli, 2008). Lamin A alterations have also been associated with suppression of the Wnt and Mitf signaling pathways in epidermal stem cells (Espada et al., 2008) and of myogenin (Myog) expression in muscle cells. Increases in H3-K9me2 and failures to hypertrimethylate H3-K4 around Myog and the redistribution of H3-K27me3 away from the pericentric heterochromatin have also been observed (Hakelien et al., 2008). Therefore, lamin A appears to be important in maintaining an open chromatin conformation and thus the sustaining of transcription, perhaps within transcriptional factories important for coodinate mitochondrial gene expression.
Patients with LMNA mutations have been classified into four classes based on clinical phenotypes (Liu and Zhou, 2008). However, LMNA mutations frequently exhibit pleiotrophy phenotypes and variable familial expressivity (Mercuri et al., 2005; Rankin et al., 2008). Class I laminopathies encompasses diseases of skeletal muscle and heart, including autosomal dominant (AD)-Emery-Dreifuss muscular dystrophy (EDMD), autosomal recessive (AR)-EDMD, AD-dilated cardiomyopathy 1A, and AD-limb girdle muscular dystrophy (LGMD). EDMD and LGMD LMNA mutations present with variable degrees of muscle weakness, cardiomyopathy, and contractures (Park et al., 2009). Cardiomyopathy patients are subject to cardiac conduction defects, sudden death, and dilated cardiomyopathy, with or without skeletal muscle weakness (Perrot et al., 2009; Wolf et al., 2008). EDMD mutations are distributed in lamin A from the N-terminal end through the Ig-like domain, but can be found elsewhere. DCM1A and LGMD1A mutations are found primarily in the coiled-coil protein domain (Liu and Zhou, 2008). Suggestive evidence that heart-muscle lamin A diseases are associated with mitochondria dysfunction comes for a boy who presented at six with delayed motor milestones, and proximal and truncal weakness. This progressed by nine years to severe, progressive cardiomyopathy necessitating heart transplant. His mother and brother had died at 31 and 8 years, respectively, of cardiomyopathy. Mitochondrial respiratory chain analysis of heart and skeletal muscle revealed a complex IV activity of 0.005 (normal being 0.014–0.034). Molecular studies failed to detect any of the known common pathogenic mtDNA mutations. Morever, nDNA studies revealed that the family harbored the common LMNA p.R644C missense mutation (Mercuri et al., 2005).
The Class II laminopathy involves AR-Charcot-Marie-Tooth (CMT) type 2B1, a peripheral neuropathy. The two LMNA mutations linked with AD-CMT2B1 are associated with mildly reduced nerve conduction, neuron demyelination, and axonal degeneration (Liu and Zhou, 2008), and both are adjacent to each other in the 2A coiled-coil domain (Liu and Zhou, 2008). Charcot-Marie-Tooth, Type 2A can also be caused by mutations in the nDNA-encoded mitochondrial fusion gene, mitofusin 2 (Zuchner et al., 2004)
Class III laminopathies encompasses the lipodystrophy syndromes and include AD-familial partial lipodystrophy, Dunnigan-type (FPLD2); AD-congenital generalized lipodystrophy type 2 (CGL2); and AR-mandibuloacral dysplasia. FPLD2 LMNA mutations are associated with loss of subcutaneous white adipose tissue (WAT) in limbs and the accumulation of WAT in the face, neck, and abdominal areas. The CGL2 LMNA mutations lead to diabetes with glucose intolerance, hepatic stenosis, hyperpigmentation, muscular atrophy, and hypertrophic cardiomyopathy (Araujo-Vilar et al., 2008; Liu and Zhou, 2008; Worman and Bonne, 2007). Women with FPLD2 from LMNA mutations can develop an array of fertility problems including polycystic ovaries, infertility, gestational diabetes, and preeclampsia (Vantyghem et al., 2008). Population variants in LMNA have also been associated with increased risk of diabetes (Duesing et al., 2008), and alterations in LMNA mRNA levels are observed in patients with diabetes and obesity (Miranda et al., 2008). FLPD mutations are often found in the Ig-like domain and the C-terminal region of the mature lamin A protein (Liu and Zhou, 2008).
A mitochondrial association with Class III laminopathies is supported by a study of cultured human fibroblast cell lines harboring six different heterozygous LMNA mutations (D47Y, L92F, L387V, R399H, L421P, and R482W) all derived from patients with insulin resistance and/or lipodystrophy. All of these cell lines were found by western blot to have a marked deficiency in the level of the mtDNA COII (COX2) protein but not the nDNA COX4 protein. These cell lines also showed increased ROS production which colocalized with the mitochondria (Caron et al., 2007).
All of these cells exhibit distorted nuclei have reduced proliferative capacity, increased tendency to generate senescent SA-β-galactosidase positive cells, and elevated prelamin A protein levels. Treatment of normal fibroblast cultures with the HIV anti-retroviral protease inhibitors indinavir and nelfinavir, which cause chemically-induced lipodystrophy syndromes, generated the same mitochondrial defects and cellular phenotypes as seen in the laminopathies (Caron et al., 2007). Furthermore, AIDS progression and drug-induced lipodystrophy have also been shown to be modulate mtDNA haplogroups (Hendrickson et al., 2008; Hendrickson et al., 2009).
Class IV laminopathies encompasses the accelerated aging syndromes, AD-Hutchinson-Gilford progeria syndrome (HGPS), AD-Werner Syndrome (WS), and AD-restrictive lethal dermopathy (Worman and Bonne, 2007). AD-HGPS individuals only live into their mid teens and exhibit an array of premature aging manifestations including ischemic heart disease, central nervous system problems, hearing loss, and insulin unresponsiveness (Korf, 2008; Merideth et al., 2008). LMNA mutations can also result in the later-onset Werner premature aging syndrome (Kudlow et al., 2007). Class IV LMNA mutations are primarily confined to the C-terminal end of the protein. The most common HGPS mutation is a synonymous mutation, p.G608G (Liu and Zhou, 2008), which exposes a cryptic splice site leading to a 150 nucleotide deletion in the transcript. As a result, 50 amino acids of the mature protein are deleted which eliminates the ZMPSTE24 proteolytic processing site for removal of the C-terminal prelamin A domain, while still retaining the C-terminal end of the lamin A protein with the Cys-Ser-Ile-Met sequence. The p.G608G mutation thus generates a novel protein, progerin, which remains attached to the ER and nuclear envelope membranes by its farnesyl group. This results in faulty assembly of the nuclear lamina, distortion of the nuclear shape, and a dominant negative gene action (Caron et al., 2007; Liu and Zhou, 2008; Scaffidi and Misteli, 2008). Since, the level of progerin has been found to correlate with the level of mitochondrial dysfunction in cells (Caron et al., 2007), the progeric laminopathies could also have a mitochondrial etiology.
In HGPS, the expression of progerin is also associated with reduced proliferative life span in cultured cells and increased telomere shortening rate. Telomeres, consist of thousands of copies of the TTAGGG repeat. They cap the ends of chromosomes and are maintained by the RNA-protein reverse transcriptase telomerase. The limited proliferative life span and rapid telomere decay of progeria and WS cells expressing porgerin can be ameliorated by introduction into these cells of the apoprotein of telomerase, TERT (Huang et al., 2008; Kudlow et al., 2008). Furthermore, lamin A has intrinsic affinities for binding DNA containing telomeric repeats (Shoeman and Traub, 1990) and telomeres are generally centrally localized within the nucleus of interphase cells (Scherthan, 2003).
WS can also be caused by mutations in the WRN helicase gene. The WRN helicase facilitates the opening of the ordered structure of the G-rich telomere sequences making it important in telomere maintenance and suppression of telomere sister-chromatid exchanges (Kudlow et al., 2007). Since LMNA mutations also cause WS, lamin A must be important in telomere decay. This creates a direct link between progeria and the mitochondrion. It is now clear that telomere decay is primarily due to mitochondrially-generated ROS which causes the oxidation and loss of telomere repeats (Parrinello et al., 2003; Passos et al., 2007; Passos and von Zglinicki, 2005; Richter et al., 2007; Richter and von Zglinicki, 2007; Saretzki et al., 2003; Saretzki et al., 2008; von Zglinicki, 2002; Yang et al., 2008). In cells expressing telomerase, increased ROS production activates a Src family tyrosine kinase located in the nuclear periphery which phosphorylates TERT. This causes TERT to be exported from the nucleus and imported into the mitochondrion where it reduces ROS production, protects mtDNA from damage, and regulates apoptosis in a Bcl-2 dependent fashion (Ahmed et al., 2008; Del Bufalo et al., 2005; Haendeler et al., 2003; Haendeler et al., 2004; Santos et al., 2004). Since non-stem cells do not express TERT, the increased telomere shortening of progeria cells must be due to increased ROS production due to mitochondrial dysfunction.
6. PERSPECTIVE
The similarity in symptoms between mitochondrial and epigenomic diseases suggests that they have a common pathophysiology. Is so, alterations in mitochondrial function will have important effects on the epigenome and alterations in the epigenome will have significant effects on mitochondrial function (Borrelli et al., 2008).
The epigenome has been hypothesized to provide the interface between the environment and the regulation of nDNA gene expression (Feinberg, 2007, 2008). The most important factors in an organism’s environment are the availability of calories and the demands made on the organism for the use of those calories. The mitochondrion lies between availability of environmental calories and their conversion to usable energy. This occurs by the conversion by the mitochondrion and glycolysis of energy rich compounds such as carbohydrates and fats into ATP, acetyl-CoA, SAM, and NADH. ATP, acetyl-CoA, SAM, and the NADH/NAD+ ratio, in turn, are the high-energy substrates that drive the modification of the epigenome. Prevalent calories increase the ATP, acetyl-CoA, SAM and NADH, causing the modification of histones, the opening of the chromatin, increased gene expression, and the stimulation of growth and reproduction. Reduced calorie levels deplete ATP, acetyl-CoA, SAM, and increase NAD+, reducing histone modification, compacting the chromatin, and reducing gene expression, growth and reproduction.
The mitochondrial genome encompasses over 1500nDNA genes plus the mtDNA genes. The nDNA mitochondrial genes plus the glycolysis genes are dispersed throughout the chromosomes. Therefore, the efficient exploitation of environmental calories requires the coordinate expression of the bioenergetic genes according to the availability of calories and the energy requirements of the organism. Procedures must then exist for regulating the expression of the nDNA bioenergetic genes in relation to calorie availability. The large number and random distribution of the bioenergetic genes would require both cis and trans regulation. The cis regulation of nDNA bioenergetic genes could occur within chromatin loops and be influenced by imprinting. The trans regulation of nDNA bioenergetic genes would require inter-chromosomal regulation by diffusible transcription factors or by the interaction of related genes through tertiary chromatin structures such as transcriptional islands and lamin A associated interactions.
Mutations in mtDNA or nDNA mitochondrial genes diminish energy production causing energy deficiency diseases. These preferentially affect the tissues most reliant on high energy flux: brain, heart, muscle, renal, and endocrine systems. Severe alterations of mitochondrial energy production should alter ATP, acetyl-CoA, SAM, and NADH levels and thus perturb the epigenome and the coordinate expression of bioenergetic genes.
Similarly, certain alterations in the epigenome should alter the coordinate expression of nDNA bioenergetic genes, thus diminishing bioenergetics and resulting in symptoms in the high energy flux tissues. Coordinate expression of bioenergetic genes could be perturbed through either cis or trans regulation systems. Alterations in the structure of chromatin loop domains should cause aberrant cis gene expression and are most apparent in imprinting diseases such as BWS, AS, PWS, etc. Alterations in the epigenome that affect trans interactions include mutations in trans acting factors such a PGC-1α, FOXOs, MeCP2, but also could be affected by alterations in the overall chromatin arrangement in the nucleus as seen in the laminopathies.
Therefore, mitochondrial diseases and many epigenomic diseases may have a common pathophysiology, bioenergetic failure. If this continues to be shown to be true, then it will have major implications for the diagnosis and treatment of the common complex diseases associated with mitochondrial and epigenomic dysfunction.
ACKNOWLEDGEMENTS
The authors would like to thank Ms. Marie T. Lott for her assistance in assembling this document. The work has been supported by NIH grants NS21328, AG24373, DK73691, AG13154, AG16573, a CIRM Comprehensive Grant RC1-00353-1, a Doris Duke Clinical Interfaces Award 2005, and an Autism Speaks High Impact Grant awarded to DCW and a CIRM Predoctoral Fellowship awarded to WF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421–426. doi: 10.1016/s0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- Ahmed S, Passos JF, Birket MJ, Beckmann T, Brings S, Peters H, Birch-Machin MA, von Zglinicki T, Saretzki G. Telomerase does not counteract telomere shortening but protects mitochondrial function under oxidative stress. Journal of Cell Science. 2008;121:1046–1053. doi: 10.1242/jcs.019372. [DOI] [PubMed] [Google Scholar]
- Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of The Cell. New York and London: Garland Science; 2002. [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nature Genetics. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Andreu AL, Bruno C, Dunne TC, Tanji K, Shanske S, Sue CM, Krishna S, Hadjigeorgiou GM, Shtilbans A, Bonilla E, DiMauro S. A nonsense mutation (G15059A) in the cytochrome b gene in a patient with exercise intolerance and myoglobinuria. Annals of Neurology. 1999a;45:127–130. doi: 10.1002/1531-8249(199901)45:1<127::aid-art20>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Andreu AL, Bruno C, Shanske S, Shtilbans A, Hirano M, Krishna S, Hayward L, Systrom DS, Brown RH, Jr, DiMauro S. Missense mutation in the mtDNA cytochrome b gene in a patient with myopathy. Neurology. 1998;51:1444–1447. doi: 10.1212/wnl.51.5.1444. [DOI] [PubMed] [Google Scholar]
- Andreu AL, Hanna MG, Reichmann H, Bruno C, Penn AS, Tanji K, Pallotti F, Iwata S, Bonilla E, Lach B, Morgan-Hughes J, DiMauro S. Exercise intolerance due to mutations in the cytochrome b gene of mitochondrial DNA. New England Journal of Medicine. 1999b;341:1037–1044. doi: 10.1056/NEJM199909303411404. [DOI] [PubMed] [Google Scholar]
- Araujo-Vilar D, Lado-Abeal J, Palos-Paz F, Lattanzi G, Bandin MA, Bellido D, Dominguez-Gerpe L, Calvo C, Perez O, Ramazanova A, Martinez-Sanchez N, Victoria B, Costa-Freitas AT. A novel phenotypic expression associated with a new mutation in LMNA gene, characterized by partial lipodystrophy, insulin resistance, aortic stenosis and hypertrophic cardiomyopathy. Clinical Endocrinology (Oxford) 2008;69:61–68. doi: 10.1111/j.1365-2265.2007.03146.x. [DOI] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- Bai RK, Leal SM, Covarrubias D, Liu A, Wong LJ. Mitochondrial genetic background modifies breast cancer risk. Cancer Research. 2007;67:4687–4694. doi: 10.1158/0008-5472.CAN-06-3554. [DOI] [PubMed] [Google Scholar]
- Baudouin SV, Saunders D, Tiangyou W, Elson JL, Poynter J, Pyle A, Keers S, Turnbull DM, Howell N, Chinnery PF. Mitochondrial DNA and survival after sepsis: a prospective study. Lancet. 2005;366:2118–2121. doi: 10.1016/S0140-6736(05)67890-7. [DOI] [PubMed] [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang MD, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur JA, Sinclair DA. Therapeutic potential of resveratrol: the in vivo evidence. Nature Reviews. Drug Discovery. 2006;5:493–506. doi: 10.1038/nrd2060. [DOI] [PubMed] [Google Scholar]
- Bechara EG, Didiot MC, Melko M, Davidovic L, Bensaid M, Martin P, Castets M, Pognonec P, Khandjian EW, Moine H, Bardoni B. A novel function for fragile X mental retardation protein in translational activation. PLoS Biology. 2009;7:e16. doi: 10.1371/journal.pbio.1000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellomo F, Piccoli C, Cocco T, Scacco S, Papa F, Gaballo A, Boffoli D, Signorile A, D'Aprile A, Scrima R, Sardanelli AM, Capitanio N, Papa S. Regulation by the cAMP cascade of oxygen free radical balance in mammalian cells. Antioxidants and Redox Signaling. 2006;8:495–502. doi: 10.1089/ars.2006.8.495. [DOI] [PubMed] [Google Scholar]
- Bjornsson HT, Brown LJ, Fallin MD, Rongione MA, Bibikova M, Wickham E, Fan JB, Feinberg AP. Epigenetic specificity of loss of imprinting of the IGF2 gene in Wilms tumors. Journal of the National Cancer Institute. 2007;99:1270–1273. doi: 10.1093/jnci/djm069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boily G, Seifert EL, Bevilacqua L, He XH, Sabourin G, Estey C, Moffat C, Crawford S, Saliba S, Jardine K, Xuan J, Evans M, Harper ME, McBurney MW. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS ONE. 2008;3:e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker LM, Habermacher GM, Jessie BC, Sun QC, Baumann AK, Amin M, Lim SD, Fernandez-Golarz C, Lyles RH, Brown MD, Marshall FF, Petros JA. North American white mitochondrial haplogroups in prostate and renal cancer. Journal of Urology. 2006;175:468–472. doi: 10.1016/S0022-5347(05)00163-1. discussion 472–473. [DOI] [PubMed] [Google Scholar]
- Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. Decoding the epigenetic language of neuronal plasticity. Neuron. 2008;60:961–974. doi: 10.1016/j.neuron.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- Brown MD, Starikovskaya E, Derbeneva O, Hosseini S, Allen JC, Mikhailovskaya IE, Sukernik RI, Wallace DC. The role of mtDNA background in disease expression: A new primary LHON mutation associated with Western Eurasian haplogroup. J. Human Genetics. 2002;110:130–138. doi: 10.1007/s00439-001-0660-8. [DOI] [PubMed] [Google Scholar]
- Brown MD, Sun F, Wallace DC. Clustering of Caucasian Leber hereditary optic neuropathy patients containing the 11778 or 14484 mutations on an mtDNA lineage. American Journal of Human Genetics. 1997;60:381–387. [PMC free article] [PubMed] [Google Scholar]
- Brown MD, Torroni A, Reckord CL, Wallace DC. Phylogenetic analysis of Leber's hereditary optic neuropathy mitochondrial DNA's indicates multiple independent occurrences of the common mutations. Human Mutation. 1995;6:311–325. doi: 10.1002/humu.1380060405. [DOI] [PubMed] [Google Scholar]
- Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nature Genetics. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- Caron M, Auclair M, Donadille B, Bereziat V, Guerci B, Laville M, Narbonne H, Bodemer C, Lascols O, Capeau J, Vigouroux C. Human lipodystrophies linked to mutations in A-type lamins and to HIV protease inhibitor therapy are both associated with prelamin A accumulation, oxidative stress and premature cellular senescence. Cell Death and Differentiation. 2007;14:1759–1767. doi: 10.1038/sj.cdd.4402197. [DOI] [PubMed] [Google Scholar]
- Carrieri G, Bonafe M, De Luca M, Rose G, Varcasia O, Bruni A, Maletta R, Nacmias B, Sorbi S, Corsonello F, Feraco E, Andreev KF, Yashin AI, Franceschi C, De Benedictis G. Mitochondrial DNA haplogroups and APOE4 allele are non-independent variables in sporadic Alzheimer's disease. Human Genetics. 2001;108:194–198. doi: 10.1007/s004390100463. [DOI] [PubMed] [Google Scholar]
- Carrozzo R, Dionisi-Vici C, Steuerwald U, Lucioli S, Deodato F, Di Giandomenico S, Bertini E, Franke B, Kluijtmans LA, Meschini MC, Rizzo C, Piemonte F, Rodenburg R, Santer R, Santorelli FM, van Rooij A, Vermunt-de Koning D, Morava E, Wevers RA. SUCLA2 mutations are associated with mild methylmalonic aciduria, Leigh-like encephalomyopathy, dystonia and deafness. Brain. 2007;130:862–874. doi: 10.1093/brain/awl389. [DOI] [PubMed] [Google Scholar]
- Chagnon P, Gee M, Filion M, Robitaille Y, Belouchi M, Gauvreau D. Phylogenetic analysis of the mitochondrial genome indicates significant differences between patients with Alzheimer disease and controls in a French-Canadian founder population. American Journal of Medical Genetics. 1999;85:20–30. doi: 10.1002/(sici)1096-8628(19990702)85:1<20::aid-ajmg6>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Chen C, Liu Y, Liu R, Ikenoue T, Guan KL, Liu Y, Zheng P. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. The Journal of Experimental Medicine. 2008;205:2397–2408. doi: 10.1084/jem.20081297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. The Journal of Biological Chemistry. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. Journal of Cell Biology. 2003a;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Fearnley IM, Peak-Chew SY, Walker JE. The phosphorylation of subunits of complex I from bovine heart mitochondria. The Journal of Biological Chemistry. 2004;279:26036–26045. doi: 10.1074/jbc.M402710200. [DOI] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003b;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolat S, Martins de Brito O, Dal Zilio B, Scorrano L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15927–15932. doi: 10.1073/pnas.0407043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia C, Coutinho AM, Diogo L, Grazina M, Marques C, Miguel T, Ataide A, Almeida J, Borges L, Oliveira C, Oliveira G, Vicente AM. Brief report: high frequency of biochemical markers for mitochondrial dysfunction in autism: no association with the mitochondrial aspartate/glutamate carrier SLC25A12 gene. Journal of Autism and Developmental Disorders. 2006;36:1137–1140. doi: 10.1007/s10803-006-0138-6. [DOI] [PubMed] [Google Scholar]
- Cremer M, Kupper K, Wagler B, Wizelman L, von Hase J, Weiland Y, Kreja L, Diebold J, Speicher MR, Cremer T. Inheritance of gene density-related higher order chromatin arrangements in normal and tumor cell nuclei. The Journal of Cell Biology. 2003;162:809–820. doi: 10.1083/jcb.200304096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispim D, Canani LH, Gross JL, Tschiedel B, Souto KE, Roisenberg I. The European-specific mitochondrial cluster J/T could confer an increased risk of insulin-resistance and type 2 diabetes: an analysis of the m.4216T > C and m.4917A > G variants. Annals of Human Genetics. 2006;70:488–495. doi: 10.1111/j.1469-1809.2005.00249.x. [DOI] [PubMed] [Google Scholar]
- Daitoku H, Yamagata K, Matsuzaki H, Hatta M, Fukamizu A. Regulation of PGC-1 promoter activity by protein kinase B and the forkhead transcription factor FKHR. Diabetes. 2003;52:642–649. doi: 10.2337/diabetes.52.3.642. [DOI] [PubMed] [Google Scholar]
- Darvishi K, Sharma S, Bhat AK, Rai E, Bamezai RN. Mitochondrial DNA G10398A polymorphism imparts maternal Haplogroup N a risk for breast and esophageal cancer. Cancer Letters. 2007;249:249–255. doi: 10.1016/j.canlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- De Benedictis G, Rose G, Carrieri G, De Luca M, Falcone E, Passarino G, Bonafe M, Monti D, Baggio G, Bertolini S, Mari D, Mattace R, Franceschi C. Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. FASEB Journal. 1999;13:1532–1536. doi: 10.1096/fasebj.13.12.1532. [DOI] [PubMed] [Google Scholar]
- Del Bufalo D, Rizzo A, Trisciuoglio D, Cardinali G, Torrisi MR, Zangemeister-Wittke U, Zupi G, Biroccio A. Involvement of hTERT in apoptosis induced by interference with Bcl-2 expression and function. Cell Death and Differentiation. 2005;12:1429–1438. doi: 10.1038/sj.cdd.4401670. [DOI] [PubMed] [Google Scholar]
- Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, Belenguer P, Pelloquin L, Grosgeorge J, Turc-Carel C, Perret E, Astarie-Dequeker C, Lasquellec L, Arnaud B, Ducommun B, Kaplan J, Hamel CP. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nature Genetics. 2000;26:207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes and Development. 2008;22:239–251. doi: 10.1101/gad.1617608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Duckles SP, Krause DN, Stirone C, Procaccio V. Estrogen and mitochondria: a new paradigm for vascular protection? Molecular Interventions. 2006;6:26–35. doi: 10.1124/mi.6.1.6. [DOI] [PubMed] [Google Scholar]
- Duesing K, Charpentier G, Marre M, Tichet J, Hercberg S, Froguel P, Gibson F. Evaluating the association of common LMNA variants with type 2 diabetes and quantitative metabolic phenotypes in French Europids. Diabetologia. 2008;51:76–81. doi: 10.1007/s00125-007-0857-z. [DOI] [PubMed] [Google Scholar]
- Eckel-Mahan K, Sassone-Corsi P. Metabolism control by the circadian clock and vice versa. Nature Structural and Molecular Biology. 2009;16:462–467. doi: 10.1038/nsmb.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeg-Olofsson O, al-Zuhair AG, Teebi AS, al-Essa MM. Rett syndrome: genetic clues based on mitochondrial changes in muscle. American Journal of Medical Genetics. 1989;32:142–144. doi: 10.1002/ajmg.1320320131. [DOI] [PubMed] [Google Scholar]
- Escobar-Henriques M, Westermann B, Langer T. Regulation of mitochondrial fusion by the F-box protein Mdm30 involves proteasome-independent turnover of Fzo1. The Journal of Cell Biology. 2006;173:645–650. doi: 10.1083/jcb.200512079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espada J, Varela I, Flores I, Ugalde AP, Cadinanos J, Pendas AM, Stewart CL, Tryggvason K, Blasco MA, Freije JM, Lopez-Otin C. Nuclear envelope defects cause stem cell dysfunction in premature-aging mice. Journal of Cell Biology. 2008;181:27–35. doi: 10.1083/jcb.200801096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans AR, Limp-Foster M, Kelley MR. Going APE over ref-1. Mutation Research. 2000;461:83–108. doi: 10.1016/s0921-8777(00)00046-x. [DOI] [PubMed] [Google Scholar]
- Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, Lambert PD, Mataki C, Elliott PJ, Auwerx J. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metabolism. 2008;8:347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- Feinberg AP. Epigenetics at the epicenter of modern medicine. JAMA. 2008;299:1345–1350. doi: 10.1001/jama.299.11.1345. [DOI] [PubMed] [Google Scholar]
- Figarella-Branger D, Pellissier JF, Scheiner C, Wernert F, Desnuelle C. Defects of the mitochondrial respiratory chain complexes in three pediatric cases with hypotonia and cardiac involvement. Journal of the Neurological Sciences. 1992;108:105–113. doi: 10.1016/0022-510x(92)90195-q. [DOI] [PubMed] [Google Scholar]
- Filipek PA. Medical Aspects of Autism. In: Volkmar FR, Klin A, Paul R, Cohen DJ, editors. Handbook of Autism and Pervasive Developmental Disorders. Chapter 520. New York: John Wiley and Sons; 2005. pp. 534–578. [Google Scholar]
- Fillano JJ, Goldenthal MJ, Rhodes CH, Marin-Garcia J. Mitochondrial dysfunction in patients with hypotonia, epilepsy, autism, and developmental delay: HEADD syndrome. Journal of Child Neurology. 2002;17:435–439. doi: 10.1177/088307380201700607. [DOI] [PubMed] [Google Scholar]
- Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- Fuchs SM, Laribee RN, Strahl BD. Protein modifications in transcription elongation. Biochimica et Biophysica Acta. 2009;1789:26–36. doi: 10.1016/j.bbagrm.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuku N, Park KS, Yamada Y, Nishigaki Y, Cho YM, Matsuo H, Segawa T, Watanabe S, Kato K, Yokoi K, Nozawa Y, Lee HK, Tanaka M. Mitochondrial Haplogroup N9a Confers Resistance against Type 2 Diabetes in Asians. American Journal of Human Genetics. 2007;80:407–415. doi: 10.1086/512202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiova G, Bartova E, Raska I, Krejci J, Kozubek S. Chromatin changes induced by lamin A/C deficiency and the histone deacetylase inhibitor trichostatin A. European Journal of Cell Biology. 2008;87:291–303. doi: 10.1016/j.ejcb.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Molecular Biology of the Cell. 2008;19:2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargus JJ. Genetic calcium signaling abnormalities in the central nervous system: seizures, migraine, and autism. Annals of the New York Academy of Sciences. 2009;1151:133–156. doi: 10.1111/j.1749-6632.2008.03572.x. [DOI] [PubMed] [Google Scholar]
- Gargus JJ, Imtiaz F. Mitochondrial energy-deficient endophenotype in autism. American Journal of Biochemistry and Biotechnology. 2008;4:198–207. [Google Scholar]
- Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO Journal. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzi D, Marelli C, Achilli A, Goldwurm S, Pezzoli G, Barone P, Pellecchia MT, Stanzione P, Brusa L, Bentivoglio AR, Bonuccelli U, Petrozzi L, Abbruzzese G, Marchese R, Cortelli P, Grimaldi D, Martinelli P, Ferrarese C, Garavaglia B, Sangiorgi S, Carelli V, Torroni A, Albanese A, Zeviani M. Mitochondrial DNA haplogroup K is associated with a lower risk of Parkinson's disease in Italians. European Journal of Human Genetics. 2005;13:748–752. doi: 10.1038/sj.ejhg.5201425. [DOI] [PubMed] [Google Scholar]
- Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. 2007;26:5420–5432. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- Goel A, Mathupala SP, Pedersen PL. Glucose metabolism in cancer. Evidence that demethylation events play a role in activating type II hexokinase gene expression. The Journal of Biological Chemistry. 2003;278:15333–15340. doi: 10.1074/jbc.M300608200. [DOI] [PubMed] [Google Scholar]
- Goetze S, Mateos-Langerak J, Gierman HJ, de Leeuw W, Giromus O, Indemans MH, Koster J, Ondrej V, Versteeg R, van Driel R. The three-dimensional structure of human interphase chromosomes is related to the transcriptome map. Molecular and Cellular Biology. 2007;27:4475–4487. doi: 10.1128/MCB.00208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondor A, Ohlsson R. Transcription in the loop. Nature Genetics. 2006;38:1229–1230. doi: 10.1038/ng1106-1229. [DOI] [PubMed] [Google Scholar]
- Goto Y, Nonaka I, Horai S. A mutation in the tRNALeu(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1990;348:651–653. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- Gottlieb E, Tomlinson IP. Mitochondrial tumour suppressors: a genetic and biochemical update. Nature Reviews. Cancer. 2005;5:857–866. doi: 10.1038/nrc1737. [DOI] [PubMed] [Google Scholar]
- Graf WD, Marin-Garcia J, Gao HG, Pizzo S, Naviaux RK, Markusic D, Barshop BA, Courchesne E, Haas RH. Autism associated with the mitochondrial DNA G8363A transfer RNA(Lys) mutation. Journal of Child Neurology. 2000;15:357–361. doi: 10.1177/088307380001500601. [DOI] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, van Steensel B. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Gulati P, Gaspers LD, Dann SG, Joaquin M, Nobukuni T, Natt F, Kozma SC, Thomas AP, Thomas G. Amino acids activate mTOR complex 1 via Ca2+/CaM signaling to hVps34. Cell Metabolism. 2008;7:456–465. doi: 10.1016/j.cmet.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas RH, Townsend J, Courchesne E, Lincoln AJ, Schreibman L, Yeung-Courchesne R. Neurologic abnormalities in infantile autism. Journal of Child Neurology. 1996;11:84–92. doi: 10.1177/088307389601100204. [DOI] [PubMed] [Google Scholar]
- Haendeler J, Hoffmann J, Brandes RP, Zeiher AM, Dimmeler S. Hydrogen peroxide triggers nuclear export of telomerase reverse transcriptase via Src kinase family-dependent phosphorylation of tyrosine 707. Molecular and Cellular Biology. 2003;23:4598–4610. doi: 10.1128/MCB.23.13.4598-4610.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haendeler J, Hoffmann J, Diehl JF, Vasa M, Spyridopoulos I, Zeiher AM, Dimmeler S. Antioxidants inhibit nuclear export of telomerase reverse transcriptase and delay replicative senescence of endothelial cells. Circulation Research. 2004;94:768–775. doi: 10.1161/01.RES.0000121104.05977.F3. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Hakelien AM, Delbarre E, Gaustad KG, Buendia B, Collas P. Expression of the myodystrophic R453W mutation of lamin A in C2C12 myoblasts causes promoter-specific and global epigenetic defects. Experimental Cell Research. 2008;314:1869–1880. doi: 10.1016/j.yexcr.2008.02.018. [DOI] [PubMed] [Google Scholar]
- Hale CM, Shrestha AL, Khatau SB, Stewart-Hutchinson PJ, Hernandez L, Stewart CL, Hodzic D, Wirtz D. Dysfunctional connections between the nucleus and the actin and microtubule networks in laminopathic models. Biophysical Journal. 2008;95:5462–5475. doi: 10.1529/biophysj.108.139428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JM, Go YM, Jones DP. Nuclear and mitochondrial compartmentation of oxidative stress and redox signaling. Annu Rev Pharmacol Toxicol. 2006;46:215–234. doi: 10.1146/annurev.pharmtox.46.120604.141122. [DOI] [PubMed] [Google Scholar]
- Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nature Reviews. Molecular Cell Biology. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- Heilstedt HA, Shahbazian MD, Lee B. Infantile hypotonia as a presentation of Rett syndrome. American Journal of Medical Genetics. 2002;111:238–242. doi: 10.1002/ajmg.10633. [DOI] [PubMed] [Google Scholar]
- Hendrickson SL, Hutcheson HB, Ruiz-Pesini E, Poole JC, Lautenberger J, Sezgin E, Kingsley L, Goedert JJ, Vlahov D, Donfield S, Wallace DC, O'Brien SJ. Mitochondrial DNA haplogroups influence AIDS progression. AIDS. 2008;22:2429–2439. doi: 10.1097/QAD.0b013e32831940bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson SL, Kingsley LA, Ruiz-Pesini E, Poole JC, Jacobson LP, Palella FJ, Bream JH, Wallace DC, O’Brien SJ. Mitochondrial DNA haplogroups influence lipoatrophy after highly active anti-retroviral therapy. Journal of Acquired Immune Deficiency Syndromes. 2009;51:111–116. doi: 10.1097/QAI.0b013e3181a324d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt IJ, Harding AE, Morgan-Hughes JA. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature. 1988;331:717–719. doi: 10.1038/331717a0. [DOI] [PubMed] [Google Scholar]
- Holt IJ, Harding AE, Petty RK, Morgan-Hughes JA. A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. American Journal of Human Genetics. 1990;46:428–433. [PMC free article] [PubMed] [Google Scholar]
- Holtzman D. Autistic spectrum disorders and mitochondrial encephalopathies. Acta Paediatrica. 2008;97:859–860. doi: 10.1111/j.1651-2227.2008.00883.x. [DOI] [PubMed] [Google Scholar]
- Hong CJ, Liou YJ, Liao DL, Hou SJ, Yen FC, Tsai SJ. Association study of polymorphisms in the mitochondrial aspartate/glutamate carrier SLC25A12 (aralar) gene with schizophrenia. Progress in Neuro-Psychopharmacology and Biological Psychiatry. 2007;31:1510–1513. doi: 10.1016/j.pnpbp.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Huang S, Risques RA, Martin GM, Rabinovitch PS, Oshima J. Accelerated telomere shortening and replicative senescence in human fibroblasts overexpressing mutant and wild-type lamin A. Experimental Cell Research. 2008;314:82–91. doi: 10.1016/j.yexcr.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, Nakada K, Honma Y, Hayashi J. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- Ivanova R, Lepage V, Charron D, Schachter F. Mitochondrial genotype associated with French Caucasian centenarians. Gerontology. 1998;44:349. doi: 10.1159/000022041. [DOI] [PubMed] [Google Scholar]
- Jager S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James DI, Parone PA, Mattenberger Y, Martinou JC. hFis1, a novel component of the mammalian mitochondrial fission machinery. The Journal of Biological Chemistry. 2003;278:36373–36379. doi: 10.1074/jbc.M303758200. [DOI] [PubMed] [Google Scholar]
- Jones DP. Disruption of mitochondrial redox circuitry in oxidative stress. Chemico-Biological Interactions. 2006a;163:38–53. doi: 10.1016/j.cbi.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Jones DP. Redefining oxidative stress. Antioxidants and Redox Signaling. 2006b;8:1865–1879. doi: 10.1089/ars.2006.8.1865. [DOI] [PubMed] [Google Scholar]
- Jones DP. Radical-free biology of oxidative stress. American Journal of Physiology. Cell Physiology. 2008;295:C849–C868. doi: 10.1152/ajpcell.00283.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MM, Manwaring N, Wang JJ, Rochtchina E, Mitchell P, Sue CM. Mitochondrial DNA haplogroups and age-related maculopathy. Archives of Ophthalmology. 2007;125:1235–1240. doi: 10.1001/archopht.125.9.1235. [DOI] [PubMed] [Google Scholar]
- Kaneda A, Wang CJ, Cheong R, Timp W, Onyango P, Wen B, Iacobuzio-Donahue CA, Ohlsson R, Andraos R, Pearson MA, Sharov AA, Longo DL, Ko MS, Levchenko A, Feinberg AP. Enhanced sensitivity to IGF-II signaling links loss of imprinting of IGF2 to increased cell proliferation and tumor risk. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20926–20931. doi: 10.1073/pnas.0710359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JS, Tian JH, Pan PY, Zald P, Li C, Deng C, Sheng ZH. Docking of axonal mitochondria by syntaphilin controls their mobility and affects short-term facilitation. Cell. 2008;132:137–148. doi: 10.1016/j.cell.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M, Neutzner A, Youle RJ. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. The Journal of Cell Biology. 2007;178:71–84. doi: 10.1083/jcb.200611064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaukonen J, Juselius JK, Tiranti V, Kyttala A, Zeviani M, Comi GP, Keranen S, Peltonen L, Suomalainen A. Role of adenine nucleotide translocator 1 in mtDNA maintenance. Science. 2000;289:782–785. doi: 10.1126/science.289.5480.782. [DOI] [PubMed] [Google Scholar]
- Kelley MR, Parsons SH. Redox regulation of the DNA repair function of the human AP endonuclease Ape1/ref-1. Antioxidants and Redox Signaling. 2001;3:671–683. doi: 10.1089/15230860152543014. [DOI] [PubMed] [Google Scholar]
- Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes and Development. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- Kemp M, Go YM, Jones DP. Nonequilibrium thermodynamics of thiol/disulfide redox systems: a perspective on redox systems biology. Free Radical Biology and Medicine. 2008;44:921–937. doi: 10.1016/j.freeradbiomed.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khusnutdinova E, Gilyazova I, Ruiz-Pesini E, Derbeneva O, Khusainova R, Khidiyatova I, Magzhanov R, Wallace DC. A mitochondrial etiology of neurodegenerative diseases: evidence from Parkinson's disease. Annals of the New York Academy of Sciences. 2008;1147:1–20. doi: 10.1196/annals.1427.001. [DOI] [PubMed] [Google Scholar]
- Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nature Neuroscience. 2007;10:1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- Korf B. Hutchinson-Gilford progeria syndrome, aging, and the nuclear lamina. New England Journal of Medicine. 2008;358:552–555. doi: 10.1056/NEJMp0800071. [DOI] [PubMed] [Google Scholar]
- Kriaucionis S, Paterson A, Curtis J, Guy J, Macleod N, Bird A. Gene expression analysis exposes mitochondrial abnormalities in a mouse model of Rett syndrome. Molecular and Cellular Biology. 2006;26:5033–5042. doi: 10.1128/MCB.01665-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudlow BA, Kennedy BK, Monnat RJ., Jr Werner and Hutchinson-Gilford progeria syndromes: mechanistic basis of human progeroid diseases. Nature Reviews. Molecular Cell Biology. 2007;8:394–404. doi: 10.1038/nrm2161. [DOI] [PubMed] [Google Scholar]
- Kudlow BA, Stanfel MN, Burtner CR, Johnston ED, Kennedy BK. Suppression of proliferative defects associated with processing-defective lamin A mutants by hTERT or inactivation of p53. Molecular Biology of the Cell. 2008;19:5238–5248. doi: 10.1091/mbc.E08-05-0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, Morrow JD, Van Remmen H, Sedivy JM, Yamasoba T, Tanokura M, Weindruch R, Leeuwenburgh C, Prolla TA. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Lane N. Oxygen: The Molecule that made the World. Oxford: Oxford University Press; 2002. [Google Scholar]
- Lane N. Power, Sex, Suicide: Mitochondria and the Meaning of Life. Oxford: Oxford University Press; 2005. [Google Scholar]
- Lee I, Salomon AR, Ficarro S, Mathes I, Lottspeich F, Grossman LI, Huttemann M. cAMP-dependent tyrosine phosphorylation of subunit I inhibits cytochrome c oxidase activity. The Journal of Biological Chemistry. 2005;280:6094–6100. doi: 10.1074/jbc.M411335200. [DOI] [PubMed] [Google Scholar]
- Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Hale CM, Panorchan P, Khatau SB, George JP, Tseng Y, Stewart CL, Hodzic D, Wirtz D. Nuclear lamin A/C deficiency induces defects in cell mechanics, polarization, and migration. Biophysical Journal. 2007;93:2542–2552. doi: 10.1529/biophysj.106.102426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepagnol-Bestel AM, Maussion G, Boda B, Cardona A, Iwayama Y, Delezoide AL, Moalic JM, Muller D, Dean B, Yoshikawa T, Gorwood P, Buxbaum JD, Ramoz N, Simonneau M. SLC25A12 expression is associated with neurite outgrowth and is upregulated in the prefrontal cortex of autistic subjects. Molecular Psychiatry. 2008;13:385–397. doi: 10.1038/sj.mp.4002120. [DOI] [PubMed] [Google Scholar]
- Li Z, Okamoto K, Hayashi Y, Sheng M. The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell. 2004;119:873–887. doi: 10.1016/j.cell.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jager S, Vianna CR, Reznick RM, Cui L, Manieri M, Donovan MX, Wu Z, Cooper MP, Fan MC, Rohas LM, Zavacki AM, Cinti S, Shulman GI, Lowell BB, Krainc D, Spiegelman BM. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- Liu B, Zhou Z. Lamin A/C, laminopathies and premature ageing. Histology and Histopathology. 2008;23:747–763. doi: 10.14670/HH-23.747. [DOI] [PubMed] [Google Scholar]
- Liu J, Cao L, Chen J, Song S, Lee IH, Quijano C, Liu H, Keyvanfar K, Chen H, Cao LY, Ahn BH, Kumar NG, Rovira II, Xu XL, van Lohuizen M, Motoyama N, Deng CX, Finkel T. Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature. 2009;459:387–392. doi: 10.1038/nature08040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu QA, Shio H. Mitochondrial morphogenesis, dendrite development, and synapse formation in cerebellum require both Bcl-w and the glutamate receptor delta2. PLoS Genetics. 2008;4 doi: 10.1371/journal.pgen.1000097. e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loat CS, Curran S, Lewis CM, Duvall J, Geschwind D, Bolton P, Craig IW. Methyl-CpG-binding protein 2 polymorphisms and vulnerability to autism. Genes, Brain, and Behavior. 2008;7:754–760. doi: 10.1111/j.1601-183X.2008.00414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr, Weissman S, Verdin E, Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Molecular and Cellular Biology. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard J. Autism: a mitochondrial disorder? Medical Hypotheses. 1998;50:497–500. doi: 10.1016/s0306-9877(98)90270-5. [DOI] [PubMed] [Google Scholar]
- Mandel H, Szargel R, Labay V, Elpeleg O, Saada A, Shalata A, Anbinder Y, Berkowitz D, Hartman C, Barak M, Eriksson S, Cohen N. The deoxyguanosine kinase gene is mutated in individuals with depleted hepatocerebral mitochondrial DNA. Nature Genetics. 2001;29:337–341. doi: 10.1038/ng746. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- Markham A, Cameron I, Franklin P, Spedding M. BDNF increases rat brain mitochondrial respiratory coupling at complex I, but not complex II. European Journal of Neuroscience. 2004;20:1189–1196. doi: 10.1111/j.1460-9568.2004.03578.x. [DOI] [PubMed] [Google Scholar]
- Marmorstein R, Trievel RC. Histone modifying enzymes: Structures, mechanisms, and specificities. Biochimica et Biophysica Acta. 2009;1789:58–68. doi: 10.1016/j.bbagrm.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matijevic T, Knezevic J, Slavica M, Pavelic J. Rett syndrome: from the gene to the disease. European Neurology. 2009;61:3–10. doi: 10.1159/000165342. [DOI] [PubMed] [Google Scholar]
- Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- Mattson MP. Glutamate and neurotrophic factors in neuronal plasticity and disease. Annals of the New York Academy of Sciences. 2008;1144:97–112. doi: 10.1196/annals.1418.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Liu D. Energetics and oxidative stress in synaptic plasticity and neurodegenerative disorders. Neuromolecular Medicine. 2002;2:215–231. doi: 10.1385/NMM:2:2:215. [DOI] [PubMed] [Google Scholar]
- McBurney MW, Yang X, Jardine K, Hixon M, Boekelheide K, Webb JR, Lansdorp PM, Lemieux M. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Molecular and Cellular Biology. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord JM. The evolution of free radicals and oxidative stress. American Journal of Medical Genetics. 2000;108:652–659. doi: 10.1016/s0002-9343(00)00412-5. [DOI] [PubMed] [Google Scholar]
- Mercuri E, Brown SC, Nihoyannopoulos P, Poulton J, Kinali M, Richard P, Piercy RJ, Messina S, Sewry C, Burke MM, McKenna W, Bonne G, Muntoni F. Extreme variability of skeletal and cardiac muscle involvement in patients with mutations in exon 11 of the lamin A/C gene. Muscle and Nerve. 2005;31:602–609. doi: 10.1002/mus.20293. [DOI] [PubMed] [Google Scholar]
- Merideth MA, Gordon LB, Clauss S, Sachdev V, Smith AC, Perry MB, Brewer CC, Zalewski C, Kim HJ, Solomon B, Brooks BP, Gerber LH, Turner ML, Domingo DL, Hart TC, Graf J, Reynolds JC, Gropman A, Yanovski JA, Gerhard-Herman M, Collins FS, Nabel EG, Cannon RO, 3rd, Gahl WA, Introne WJ. Phenotype and course of Hutchinson-Gilford progeria syndrome. New England Journal of Medicine. 2008;358:592–604. doi: 10.1056/NEJMoa0706898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne JC, Denu JM. The Sirtuin family: therapeutic targets to treat diseases of aging. Current Opinion in Chemical Biology. 2008;12:11–17. doi: 10.1016/j.cbpa.2008.01.019. [DOI] [PubMed] [Google Scholar]
- Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, Bemis JE, Xie R, Disch JS, Ng PY, Nunes JJ, Lynch AV, Yang H, Galonek H, Israelian K, Choy W, Iffland A, Lavu S, Medvedik O, Sinclair DA, Olefsky JM, Jirousek MR, Elliott PJ, Westphal CH. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda M, Chacon MR, Gutierrez C, Vilarrasa N, Gomez JM, Caubet E, Megia A, Vendrell J. LMNA mRNA expression is altered in human obesity and type 2 diabetes. Obesity. 2008;16:1742–1748. doi: 10.1038/oby.2008.276. [DOI] [PubMed] [Google Scholar]
- Mishmar D, Ruiz-Pesini EE, Golik P, Macaulay V, Clark AG, Hosseini S, Brandon M, Easley K, Chen E, Brown MD, Sukernik RI, Olckers A, Wallace DC. Natural selection shaped regional mtDNA variation in humans. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohlke KL, Jackson AU, Scott LJ, Peck EC, Suh YD, Chines PS, Watanabe RM, Buchanan TA, Conneely KN, Erdos MR, Narisu N, Enloe S, Valle TT, Tuomilehto J, Bergman RN, Boehnke M, Collins FS. Mitochondrial polymorphisms and susceptibility to type 2 diabetes-related traits in Finns. Human Genetics. 2005;118:245–254. doi: 10.1007/s00439-005-0046-4. [DOI] [PubMed] [Google Scholar]
- Muchir A, Pavlidis P, Decostre V, Herron AJ, Arimura T, Bonne G, Worman HJ. Activation of MAPK pathways links LMNA mutations to cardiomyopathy in Emery-Dreifuss muscular dystrophy. Journal of Clinical Investigation. 2007;117:1282–1293. doi: 10.1172/JCI29042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nature Genetics. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Hirose S. Regulation of mitochondrial morphology by USP30, a deubiquitinating enzyme present in the mitochondrial outer membrane. Molecular Biology of the Cell. 2008;19:1903–1911. doi: 10.1091/mbc.E07-11-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Kimura Y, Tokuda M, Honda S, Hirose S. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Reports. 2006;7:1019–1022. doi: 10.1038/sj.embor.7400790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naviaux RK. Mitochondrial control of epigenetics. Cancer Biology and Therapy. 2008;7:1191–1193. doi: 10.4161/cbt.7.8.6741. [DOI] [PubMed] [Google Scholar]
- Neutzner A, Youle RJ. Instability of the mitofusin Fzo1 regulates mitochondrial morphology during the mating response of the yeast Saccharomyces cerevisiae. The Journal of Biological Chemistry. 2005;280:18598–18603. doi: 10.1074/jbc.M500807200. [DOI] [PubMed] [Google Scholar]
- Niemi AK, Hervonen A, Hurme M, Karhunen PJ, Jylha M, Majamaa K. Mitochondrial DNA polymorphisms associated with longevity in a Finnish population. Human Genetics. 2003;112:29–33. doi: 10.1007/s00439-002-0843-y. [DOI] [PubMed] [Google Scholar]
- Nishigaki Y, Yamada Y, Fuku N, Matsuo H, Segawa T, Watanabe S, Kato K, Yokoi K, Yamaguchi S, Nozawa Y, Tanaka M. Mitochondrial haplogroup N9b is protective against myocardial infarction in Japanese males. Human Genetics. 2007;120:827–836. doi: 10.1007/s00439-006-0269-z. [DOI] [PubMed] [Google Scholar]
- Nishino I, Spinazzola A, Hirano M. Thymidine phosphorylase gene mutations in MNGIE, a human mitochondrial disorder. Science. 1999;283:689–692. doi: 10.1126/science.283.5402.689. [DOI] [PubMed] [Google Scholar]
- Oliveira G, Ataide A, Marques C, Miguel TS, Coutinho AM, Mota-Vieira L, Goncalves E, Lopes NM, Rodrigues V, Carmona da Mota H, Vicente AM. Epidemiology of autism spectrum disorder in Portugal: prevalence, clinical characterization, and medical conditions. Developmental Medicine and Child Neurology (London) 2007;49:726–733. doi: 10.1111/j.1469-8749.2007.00726.x. [DOI] [PubMed] [Google Scholar]
- Palmieri L, Alberio S, Pisano I, Lodi T, Meznaric-Petrusa M, Zidar J, Santoro A, Scarcia P, Fontanesi F, Lamantea E, Ferrero I, Zeviani M. Complete loss-of-function of the heart/muscle-specific adenine nucleotide translocator is associated with mitochondrial myopathy and cardiomyopathy. Human Molecular Genetics. 2005;14:3079–3088. doi: 10.1093/hmg/ddi341. [DOI] [PubMed] [Google Scholar]
- Palmieri L, Papaleo V, Porcelli V, Scarcia P, Gaita L, Sacco R, Hager J, Rousseau F, Curatolo P, Manzi B, Militerni R, Bravaccio C, Trillo S, Schneider C, Melmed R, Elia M, Lenti C, Saccani M, Pascucci T, Puglisi-Allegra S, Reichelt KL, Persico AM. Altered calcium homeostasis in autism-spectrum disorders: evidence from biochemical and genetic studies of the mitochondrial aspartate/glutamate carrier AGC1. Molecular Psychiatry ePub ahead of print. 2008 doi: 10.1038/mp.2008.63. http://dx.doi.org/10.1038/mp.2008.1063. [DOI] [PubMed] [Google Scholar]
- Papa S, Scacco S, Sardanelli AM, Petruzzella V, Vergari R, Signorile A, Technikova-Dobrova Z. Complex I and the cAMP cascade in human physiopathology. Bioscience Reports. 2002;22:3–16. doi: 10.1023/a:1016004921277. [DOI] [PubMed] [Google Scholar]
- Park YE, Hayashi YK, Goto K, Komaki H, Hayashi Y, Inuzuka T, Noguchi S, Nonaka I, Nishino I. Nuclear changes in skeletal muscle extend to satellite cells in autosomal dominant Emery-Dreifuss muscular dystrophy/limb-girdle muscular dystrophy 1B. Neuromuscular Disorders. 2009;19:29–36. doi: 10.1016/j.nmd.2008.09.018. [DOI] [PubMed] [Google Scholar]
- Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nature Cell Biology. 2003;5:741–747. doi: 10.1038/ncb1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passos JF, Saretzki G, Ahmed S, Nelson G, Richter T, Peters H, Wappler I, Birket MJ, Harold G, Schaeuble K, Birch-Machin MA, Kirkwood TB, von Zglinicki T. Mitochondrial dysfunction accounts for the stochastic heterogeneity in telomere-dependent senescence. PLoS Biology. 2007;5:e110. doi: 10.1371/journal.pbio.0050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passos JF, von Zglinicki T. Mitochondria, telomeres and cell senescence. Experimental Gerontology. 2005;40:466–472. doi: 10.1016/j.exger.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Pearson KJ, Baur JA, Lewis KN, Peshkin L, Price NL, Labinskyy N, Swindell WR, Kamara D, Minor RK, Perez E, Jamieson HA, Zhang Y, Dunn SR, Sharma K, Pleshko N, Woollett LA, Csiszar A, Ikeno Y, Le Couteur D, Elliott PJ, Becker KG, Navas P, Ingram DK, Wolf NS, Ungvari Z, Sinclair DA, de Cabo R. Resveratrol delays age-related deterioration and mimics transcriptional aspects of dietary restriction without extending life span. Cell Metabolism. 2008;8:157–168. doi: 10.1016/j.cmet.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedram A, Razandi M, Wallace DC, Levin ER. Functional estrogen receptors in the mitochondria of breast cancer cells. Molecular Biology of the Cell. 2006;17:2125–2137. doi: 10.1091/mbc.E05-11-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekovic V, Harborth J, Broers JL, Ramaekers FC, van Engelen B, Lammens M, von Zglinicki T, Foisner R, Hutchison C, Markiewicz E. Nucleoplasmic LAP2alpha-lamin A complexes are required to maintain a proliferative state in human fibroblasts. Journal of Cell Biology. 2007;176:163–172. doi: 10.1083/jcb.200606139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekovic V, Hutchison CJ. Adult stem cell maintenance and tissue regeneration in the ageing context: the role for A-type lamins as intrinsic modulators of ageing in adult stem cells and their niches. Journal of Anatomy. 2008;213:5–25. doi: 10.1111/j.1469-7580.2008.00928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrot A, Hussein S, Ruppert V, Schmidt HH, Wehnert MS, Duong NT, Posch MG, Panek A, Dietz R, Kindermann I, Bohm M, Michalewska-Wludarczyk A, Richter A, Maisch B, Pankuweit S, Ozcelik C. Identification of mutational hot spots in LMNA encoding lamin A/C in patients with familial dilated cardiomyopathy. Basic Research in Cardiology. 2009;104:90–99. doi: 10.1007/s00395-008-0748-6. [DOI] [PubMed] [Google Scholar]
- Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders WD, Hosseini SH, Marshall FF, Wallace DC. mtDNA mutations increase tumorigenicity in prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poling JS, Frye RE, Shoffner J, Zimmerman AW. Developmental regression and mitochondrial dysfunction in a child with autism. Journal of Child Neurology. 2006;21:170–172. doi: 10.2310/7010.2006.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons R, Andreu AL, Checcarelli N, Vila MR, Engelstad K, Sue CM, Shungu D, Haggerty R, de Vivo DC, DiMauro S. Mitochondrial DNA abnormalities and autistic spectrum disorders. Journal of Pediatrics. 2004;144:81–85. doi: 10.1016/j.jpeds.2003.10.023. [DOI] [PubMed] [Google Scholar]
- Procaccio V, Wallace DC. Late-onset Leigh syndrome in a patient with mitochondrial complex I NDUFS8 mutations. Neurology. 2004;62:1899–1901. doi: 10.1212/01.wnl.0000125251.56131.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raby BA, Klanderman B, Murphy A, Mazza S, Camargo CA, Jr, Silverman EK, Weiss ST. A common mitochondrial haplogroup is associated with elevated total serum IgE levels. The Journal of Allergy and Clinical Immunology. 2007;120:351–358. doi: 10.1016/j.jaci.2007.05.029. [DOI] [PubMed] [Google Scholar]
- Ramoz N, Reichert JG, Smith CJ, Silverman JM, Bespalova IN, Davis KL, Buxbaum JD. Linkage and association of the mitochondrial aspartate/glutamate carrier SLC25A12 gene with autism. American Journal of Psychiatry. 2004;161:662–669. doi: 10.1176/appi.ajp.161.4.662. [DOI] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai S, Bass J. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin J, Auer-Grumbach M, Bagg W, Colclough K, Nguyen TD, Fenton-May J, Hattersley A, Hudson J, Jardine P, Josifova D, Longman C, McWilliam R, Owen K, Walker M, Wehnert M, Ellard S. Extreme phenotypic diversity and nonpenetrance in families with the LMNA gene mutation R644C. American Journal of Medical Genetics. Part A. 2008;146A:1530–1542. doi: 10.1002/ajmg.a.32331. [DOI] [PubMed] [Google Scholar]
- Rathmell JC, Newgard CB. Biochemistry. A glucose-to-gene link. Science. 2009;324:1021–1022. doi: 10.1126/science.1174665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razmara A, Duckles SP, Krause DN, Procaccio V. Estrogen suppresses brain mitochondrial oxidative stress in female and male rats. Brain Research. 2007;1176:71–81. doi: 10.1016/j.brainres.2007.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razmara A, Sunday L, Stirone C, Wang XB, Krause DN, Duckles SP, Procaccio V. Mitochondrial effects of estrogen are mediated by estrogen receptor alpha in brain endothelial cells. Journal of Pharmacology and Experimental Therapeutics. 2008;325:782–790. doi: 10.1124/jpet.107.134072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter T, Saretzki G, Nelson G, Melcher M, Olijslagers S, von Zglinicki T. TRF2 overexpression diminishes repair of telomeric single-strand breaks and accelerates telomere shortening in human fibroblasts. Mechanisms of Ageing and Development. 2007;128:340–345. doi: 10.1016/j.mad.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Richter T, von Zglinicki T. A continuous correlation between oxidative stress and telomere shortening in fibroblasts. Experimental Gerontology. 2007;42:1039–1042. doi: 10.1016/j.exger.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose G, Passarino G, Carrieri G, Altomare K, Greco V, Bertolini S, Bonafe M, Franceschi C, De Benedictis G. Paradoxes in longevity: sequence analysis of mtDNA haplogroup J in centenarians. European Journal of Human Genetics. 2001;9:701–707. doi: 10.1038/sj.ejhg.5200703. [DOI] [PubMed] [Google Scholar]
- Ruiz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace DC. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science. 2004;303:223–226. doi: 10.1126/science.1088434. [DOI] [PubMed] [Google Scholar]
- Ruiz-Pesini E, Wallace DC. Evidence for adaptive selection acting on the tRNA and rRNA genes of the human mitochondrial DNA. Human Mutation. 2006;27:1072–1081. doi: 10.1002/humu.20378. [DOI] [PubMed] [Google Scholar]
- Saada A, Shaag A, Mandel H, Nevo Y, Eriksson S, Elpeleg O. Mutant mitochondrial thymidine kinase in mitochondrial DNA depletion myopathy. Nature Genetics. 2001;29:342–344. doi: 10.1038/ng751. [DOI] [PubMed] [Google Scholar]
- Samaco RC, Hogart A, LaSalle JM. Epigenetic overlap in autism-spectrum neurodevelopmental disorders: MECP2 deficiency causes reduced expression of UBE3A and GABRB3. Human Molecular Genetics. 2005;14:483–492. doi: 10.1093/hmg/ddi045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JH, Meyer JN, Skorvaga M, Annab LA, Van Houten B. Mitochondrial hTERT exacerbates free-radical-mediated mtDNA damage. Aging Cell. 2004;3:399–411. doi: 10.1111/j.1474-9728.2004.00124.x. [DOI] [PubMed] [Google Scholar]
- Saretzki G, Murphy MP, von Zglinicki T. MitoQ counteracts telomere shortening and elongates lifespan of fibroblasts under mild oxidative stress. Aging Cell. 2003;2:141–143. doi: 10.1046/j.1474-9728.2003.00040.x. [DOI] [PubMed] [Google Scholar]
- Saretzki G, Walter T, Atkinson S, Passos JF, Bareth B, Keith WN, Stewart R, Hoare S, Stojkovic M, Armstrong L, von Zglinicki T, Lako M. Downregulation of multiple stress defense mechanisms during differentiation of human embryonic stem cells. Stem Cells. 2008;26:455–464. doi: 10.1634/stemcells.2007-0628. [DOI] [PubMed] [Google Scholar]
- Saxena R, de Bakker PI, Singer K, Mootha V, Burtt N, Hirschhorn JN, Gaudet D, Isomaa B, Daly MJ, Groop L, Ardlie KG, Altshuler D. Comprehensive association testing of common mitochondrial DNA variation in metabolic disease. American Journal of Human Genetics. 2006;79:54–61. doi: 10.1086/504926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nature Cell Biology. 2008;10:452–459. doi: 10.1038/ncb1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla RC. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochimica et Biophysica Acta. 2002;1576:1–14. doi: 10.1016/s0167-4781(02)00343-3. [DOI] [PubMed] [Google Scholar]
- Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radical Biology and Medicine. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- Scherthan H. Knockout mice provide novel insights into meiotic chromosome and telomere dynamics. Cytogenetic and Genome Research. 2003;103:235–244. doi: 10.1159/000076809. [DOI] [PubMed] [Google Scholar]
- Schneider R, Grosschedl R. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes and Development. 2007;21:3027–3043. doi: 10.1101/gad.1604607. [DOI] [PubMed] [Google Scholar]
- Schriner SE, Linford NJ, Martin GM, Treuting P, Ogburn CE, Emond M, Coskun PE, Ladiges W, Wolf N, Van Remmen H, Wallace DC, Rabinovitch PS. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–1911. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- Schuman E, Chan D. Fueling synapses. Cell. 2004;119:738–740. doi: 10.1016/j.cell.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, Leotta A, Pai D, Zhang R, Lee YH, Hicks J, Spence SJ, Lee AT, Puura K, Lehtimaki T, Ledbetter D, Gregersen PK, Bregman J, Sutcliffe JS, Jobanputra V, Chung W, Warburton D, King MC, Skuse D, Geschwind DH, Gilliam TC, Ye K, Wigler M. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadel GS. Expression and maintenance of mitochondrial DNA: new insights into human disease pathology. American Journal of Pathology. 2008;172:1445–1456. doi: 10.2353/ajpath.2008.071163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoeman RL, Traub P. The in vitro DNA-binding properties of purified nuclear lamin proteins and vimentin. The Journal of Biological Chemistry. 1990;265:9055–9061. [PubMed] [Google Scholar]
- Shoffner JM, Brown MD, Torroni A, Lott MT, Cabell MR, Mirra SS, Beal MF, Yang C, Gearing M, Salvo R, Watts RL, Juncos JL, Hansen LA, Crain BJ, Fayad M, Reckord CL, Wallace DC. Mitochondrial DNA variants observed in Alzheimer disease and Parkinson disease patients. Genomics. 1993;17:171–184. doi: 10.1006/geno.1993.1299. [DOI] [PubMed] [Google Scholar]
- Shoffner JM, Lott MT, Lezza AM, Seibel P, Ballinger SW, Wallace DC. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNALys mutation. Cell. 1990;61:931–937. doi: 10.1016/0092-8674(90)90059-n. [DOI] [PubMed] [Google Scholar]
- Shulman AI, Mangelsdorf DJ. Retinoid x receptor heterodimers in the metabolic syndrome. New England Journal of Medicine. 2005;353:604–615. doi: 10.1056/NEJMra043590. [DOI] [PubMed] [Google Scholar]
- Smiraglia DJ, Kulawiec M, Bistulfi GL, Gupta SG, Singh KK. A novel role for mitochondria in regulating epigenetic modification in the nucleus. Cancer Biology and Therapy. 2008;7:1182–1190. doi: 10.4161/cbt.7.8.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Molecular Biology of the Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BC, Denu JM. Chemical mechanisms of histone lysine and arginine modifications. Biochimica et Biophysica Acta. 2009;1789:45–57. doi: 10.1016/j.bbagrm.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M, Spence MA, Flodman P. Nuclear and mitochondrial genome defects in autisms. Annals of the New York Academy of Science. 2008;1151:102–132. doi: 10.1111/j.1749-6632.2008.03571.x. [DOI] [PubMed] [Google Scholar]
- Sonoda J, Mehl IR, Chong LW, Nofsinger RR, Evans RM. PGC-1beta controls mitochondrial metabolism to modulate circadian activity, adaptive thermogenesis, and hepatic steatosis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:5223–5228. doi: 10.1073/pnas.0611623104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Pei L, Evans RM. Nuclear receptors: decoding metabolic disease. FEBS Letters. 2008;582:2–9. doi: 10.1016/j.febslet.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelbrink JN, Li FY, Tiranti V, Nikali K, Yuan QP, Tariq M, Wanrooij S, Garrido N, Comi G, Morandi L, Santoro L, Toscano A, Fabrizi GM, Somer H, Croxen R, Beeson D, Poulton J, Suomalainen A, Jacobs HT, Zeviani M, Larsson C. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nature Genetics. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- Splawski I, Yoo DS, Stotz SC, Cherry A, Clapham DE, Keating MT. CACNA1H mutations in autism spectrum disorders. The Journal of Biological Chemistry. 2006;281:22085–22091. doi: 10.1074/jbc.M603316200. [DOI] [PubMed] [Google Scholar]
- Stewart CL, Kozlov S, Fong LG, Young SG. Mouse models of the laminopathies. Experimental Cell Research. 2007;313:2144–2156. doi: 10.1016/j.yexcr.2007.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirone C, Duckles SP, Krause DN, Procaccio V. Estrogen increases mitochondrial efficiency and reduces oxidative stress in cerebral blood vessels. Molecular Pharmacology. 2005;68:959–965. doi: 10.1124/mol.105.014662. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Su H, Fan W, Coskun PE, Vesa J, Gold JA, Jiang YH, Potluri P, Procaccio V, Acab A, Weiss JH, Wallace DC, Kimonis VE. Mitochondrial dysfunction in CA1 hippocampal neurons of the UBE3A deficient mouse model for Angelman syndrome. Neuroscience Letters. 2009 doi: 10.1016/j.neulet.2009.06.079. ePub ahead of print http://dx.doi.org/10.1016/j.neulet.2009.1006.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Molecular Cell. 2006;23:207–217. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Gong J, Zhang J, Yamada Y, Borgeld HJ, Yagi K. Mitochondrial genotype associated with longevity and its inhibitory effect on mutagenesis. Mechanisms of Ageing and Development. 2000;116:65–76. doi: 10.1016/s0047-6374(00)00149-4. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Gong JS, Zhang J, Yoneda M, Yagi K. Mitochondrial genotype associated with longevity. Lancet. 1998;351:185–186. doi: 10.1016/S0140-6736(05)78211-8. [DOI] [PubMed] [Google Scholar]
- Technikova-Dobrova Z, Sardanelli AM, Speranza F, Scacco S, Signorile A, Lorusso V, Papa S. Cyclic adenosine monophosphate-dependent phosphorylation of mammalian mitochondrial proteins: enzyme and substrate characterization and functional role. Biochemistry. 2001;40:13941–13947. doi: 10.1021/bi011066p. [DOI] [PubMed] [Google Scholar]
- Tini M, Benecke A, Um SJ, Torchia J, Evans RM, Chambon P. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Molecular Cell. 2002;9:265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- Tong J, Schriner SE, McCleary D, Day BJ, Wallace DC. Life extension through neurofibromin mitochondrial regulation and antioxidant therapy for Neurofibromatosis-1 in Drosophila melanogaster. Nature Genetics. 2007;39:476–485. doi: 10.1038/ng2004. [DOI] [PubMed] [Google Scholar]
- Torroni A, Petrozzi M, D'Urbano L, Sellitto D, Zeviani M, Carrara F, Carducci C, Leuzzi V, Carelli V, Barboni P, De Negri A, Scozzari R. Haplotype and phylogenetic analyses suggest that one European-specific mtDNA background plays a role in the expression of Leber hereditary optic neuropathy by increasing the penetrance of the primary mutations 11778 and 14484. American Journal of Human Genetics. 1997;60:1107–1121. [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Stepien G, Hodge JA, Wallace DC. Neoplastic transformation is associated with coordinate induction of nuclear and cytoplasmic oxidative phosphorylation genes. The Journal of Biological Chemistry. 1990;265:20589–20593. [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, Tornell J, Jacobs HT, Larsson NG. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Udar N, Atilano SR, Memarzadeh M, Boyer DS, Chwa M, Lu S, Maguen B, Langberg J, Coskun P, Wallace DC, Nesburn AB, Khatibi N, Hertzog D, Le K, Hwang D, Kenney MC. Mitochondrial DNA haplogroups associated with age-related macular degeneration. Investigative Ophthalmology and Visual Science. 2009;50:2966–2974. doi: 10.1167/iovs.08-2646. [DOI] [PubMed] [Google Scholar]
- van der Walt JM, Dementieva YA, Martin ER, Scott WK, Nicodemus KK, Kroner CC, Welsh-Bohmer KA, Saunders AM, Roses AD, Small GW, Schmechel DE, Murali Doraiswamy P, Gilbert JR, Haines JL, Vance JM, Pericak-Vance MA. Analysis of European mitochondrial haplogroups with Alzheimer disease risk. Neuroscience Letters. 2004;365:28–32. doi: 10.1016/j.neulet.2004.04.051. [DOI] [PubMed] [Google Scholar]
- van der Walt JM, Nicodemus KK, Martin ER, Scott WK, Nance MA, Watts RL, Hubble JP, Haines JL, Koller WC, Lyons K, Pahwa R, Stern MB, Colcher A, Hiner BC, Jankovic J, Ondo WG, Allen FH, Jr, Goetz CG, Small GW, Mastaglia F, Stajich JM, McLaurin AC, Middleton LT, Scott BL, Schmechel DE, Pericak-Vance MA, Vance JM. Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. American Journal of Human Genetics. 2003;72:804–811. doi: 10.1086/373937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Goethem G, Dermaut B, Lofgren A, Martin JJ, Van Broeckhoven C. Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions. Nature Genetics. 2001;28:211–212. doi: 10.1038/90034. [DOI] [PubMed] [Google Scholar]
- Vantyghem MC, Vincent-Desplanques D, Defrance-Faivre F, Capeau J, Fermon C, Valat AS, Lascols O, Hecart AC, Pigny P, Delemer B, Vigouroux C, Wemeau JL. Fertility and obstetrical complications in women with LMNA-related familial partial lipodystrophy. The Journal of Clinical Endocrinology and Metabolism. 2008;93:2223–2229. doi: 10.1210/jc.2007-2521. [DOI] [PubMed] [Google Scholar]
- Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zglinicki T. Oxidative stress shortens telomeres. Trends in Biochemical Sciences. 2002;27:339–344. doi: 10.1016/s0968-0004(02)02110-2. [DOI] [PubMed] [Google Scholar]
- Wagner BK, Kitami T, Gilbert TJ, Peck D, Ramanathan A, Schreiber SL, Golub TR, Mootha VK. Large-scale chemical dissection of mitochondrial function (Erratum in Nat Biotechnol 2008 Jul 26(7):831; comment in Nat Biotechnol 2008 Mar 26(3):294–6) Nature Biotechnology. 2008;26:343–351. doi: 10.1038/nbt1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Structure and evolution of organelle genomes. Microbiological Reviews. 1982;46:208–240. doi: 10.1128/mr.46.2.208-240.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Mitochondria and Cancer: Warburg Address. Cold Spring Harbor Symposia on Quantitative Biology. 2005a;70:363–374. doi: 10.1101/sqb.2005.70.035. [DOI] [PubMed] [Google Scholar]
- Wallace DC. The mitochondrial genome in human adaptive radiation and disease: On the road to therapeutics and performance enhancement. Gene. 2005b;354:169–180. doi: 10.1016/j.gene.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annual Review of Genetics. 2005c;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Why do we have a maternally inherited mitochondrial DNA? Insights from Evolutionary Medicine. Annual Review of Biochemistry. 2007;76:781–821. doi: 10.1146/annurev.biochem.76.081205.150955. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondria as chi. Genetics. 2008;179:727–735. doi: 10.1534/genetics.104.91769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Brown MD, Lott MT. Mitochondrial DNA variation in human evolution and disease. Gene. 1999;238:211–230. doi: 10.1016/s0378-1119(99)00295-4. [DOI] [PubMed] [Google Scholar]
- Wallace DC, Lott MT, Brown MD, Kerstann K. Mitochondria and Neuro-Ophthalmological Diseases. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic and Molecular Basis of Inherited Disease. vol. II. New York: McGraw-Hill; 2001. pp. 2425–2512. [Google Scholar]
- Wallace DC, Lott MT, Procaccio V. Mitochondrial Genes in Degenerative Diseases, Cancer and Aging. In: Rimoin DL, Connor JM, Pyeritz RE, Korf BR, editors. Emery and Rimoin's Principles and Practice of Medical Genetics. Chapter 13. 5th Edition. vol. Volume 1. Philadelphia, PA: Churchill Livingstone Elsevier; 2007. pp. 194–298. [Google Scholar]
- Wallace DC, Ruiz-Pesini E, Mishmar D. mtDNA variation, climatic adaptation, degenerative diseases, and longevity. Cold Spring Harbor Symposia on Quantitative Biology. 2003;68:479–486. doi: 10.1101/sqb.2003.68.471. [DOI] [PubMed] [Google Scholar]
- Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, Lezza AM, Elsas LJ, Nikoskelainen EK. Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science. 1988a;242:1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- Wallace DC, Zheng X, Lott MT, Shoffner JM, Hodge JA, Kelley RI, Epstein CM, Hopkins LC. Familial mitochondrial encephalomyopathy (MERRF): Genetic, pathophysiological, and biochemical characterization of a mitochondrial DNA disease. Cell. 1988b;55:601–610. doi: 10.1016/0092-8674(88)90218-8. [DOI] [PubMed] [Google Scholar]
- Wang YX, Zhang CL, Yu RT, Cho HK, Nelson MC, Bayuga-Ocampo CR, Ham J, Kang H, Evans RM. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biology. 2004;2:e294. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nature Genetics. 2009;49:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann FR, Siemen D, Mawrin C, Horn TF, Dietzmann K. The neurotrophin receptor TrkB is colocalized to mitochondrial membranes. International Journal of Biochemistry and Cell Biology. 2006;38:610–620. doi: 10.1016/j.biocel.2005.10.024. [DOI] [PubMed] [Google Scholar]
- Wijnen H. Circadian rhythms. A circadian loop asSIRTs itself. Science. 2009;324:598–599. doi: 10.1126/science.1174132. [DOI] [PubMed] [Google Scholar]
- Wolf CM, Wang L, Alcalai R, Pizard A, Burgon PG, Ahmad F, Sherwood M, Branco DM, Wakimoto H, Fishman GI, See V, Stewart CL, Conner DA, Berul CI, Seidman CE, Seidman JG. Lamin A/C haploinsufficiency causes dilated cardiomyopathy and apoptosis-triggered cardiac conduction system disease. Journal of Molecular and Cellular Cardiology. 2008;44:293–303. doi: 10.1016/j.yjmcc.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- Worman HJ, Bonne G. "Laminopathies": a wide spectrum of human diseases. Experimental Cell Research. 2007;313:2121–2133. doi: 10.1016/j.yexcr.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Wyrick JJ, Parra MA. The role of histone H2A and H2B post-translational modifications in transcription: A genomic perspective. Biochimica et Biophysica Acta. 2009;1789:37–44. doi: 10.1016/j.bbagrm.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Xiao H, Hasegawa T, Isobe K. p300 collaborates with Sp1 and Sp3 in p21(waf1/cip1) promoter activation induced by histone deacetylase inhibitor. The Journal of Biological Chemistry. 2000;275:1371–1376. doi: 10.1074/jbc.275.2.1371. [DOI] [PubMed] [Google Scholar]
- Yang C, Przyborski S, Cooke MJ, Zhang X, Stewart R, Anyfantis G, Atkinson SP, Saretzki G, Armstrong L, Lako M. A key role for telomerase reverse transcriptase unit in modulating human embryonic stem cell proliferation, cell cycle dynamics, and in vitro differentiation. Stem Cells. 2008;26:850–863. doi: 10.1634/stemcells.2007-0677. [DOI] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Yoon Y, Krueger EW, Oswald BJ, McNiven MA. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Molecular and Cellular Biochemistry. 2003;23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Leotta A, Kustanovich V, Lajonchere C, Geschwind DH, Law K, Law P, Qiu S, Lord C, Sebat J, Ye K, Wigler M. A unified genetic theory for sporadic and inherited autism. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12831–12836. doi: 10.1073/pnas.0705803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nature Genetics. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Yao J, Johns T, Fu K, De Bie I, Macmillan C, Cuthbert AP, Newbold RF, Wang J, Chevrette M, Brown GK, Brown RM, Shoubridge EA. SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nature Genetics. 1998;20:337–343. doi: 10.1038/3804. [DOI] [PubMed] [Google Scholar]
- Zuchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, Parman Y, Evgrafov O, Jonghe PD, Takahashi Y, Tsuji S, Pericak-Vance MA, Quattrone A, Battaloglu E, Polyakov AV, Timmerman V, Schroder JM, Vance JM. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nature Genetics. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]