Abstract
Mitochondrial dysfunction has been linked to a wide range of degenerative and metabolic diseases, cancer, and aging. All these clinical manifestations arise from the central role of bioenergetics in cell biology. Although genetic therapies are maturing as the rules of bioenergetic genetics are clarified, metabolic therapies have been ineffectual. This failure results from our limited appreciation of the role of bioenergetics as the interface between the environment and the cell. A systems approach, which, ironically, was first successfully applied over 80 years ago with the introduction of the ketogenic diet, is required. Analysis of the many ways that a shift from carbohydrate glycolytic metabolism to fatty acid and ketone oxidative metabolism may modulate metabolism, signal transduction pathways, and the epigenome gives us an appreciation of the ketogenic diet and the potential for bioenergetic therapeutics.
Keywords: mitochondria, oxidative phosphorylation, mitochondrial disorders, redox state, mitochondrial therapies
INTRODUCTION
The role of energetic dysfunction in a broad range of metabolic and degenerative diseases, cancer, and aging is becoming increasingly apparent (1–3). Whereas the identification of mutations in mitochondrial DNA (mtDNA) genes 20 years ago (4–7) proved that energy deficiency could cause degenerative diseases, the pathophysiology of energy-deficiency diseases remains poorly understood. Consequently, although genetic therapies targeting energy-production genes have advanced in sophistication and potential effectiveness, metabolic therapies to ameliorate the symptoms of affected individuals have been slow to develop and disappointing in their effectiveness.
The disappointing progress in developing energetic medicines is the direct result of our very limited understanding of energy biology and the interrelationship between bioenergetics and environmental influences. Therefore, to advance energetic therapeutics, we must place mitochondrial diseases and their therapeutics in the broader context of organismal and cellular bioenergetics. This endeavor should not be confined to a few rare genetic diseases, as it may prove to be central to our understanding and treatment of a broad range of common metabolic and degenerative clinical problems.
At present, our knowledge of energetic diseases stems from the past 20 years of studies on mitochondrial bioenergetics and diseases. Mitochondrial diseases are heterogeneous and often multisystemic. Because the mitochondrion provides much of the energy for the cell, mitochondrial disorders preferentially affect tissues with high-energy demands such as the brain, muscle, heart, and endocrine system, although any organ can be affected. Thus, energetic defects have been implicated in forms of blindness, deafness, movement disorders, dementias, cardiomyopathy, myopathy, renal dysfunction, and aging. Moreover, because the mitochondria lie at the interface between environmental calories and organ energetic demands, the mitochondrion is the likely mediator of the common metabolic disorders. As a consequence, mitochondrial dysfunction is probably central to diabetes, obesity, cardiovascular disease, and cancer.
Mitochondrial diseases can be caused by genetic defects in any mtDNA or nuclear DNA (nDNA) gene that encodes a mitochondrial protein or structural RNAs. Although mitochondrial diseases had been assumed to be rare, epidemiological studies have concluded that the frequency of mtDNA diseases alone is on the order of 1 in 5000 (8, 9), and known pathogenic mtDNA mutations have been detected in the cord blood of 1 in 200 live births (10). Furthermore, nDNA mutations affecting mitochondrial genes may be even more common than mtDNA mutations (3). Thus, the genetic burden of energy dysfunction is likely to be enormous. Because the mitochondria are believed to generate the majority of our energy, understanding the pathophysiology of rare and common mitochondrial diseases and the development of energetic therapies must start with the mitochondrion.
MITOCHONDRIAL BIOENERGETICS AND GENETICS
The mitochondria perform four central functions in the cell that are relevant to the pathophysiology of disease: They (a) provide the majority of the cellular energy in the form of ATP, (b) generate and regulate reactive oxygen species (ROS), (c) buffer cytosolic calcium (Ca2+), and (d) regulate apoptosis through the mitochondrial permeability transition pore (mtPTP) (1).
Mitochondrial Energetics
Energetics in animals is based on the availability of reducing equivalents (hydrogen), consumed as carbohydrates and fats, that react with oxygen to generate water via mitochondrial oxidative phosphorylation (OXPHOS) (Figure 1). Glucose is cleaved into pyruvate via glycolysis within the cytosol, reducing cytosolic NAD+ to NADH (reduced nicotinamide adenine nucleotide). The pyruvate then enters the mitochondrion via pyruvate dehydrogenase (PDH), resulting in mitochondrial acetyl-CoA, NADH + H+, and CO2. The acetyl-CoA then enters the tricarboxylic acid (TCA) cycle, which strips the hydrogens from the organic acids, thereby generating NADH + H+. Fatty acids are oxidized entirely within the mitochondrion by beta oxidation, generating mitochondrial acetyl-CoA, NADH + H+, and FADH2; the latter is contained in the electron transfer factor. Two electrons are transferred from NADH + H+ to NADH dehydrogenase (complex I) or from FADH2-containing enzymes such as the electron transfer factor dehydrogenase or succinate dehydrogenase (complex II) to reduce ubiquinone [coenzyme Q10 (CoQ)] to ubisemiquinone, and then to ubiquinol. The electrons from ubiquinol are then transferred down the remainder of the electron transport chain (ETC) successively to complex III (bc1 complex), cytochrome c, complex IV [cytochrome c oxidase (COX)], and finally to oxygen () to yield H2O.
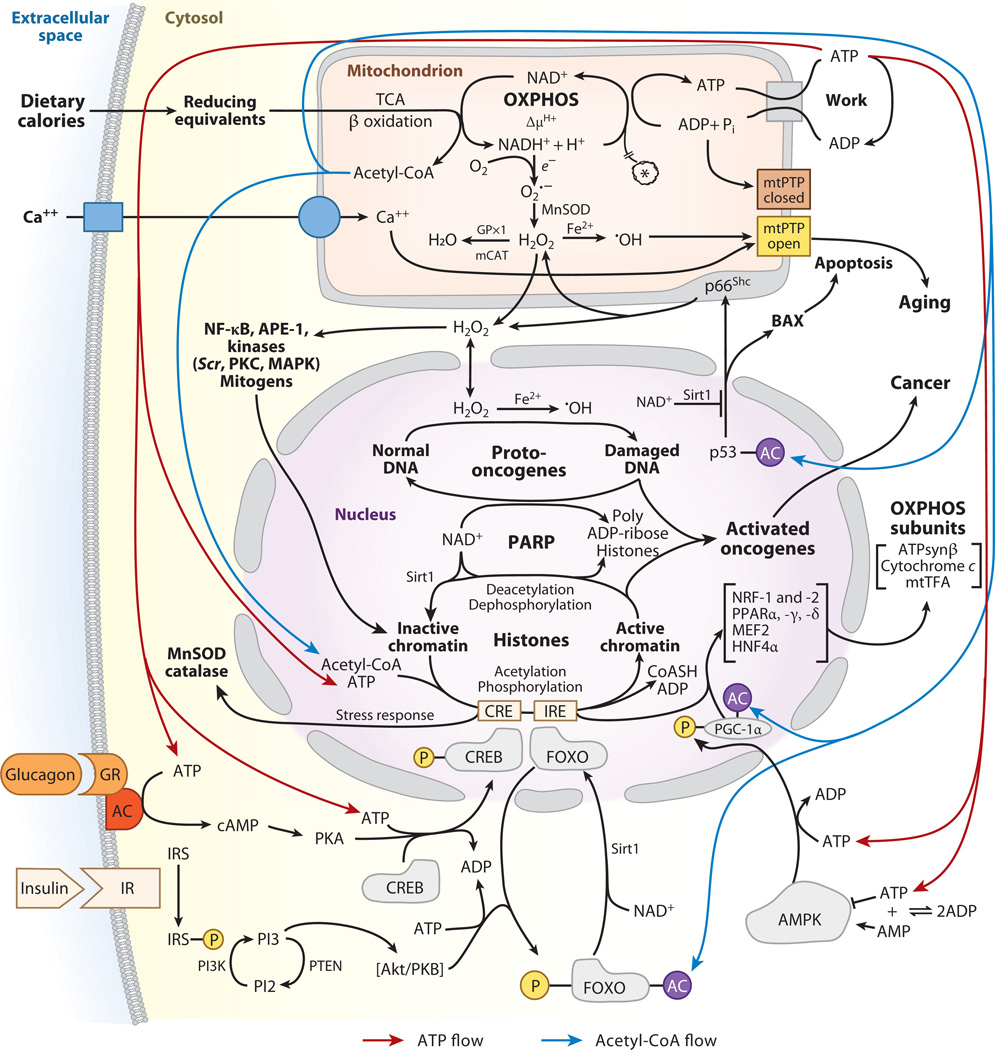
Figure 1.
Bioenergetic regulation of cellular physiology and gene expression. Dietary calories enter the cell as reducing equivalents. Carbohydrates are processed through glycolysis to generate cytosolic pyruvate and NADH (reduced nicotinamide adenine nucleotide). The pyruvate then enters the mitochondrion, is processed through pyruvate dehydrogenase, and is converted to acetyl-CoA, CO2, and NADH. Fatty acids and ketone bodies enter the mitochondrion directly, where they generate acetyl-CoA and mitochondrial NADH. NADH can be oxidized within the mitochondrion by the electron transport chain to generate an inner-membrane electrochemical gradient (ΔP = Δψ + ΔµH+). This ΔP can then be used to generate ATP by the ATP synthase. The ATP is exported to the cytosol by the adenine nucleotide translocator (ANT) to energize work. Excess mitochondrial reducing equivalents can be transferred to O2 to generate superoxide anion (O2·−). O2·− is converted to hydrogen peroxide (H2O2) by manganese superoxide dismutase (MnSOD). H2O2 can diffuse out of the mitochondrion into the cytosol and nucleus. Further reduction of H2O2 results in hydroxyl radical (·OH). The mitochondrial permeability transition pore (mtPTP) senses mitochondrial energy decline, reactive oxygen species (ROS) production, altered oxidation-reduction (redox) state, and increased Ca2+. When activated, it opens a channel in the inner membrane, collapses ΔP, and induces apoptosis. Carbohydrate calories, in the form of glucose, are monitored by the pancreatic islet cells. High serum glucose elicits the secretion of insulin, which binds to the insulin receptors of target cells. This activates the phosphatidylinositol 3 kinase (PI3K) pathway to activate Akt protein kinase B (PKB). Akt phosphorylates the forkhead box, subgroup O (FOXO) transcription factors, barring them from the nucleus and their binding to insulin response elements (IREs). IREs are upstream of the mitochondrial transcription factor coactivator, peroxisome proliferator–activated receptor gamma coactivator 1 alpha (PGC-1α). In the absence of FOXO binding to IRE, PGC-1α transcription is reduced and mitochondrial biogenesis and oxidative phosphorylation (OXPHOS) decline, shifting metabolism toward glycolysis. When carbohydrates are limiting, serum glucose declines; insulin secretion diminishes; and the FOXOs become dephosphorylated and enter the nucleus, where they induce PGC-1α, upregulating mitochondrial biogenesis and OXPHOS. Furthermore, low glucose activates the pancreatic alpha cells to secrete glucagon. Glucagon binds to glucocorticoid receptors (GR) on target cells, activating adenylate cyclase (AC). cAMP activates protein kinase A (PKA) to phosphorylate cAMP response element binding (CREB), and phospho-CREB enters the nucleus, where it binds to cAMP response elements (CREs). One CRE is upstream of PGC-1α, resulting in its increased expression and the induction of mitochondrial biogenesis. Mitochondrial energetics (ATP and acetyl-CoA), redox status, and ROS also regulate cytosolic signal transduction pathways and the epigenome. Mitochondrial acetyl-CoA generated from pyruvate or fatty acids and ketones is converted to citrate. The citrate either drives the tricarboxylic acid (TCA) cycle to generate ATP or is exported to the cytosol and cleaved back to acetyl-CoA. Elevated cytosolic ATP and acetyl-CoA produced when calories are abundant can stimulate the phosphorylation and acetylation of histones, opening chromatin and stimulating transcription, growth, and cell replication. Diminished calories have the opposite effect. High acetyl-CoA also drives the acetylation and inactivation of the FOXOs and PGC-1α, shifting cellular metabolism away from OXPHOS and toward glycolysis. Glycolysis also causes the reduction of NAD+ to NADH, but oxidation of fatty acids and ketones reduces mitochondrial NAD+ to NADH but not cytosolic NAD+. The cytosolic and nuclear protein deacetylase, Sirt1, requires NAD+ as a coreactant and cannot use NADH. Therefore, during active glycolysis Sirt1 is inhibited, the FOXOs and PGC-1α remain acetylated, and the cell is biased toward glycolysis. However, during fatty acid and ketone oxidation, cytosolic NAD+ remains oxidized, the Sirt1 deacetylates the FOXOs and PGC-1α, and OXPHOS is induced. Mitochondrial H2O2 is also an important agent in signal transduction, activating an array of kinases and other signaling molecules. However, excessive H2O2 production can damage cells and can act as a mutagen, potentially activating nuclear DNA (nDNA) oncogenes or inactivating tumor-suppressor genes. Abbreviations: AMPK, AMP-activated protein kinase; APE-1, apurinic/apyrimidinic endonuclease factor 1; HNF4α, hepatocyte nuclear factor 4 alpha; IR, insulin receptor; IRS, insulin receptor substrate; MAPK, mitogen-activated protein kinase; mCAT, mitochondrially targeted catalase; MEF2, myocyte-enhancing factor 2; mtTFA, mitochondrial transcription factor A; NF-κB, nuclear factor kappa B; NRF, nuclear regulatory factor; PTEN, phosphatase and tensin homolog.
The energy that is released as the electrons flow down the ETC is used to pump protons out across the mitochondrial inner membrane through complexes I, III, and IV. This creates a proton electrochemical gradient (ΔP = ΔΨ + ΔµH+), a capacitor that is acidic and positive in the intermembrane space and negative and alkaline on the matrix side. The potential energy stored in ΔP is used for multiple purposes: (a) to import proteins and Ca2+ into the mitochondrion, (b) to generate heat, and (c) to synthesize ATP within the mitochondrial matrix. The energy to convert ADP + Pi to ATP comes from the flow of protons through the ATP synthetase (complex V) back into the matrix. Matrix ATP is then exchanged for cytosolic ADP by the inner-membrane adenine nucleotide translocators (ANTs) (Figure 1) (11).
The efficiency with which dietary reducing equivalents are converted to ATP by OXPHOS is known as the coupling efficiency. This is determined by the efficiency with which protons are pumped out of the matrix by complexes I, III, and IV and by the efficiency with which proton flux through complex V is converted to ATP. The uncoupler drug 2,4-dinitrophenol and the nDNA-encoded uncoupler proteins 1, 2, and 3 render the mitochondrial inner membrane “leaky” for protons. This short-circuits the mitochondrial inner-membrane capacitor; uncouples electron transport from ATP synthesis; and causes the ETC to run at its maximum rate, thereby dissipating the energy as heat.
Variation in mitochondrial proteins has been proposed to alter the OXPHOS coupling efficiency, thus altering the proportion of the calories burned by the mitochondrion that are allocated to ATP generation versus heat production. Alterations in the coupling efficiency can influence ROS generation, modulating Ca2+ uptake, and predilection to apoptosis. Such physiological changes permitted our ancestors to adapt to a range of new environments (12–15).
OXPHOS Complexes
The mitochondrion is assembled from genes encoded in both the nDNA and the mtDNA. OXPHOS complex I is assembled from 45 polypeptides, of which seven (ND1, -2, -3, -4, -4L, -5, and -6) are encoded by the mtDNA; complex II from four nDNA polypeptides; complex III from 11 polypeptides, of which one (cytochrome b) is encoded by the mtDNA; complex IV from 13 polypeptides, of which three (COI, -II, and -III) are from the mtDNA; and complex V from approximately 16 polypeptides, of which two (ATP6 and -8) are from the mtDNA. Of the five complexes, only complexes I, III, IV, and V transport protons, and they also retain mtDNA-encoded polypeptides. Because all of the proton-transporting complexes share a common electrochemical gradient (ΔP) that is central to the energetics of the cell, the proton transport of all four complexes must be balanced to avoid one of the complexes short-circuiting the common capacitor and negating the energy-generating capacity of the other complexes. Consequently, the major electrical components of the four proton-pumping complexes must coevolve. This is accomplished through the maintenance of all these proteins on a single piece of DNA inherited from only one parent, the mother in most animals. As a result, protein genes from different mtDNA maternal lineages with different coupling efficiencies cannot be mixed by recombination. Hence, all 13 mtDNA polypeptide genes are selected as a single unit, and only combinations of variants that optimally manage and utilize ΔP relative to environmental energy supplies and demands survive (11).
Each of the ETC complexes incorporates multiple electron carriers. Complexes I and II utilize flavins and iron-sulfur (Fe-S) centers. The TCA enzyme aconitase also utilizes an Fe-S group, which renders it sensitive to oxidative stress. Complex III encompasses an Fe-S center plus cytochromes b and c1. Complex IV encompasses two Cu centers plus cytochromes a and a3.
Mitochondrial Genetics
Although animal mtDNA encodes 13 core OXPHOS polypeptides plus the 22 transfer RNA (tRNA) and 12S and 16S ribosomal RNA (rRNA) genes for mitochondrial protein synthesis, the nDNA encodes the remaining 80% of the OXPHOS protein genes. In addition, the nDNA encodes all of the genes for the mitochondrial metabolic pathways and all of the enzymes required for mitochondrial biogenesis, including mtDNA polymerase gamma (POLG), RNA polymerase, mtDNA transcription factors, ribosomal proteins, etc. Therefore, the mitochondrial genome encompasses approximately 1500 nDNA-encoded mitochondrial genes plus 37 mtDNA genes (1, 3, 11, 16).
It is because the mitochondrial genome is dispersed between the uniparentally inherited mtDNA and the biparentally inherited nDNA that the genetics of energy metabolism is so complex. The mtDNA is a double-stranded, closed-circular molecule of 16,569 nucleotide pairs. In mammals, the two strands differ in their distribution of Gs and Cs, resulting in a C-rich light (L) strand and a G-rich heavy (H) strand. The gene arrangement of all mammals is conserved, and in addition to the structural genes the mtDNA contains a control region that encompasses the promoters for both H- and L-strand transcription plus the origin of H-strand replication. The origin of L-strand replication is located two-thirds of the way around the mtDNA in a clockwise direction. All of the rRNA and polypeptide genes are located on the H-strand, except for the ND6 gene, which is located on the L-strand. The tRNAs punctuate the genes, and the mature transcripts are generated from the polycistronic transcripts by cleavage of the tRNAs from the transcript, followed by polyadenylation of the mRNAs and rRNAs (17–20). The mtDNA genes have a much higher mutation rate than do the nDNA genes (21, 22). One likely reason for the high mtDNA mutation rate is the proximity of mtDNA to mitochondrial ROS production.
Each mammalian cell contains hundreds of mitochondria and thousands of mtDNAs. When a mutation arises in a mtDNA, it creates a mixed population of normal and mutant mtDNAs, resulting in a state known as heteroplasmy. When a heteroplasmic cell divides, the two types of mtDNAs are randomly distributed into the daughter cells, resulting in genetic drift toward either pure mutant or wild type. Over time, this replicative segregation results in segregation of the mutant mtDNAs into pure mutant or normal populations, resulting in a state termed homoplasmy (11). As the percentage of mutant mtDNAs increases, mitochondrial energetic function declines. When energy output is insufficient for normal tissue function, a threshold is crossed, symptoms appear, and apoptosis or necrosis may be initiated (3, 23).
Human mtDNAs are strictly maternally inherited (24). Only one human exception—in an individual who developed mitochondrial myopathy—has been reported. His muscle tissue was found to have some paternal mtDNA containing a deleterious mutation (25).
MITOCHONDRIAL GENETIC DISEASES
A broad spectrum of complex clinical phenotypes has been linked to mutations in nDNA and mtDNA mitochondrial genes. Very different gene mutations can cause a similar range of phenotypes, mutations in the same gene can give a range of different phenotypes, and the same mtDNA mutation at different levels of heteroplasmy can result in totally different phenotypes. Some mtDNA mutations, such as those causing Leber hereditary optic neuropathy (LHON) (5), yield stereotypic phenotypes, whereas other mtDNA mutations, such as the tRNA mutations causing myoclonic epilepsy and ragged red fiber (MERRF) and the mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS) syndromes, can result in highly variable multisystem diseases (3, 6, 7).
Mitochondrial DNA Diseases
Clinically relevant mtDNA variants fall into three classes: (a) recent deleterious mutations that result in maternally transmitted disease, (b) ancient adaptive variants that predispose individuals to disease in different environments, and (c) age-related accumulation of somatic mtDNA mutations that erode function and provide the aging clock (1). Pathogenic mtDNA mutations include both rearrangement mutations and base-substitution mutations.
Rearrangement mutations can be either de novo deletion mutations or maternally transmitted insertion mutations that are unstable and generate deletion mutations in postmitotic cells. Most deletion mutations remove at least one tRNA and thus affect protein synthesis (20). mtDNA-rearrangement syndromes are invariably heteroplasmic and can result in a range of clinical manifestations and severities. The mildest mtDNA-rearrangement phenotype is maternally inherited type 2 diabetes and deafness, thought to be caused by inheritance of an mtDNA-duplication mutation. The next most severe group of mtDNA rearrangement diseases comprises chronic progressive external ophthalmoplegia (CPEO) and the Kearns-Sayre syndrome (KSS), associated with ophthalmoplegia, ptosis, and mitochondrial myopathy with ragged red fibers (RRFs). RRFs are often associated with COX-negative and succinate dehydrogenase–hyperactive fiber zones in skeletal muscle that contain high concentrations of the rearranged mtDNAs (1, 20, 26, 27). The most severe mtDNA-rearrangement disease is the Pearson marrow/pancreas syndrome. Affected patients develop pancytopenia early in life and become transfusion dependent (28). If they survive the pancytopenia, they progress to KSS (29). Differences in mtDNA-rearrangement phenotypes appear to stem from differences between insertions and deletions, the diversity of tissues that contain the rearrangement, and the percentage of mtDNAs harboring the rearrangement in each tissue (20, 27).
Base-substitution mutations can alter either polypeptide genes (polypeptide mutations) or rRNAs and tRNAs (protein-synthesis mutations). Pathogenic polypeptide mutations encompass a broad spectrum of multisystem diseases, including LHON (5), Leigh syndrome (30), and mitochondrial myopathy (31–33), etc. Mitochondrial tRNA and rRNA protein-synthesis mutations can result in multisystem diseases such as MERRF (6, 7), MELAS (34), encephalomyopathy, mitochondrial myopathy and exercise intolerance, CPEO and KSS, gastrointestinal syndrome, dystonia, diabetes, deafness, cardiomyopathy, renal failure, Alzheimer disease, Parkinson disease, etc. (3).
Both somatic and germline mtDNA mutations have been reported in cancers including renal adenocarcinomas, colon cancer cells, head and neck tumors, astrocytic tumors, thyroid tumors, breast tumors, prostate tumors, etc. (35, 36). Mitochondrial ROS production appears to be an important component of carcinogenesis. Introduction of the pathogenic human mtDNA ATP6 8993T>G missense mutation into prostate cancer cells increased mitochondrial ROS production and prostate tumor growth (37), and the presence of the mtDNA ND6 13997G>A mutation increased the metastatic potential of mouse cancer cell lines in association with increased mitochondrial ROS (38).
mtDNA lineages descended from a single founder mtDNA have accumulated sequential mutations along radiating mtDNA lineages, giving rise to maternal lineages encompassing groups of related mtDNA haplotypes (haplogroups). Haplogroups have generally been found to be associated with specific indigenous populations located in specific geographic regions. The human mtDNA sequence is highly variable, and approximately one-quarter to one-third of mtDNA polypeptide and structural RNA-sequence variants found in the general population appear to be functionally important. These observations suggest that functional mtDNA variants that altered mitochondrial coupling efficiency permitted individuals with that mtDNA to adapt to and multiply within new energetic environments. Natural selection then enriches for that mtDNA and all of its descendants that carried the beneficial variant to create a haplogroup specific for that region (12–15). Adaptive mtDNA haplogroups still correlate with the geographic distribution of indigenous people today. Two-thirds to three-quarters of all African mtDNAs belong to macrohaplogroup L; all European mtDNAs belong to macrohaplogroup N, encompassing the European haplogroups H, I, J, Uk, T, U, V, W, and X. Asian mtDNAs belong to either macrohaplogroup M or macrohaplogroup N. Of all of the Asian haplogroups, only A, C, and D became enriched in northeastern Siberia and crossed into the Americas on the Bering land bridge to found the Native American populations. These Native American haplogroups were subsequently joined by haplogroups B and X (1, 39).
These ancient adaptive mtDNA haplogroups still influence individual predisposition to a wide spectrum of common diseases. The first evidence that mtDNA haplogroups could modify disease predisposition was the discovery that European haplogroup J increases penetrance of the milder LHON mutations (40–43). It was subsequently shown that haplogroup H correlates with reduced risk of age-related macular degeneration and that haplogroups J and U are associated with increased risk of macular degeneration as well as increased drusen levels and retinal pigment abnormalities (44, 45). Haplogroups H and H-nt 4336 are associated with increased risk, and haplogroups J and Uk with decreased risk, for developing Parkinson disease (46–48). The haplogroup H-nt 4336 sublineage is also associated with increased Alzheimer disease risk, and haplogroups U and T are associated with decreased risk in certain contexts (49–52). Haplogroup J has been correlated with longevity in Europeans (53–56), and D has been correlated with longevity in Asians (57, 58). Haplogroup J has been associated with increased risk of diabetes in certain European descent populations (59–61), whereas haplogroup N9a is protective of diabetes, metabolic syndrome, and myocardial infarction in Asians (62–64). Haplogroup U has been associated with increased serum immunoglobin E levels (65), and haplogroup H has been associated with protection against sepsis (66). Haplogroups J and U5a are associated with more rapid progression of acquired immune deficiency syndrome, whereas H3, Uk, and IWX retard this disease’s progression (67). Haplogroup H is protective against lipodystrophy associated with highly active antiretroviral therapy, but haplogroup T may increase risk of lipodystrophy (68). Finally, various haplogroups have been correlated with altered risk for particular cancers (35, 36, 69–72). Therefore, mtDNA functional variation modulates predisposition to a wide range of metabolic and degenerative diseases as well as to cancer and longevity.
Mitochondrial Diseases of Nuclear DNA Mutations
Mutations in nDNA-encoded OXPHOS genes have also been linked to a variety of multisystem disorders ranging from lethal childhood Leigh syndrome to predisposition to depression (Table 1). These include mutations in structural and assembly genes of the respiratory complexes (73, 74), mutations in mitochondrial biogenesis genes such as POLG (75, 76), mutations in nucleoside and nucleotide metabolism (77–82), and mutations in genes involved in mitochondrial fission and fusion (83, 84).
Table 1.
Examples of nuclear DNA mutations that cause mitochondrial diseasesa
| Gene name | Function | Chromosome | Inheritance | Clinical phenotype | Reference(s) |
|---|---|---|---|---|---|
| NDUFS8 | Complex I | 11q13 | AR | Leigh syndrome | 73 |
| SURF1 | Complex IV assembly | 9q34 | AR | Leigh syndrome | 74 |
| C10ORF2 (PEOA3) | Twinkle helicase, mtDNA stability | 10q24 | AD | AD-PEO, SANDO syndrome | 75 |
| POLG (PEOA1) | Polymerase gamma mtDNA replication, mtDNA stability | 15q25 | AD-AR | Alpers syndrome, AD and AR-PEO, SANDO syndrome, SCAE | 76 |
| ANT1 (PEOA2) | Adenine nucleotide translocator isoform 1, mtDNA stability | 4q35 | AD-AR | PEO, multiple mtDNA deletions | 77, 78 |
| DGUOK | Deoxyguanosine kinase, mitochondrial dNTP pool maintenance | 2p13 | AR | Hepatocerebral mtDNA depletion syndrome | 79 |
| TK2 | Thymidine kinase, mitochondrial dNTP pool maintenance | 16q22 | AR | Myopathic mtDNA depletion | 80 |
| SUCLA2 | Succinate-CoA ligase | 13q12 | AR | Leigh-like, dystonia, mild methylmalonic aciduria, mtDNA depletion | 81 |
| ECGF1 | Thymidine phosphorylase, mtDNA stability | 22q13 | AR | MNGIE, multiple mtDNA deletions, depletion | 82 |
| OPA1 | Dynamin-related protein | 3q28-q29 | AD | AD–optic atrophy | 83 |
| MFN2 | Mitofusin, mitochondrial fusion | 1p36-p35 | AD | Charcot-Marie-Tooth | 84 |
Abbreviations: AD, autosomal dominant; AR, autosomal recessive; dNTP, deoxyribonucleotide triphosphate; MNGIE, myoneurogastrointestinal encephalopathy; mtDNA, mitochondrial DNA; PEO, progressive external ophthalmoplegia; SANDO, sensory ataxic neuropathy, dysarthria, and ophthalmoparesis; SCAE, spinocerebellar ataxia with epilepsy.
MITOCHONDRIAL GENETIC THERAPIES
As our understanding of mitochondrial genetics has improved, approaches for the genetic therapy of mitochondrial diseases have also advanced. The modification of nDNA-encoded mitochondrial genes has drawn heavily on prior developments in the gene therapy of other Mendelian disorders. However, new gene-therapy approaches are required for mtDNA diseases because mutant mtDNA can be present in thousands of copies per cell, with the phenotype depending on the percentage of mutant and normal mtDNAs and each tissue’s threshold expression.
Gene Therapy of Nuclear DNA Mitochondrial Mutations
Because many chromosomal mutations are recessive, introduction of a single functional gene into the nDNA should restore the missing biochemical function. In the case of recessive ANT1 cardiomyopathy, adenoassociated virus (AAV) has been used to transduce the heart-muscle isoform of Ant1 into the skeletal muscle of mice in which the Ant1 gene has been genetically inactivated. Ant1−/− mice develop mitochondrial myopathy with RRFs and proliferation of abnormal mitochondria, but without external ophthalmoplegia, together with progressive cardiomyopathy (85, 86). Exactly the same phenotype has been reported for a patient harboring a homozygous ANT1 null mutation (77). The skeletal muscle of mice transduced with the AAV-Ant1 virus showed a 25% increase in muscle enzyme and ATP levels as well as a strong reversal of the histological abnormalities associated with the disease (86). Therefore, gene transduction of nDNA-encoded mitochondrial genes should be a feasible strategy for recessive mutations.
Gene Therapy of Mitochondrial DNA Mutations
To address mtDNA diseases, we could influence the percentage of mutant and normal mtDNAs and/or change the structure and function of the mitochondrial gene. Therefore, investigators are addressing mtDNA mutations with three different approaches: (a) import of normal mtDNA polypeptides into the mitochondrion to complement the mtDNA defect, (b) reduction of the proportion of mutant mtDNAs (heteroplasmy shifting), and (c) direct medication of the mtDNA.
Mitochondrial import polypeptides: allotopic versus xenotopic
Of the more than 200 pathogenic mtDNA mutations that have been published, approximately one-third are located in mtDNA-encoded polypeptide genes (3, 87). For treatment of polypeptide mutations, one approach under development is to isolate a normal copy of the mutant mtDNA polypeptide gene, adjust its genetic code to that of the nDNA, introduce appropriate transcriptional regulatory elements and an amino-terminal mitochondrial targeting peptide, and introduce the resulting allotopic gene into the nDNA. It is hoped that a sufficient amount of the allotopic protein would be introduced into the mitochondrion to overwhelm the high level of mtDNA mutant protein, partially correcting the biochemical defect and leading to suppression of the pathological symptoms.
Allotopic expression has already been attempted with some success in cultured cells. The human ATP8 gene has been reengineered for allotopic expression and successfully introduced into the nucleus, and its protein has been transported back into the mitochondrion (88). The mtDNA ATP6 gene has also been reengineered and introduced into the nucleus, and it has reportedly been able to biochemically rescue human transmitochondrial cybrids harboring the homoplasmic mtDNA 8993T>G mutation (89). Allotopic expression of the mtDNA ND4 gene has also been reported to restore biochemical function to cybrids harboring the ND4 11778G>A missense mutation responsible for LHON. The aldehyde dehydrogenase–targeting peptide was used to direct the reengineered ND4 protein to the mitochondria, thereby partially restoring mitochondrial ATP production (90).
A large proportion of pathogenic mtDNA mutations alter the structure and function of mtDNA tRNAs. Although it is thought that all or virtually all of the mammalian mtDNA tRNAs are encoded by the mtDNA, in other organisms a portion of the mitochondrial tRNAs are encoded by nDNA genes and imported into the mitochondrion. Therefore, mtDNA tRNA mutations may be complemented by introduction of a xenotopic nDNA-encoded yeast mitochondrial tRNA into the mammalian nucleus and having the yeast tRNA imported into the mitochondrion to complement the defect. This approach has been attempted for the treatment of human cells harboring the tRNALys nucleotide 8344A>G mutation with the yeast tRNALys nDNA gene. This partially restored the mitochondrial dysfunction associated with the mitochondrial protein-synthesis defect (91). Similarly, the Leishmania mitochondrial RNA import complex has been used to introduce, through a caveolin-1-dependent pathway, the human cytosolic tRNALys into human cybrids harboring the tRNALys 8344A>G mtDNA mutation. Significant restoration of mitochondrial function was observed (92).
The yeast Saccharomyces cerevisiae NDI1 gene encodes a single polypeptide NADH oxidase, which performs the NADH-CoQ oxidation-reduction (redox) reaction without pumping protons. The introduction of NDI1 into the nDNA of cell lines harboring LHON mtDNA complex I mutations through use of an AAV vector has been reported to restore electron transport activity, although the mitochondria lacked one proton-pumping (coupling) site. The AAV-NDI1 vector was found to transduce mouse brain and skeletal muscle (93). AAV-NDI1 transduction has also been shown to be neuroprotective when injected into the substantia nigra of mice treated with the complex I inhibitor, rotenone, which results in a Parkinson-like disease in rats. NDI1 transduction reduced cell death and oxidative damage of the dopaminergic neurons (94).
AAV transduction has also been used to induce a disease similar to LHON. The retina and optic nerve were transduced with viruses carrying ribozymes that destroy the mRNAs for either mitochondrial manganese superoxide dismutase (MnSOD; Sod2) (95) or the nDNA-encoded NDUFA1 subunit of complex I (96). The loss of optic nerve cells resulting from NDUFA1 ribozyme–transduced cells could then be partially ameliorated by a secondary transduction of the retina with an AAV carrying the human Sod2. The AAV-Sod2 transduction was found to reduce apoptosis in the retinal ganglion cells and the accompanying degeneration of the optic nerve (97).
The allotopic expression of the cyanide-insensitive alternative oxidase (AOX) from the ascidian Ciona intestinalis has been used to confer significant cyanide-insensitive COX activity in cultured cells. Transduction of this enzyme into the nucleus both bypassed a patient’s COX defects and prevented overreduction of the mitochondrial quinone pool responsible for ROS, thus acting as an antioxidant (98). Moreover, transforming mouse mtDNA-deficient (ρ°) cells with the S. cerevisiae NADH dehydrogenase gene (Ndi1) and the Emericella nidulans Aox gene restored NADH/CoQ plus CoQ oxidase activity, relieving the ρ° cell’s dependency on exogenous pyruvate and uridine (99).
Heteroplasmic shifting
Because the threshold proportion of mtDNAs necessary to result in a clinically relevant biochemical defect can occur over a narrow range (generally a few percent), various approaches have been used to shift the proportion of mutant mtDNAs to below the clinical expression threshold. This heteroplasmic shifting strategy has been achieved in cultured cells containing a mixture of wild-type and deleted mtDNA molecules when the heteroplasmic cells were maintained in a ketogenic medium. This medium lacked glucose but contained either beta-hydroxybutyrate or acetoacetate, or both. This obligatory oxidative medium resulted in an increase in wild-type mtDNAs from 13% to 22% within 5 days (100). Similarly, in cybrid clones heteroplasmic for the ATP6 8993T>G mutation associated with a L156R amino acid substitution, growth in medium containing the ATP synthase inhibitor oligomycin to accentuate the ATP synthase defect and galactose to enhance the cellular dependency on mitochondrial OXPHOS caused the wild-type mtDNAs to increase from 16% to 22% (101).
A more direct approach has been to introduce into the nDNA of a heteroplasmic cell, a mitochondrially targeted restriction endonuclease that recognizes and cleaves the mtDNAs harboring the pathogenic mutation but spares the mtDNAs with the normal sequence. Through this approach, cells heteroplasmic for the ATP6 8993T>G mutation have been shifted toward the wild-type 8993T mtDNA via a mitochondrially targeted Sma1 restriction endonuclease (102). A similar approach has been used in vivo to shift the mtDNA heteroplasmy level in muscle and brain of NZB/BALB heteroplasmic mice through the introduction of a transgene that targets the ApaLI restriction endonuclease to the mitochondria. The BALB mtDNA has an ApaLI restriction site that the NZB mtDNA lacks (103).
Direct modifications of the mitochondrial DNA
Attempts to directly correct mtDNA mutations through the introduction of exogenous DNA into the mitochondrion have not as yet been routinely successful. In an alternative approach, protein nucleic acids (PNAs) have been introduced into cellular mitochondria in an effort to inhibit the replication of the mutant mtDNAs. PNAs substitute a peptide backbone for the sugar phosphate background of nucleic acids. Because peptide bonds are not charged, the PNAs bind with a much higher affinity to complementary nucleic acid strands than to regular nucleic acid strands. PNAs have been synthesized to be complementary either to the tRNALys nucleotide 8344A>G mutation or to the breakpoint junction associated with the common 4977 nucleotide deletion. The PNAs are taken up by cells and inhibit the replication of the mutant mtDNA between 75% and 80% without affecting the replication of the wild type. However, in vivo efficacy of this approach has yet to be established (104, 105).
The permeability of PNAs to mitochondria has also been used to introduce oligonucleotides into the mitochondrial matrix. Oligonucleotides complementary to the mtDNA have been annealed to complementary PNAs, which in turn were linked to synthetically synthesized N-terminal mitochondrial-targeting sequences. These conjugates were then introduced into cells by various procedures, including streptolysin permeabilization. Once the conjugate was inside the cytoplasm, it was selectively imported into the mitochondrial matrix (106). Assuming that an oligonucleotide with the wild-type sequence can dissociate from the PNA and anneal to the single-stranded mtDNA replication intermediate, the oligonucleotide containing the wild-type base could act as a primer and replace the mutation in the replicating strand. However, this conjecture has not yet been shown to be feasible.
Stem Cell and Organ Transplantation Therapy of Mitochondrial Disorders
As an alternative to correcting a mutated nDNA or mtDNA mitochondrial gene, it may be possible to replace the cells of the affected mutant tissue with stem cells. Such replacement has been achieved for mitochondrial neurogastrointestinal encephalomyopathy (MNGIE), an autosomal recessive progressive and fatal disorder resulting from mutations in the thymidine phosphorylase gene (TYMP). Absence of the TYMP gene results in the accumulation of toxic levels of thymidine and deoxyuridine in blood. The introduction of allogeneic stem cells, which produce the enzyme and release it into the blood stream, may then establish the enzyme-producing cells in various organs and eliminate the toxic serum metabolite. In one MNGIE patient, this approach resulted in the nearly complete reduction of the toxic blood substrates, as well as significant clinical improvement (107).
Organ transplantation is another approach for correcting specific mitochondrial disease pathologies. This approach has been successful in treating certain mitochondrial diseases, including cardiomyopathy (108) and liver failure (109).
Prenatal Diagnosis, Preimplantation Genetics, and Germline Gene Therapy
Although the above approaches show promise for most patients, women harboring a deleterious mtDNA mutation have a very substantial risk of bearing children with debilitating mitochondrial disease. Because the transmission of a pathogenic mtDNA mutation is stochastic, the mtDNA genotype of chorionic villi or amniocytes may not provide a reliable estimate of the mtDNA mutational load in the target tissues (110, 111). Although the precise genotype of a fetus for the MELAS 3243A>G mutation cannot be deduced, the demonstration that fetal cells have very low mutant mtDNA has been considered reassuring (112). By contrast, placental and amniotic mutation loads have been found to reflect fetal tissue heteroplasmy levels for the ATP6 8993T>G mutation, potentially enabling limited prenatal predictions for this mutation (113).
Preimplantation genetic diagnosis appears to be a promising new approach for the mtDNA-inheritance dilemma. The oocytes of the mtDNA mutation carrier (the mother) could be fertilized in vitro, and either a polar body or a blastomere could be collected and the mtDNA heteroplasmic genotype determined. The embryos with the lowest proportion of mutant mtDNAs could then be selected for implantation (114, 115). Adoption or oocyte donation provide other reproductive alternatives for families with maternally inherited mtDNA disease (116).
A more direct, albeit more complex, approach to eliminating maternally inherited pathogenic mutations may be germline mtDNA therapy. One proposal is to fertilize the mother’s mutant oocytes in vitro, then remove the zygote or early blastomere nucleus by micropipette and inject it into an enucleated oocyte from a woman with normal mtDNA. Implantation of this nuclear transplantation embryo into the mother’s uterus should result in child(ren) with the nDNA from a normal conception by the parents, but with nonmaternal and thus normal mtDNA. If the recipient oocyte were donated by the father’s sister, the mtDNA lineage would simply switch from the maternal to the paternal side (114, 117). Because some of the mother’s mitochondria might be transmitted along with the nucleus during the nuclear transplantation, creating an undesirable heteroplasmy, any residual maternal mitochondria and mtDNAs might be eliminated by rhodamine 6G treatment (118).
MITOCHONDRIAL AND CELLULAR BIOENERGETICS
Although these genetic approaches could help ameliorate the symptoms of some patients with nDNA-encoded mitochondrial diseases and could assist with the reproductive decisions of women harboring deleterious heteroplasmic mtDNA mutations, they provide little hope of relieving the debilitating symptoms of most patients with mitochondrial disease. What is required is an effective metabolic or pharmacological treatment that ameliorates these symptoms. Although numerous case reports and a limited number of clinical trials on putative mitochondrial therapies have been conducted, marked reproducible clinical improvement has been difficult to verify.
In a recent review on treatment of mitochondrial disorders, the so-called Cochrane collaboration concluded that no clear benefit from the main components of the mitochondrial therapies could be scientifically established. The Cochrane collaboration judged only seven clinical trials to be scientifically valid. These trials encompassed a variety of compounds such as CoQ, creatine monohydrate, dichloroacetate, and dimethyglycine, but they used limited sample sizes of 5 to 30 patients (119, 120).
Beyond these controlled clinical trials, there is an extensive literature comprising open-label or retrospective studies and single case reports. However, these reports often present conflicting results. Thus, the development and testing of metabolic therapies have been plagued by the limited availability of groups of well-characterized patients with similar molecular defects and by our imperfect understanding of the pathophysiology of mitochondrial diseases.
Given that mitochondrial dysfunction involves a perturbation of metabolism, why have metabolic and pharmacological treatments not been more effective? We believe that the answer lies in the integrated network of interactions within the human cell, in which mitochondrial bioenergetics is involved. A single energetic defect can have many different consequences. Thus, use of a single metabolite such as ascorbate (vitamin C) or tocopherol (vitamin E) to perturb the system may have only a transient effect, given that the other components of the network ultimately counteract the therapeutic intervention. Furthermore, the varying energetic defects in different patients perturb the network in different ways. Hence, the same therapeutic approach may be beneficial for one individual and deleterious for another.
Therefore, to develop effective metabolic and pharmacological therapies, we must understand the energetics network. Then we could develop therapeutic strategies to address the various ramifications of each bioenergetic defect, thus permitting coordinated targeting of multiple different dysfunctional consequences of the genetic mutation.
Energetic Regulation
To understand mitochondrial bioenergetics, we must start with the source of our energy: the dietary calories that we consume. The environmental calories consumed determine the availability of reducing equivalents for the mitochondrial redox reactions (Figure 1).
In the presence of excess calories, cells are programmed to actively catabolize the available calories and use the energy for cellular growth, maintenance, repair, and—in the case of proliferative cells—to replicate their DNA and divide. We have hypothesized that the energetic status of the cell is communicated to the nucleus by the modification of nuclear chromatin through phosphorylation via ATP, through acetylation via acetyl-CoA, and through methylation via S-adenosylmethionine—all high-energy intermediates generated via glycolysis and mitochondrial metabolic and OXPHOS pathways. Thus, when calories are abundant, chromatin histones become phosphorylated by kinases and acetylated by histone acetylases, which reduce their affinity for DNA and permit active gene expression. In the absence of sufficient calories, ATP and acetyl-CoA levels decline, histone phosphorylation and acetylation are reduced, gene expression is repressed, and growth declines and ultimately stops.
In multicellular organisms, this system has become modified such that when carbohydrates in the diet are abundant, cellular metabolism shifts toward glycolysis with the excess calories being stored as fat in the white and brown adipose tissues. When carbohydrate supply is limited, energy must be generated through oxidation of fatty acids by mitochondrial beta oxidation. The shift to ward a more glycolytic metabolism is achieved because high serum glucose stimulates pancreatic beta cell insulin secretion, which acts on target cells through phosphatidylinositol 3 kinase (PI3K) and Akt to phosphorylate and inactivate the forkhead box, subgroup O (FOXO) transcription factors. Inactivation of the FOXOs suppresses the expression of the mitochondrial gene cotranscriptional activator, peroxisome proliferator–activated receptor (PPAR) gamma coactivator 1 alpha (PGC-1α), thus downregulating mitochondrial OXPHOS and antioxidant defenses. In the high-carbohydrate state, acetyl-CoA is abundant and glycolysis generates excess cytosolic NADH. This process results in the acetylation and partial inactivation of the FOXOs and PGC-1α, which are stable because deacetylation by the protein deacetylase Sirt1 requires NAD+ as a substrate and Sirt1 cannot use NADH. When carbohydrates are limited, insulin secretion declines, the FOXOs become dephosphorylated, and PGC-1α transcription increases. Moreover, the pancreatic alpha cells secrete glucagon-activating adenylylcyclase in the target tissues. The resulting cAMP stimulates protein kinase A to phosphorylate the cAMP response element binding protein (CREB), which also increases the transcription of PGC-1α. Moreover, because oxidation of fatty acids and of the liver-generated ketone bodies (beta-hydroxybutyrate and acetoacetate) can only occur within the mitochondrion, the cytosolic NADH/NAD+ ratio remains low. The high proportion of cytosolic NAD+ then stimulates Sirt1 to deacetylate the FOXOs and PCG-1α, thus enhancing mitochondrial biogenesis, OXPHOS activity, and the antioxidant enzyme level. These changes facilitate the extraction of the calories from stored fats and circulating ketones (Figure 1) (11, 121).
The induction of mitochondrial biogenesis can be further stimulated by fasting and starvation through the action of AMP-activated protein kinase (AMPK). When ATP is limited, the increased AMP activates AMPK, which phosphorylates and further activates PGC-1α to up-regulate mitochondrial biogenesis.
As mitochondrial OXPHOS increases, hydrogen peroxide (H2O2) production also increases. The mitochondrial H2O2 can then diffuse out of the mitochondrion and into the nucleus, changing the nuclear redox potential and altering the redox status of transcription factors. In potentially mitotic cells, the mitochondrial H2O2 can activate immediate early transcription and induce the replication cycle (Figure 1). This mitogenic character of subtoxic levels of H2O2 has been well documented in cultured mammalian cells (122).
Reactive Oxygen Species and Reactive Nitrogen Species
The mitochondria generate much of the endogenous cellular ROS through OXPHOS. Under normal physiological conditions, ROS production is highly regulated, at least in part, by complex I (123–127). However, when the ETC is inhibited by an OXPHOS gene mutation or becomes highly reduced from excessive calorie consumption relative to exercise level, then the ETC electron carriers accumulate excessive electrons, which can be passed directly to O2 to generate superoxide anion (O2·−). The O2·− generated by complex I is released into the mitochondrial matrix, where it is converted to H2O2 by MnSOD (the Sod2 gene). The O2·− generated from complex III is released into the mitochondrial intermembrane space, where it is converted to H2O2 by copper/zinc superoxide dismutase (Cu/ZnSOD; Sod1) located in the intermembrane space and cytosol. Mitochondrial H2O2 can then diffuse into the nucleuscytosol. If H2O2 encounters a reduced transition metal or is mixed with O2·−, the H2O2 can be further reduced to hydroxyl radical (OH·), the most potent oxidizing agent of the ROS (Figure 1). ROS can damage cellular proteins, lipids, and nucleic acids. Hence, excessive mitochondrial ROS production can exceed the antioxidant defenses of the cell, and the cumulative damage can ultimately destroy the cell by necrosis or apoptosis.
Reactive nitrogen species (RNS) can be generated by mitochondria. At high oxygen tensions, COX (complex IV) uses O2 as its terminal electron acceptor. However, at low oxygen tension, complex IV can reduce nitrite to nitric oxide (NO). In yeast, the mitochondrially produced NO then induces an alternative nuclear complex IV subunit, COX5b, which enhances complex IV NO production (128, 129). NO is also generated from arginine and O2 by the three types of nitric oxide synthase (NOS): neuronal, inducible, and endothelial (130–132).
NO is a vasodilator. Hence, NO production by complex IV provides an oxygen-based feedback loop. When oxygen tension is low, complex IV switches to NO synthesis. NO diffuses out of the mitochondrion and cell, causing relaxation of the vascular smooth muscle cells that results in vasodilation (133). This in turn increases tissue oxygenation. However, this system can be compromised by genetic defects that inhibit the ETC and increase ROS production, given that mitochondrial O2·− reacts with NO to generate peroxynitrite, which is inactive as a vasodilator. This may be the reason for the stroke-like episodes associated with the MELAS mtDNA tRNALeu(UUR) nucleotide 3243A>G mutation (34), leading to the proposal that MELAS strokes could be treated with arginine supplementation (134).
Peroxynitrite is a potential reactant that can oxidize reduced groups such as Fe-S centers and sulfhydryls. Peroxynitrite also reacts with tyrosines in proteins to produce nitrotyrosines (nitrosylation), which can affect protein structure and function (135).
The Mitochondrial Permeability Transition Pore
The mitochondria contain a self-destruct system, the mtPTP. The mtPTP is activated when the biochemical health of the mitochondria and cell declines, specifically when mitochondrial energy production declines, ROS generation increases, and excessive Ca2+ is released into the cytosol and taken up by the mitochondria. When the mtPTP is activated, it opens a channel in the mitochondrial inner membrane, short-circuits ΔP, and initiates programmed cell death (apoptosis). Activation of the mtPTP can be inhibited by cyclosporin A (1).
The opening of the mtPTP and the collapse of ΔP are associated with the release of cytochrome c, procaspase-9, apoptosis-initiating factor, and endonuclease G from the mitochondrial intermembrane space into the cytosol. The former two interact with cytosolic Apaf1 to form the apoptosome, activating the caspase cascade that degrades the cellular proteins. The latter two are transported to the nucleus and degrade the chromatin. Cells with defective mitochondria are thus degraded from the inside out, stopping the release of the mitochondrial bacteria–like antigens into the circulation (1).
Mitochondrial and Cellular Redox Biology
Mitochondrial redox reactions are generally considered in the context of generating ATP by OXPHOS. However, mitochondrial redox chemistry is of profound importance to the redox balance of the entire cell and is central to the signal transduction pathways that regulate nuclear gene expression (11, 136, 137). As a result, the use of therapeutics from the perspective of increasing ATP production by feeding more electrons into the ETC when the ETC is inhibited by a genetic defect simply increases the cellular reduction state and increases ROS production. Similarly, attempting to limit mitochondrial ROS production by providing excessive antioxidant compounds may disrupt ROS signaling pathways essential for cell growth. For these reasons, attempts to treat mitochondrial diseases by metabolic supplementation without consideration of the complex network of cellular redox reactions have generally resulted in either disappointing or counterproductive results. Therefore, for us to address mitochondrial metabolic therapy, it is essential that we first discuss the cellular redox chemistry that would be influenced by the treatments.
Redox regulation of the epigenome
The direct link between the mitochondrial redox state and the regulation of cellular energy metabolism is evident from the biology of the sirtuins. Mammals possess seven sirtuins: Sirt1 and -2 located in the nucleus and cytosol; Sirt3, -4, and -5 in the mitochondrion; and Sirt6 and -7 in the nucleus (121). Sirt1 regulates both mitochondrial biogenesis, through the FOXOs and PGC-1α, and mitophagy, through deacetylation of members of the Agt family of proteins (138). Sirt3 deacetylates various OXPHOS proteins and also modulates the mitochondrial metabolic rate (139).
However, redox signaling’s role in regulation of the epigenome is much more pervasive and more ancient than that of the sirtuins. Energy, in the form of reducing equivalents, flows through biological systems from reduced (calorie intake) to oxidized (oxygen). Thus, the use of oxygen as the terminal electron acceptor makes a wide range of redox potentials available to the eukaryotic cell, permitting a broad range of protein modulation through alterations in thiol/disulfide ratios, ROS oxidation, and RNS nitrosylation.
Redox control systems
ROS participate in the activation of a wide spectrum of tyrosine and serine/threonine kinases including Src kinase, protein kinase C (PKC), mitogen-activated protein kinase (MAPK), receptor tyrosine kinases, etc. (123–127). Moreover, ROS also modulate the activity of phosphatases such as protein tyrosine phosphatase 1B, Src homology 2–domain-containing tyrosine phosphatase 2, and phosphatase and tensin homolog, which modulate phosphoprotein levels (140). Mitochondrial and cellular redox states also determine the redox ratios of the nucleus-cytosolic and mitochondrial NADPH/NADP+, nucleus-cytosolic and mitochondrial NADH/NAD+, thioredoxins 1 and 2 (SH)2/SS [Trx1(SH)2/SS and Trx2(SH)2/SS, where SH stands for thiol and SS for disulfide], gamma-glutamylcysteinylglycine [reduced glutathione/oxidized glutathione (GSH/GSSG)], and cysteine/cystine (CyS/CySS). These eight key redox nodes coordinate the regulation of a broad range of processes. They are also coupled to each other and to a wide range of redox-sensitive proteins.
Within the mitochondrion, the NADPH/NADP+ redox state is coupled to that of Trx2(SH)2/SS and to GSH/GSSG by thioredoxin reductase 2Oxd/Red (TrxR2Oxd/Red) and glutathione reductaseOxd/Red (GSHROx/Red). Similarly, in the cytosol NADPH/NADP+ is coupled to Trx1(SH)2/SS and to GSH/GSSG by TrxR2Oxd/Red and by GSHROx/Red (127, 140–143).
The above redox control nodes regulate ROS levels and alter the thiol/disulfide redox status of multiple proteins. Consequently, changes in the redox potential of the control nodes provide a powerful system through which a wide range of signal transduction pathways and transcription factors can be coordinated (127, 140–143).
The importance of these redox control nodes for cell growth, differentiation, and death is clear from the consequences of inactivation of the associated genes in the mouse. Glutathione is synthesized in the cytosol by the condensation of cysteine and L-glutamate to generate gamma-glutamylcysteine, catalyzed by gamma-glutamylcysteine synthetase, the rate-limiting step. Glutathione synthesis is completed by the addition of glycine to the C terminus by glutathione synthetase. Mice that have been mutated to lack the gamma-glutamylcysteine synthetase [also known as glutamylcysteine ligase (GCL)] catalytic subunit (Gclc) experience early embryonic lethality. The mutant embryos show increased apoptosis, and the Gclc null blasto-cyst cells cannot grow in a GSH-free medium unless supplemented with N-acetylcysteine (144, 145). Liver-specific inactivation of mouse Gclc results in hepatic steatosis, inflammation, increased lipid peroxidation, and marked changes in mitochondrial morphology and function (146). Mice lacking the GCL modifier subunit (Gclm) are viable and fertile, but they have reduced GSH levels, and their Gclm null fetal fibroblasts are more sensitive to oxidative stress (147).
The importance of the thioredoxin redox system for cell survival, growth, and differentiation is demonstrated by the genetic inactivation of the mouse Trx1(SH)2/SS, Trx2(SH)2/SS, TrxR1Oxd/Red, and TrxR2Oxd/Red genes. Targeted disruption of the Trx1(SH)2/SS gene resulted in early embryonic lethality (148). The absence of mitochondrial Trx2(SH)2/SS in mouse also causes early embryonic lethality associated with massive apoptosis in the embryo and in nonviable embryonic fibroblasts (149).
Targeted depletion of TrxR1Oxd/Red results in early embryonic lethality and nonviable embryonic fibroblasts, although cardiomyocytes are not affected (150). The inactivation of the mitochondrial TrxR2Oxd/Red in mouse also causes early embryonic lethality, with increased liver apoptosis, reduced hematopoietic differentiation, heart defects with abnormal heart mitochondria, and slow-growing embryonic fibroblasts (151).
The critical role of the thiol/disulfide redox control systems stems from the fact that many proteins harbor redox-sensitive cysteines. These thiols can become oxidized to disulfides by reacting with another cysteine thiol within the protein to generate a disulfide bridge, or with a free cysteine to generate protein-CyS-SCy (cysteinylation) or with the thiol in glutathione to generate protein-CyS-SG (glutathionylation). These thiol-disulfide reactions are mediated by the redox status of 2GSH/GSSG, 2CyS/CySS, Trx1(SH)2/SS, and Trx2(SH)2/SS. The redox potential of each of these redox control nodes is a function of the steady-state redox status of the couples and the concentrations of their reactants and products: Eh = Eo + RT/NF ln[SS/(SH)2], where Eh and Eo stand for actual and steady-state redox potential, respectively. Hence, each redox control node can have different redox potentials and act on different proteins to regulate their redox state and thus activity (127, 140–142). Factors that modulate these redox nodes include the availability of reducing equivalents (calories), oxidative stress, and intracellular and extracellular environmental influences (Figure 2).
Figure 2.
Oxidation-reduction (redox) regulation of the epigenome. The cellular redox system is coordinated by eight redox control nodes: mitochondrial and cytosolic NADPH/NADP+, mitochondrial and nucleus-cytosolic NADH/NAD+, thioredoxins 1 and 2 (SH)2/SS [Trx1(SH)2/SS and Trx2(SH)2/SS, where SH stands for thiol and SS stands for disulfide], reduced glutathione/oxidized glutathione (GSH/GSSG), and cysteine/cystine (CyS/CySS). In the mitochondrion, a small portion of the electron transport chain (ETC) electrons are directly transferred to O2 to generate superoxide anion (O2·−), which is converted to hydrogen peroxide (H2O2) by manganese superoxide dismutase (MnSOD) in the matrix or by copper/zinc superoxide dismutase (Cu/ZnSOD) in the cytosol. The mitochondrial H2O2 level is regulated by the GSH-driven glutathione peroxidases 1 and 4 (Gpx1 and -4), and by the Trx2(SH)2-driven peroxiredoxins 3 and 5 (Prx3 and -5). Both GSH and Trx2(SH)2 are reduced from NADPH, which is generated from NADH by the mitochondrial nicotinamide nucleoside transhydrogenase (NNT). H2O2 can diffuse into the cytosol-nucleus, where it can be detoxified by peroxisomal catalase. H2O2 can also be converted to hydroxyl radical (OH·) when it encounters a reduced transition metal. Reactive oxygen species (ROS), O2·−, H2O2, and OH· can alter redox potentials of Trx1(SH)2/SS, Trx2(SH)2/SS, GSH/GSSG, and CyS/CySS, which regulate the activities of redox-sensitive proteins containing cysteines. ROS can also react with the guanine-cytosine (GC)-rich chromosomal telomere repeats, causing their premature shortening. On the plasma membrane, NADPH oxidases (NOX) can also generate ROS to produce a bactericidal oxidative burst or to stimulate cell growth or mediate cell death. The cytosolic and nuclear Trx1(SH)2/SS can act through apurinic/apyrimidinic endonuclease/redox factor 1 (APE-1/ Ref1Red/Ox) to regulate activator protein 1 (AP1, c-Jun), hypoxia-inducible factor 1 (HIF-1), nuclear factor–E2 related factor 2 (Nrf2), nuclear factor kappa B (NF-κB), p53, glucocorticoid receptor (GR), and estrogen receptor (ER). AP1 regulates DNA replication and cell growth, HIF-1 modulates glycolytic and oxidative energy metabolism, Nrf2 regulates cellular stress response genes, NF-κB regulates cytokines production and the inflammatory response, and p53 modulates cell death. Trx1(SH)2/SS also regulates the activity of the stem cell pluripotency factor, Oct4. GSH/GSSG maintain the redox potential of the endoplasmic reticulum for the proper processing of secreted proteins. CyS/CySS play an important role in the lysosomal hydrolysis of cysteine-rich proteins containing disulfide bridges. Extracellular CyS/CySS and GSH/GSSG can also regulate cell growth and proliferation through epidermal growth factor receptor (EGFR) and mitogen-activated protein kinase (MAPK). Abbreviations: AMPK, AMP-activated protein kinase; ANT, adenine nucleotide translocator; COX, cytochrome c oxidase; CREB, cAMP response element binding; dNDP, deoxynucleoside diphosphate; EPO, erythropoietin; FOXO, forkhead box, subgroup O; HO, heme oxygenase; IGF, insulin-like growth factor; LON, mitochondrial LON protease; mTORC, mammalian target of rapamycin; NADH, reduced nicotinamide adenine nucleotide; NDP, nucleoside diphosphate; OAA, oxaloacetic acid; OXPHOS, oxidative phosphorylation; PDK, pyruvate dehydrogenase kinase; PGC-1β, peroxisome proliferator–activated receptor gamma coactivator 1 beta; PHD, prolylhydroxylase domain protein; PKA, protein kinase A; PrSS, protein disulphide; PrSH, protein thiol; TGF, transforming growth factor; TGFR, TGF receptor; TNF, tumor necrosis factor; TNFR, TNF receptor; TSC, tuberous sclerosis protein; VEGF, vascular endothelial growth factor; VHL, von Hippel–Lindau tumor suppressor.
Each cellular compartment maintains discrete redox nodes at the potential necessary to perform that compartment’s function. GSH/GSSG redox control nodes are found in the mitochondrion, peroxisome, cytosol, endoplasmic reticulum, lysosome, nucleus, and extracellular space. CyS/CySS nodes are important in the cytosol, lysosome, and extracellular space. Trx2(SH)2/SS control nodes function exclusively in the mitochondrion, whereas Trx1(SH)2/SS control nodes function in the cytosol and the nucleus (Figure 2). The overall hierarchy of redox control node potentials from reduced to oxidized is: mitochondria, cytosol, nucleus, endoplasmic reticulum, extracellular space. This hierarchy follows from the flow of reducing equivalents into the mitochondria via the ETC and into the cytosol through glycolysis, the pentose phosphate shunt, and the mitochondria.
Within the mitochondrion, reducing equivalents are maintained on the soluble carriers, NAD+ and NADP+. Hence, the concentration and redox status of these electron carriers set the upper limits of electronegativity for the pathways to which they contribute electrons. The redox status of the mitochondrial redox couples has been estimated to be (a) −405 mV for NADPH/NADP+, (b) −340 to −360 mV for Trx2(SH)2/SS, (c) −300 mV for GSH/GSSG, and (d) −250 mV for NADH/NAD+ (140–142, 152, 153). In the cytosol, the potentials of the redox couples have been estimated to be (a) −393 mV for NADPH/NADP+, (b) −280 mV for Trx1(SH)2/SS, (c) from −260 to −170 mV for GSH/GSSG, and (d) −160 mV for CyS/CySS. In the nucleus, the redox status of Trx1(SH)2/SS is −300 mV. The redox state of the endoplasmic reticulum is set by GSH/GSSG at −189 mV. The plasma (extra-cellular space) redox control nodes for healthy individuals have been estimated to be −140 mV for GSH/GSSG and −80 mV for CyS/CySS (Figure 2) (140–142, 152, 153).
In the mitochondrion (Figure 2), reducing equivalents from dietary calories are converted to NADH and fed into the ETC through complex I. Because the redox potential of NADH/NAD+ within the mitochondrion is −250 mV, most of the electrons from NADH flow down the ETC to O2 at +1200 mV to generate H2O. However, approximately 1%–5% of the electrons of the ETC are transferred directly to O2 to generate O2·−. The chemical affinity of O2 for electrons is unchanged by oxygen tension, and the KM (Michaelis constant) for O2 by complex IV places a lower limit on the reduction of O2 by complex IV. As a result, as O2 concentration declines, the ETC becomes reduced and mitochondrial ROS production increases.
Mitochondrial H2O2 levels are regulated by the glutathione peroxidases and the peroxiredoxins. These in turn derive their reducing potential from NADPH. Generation of NADPH from NADH requires additional reducing potential. This is provided by the mitochondrial nicotinamide nucleoside transhydrogenase (NNT), which uses the mitochondrial inner-membrane ΔP to provide the necessary energy to transfer the reducing equivalents from NADH to NADP+, increasing the reducing potential from −250 mV (NADH/NAD+) to −405 mV (NADPH/NADP+). NADPH has sufficient reducing potential to convert GSSG to 2GSH via the mitochondrial GSHROx/Red and also to convert Trx2(SS) to Trx2(SH)2 via the mitochondrial TrxR2Red/Ox (Figure 2).
The concentration of peroxide within the mitochondrial matrix is regulated by the GSH-driven glutathione peroxidases (Gpxs), specifically Gpx1 and -4, and by the Trx2(SH)2/SS-driven peroxiredoxins (Prxs) Prx3 and -5. The mitochondrial Gpx system reduces H2O2 to 2H2O by a 2e− transfer from 2GSH to produce GSSG. Because glutathione cannot be synthesized by the mitochondrion, it must be imported from the cytosol by the mitochondrial inner-membrane dicarboxylate and 2-oxoglutarate transporters (127, 141). Trx2(SH)2/SS, acting through mitochondrial Prx3, can reduce a variety of mitochondrial peroxides (H2O2, alkyl hydroperoxide, and peroxynitrite) (Figure 2). Inactivation of Prx3 in the mouse results in animals that are grossly normal but whose macrophages show significantly higher ROS production. However, upon lipopolysaccharide (LPS) exposure, the mutant mice develop severe injury to lung DNA and proteins (154). Thus, mitochondrial H2O2 production is directly regulated by the redox potential of the NADPH/NADP+ couple, which in turn is maintained by the flow of reducing equivalents through the mitochondrial ETC and the ΔP. The Trx2(SH)2/SS redox state can also stabilize the mtPTP (140, 155). Finally, the mitochondrial GSH/GSSG redox control node acts through glutaredoxin 2 (Grx2) to regulate the function of a variety of proteins through their thiols (156).
Mitochondrial NADH/NAD+ redox levels are coupled to the cytosol through the mitochondrial inner-membrane NADH/NAD+ shuttle systems. The malate-oxaloacetate shuttle involves the aspartate-glutamate carrier and the malate-alpha-ketoglutarate carrier linked to cytosolic and mitochondrial malate dehydrogenase and aspartate aminotransferase (Figures 2 and 3). Although the redox statuses of the mitochondria and cytosol are coupled, the redox potential of the two NADH/NAD+ systems can differ due to differences in concentrations of NAD+ in the two compartments and differences in the activities of the carriers.
Figure 3.
Metabolic, redox, and epigenomic influences of the ketogenic diet. Calories can enter the cell as glucose or as ketones [beta-hydroxybutyrate (BOHB) and acetoacetate (AcAc)]. Glucose is processed through the cytosolic glycolytic pathway to generate cytosolic NADH (reduced nicotinamide adenine nucleotide) and pyruvate. Pyruvate can enter the mitochondrion and be processed into acetyl-CoA, CO2, and mitochondrial NADH. Ketones are directly metabolized by the mitochondrion to generate acetyl-CoA and mitochondrial NADH, leaving cytosolic NAD+ oxidized. Ketone body–derived acetyl-CoA is condensed with oxaloacetic acid (OAA) to generate citrate. In inhibitory neurons, citrate is converted to alpha-ketoglutarate (α-KG), then glutamate (Glu), and then gamma-aminobutyric acid (GABA). Increased GABA inhibits neuronal excitation, suppressing seizures. Increased acetyl-CoA also drives the tricarboxylic acid (TCA) cycle to generate mitochondrial NADH, which is oxidized by the electron transport chain (ETC) to increase ΔP and drive ATP synthesis. The elevated ATP drives the neuron plasma membrane K+ and Ca2+ pumps, thereby stabilizing the membrane potential, decreasing depolarization, and inhibiting seizures. The increased ΔP also drives the mitochondrial innermembrane nicotinamide nucleotide transhydrogenase (NNT) to convert NADH to NADPH. The elevated NADPH drives the reduction of mitochondrial oxidized glutathione (GSSG) to reduced glutathione (GSH), protecting the mitochondria against oxidative stress. Mitochondrial citrate can be exported to the cytosol, where it is cleaved by ATP-citrate lyase to generate cytosolic acetyl-CoA and OAA. The OAA is reduced by malic dehydrogenase to malate, and the malate is oxidized to pyruvate, generating NADPH. The cytosolic NADPH can then reduce cytosolic GSSG to GSH, protecting the cytosol against oxidative stress. Cytosolic acetyl-CoA can drive histone acetylation by histone acetylases (HATs), opening chromatin and activating transcription of energetic genes. Histone deacetylases (HDACs) can reverse this acetylation, inhibiting transcription of energetic genes. HDAC inhibitors such as valproate keep the chromatin open and energy production high. However, valproate treatment of patients with severe energy deficits may drive energy production beyond the capacity of the impaired bioenergetic system, resulting in failure and pathology (306). Oxidation of ketones by the mitochondria leaves cytosolic NAD+ oxidized. This activates Sirt1 to deacetylate the forkhead box, subgroup O (FOXO) and peroxisome proliferator–activated receptor gamma coactivator 1 alpha (PGC-1α) transcription factors, resulting in the induction of mitochondrial biogenesis, oxidative phosphorylation (OXPHOS) and ATP production. Abbreviations: AcAcCoA, acetoacetyl-CoA; Suc-CoA, succinyl-CoA; CoASH, coenzyme A; Asp, aspartate; Gln, glutamine; SSA, succinic semialdehyde; Ac-histone, acetylated histone; FAD, flavin adenine dinucleotide; OAADPr, O-acetyl-ADP-ribose.
The nucleus and cytosol also rely on the NADPH/NADP+ redox couple, with its redox potential of −393 mV, to reduce Trx1(SH)2/SS. However, the cytosolic NADPH/NADP+ redox potential is physically separated from the mitochondrial NADPH/NADP+ redox couple. Hence, the cytosolic NADPH must be generated within the cytosol. One of the processes by which mitochondrial redox potential can be transferred to the cytosol is via the export of mitochondrial citrate. Within the mitochondrion, acetyl-CoA is condensed with oxaloacetate by citrate synthase to generate citrate. The citrate can then be exported to the cytosol by the citrate carrier. In the cytosol, citrate can be cleaved by ATP-citrate lyase to generate acetyl-CoA and oxaloacetate. The oxaloacetate can then be reduced to malate by malate dehydrogenase and NADH. Malate is then converted to pyruvate by malic enzyme, reducing NADP+ to NADPH in the process. The pyruvate can either return to the mitochondrion to generate acetyl-CoA or be reduced to lactate in the cytosol by lactate dehydrogenase via NADH (Figures 2 and 3). The generation of NADPH via malic enzyme was recently found to be central to cancer cell metabolism, mediated by the c-myc induction of the glutaminolysis pathway (157–159). Therefore, mitochondrial redox reactions play an important role in maintaining the cytosolic NADPH/NADP+ redox ratio.
Cytosolic NADPH can also be generated by the pentose shunt. Glucose is phosphorylated to glucose-6-phosphate by ATP and hexokinase. Glucose-6-phosphate dehydrogenase then converts glucose-6-phosphate + H2O + 2NADP+ to ribose-5-phosphate + 2NADPH + CO2 + 2H+.
In the nucleus and cytosol, reducing equivalents flow from NADPH/NADP+ through GSHROx/Red and TrxR1Red/Ox to regulate the cytosolic GSH/GSSG and Trx1(SH)2/SS ratios. The GSH/GSSG redox potential regulates (a) Gpx1 and Gpx3 to control peroxides and (b) Grx1 to regulate the redox state of proteins (Figure 2). Grx1-deficient mice are prone to myocardial injury as a result of ischemic reperfusion insult and to hyperoxia-induced lung injury, and Grx1 null myocyte-enhancing factors are sensitive to paraquat but are more resistant to tumor necrosis factor alpha (TNFα) and actinomycin D–induced apoptosis (160).
The Trx1(SH)2/SS redox potential acts through Prx1 and -2 to reduce peroxides (Figure 2). Prx1 mutant mice have shortened life spans due to hemolytic anemia, cancer, and reduced natural killer cell number and function. Also Prx1 null myocyte-enhancing factors proliferate slowly and are more sensitive to oxidative stress (161). Mice lacking Prx2 generate markedly increased levels of ROS and are more susceptible to paraquat-induced oxidative stress. They develop splenomegaly and hemolytic anemia, their macrophages are more susceptible to oxidative stress, and their hearts are sensitive to ischemia-reperfusion injury (162–164).
Trx1(SH)2/SS redox potential also regulates apoptosis signal–regulating kinase (ASK1). ASK1 is a MAPK that activates the c-Jun N-terminal kinase (JNK) and p38 MAPK pathways and is required for TNFα-induced apoptosis. Under reducing conditions, ASK1 is complexed with Trx(SH)2. However, under oxidizing conditions presumably generated by ROS and GSSG, Trx(SH)2 is converted to the oxidized form [Trx1(SS)], and ASK1 is released. The resulting activated ASK1 then initiates the stress-response system and apoptosis (165, 166). Because TNFα preferentially oxidizes the mitochondrial Trx2(SH)2/(SS), rather than the nucleus-cytosol Trx1(SH)2/(SS) (167), it is possible that ASK1 interacts with both Trx2(SH)2 and Trx1(SH)2 (140, 141).
The Trx1(SH)2/SS redox potential also acts through the bifunctional protein apurinic/apyrimidinic endonuclease/redox factor 1 (APE-1/ Ref1Red/Ox). Ref1Red/Ox, in turn, regulates activator protein 1 (AP1, c-Jun), nuclear factor–E2 related factor 2 (Nrf2), nuclear factor kappa B (NF-κB), p53, glucocorticoid receptor, estrogen receptor, and hypoxia-inducible factor 1 alpha (HIF-1α) by maintaining conserved cysteine residues in the reduced form required for DNA binding. AP1 is also activated by oxidant induction of JNK, which phosphorylates critical domains in the c-Jun transactivation domain. Activation of the AP1 system directly regulates an array of genes involved in cell growth and development (Figure 2). Nrf2, which regulates stress-response genes, is maintained in an inactive state in the cytosol in a complex with the Kelch-like ECH-associated protein 1 (Keap 1). Oxidation of a key cysteine in Keap 1 releases Nrf2 to migrate to the nucleus, where it binds to its antioxidant-response elements. This process induces a wide range of antioxidant and stress-response genes, including many of those for glutathione biology (Figure 2). NF-κB is also activated by ROS. Physiological stimuli such as TNFα binding to its receptor increase ROS production, which activates NF-κB. NF-κB then enters the nucleus to induce cytokine and inflammatory gene expression (Figure 2) (127, 140, 141, 152, 168). The capacity of the stem cell transcription factor Oct-4 to bind to DNA is also regulated by oxidation and is associated with the binding of Trx to cysteines in the POU domain of the protein. Overexpression of Trx(SH)2/SS increases the transcriptional activity of Oct-4, whereas expression of an inactive Trx(SH)2/SS inhibits its transcriptional activity (169).
Studies of the Trx1(SH)2/SS, GSH/GSSG, and CyS/CySS redox control nodes of the cytosol have revealed that the GSH/GSSG redox state can vary depending on the cell status and environmental conditions. The human colon carcinoma cell line, HT29, actively proliferates when the cellular GSH/GSSG redox ratio is −260 mV, but when induced to differentiate through use of butyrate or isothiocyanate, the redox state shifts to −200 mV (170). Exposure of HT29 cells to an extracellular CyS/CySS ratio of −75 mV activates NF-κB and induces mitochondrial apoptosis (142). Exposure of murine lung fibroblasts to a CyS/CySS redox potential of −46 mV stimulates their proliferation and expression of fibronectin, NF-κB, and transforming growth factor beta 1 (TGFβ1) (171). Different cell types can respond to the same redox signal differently, presumably due to differences in the signal transduction pathways. Thus, exposing cells to external redox signals can cause the cells to grow, differentiate, or die by changing their internal cellular redox status.
Trx1(SH)2/(SS) is also an important redox control node for the nucleus. However, its redox potential in the nucleus is less variable than that of cytosolic Trx1(SH)2/(SS), remaining close to −300 mV (Figure 2) (141, 142). The redox potential of the endoplasmic reticulum is maintained by GSH/GSSG and is relatively oxidized. This is important in the processing of secreted proteins (Figure 2) (141). The redox status of the peroxisome has not been determined. However, the peroxisome encompasses a variety of active redox reactions and antioxidant enzymes such as catalase (141).
In the lysosome, the potential of the CyS/CySS redox control node is thought to play an important role in the lysosomal hydrolysis of cysteine-rich proteins containing disulfide bridges (152). The leftover cystine can be transported out of the lysosome by cystinosin (CTNS), a proton-cystine symporter. Cysteine may be transported out of the lysosome by the e and f lysosomal transport systems and can be actively taken up by the lysosome via the cysteine transporter (172). In the lysosome, cysteine may act as a reductant to counteract the increased oxidation state of accumulated cystine (Figure 2). Cystine can also be taken up by the cell through the plasma membrane transport system CG (X−GC) carrier, and in the cytosol it is reduced to cysteine by GSH. In cystinosis patients who lack CTNS (173), cystine accumulates in the lysosomes and ultimately precipitates as crystals. The exceedingly high concentration of cystine in the cystinosis lysosomes may result from a combination of cystine generated from protein degradation and from the import of cytosolic cysteine, which balances the lysosomal redox state. The resulting depletion of the cytosolic cysteine could inhibit cytosolic glutathione synthesis and deplete cellular GSH, thus increasing cellular oxidative stress, mitochondrial dysfunction (174– 178), and apoptosis (167, 179–181), all of which are seen in cystinosis cells and patients. Inactivation of Ctns in mice also results in lysosomal development of cystine crystals, as well as other aspects of the cystinosis phenotype (182). Cystinosis in human and mouse can be treated by cysteamine. Cysteamine is imported into the lysosome, where it generates a cysteine-cysteamine mixed disulfide; the mixed disulfide is then exported from the lysosome by the lysine transporter (183).
The redox status of the extracellular space (plasma) is determined by the GSH/GSSG and CyS/CySS redox control nodes. In normal individuals, the redox potential of plasma GSH/GSSG is approximately −140 mV, and that for CyS/CySS is approximately −80 mV (Figure 2) (184).
Nox reactive oxygen species
Because mitochondrially generated H2O2 was probably one of the first molecules used for signal transduction between the protomitochondrion and the proto-nucleus-cytosol, it would have been employed for regulating development in multicellular organisms. Hence, ROS signaling would have become important for cellular growth and differentiation in tissues and organs, even in contexts wherein the mitochondrial redox state could not change. Consequently, an alternative method for developing O2·−, the Nox and Doux system, evolved. The Nox proteins may have evolved early in multicellular organisms, as they have been found in chordates, plants, fungi, and myxomycetes (185).
The human genome contains seven members of the Nox family: Nox1–5 and Doux1–2. The Nox enzymes use NADPH and H+ to reduce O2 to O2·−. The O2·− is then converted to H2O2 by Cu/ZnSOD. Because the Nox/Doux enzymes are transplasma or vacuolar membrane proteins, the Cu/ZnSOD required to convert Nox O2·− to H2O is probably the extracellular SOD isoform (Sod3). The Douxs differ from the Noxs in having an additional peroxidase-like ectodomain.
The best-characterized Nox is Nox2 (also known as gp91phox), the enzyme responsible for the bactericidal oxidative burst generated within phagocytic vesicles of macrophages. The phagocyte NADPH oxidase is a multi-subunit enzyme encompassing gp91phox. The membrane-bound Nox2 polypeptide encompasses two hemes, a flavin adenine dinucleotide, and an NADPH-binding site. Nox2 is tightly associated with a second membrane subunit, p22phox, and together these two contain all of the catalytic sites. Activation of Nox2 also requires four soluble cytosolic polypeptides: p40phox, p47phox, p67phox, and the small GTPase Rac. Rac is complexed with the cytosolic RhoGDI. Once activated, Rac is released, and the four cytosolic subunits bind to gp91phox + p22phox. NADPH + H+ are then oxidized on the cytosolic side of the membrane; the electrons are transferred through the membrane and used to reduce O2 to O2·− on the outside of the membrane (Figure 2) (185).
Whereas Nox2 is important in innate immunity, the other Noxs are found in different tissues and have different functions. Nox1 is found in lung, vascular endothelium, intestinal epithelium, stomach, colon, uterus, pancreas, and prostate. It is induced in a number of tumors and can stimulate cellular replication through ROS production (186). Nox3 is important in inner-ear octoconia formation, Nox4 is important in kidney, and Nox5 is expressed in T- and B-lymphocytes (185).
Rac, the regulator of Nox1, is a member of the Ras family of small GTPases, which regulate ROS production in response to various stimuli. Evidence suggests that Ras-driven ROS production occurs through both Nox1 activation and mitochondrial ROS production. The Ras GTPases can activate PI3K, which generates lipid products to activate GTPase-exchange factors. These factors in turn activate Rac, which stimulates Nox1 to generate ROS (187). Rac can also stimulate NOS, and NO can react with cysteines to regulate protein function through S-nitrosylation (188).
Rac has also been shown to connect ROS production with cytoskeletal remodeling in cultured podocytes. Angiotensin II activation of its type 1 receptor activates Rac, which stimulates Nox1 to generate ROS. The increased ROS stimulates the G-coupled receptor kinase 2, which then phosphorylates the ezrin/radixin/moesin proteins. This process promotes cortical F-actin remodeling, thus tying ROS directly to cytoskeletal organization. TNFα-induced ezrin/radixin/moesin protein phosphorylation in endothelial cells has also been associated with cytoskeletal changes (189). Thus, ROS is directly linked to the regulation of the cytoskeleton.
TNF interaction with tumor necrosis factor receptor 1 (TNFR1) has been found to activate the receptor-interacting protein kinase (RIP1). RIP1 is known to activate NF-κB and MAPK, but it also stimulates the formation of a complex including Nox1, tumor necrosis factor receptor–associated death domain protein, Nox organizing protein 1, and Rac, resulting in ROS production and necrosis. RIP1 has also been reported to move to the mitochondrion and reduce the interaction between ANT and cyclophilin D (190, 190). The Src tyrosine kinase has also been shown to increase cellular ROS production by phosphorylating the RAC GTPase-exchange factor Vav2. Phosphorylated Vav2 converts the inactive Rac-GDP to the active Rac-GTP, which in turn activates Nox1 to generate ROS (192).
The chronic production of ROS by the Noxs can result in the accumulation of cellular oxidative damage. Hence, ROS from both the Nox/Doux proteins and the mitochondria may contribute to age-related diseases (193). However, mitochondrial O2·− generation must be much more important in cell toxicity than the Noxs, given that inactivation of the Sod2 gene is perinatal lethal (194, 195) and that inactivation of Sod1 (196) or Sod3 is fully compatible with life (197).
The importance of the Nox1 activation and ROS production in the action of such diverse signaling systems as Src and Ras induction of growth and TNF induction of cell death demonstrates the importance of ROS as a fundamental signal transduction system. Therefore, the Nox system confirms the importance of the mitochondrion and associated redox systems in regulation of the eukaryotic epigenome.
Reactive oxygen species, hypoxia, and stress
The regulation of cellular ROS production levels by the mitochondria and the NADPH oxidases can have a range of effects, depending on the level of ROS production and the regulatory proteins expressed in the cells involved (122–127, 140). One of the key regulatory circuits responding to changes in oxygen tension is the HIF-1 system. HIF-1 is composed of two subunits, HIF-1α and HIF-1β. Both are synthesized constitutively, but the turnover of HIF-1α is regulated by O2 concentration. At high O2 levels, prolines 402 and/or 562 are hydroxylated by prolylhydroxylase domain protein 2 (PHD2). Hydroxylation promotes the binding of the von Hippel–Lindau protein, leading to ubiquitination and degradation of HIF-1α. Hence, under normoxic conditions, HIF-1 is inactivated. Under hypoxic conditions, by contrast, O2, which is a substrate for PHD2, becomes limiting. Moreover, the reduced O2 stimulates complex III of the ETC to produce ROS, which reacts with the Fe(II) in the PHD2 catalytic center, thereby causing its inactivation. This reaction stops the hydroxylation of HIF-1α by PHD2, blocks von Hippel–Lindau protein binding, and stabilizes HIF-1α and thus HIF-1 (Figure 2).
Stabilized HIF-1 binds to the core DNA target (5′-RCGTG-3′) in a large number of genes and activates the genes permitting adaptation to low oxygen tension. Selected genes will be induced, such as genes that enhance glycolysis, including glucose transporters and glycolytic genes such as lactate dehydrogenase (LDH); genes to increase vascularization, including erythropoietin (Epo) and vascular endothelial growth factor (VEGF); and a variety of others (Figure 2) (198–200). Induction of HIF-1 also induces PDH kinase 1 (PDK1), which phosphorylates the E1α subunit of PDH, resulting in PDH inactivation. Inactivation of PDH blocks the metabolism of pyruvate to acetyl-CoA, thus inhibiting the flow of reducing equivalents through the TCA cycle to NADH and the mitochondrial ETC, the resulting reduced electron density of the ETC reduces mitochondrial ROS production, and the cytosolic pyruvate and NADH are converted to lactate by LDH (Figure 3) (199, 201–203). Activated HIF-1 also induces synthesis of the complex IV COX4-2 subunit as well as the mitochondrial LON protease. The LON protease in turn degrades the normoxic subunit, COX4-1. As a result, the kinetics of complex IV is altered to sustain ATP production at low oxygen tension (200, 204). HIF-1 also inhibits c-myc transcription through induction of MXI-1 and inactivates the c-myc protein. Because c-myc drives the expression of PGC-1β, which induces mitochondrial biogenesis in a variety of tissues (205), inhibition of c-myc transcription and inactivation of c-myc protein suppress mitochondrial biogenesis. Lastly, HIF-1 activation induces the expression of mitochondrial autophagy protein, BNIP3. BNIP3 antagonizes the binding of Beclin-1 to Bcl2 or Bclx, thus releasing Beclin-1 to initiate mitochondrial autophagy (mitophagy). This release leads to a rapid reduction in mitochondrial number and function (206). Thus, hypoxic activation of HIF-1, mediated by mitochondrial ROS production, reduces mitochondrial function in four ways: COX4 subunit switching, PDH inactivation by phosphorylation, inhibition of mitochondrial biogenesis by suppression of c-myc, and increase of mitochondrial degradation by mitophagy (207).
In addition to O2 tension, other environmental factors can alter the redox status of the mitochondrial and nucleus-cytosol control nodes. Certain heavy metals selectively alter the redox status of Trx1(SH)2/SS and Trx2(SH)2/SS. Exposure of HeLa cells to Cu, Fe, and Ni results in significant oxidation of the GSH/GSSG redox node, with relatively little effect on Trx1(SH)2/SS and Trx2(SH)2/SS redox states. By contrast, HeLa exposure to As and Hg has little effect on the GSH/GSSG redox state and partially increases the oxidation state of the cytosolic Trx1(SH)2/SS (20 to 40 mV), but it markedly increases the oxidation state of the mitochondrial Trx2(SH)2/SS (>60 mV). Cu, Fe, and Ni contribute to the generation of OH· through Fenton-type reactions, whereas As and Hg disrupt mitochondrial redox homeostasis (208).
Bacterial LPS (Escherichia coli O111:B6) injected intraperitoneally into mice is cleared by the liver, which responds with the synthesis and release of inflammatory cytokines such as TNFα and interleukin-1β. These cytokines act on the lung, resulting in acute injury. Within 2 h, plasma CyS levels decline and CySS levels increase, resulting in overall oxidation of the CyS/CySS couple by 10 mV. During the same period, the lung fluid CyS/CySS couple is oxidized by 50 mV. Because cysteine is more prone to H2O2 oxidation than glutathione, the changes in plasma CyS/CySS redox state may reflect acute increases in ROS production (209). The relationship between LPS, TNFα, and oxidative stress is demonstrated in human macrophages derived from the THP-1 leukemia cell line. LPS binds to the Toll-like receptors, activating their tyrosine kinase activity, which in turn activates NADPH oxidase to produce ROS. The ROS then activates the ROS-sensitive PKC, PI3K, MAPK, and tyrosine kinases, leading to induction of HIF-1α transcription. The increased ROS also activate mammalian target of rapamycin and S6 kinase to increase HIF-1α protein synthesis. Finally, the increased HIF-1 induces transcription of inflammation and wound-healing genes. The fact that activation of Nox increases extracellular O2·− production could partially explain the rapid oxidation of the plasma CyS/GySS pool (210). Thus, exposure to LPS results in rapid oxidation of both cytoplasmic and plasma redox pools.
Reactive oxygen species, telomeres, and replicative senescence
The importance of redox status for mammalian cell growth and differentiation has been repeatedly demonstrated by the effects of growing cells in different oxygen concentrations. Traditionally, mammalian cells have been cultured in air containing 21% O2. However, arterial blood has an oxygen concentration of approximately 12%, tissue oxygen levels are approximately 3%, and the mean oxygen level in the brain is approximately 1.5% (211). Growth of mammalian cells in ambient O2 has been shown to inhibit the differentiation of stem cells (212–216). For example, the differentiation of cultured mouse myogenic C2C12 cells into myotubes at 20% O2 is enhanced by addition of the ROS trapping agent phenyl-N-tert-butylnitrone and inhibited by addition of H2O2. Skeletal muscle myoblasts isolated from Gpx1−/− mice also show impaired myotube formation (215).
An important DNA target of ROS damage is the chromosomal telomere. Telomeres cap the ends of chromosomes and consist of thousands of copies of the TTAGGG repeat. Telomeres can be eroded during cell replication because unidirectional DNA replication necessitates the use of Okazaki fragments to initiate lagging strand replication. Consequently, lagging strand replication is always tens of nucleotides short of the chromosomal end. Telomere erosion is countered by telomerase, which uses an RNA template to extend the telomere repeats, thus compensating for the loss of repeats of the lagging stand. Telomerase consists of the RNA template and the reverse transcriptase apoprotein (TERT). Expression of the TERC gene for the RNA template is constitutive, but expression of the TERT gene is regulated, as the template is high in immortal germ line cells, pluripotent stem cells, and cancer cells but low or absent on terminally differentiated cells. TERT and TERC mutations are associated with dyskeratosis congenital and pulmonary fibrosis (217).
The ends of chromosomes are capped by a telomere loop (t-loop) and a six-protein complex, shelterin. The shelterin complex blocks the activation of the DNA-repair protein ATM, which is activated by double-strand breaks, and/or ATR, which is activated by single-strand breaks, that would be initiated by free telomere ends. Activated ATM or ATR phosphorylates Chk1 and Chk2, which enforce G1-S and G2-M arrest. They also activate p53 to induce p21, inhibit Cdk, and block the cell cycle (218). When sufficient telomere repeats are lost, the t-loop does not form. This activates the DNA-damage system, thereby activating p53 and causing cell-cycle arrest and cell death by apoptosis (219, 220).
Although telomerase is essential for maintaining telomeres during normal replication, telomeres are sufficiently long that many rounds of DNA replication would be required for incomplete lagging strand replication to erode the telomeres sufficiently to destabilize the t-loop and cause cell arrest and death. Yet telomere erosion is relatively rapid in telomerase-deficient cells such as human diploid fibroblasts. Moreover, if end replication were the primary reason for telomere erosion, then postmitotic cell telomeres would be completely stable. This discrepancy was resolved upon the discovery that the rate of telomere decay correlated with the oxygen tension at which cells were grown (221). It is now clear that telomere decay is primarily due to mitochondrially generated ROS, which cause oxidative damage in the telomere repeats (222–229).
The direct connection between telomere stability and mitochondrial ROS production was revealed following the discovery that, when ROS production increases in cells expressing telomerase, TERT is induced to translocate out of the nucleus and into the mitochondrion (230). Apparently, the increased ROS activates a Src family tyrosine kinase, located in the nuclear periphery, that phosphorylates tyrosine 707 of TERT. Tyrosine 707 signals, through a Ran GTPase–dependent process, for TERT to be exported out through the nuclear pores (231, 232). Once the TERT protein is in the cytosol, an N-terminal mitochondrial targeting peptide directs it into the mitochondrion (230). Within the mitochondrion, TERT reduces mitochondrial ROS production, protects mtDNA from damage, and regulates apoptosis in a Bcl-2-dependent fashion. Although mitochondrial TERT protects the mitochondrion and mtDNA, it does not protect the telomeres. Hence, oxidative damage to the telomeres results in rapid loss of blocks of telomere repeats (232–234). Therefore, the replicative life span of human diploid fibroblasts, the Hayflick limit (219, 220), is presumably due to mitochondrial ROS production and not the end-replication problem.
CyS/CySS and GSH/GSSG in health
Extracellular redox status, mediated by exogenous CyS/CySS and GSH/GSSG ratios, can modulate critical cysteines in extracellular signal transduction molecules (143). These extracellular signals can then modify intracellular redox control nodes to regulate cellular growth, differentiation, and death (127, 140–143).
Growth of human colon cancer Caco-2 cells increases twofold as the CyS/CsSS redox potential is reduced from 0 mV to −150 mV. This growth occurs because exposure of Caco-2 cells to the CyS/CySS −150 mV reducing environment activates extracellular metalloproteinases that contain redox-sensitive cysteines. The activated metalloproteinases cleave and release cell surface–tethered TGFα. This binds to and activates the epidermal growth factor receptor (EGFR) tyrosine kinase. Activation of the EGFR results in an 80% increase in phosphorylation of EGFR and an increase in phosphorylation of p44/42 MAPK, which activates c-myc and CREB, promoting cell growth and proliferation (Figure 2) (235, 236).
Conversely, human plasma GSH/GSSG and CyS/CySS ratios provide a window into an individual’s cellular health. In normal individuals, the plasma redox status for both CyS/CySS and GSH/GSSG declines with age. The CyS/CySS redox ratio declines continuously at approximately 0.2 mV per year, starting at −80 mV at age 18 and declining through age 93. GSH/GSSG redox status remains stable at approximately −140 mV until age 45, after which it declines at roughly 0.7 mV per year. Thus, the range of CyS/CySS and GSH/GSSG redox potentials for healthy individuals is −80 to −62 mV and −140 to −119 mV, respectively. By contrast, individuals with a variety of nutritional deficiencies or degenerative conditions have much more oxidized CyS/CySS and GSH/GSSG redox potentials on the order of −20 mV and −80 mV, respectively. This has been observed in individuals who are depleted of antioxidants or dietary cysteine, who abuse alcohol or smoke, or who have type 2 diabetes or myocardial perfusion defects—all of which impinge on mitochondrial function (142).
MITOCHONDRIAL METABOLIC THERAPEUTIC APPROACHES
Realization of the complex interactions between (a) mitochondrial redox biology and energetics and (b) cellular signaling and nuclear gene expression provides some appreciation for why treatment of patients with single mitochondrial metabolites or cofactors has yielded inconsistent results.
Metabolic Therapeutics
Current pharmacological or metabolic therapeutics have focused on increasing mitochondrial ATP production, reducing the mitochondrial ROS produced, modifying Ca2+ uptake and toxicity, stabilizing the mtPTP, and inducing mitochondrial biogenesis (Table 2). Ironically, one of the oldest therapeutic approaches—fasting and the ketogenic diet—remains the most promising treatment for mitochondrial defects. Hence, understanding the manifold potential actions of fasting and the ketogenic diet may point the way toward more comprehensive mitochondrial therapies.
Table 2.
Metabolic and pharmacological agents used to treat mitochondrial diseasea
| Agent | Indication | Mechanism | Patient dose (per day) | Reference(s) |
|---|---|---|---|---|
| Cofactors that increase the production of ATP | ||||
| CoQ | Mitochondrial disorders | Transfers electrons from complex I and II to complex III | Variable, 5 mg kg−1 | 307, 308 |
| Idebenone | Mitochondrial disorders | Transfers electrons from complex I and II to complex III | Variable; child, 5 mg kg−1 day−1; adult, 90 mg day−1 (up to 225 mg day−1) | 237, 309 |
| Succinate | Complex I deficiency | Bypasses complex I | 6 g day−1 | 244 |
| Dimethylglycine | Mitochondrial disorders | Transfers electrons to CoQ | 5 g kg−1 if > 33kg, 50 mg kg−1 if < 33 kg | 310 |
| Other cofactors and vitamin or metabolic supplements | ||||
| Thiamine (vitamin B1) | PDH deficiency | Cofactor of PDH | Variable: child, 5 mg day−1; adult, 90 mg day−1 (up to 225 mg day−1) | 249, 250 |
| Riboflavin (vitamin B2) | Complex I and II deficiency | Increases complex I and II activities | 100 mg day−1 | 289 |
| Nicotinamide (vitamin B3) | MELAS | Increases nicotinamide adenine dinucleotide | 50 mg kg−1 day−1 | 311 |
| Dichloroacetate | Severe lactic acidosis | Reduces lactic acidosis, stimulates PDH | 50 mg kg−1 | 120, 256, 312, 313 |
| Creatine monohydrate | Mitochondrial myopathy | Increases muscle phosphocreatine | 10–20 g day−1 | 314, 315 |
| Carnitine | Carnitine deficiency | Increases carnitine level | 3 g day−1 | 258 |
| L-arginine | MELAS | Increases nitric oxide production | 0.15–0.3 g kg−1 day−1 | 258 |
| Reactive oxygen species scavengers and mitochondrial antioxidants | ||||
| CoQ/idebenone | Mitochondrial disorders | Act as antioxidant | Variable; child, 5 mg kg−1 day−1; adult, 90 mg day−1 (up to 225 mg/day) | 237, 307–309 |
| Vitamin E | Mitochondrial disorders | Antioxidant | 50 µg day−1 | – |
| Vitamin C (ascorbate)/vitamin K3 (menadione) | Mitochondrial disorders | Antioxidants, bypass complex III | 4 g day−1/40 mg day−1 | 245–247 |
| Lipoic acid | Pyruvate dehydrogenase deficiency | Acts as antioxidant, stimulates PDH | 600 mg day−1 | 261 |
| Triacetyluridine | Mitochondrial disorders | De novo pyrimidine biosynthesis | Animal models | 255 |
| Modulation of the mtPTP | ||||
| Cyclosporin A | – | Inhibits the mtPTP | In vitro studies | 255 |
| Nortriptyline | – | Inhibits the mtPTP | Animal models | 269 |
| Regulation of mitochondrial biogenesis | ||||
| Sirtuin analogs | Mitochondrial disorders | Increase mitochondrial biogenesis | Animal models | 273 |
| Bezafibrate | Mitochondrial disorders | Increases mitochondrial biogenesis | In vitro studies, animal models | 276 |
| Exercise therapy and diet | ||||
| High-fat diet | Mitochondrial disorders | Increases electron transfer | 50%–60% calories | 289 |
| Ketonic diet | Intractable seizures, mitochondrial disorders | Bypasses glycolytic pathway | 4:1 lipid:nonlipid ratio | 100, 290 |
| Endurance exercise | Mitochondrial myopathy | Aerobic training induces OXPHOS | – | 280, 281 |
| Resistance exercise | Mitochondrial myopathy | Activation and proliferation of satellite cells in muscle, shifting of heteroplasmy | – | 282, 283 |
Abbreviations: CoQ, coenzyme CoQ10; MELAS, mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes; mtPTP, mitochondrial permeability transition pore; OXPHOS, oxidative phosphorylation; PDH, pyruvate dehydrogenase.
Neutraceuticals to enhance mitochondrial function
A variety of metabolic strategies to increase mitochondrial energy production have been attempted. However, to be effective, these therapies must be tailored to the patient’s specific mitochondrial defect. In contrast, treatments of patients with so-called mitochondrial cocktails have met with mixed results.
Coenzyme Q10 and analogs
CoQ is the most commonly recommended metabolic therapy for patients with putative mitochondrial disease (Table 2). Its advantages are that it is generally nontoxic and that it may both increase electron entry into the mitochondrial ETC and function as an antioxidant. Numerous open-label clinical trials have been performed with CoQ with varying success.
In instances where the patient has a primary defect in CoQ synthesis, CoQ supplementation can largely alleviate symptoms. However, primary CoQ deficiency resulting from defects in one of the several steps involved in the CoQ biosynthesis is rare (237). Primary CoQ deficiency syndromes are also heterogeneous, presenting with encephalomyopathy, Leigh syndrome, severe multisystem infantile disease, cerebellar ataxia, or pure myopathies (238). Fortunately, as soon as an accurate diagnosis has been made, the patient is very likely to benefit from CoQ supplementation.
CoQ and idebenone, a short-chain quinone that readily crosses the blood-brain barrier, have also been used to treat a wide spectrum of other mitochondrial diseases, but with varying degrees of success (239, 240). High doses have been used to treat Alzheimer disease and Friedreich’s ataxia (238). Friedreich’s ataxia is an autosomal recessive disease that manifests with hereditary ataxia in association with diabetes mellitus and hypertrophic cardiomyopathy. Idebenone has been reported to counteract the cardiomyopathy of Friedreich’s ataxia patients (241, 242), and high doses of this quinone have also been reported to afford some neurological protection (243). However, Friedreich’s ataxia patients need to be treated as early as possible to slow down the disease process before definitive mitochondrial damage occurs.
Succinate plus CoQ have been used to treat a CPEO patient with respiratory failure that was predominantly due to a complex I defect. The logic of this therapy was that reducing equivalents could be introduced into the ETC via succinate through complex II, thus bypassing the complex I block. The CoQ would then enhance the transfer of the electrons from complex II to complex III (244).
A similar bypass strategy has been reported for the treatment of a complex III defect in a patient harboring a cytochrome b nonsense mutation. This patient was treated with ascorbate (vitamin C) plus menadione; the logic was that the menadione would transfer reducing equivalents directly to complex IV (245, 246). This treatment significantly improved the patient’s muscle strength and mitochondrial energy production, as assessed by 31P magnetic resonance spectroscopy (247).
Other cofactor and vitamin supplements
Various other combinations of vitamins and metabolic supplements have also been used to treat mitochondrial disease patients; see Table 2 (240, 248). Thiamine has been used with success in children with PDH deficiency due to alterations within the thiamine binding site of the X-linked PDH-E1α subunit gene (249). Also, an adult patient with peripheral neuropathy and optic neuropathy associated with a PDH deficiency showed improvement in neurological symptoms and visual acuity with 1 g day−1 of thiamine (250). A thiamine deficiency can cause severe neurological degenerative disease and thus is a useful model of neurodegenerative disease (251).
Primary carnitine deficiency due to defects in the carnitine transporter, OCTN2, can present with hypoketotic hypoglycemia and hepatic encephalopathy or with skeletal and cardiac myopathy. This disease is effectively treated with free carnitine (252). Carnitine levels are also frequently decreased in patients with other mitochondrial disorders, due to the excretion of unoxidized acylcarnitines. These patients may also benefit from oral L-carnitine supplementation at 3 g day−1 of free carnitine (253).
Uridine
OXPHOS is necessary for de novo pyrimidine biosynthesis because dihydroorotate CoQ oxidoreductase (dihydroorate dehydrogenase) uses the ETC as an electron sink via CoQ. Therefore, patients with mitochondrial defects can become pyrimidine deficient and may benefit from exogenous uridine (254). Evidence that this approach may be clinically beneficial comes from rodents poisoned with MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine) that were partially protected by treatment with triacetyluridine, the bioavailable form of uridine. MPTP is a dominanergic neuron-specific complex I inhibitor that kills cells in the basal ganglia, resulting in movement disorders in humans and rodents (255).
Dichloroacetate
Dichloroacetate has been proposed as a treatment for mitochondrial diseases because it inhibits PDK. PDK inactivates PDH through phosphorylation of the E1α subunit. The resulting activation of PDH should drive pyruvate into the mitochondrion, increasing mitochondrial acetyl-CoA and NADH levels and potentially increasing energy production. However, when patients who have severe neonatal lactic acidosis due to PDH deficiency or who harbor the 3243 A>G mutation were treated with dichloroacetate, only limited success in ameliorating clinical symptoms was observed (120, 256, 257). Moreover, a high incidence of drug-induced peripheral neuropathy was reported in a group of 3243A>G patients treated with dichloroacetate (120).
L-arginine
L-arginine has been proposed as a treatment for MELAS 3243A>G syndrome patients during the acute stroke phase. The logic of this approach is that the stroke-like episodes of MELAS syndrome are the result of mitochondrial production of O2·−, which destroys NO in vascular endothelial and smooth muscle cells. Because NO is a vasodilator and the vasoconstrictor is a peptide, loss of NO results in chronic vasoconstriction and transient ischemia. As the substrate of NOS, L-arginine should stimulate NOS to generate more NO and alleviate the vasoconstriction. L-arginine is administrated either orally or by intravenous injections between crises at 0.5–0.3 g kg−1 day−1. Although preliminary reports indicate suppression of the stroke-like episodes with L-arginine therapy (258), these claims need to be validated by a standardized clinical trial.
Reactive oxygen species scavengers and mitochondrial antioxidants
Mitochondrial oxidative damage could be ameliorated by natural (lipoic acid, vitamins C and E, CoQ, etc.) or experimental synthetic (MnTDEIP, EUK134, etc.) antioxidants. These compounds either provide an electron sink (vitamins C and E and CoQ) or catalytically degrade the ROS (MnTDEIP, EUK134, etc.).
It is generally believed that CoQ has significant antioxidant properties (239). Although vitamins C and E should have a similar capacity, they have given mixed results in both individual patients and broad-based epidemiological studies (240). These discrepancies may relate to the action of the various compounds on the different cellular compartments (Figure 2) (136). To enhance efficacy, these compounds have been covalently linked to the lipophilic triphenylphosphonium cation. The positive charge of this cation is electrostatically attached to the negatively charged mitochondrial matrix, directing the antioxidant to this compartment. This approach has proven promising in model-system studies (259, 260).
Lipoic acid, a coenzyme of the PDH complex, is also an antioxidant. A lipoic acid dose of 600 mg day−1 has been shown to decrease several markers of oxidative stress in plasma, including low-density lipoprotein oxidation and urinary isoprostanes in healthy subjects (261).
Metabolic antioxidants have the advantage that they are relatively nontoxic, perhaps because the cell brings them back into redox homeostasis with the rest of the cellular compartments, thus minimizing perturbation of existing cellular signaling networks (Figure 2). However, they have the disadvantage of being only transiently effective at compensating for the severe mitochondrial ROS production that results from mitochondrial OXPHOS defects.
The efficacy of antioxidant therapy can be greatly enhanced by introducing catalytic antioxidants into the mitochondrial matrix. This has been demonstrated by creating a transgenic mouse in which the normally peroxisomal catalase was redirected into the mitochondrial matrix by removal of the C-terminal peroxisomal targeting sequence and addition of an N-terminal mitochondrial targeting sequence. The resulting transgenic mice have an increased life span and reduced mtDNA somatic mutation rate, and they are also more resistant to oxidative challenge and to cardiomyopathy (262).
This benefit has been observed in mouse model systems of mitochondrial disease. These mice were treated with catalytic chemical antioxidants, which remove ROS without themselves being destroyed. One class of these compounds is MnSOD and catalase mimetics. Intraperitoneal injection of MnTBAP [Mn(III)tetrakis (4-benzoic acid) porphyrin chloride] into mice deficient in the MnSOD rescued the mice from their lethal neonatal cardiomyopathy. Also, the MnTBAP partially rescued the mouse tissues from the superoxide destruction of the Fe-S center–containing enzymes, including mitochondrial aconitase and complexes I, II, and III. However, MnTBAP does not readily cross the blood-brain barrier, and the MnTBAP-treated animals developed movement disorders in association with a spongiform encephalopathy (263). Treatment of the MnSOD-deficient animals with the salen Mn complexes EUK-8, -134, and -189, which do cross the blood-brain barrier, protected these animals from both the cardiomyopathy and the spongiform encephalomyopathy (264).
Although treatment of cells and animals with mitochondrially targeted antioxidants may appear to be a promising approach for treatment of a wide variety of clinical problems associated with excessive mitochondrial ROS production, the complete elimination of mitochondrial ROS production could disrupt critical cellular redox signal transduction systems and thus be deleterious (Figure 2). Because mitochondrial ROS is an important mitogen, highly effective catalytic antioxidants may inhibit embryonic development and thus act as a teratogen or inhibit adult stem cell replication, leading to long-term toxicity. Hence, more information about the role of mitochondrial ROS and redox in the cellular signal transduction pathways will be required before optimal therapeutic doses of antioxidant can be selected.
Modulation of the mitochondrial permeability transition pore
Although the accumulation of mtDNA damage that leads to the erosion of mitochondrial energy production has been proposed to be the pathophysiological basis of mitochondrial disease and aging, it is the loss of cells due to necrosis and apoptosis that leads to organ failure, clinical symptoms, and ultimately death. This cell death is precipitated by the activation of the mtPTP and initiation of apoptosis (1). Therefore, blocking of the activation of the mtPTP should prolong the life of the patient’s cells, slow the rate of onset, and diminish the severity of the symptoms (265).
It has long been known that activation of the mtPTP can be inhibited by cyclosporin A. However, cyclosporin A is also an immunosuppressant, so it is not a feasible drug for long-term treatment of mitochondrial disease patients. Analogs of cyclosporin A, now in development, inhibit the mtPTP but lack its immunosuppressive activity. These analogs show great promise for treating mitochondrial patients (265). A particularly exciting demonstration of the potential power of these mtPTP inhibitors is in the treatment of Bethlem myopathy and Ullrich congenital muscular dystrophy. Mutations in the collagen VI gene result in a progressive myopathy. Surprisingly, the clinical features of these diseases can be markedly reduced simply by treatment with mtPTP inhibitors (266, 267), which not only demonstrates the value of modulating the mtPTP in therapeutics but also indicates that mitochondrial dysfunction and the activation of the mtPTP are relevant to a much wider range of clinical problems than simple mitochondrial disorders.
Cyclosporin A exposure and Bcl-2 overexpression have also been shown to stabilize the mtPTP in cybrid cell lines carrying LHON mtDNA mutations that cause complex I defects (268). Nortriptyline, a tricyclic antidepressant, was found to be protective in a mouse model of amyotrophic lateral sclerosis. It was shown to inhibit the activation of the mtPTP and thus to delay the onset of symptoms and increase the life span of the mice (269). Therefore, modulation of the mtPTP may have broad implications for treating mitochondrial disease.
Modulating Ca2+ toxicity
Increases in cytosolic Ca2+ are buffered by the mitochondrial uptake of the Ca2+ at the expense of the mitochondrial inner-membrane ΔP. Mitochondrial Ca2+, in turn, is a major regulator of TCA enzyme activity and ATP production (270). Mitochondrial defects can reduce the capacity of the mitochondria to take up Ca2+, and the reduced mitochondrial ATP can diminish the activity of ion transporters, including Ca2+ pumps. Decreased ΔP and increased cytosolic Ca2+ could then increase the probability of mtPTP activation and apoptosis. Therefore, perturbation of the mitochondrial regulation of Ca2+ could also contribute to the pathophysiology of mitochondrial disease.
Transmitochondrial cybrid cell lines harboring the pathogenic mtDNA MERRF tRNAlys 8344 A>G mutation, but not the mtDNA NARP 8993A>G mutation, show aberrations in cellular Ca2+ signaling (271). Hence, loss of Ca2+ homeostasis should be addressed in certain mitochondrial disorders.
Regulation of mitochondrial biogenesis
Caloric restriction has been shown to increase mitochondrial biogenesis and longevity in a variety of animal models; it acts in part through the sirtuins, such as Sirt1 (272, 273). Therefore, mitochondrial energy output may be enhanced in patients through activation of Sirt1 by use of resveratrol or its analogs (SRT1720, SRT2183, etc.). The success of this approach has already been reported for several age-related diseases, including type 2 diabetes (274).
Resveratrol, a Sirt1 activator extracted from grape skins, induces mitochondrial biogenesis and aerobic capacity in mice through deacetylation and activation of PGC-1α. These processes protect the mice against diet-induced obesity and insulin resistance (273). Moreover, upregulation of PGC-1 coactivators stimulates mitochondrial OXPHOS in cells harboring mtDNA mutations and thus may provide therapeutic benefits for mtDNA disorders (275).
Overexpression of PGC-1α has also been found to delay the onset of symptoms and increase the life expectancy in a mouse model of muscle COX deficiency, generated by the muscle-specific inactivation of COX10. Bezafibrate, a known activator of the PPAR/PGC-1α pathway, had a similar effect (276). Bezafibrate has also been shown to increase several mitochondrial functions, including enzyme activities and protein levels in mitochondrial defective cell lines (277, 278). Therefore, modulation of the PPAR/PGC-1α pathway and administration of bezafibrate represent promising new therapeutic strategies for the treatment of patients with mitochondrial disorders.
Modulation of mitochondrial turnover
Manipulation of mitochondrial autophagy (mitophagy) may also be an important therapeutic approach. Mitophagy removes damaged or mutated mitochondria (279). It has been hypothesized that the accumulation of somatic mtDNA mutations observed during the aging process may be due to a decline of the mitophagy process. However, excessive mitophagy could deplete mitochondrial levels and thus be deleterious. Approaches that may permit beneficial modulation of autophagy include treatment with PI3K antagonists and agonists (279) or treatment with Sirt1 agonists (138).
Exercise therapy and diet
Mitochondrial diseases are generally associated with exercise intolerance, and exercise training is beneficial to the treatment of mitochondrial diseases. Muscle deconditioning in response to inactivity and unloading is common in patients with mitochondrial disease. Promotion of aerobic training with moderate exercise can attenuate or avoid this deconditioning and thus be beneficial to patients (280).
Exercise-based treatment strategies can be subdivided into two categories: endurance exercise and resistance exercise. Endurance training is known to induce OXPHOS and antioxidant capacity and is expected to modulate exercise intolerance in patients. The effects of aerobic training in a group of 20 patients carrying well-characterized mtDNA molecular defects were investigated over a 12-week period, and the approach had beneficial effects (281). In another study (280) of eight patients with single large-scale mtDNA deletions, the subjects showed significant improvement after 14 weeks of aerobic training. Oxygen utilization and skeletal muscle oxygen extraction, peak capacity to do work, and submaximal exercise tolerance were all increased. However, detraining resulted in a loss of these physiological benefits, whereas another round of 14 weeks of training for a subset of patients maintained those individuals’ beneficial adaptations (280). Both studies showed that aerobic training is well tolerated and safe for patients, but the investigators observed no change in mtDNA copy number.
Resistance exercise activates the proliferation of satellite cells in skeletal muscle as a consequence of muscle fiber injury. The level of mutant mtDNA in satellite cells is generally lower than in mature muscle fibers. Hence, fusion of satellite cells shifts the heteroplasmic level toward the wild-type mtDNAs, leading to improvement in patient OXPHOS capacity.
In a case study of a patient harboring a tRNA mutation, significant changes in mutant load and muscle morphology were induced by either eccentric or concentric resistance exercise training (282). In another recent study, a group of eight patients carrying large-scale mtDNA deletions underwent resistance exercise testing during 12 weeks; the patients showed significant therapeutic benefits and a reduced proportion of mutant mtDNAs (283).
As an alternative, although riskier, approach to exercise, skeletal muscle may be partially damaged by bupivacaine injection. This damage could induce satellite cells with reduced mtDNA mutant loads to fuse with the muscle fibers and reduce heteroplasmy (284).
Ketogenic diet
Although each of these approaches has been reported to be of some benefit for mitochondrial disease patients, several of the affected pathways appear to be activated by the ketogenic diet. Therefore, in acquiring an understanding of the mechanisms of action of the ketogenic diet, we could obtain insights into how to design a more systematic approach to treating bioenergetic disease (Figure 3).
Since the 1920s, fasting and the ketogenic diet have been known to be one of the most effective approaches for treatment of intractable seizures in children (285). Ketone bodies (beta-hydroxybutyrate and acetoacetate) are generated by the liver during fasting by beta oxidation of fatty acids. Generally, the brain uses glucose as its main energy source, with a ratio of glucose to ketones of approximately 97:3. However, during short-term fasting this ratio can shift to ~70:30, and during starvation it can shift to ~30:70 (286). Caloric restriction has also been shown to prolong life span in several organisms in association with reduced oxidative stress and increased antioxidant enzymes (272). This is the result of shifting metabolism from a more glucose-based glycolytic metabolism to a more fatty acid oxidation–based mitochondrial metabolism (1, 11).
In addition to preventing seizures in children, ketone treatment in adults has been found to (a) prevent symptoms of experimental hypoglycemia, including anxiety, fainting, hunger, irritability, and tremor, and (b) inhibit astrocytoma tumor growth and vascularization (286). Beta-hydroxybutyrate also protects cultured mesencephalic neurons from MPP+ (1-methyl-4-phenylpyridinium) toxicity and hippocampal neurons from amyloid beta 42 toxicity (287).
Generally, the ketogenic diet is composed of > 90% fat by weight; is low in carbohydrates but adequate in protein, vitamins, and minerals; and is typically calorie restricted by 10%–25% (288). The brains of rats that were fasting or on a ketogenic diet were found to have a seven- to eightfold increase in the uptake of ketones (286). A high-fat diet enhanced with vitamins and CoQ is well tolerated. It has been reported to both provide short-term benefits (289) and improve the outcomes of mitochondrial disorders and intractable epilepsy patients (290).
Although the efficacy of the ketogenic diet is well established for treating childhood seizures that are not responsive to antiepileptic drugs, the mechanism by which ketones ameliorate seizure activity is still actively debated. Current observations may be summarized and understood by a review of the metabolic pathways surrounding beta-hydroxybutyrate and acetoacetate metabolism that are centered largely within the mitochondrion (Figure 3). Beta-hydroxybutyrate and acetoacetate are taken up from the blood and are interconverted through beta-hydroxybutyrate dehydrogenase. Within the mitochondrion, beta-hydroxybutyrate and acetoacetate are converted to acetyl-CoA, which condenses with oxaloacetate to generate citrate; the excess of acetyl-CoA depletes oxaloacetate. The citrate is then converted to alpha-ketoglutarate by the TCA cycle, and alpha-ketoglutarate is converted to glutamate by transfer of an amino group from aspartate, which regenerates oxaloacetate. Because oxaloacetate is depleted, glutamate synthesis is favored. The brain glutamate can then be decarboxylated to generate gamma-aminobutyric acid (GABA), an inhibitory neurotransmitter that can suppress neuronal hyperactivity generated by excitatory neurons. The relevance of this mechanism for ketonic diet action has been demonstrated by extensive metabolic studies (291–298) and by the action of the antiepileptic drug vigabatrin, which inhibits the GABA-degrading enzyme GABA transaminase (294).
Increased production of GABA depletes the excitatory neurotransmitter, glutamate, whose release is reduced in cultured glutamatergic neurons exposed to beta-hydroxybutyrate relative to glucose (299). Moreover, in a pentylenetetrazole-induced rat seizure system, a 24-h low-glucose and low-calorie exposure was sufficient to reduce seizure activity. This reduction was associated with increased brain alanine generation and its export across the blood-brain barrier. Because alanine is generated from glucose-derived pyruvate by the transamination from glutamate, the export of brain alanine converts the excitatory neurotransmitter glutamate to alpha-ketoglutarate, thereby reducing neuronal excitation (Figure 3) (300).
In addition to enhancing GABA production, the increased acetyl-CoA generated from beta-hydroxybutyrate and acetoacetate also drives the TCA cycle (Figure 3). This increases mitochondrial NADH plus H+ and FADH2 production, fueling the ETC, increasing ΔP, and stimulating mitochondrial ATP synthesis by the H+-translocating ATP synthase. The increased ATP is then exported to the cytosol to drive the neuronal Na+ and Ca+ pumps, thus raising the neuronal plasma membrane resting potential and inhibiting the synchronous neuronal discharge characteristic of epilepsy (285). Because beta-hydroxybutyrate is more energy rich than pyruvate (the former has a 31% high caloric density per C2 unit), it produces more ATP (285). Moreover, in the heart, ketone metabolism increases the acetyl-CoA content 9- to 18-fold and citrate levels two- to fivefold; decreases succinate, oxaloacetate, and aspartate two- to threefold; increases the reduction status of NAD+ together with the oxidation status of CoQ; and increases heart hydraulic efficiency by 25% relative to pyruvate (285, 301). The ketogenic diet also increases hippocampal mitochondrial density and ATP levels in a Aldh5a1−/− mouse seizure model, in association with reduced seizures (302). In the hippocampus, the ketogenic diet also results in the coordinated induction of energy metabolism gene transcripts, inducing 33 of the 34 energy metabolism genes (including 21 genes involved in OXPHOS) and 39 of the 42 genes encoding mitochondrial proteins. A ketogenic diet also results in a 46% increase in the mitochondrial density in the hippocampus of animals and increases the phosphocreatine-to-creatine ratio (288).
Ketones have also been reported to reduce cellular and mitochondrial oxidative stress, to increase antioxidant scavenging molecules, and to stabilize the mtPTP. Studies on cultured rat neonatal cortical neurons revealed that beta-hydroxybutyrate and acetoacetate prevented neuronal cell death induced by glutamate. Ketones also significantly reduced mitochondrial production of ROS and associated cytotoxic effects in association with increased ETC oxidation of NADH (303). Rats maintained on a ketogenic diet were found to have a twofold increase in hippocampal mitochondrial GSH levels, which increased the GSH/GSSG redox potential values from −230 mV for control animals to −247 mV for ketogenic diet–fed animals. This finding correlated with a 1.3-fold increase in glutamate cysteine ligase, a 1.9-fold increase in its catalytic subunit, and a 1.6-fold increase in its modulatory subunit. Similarly, the mitochondria-specific coenzyme A/coenzyme A glutathione disulfide redox ratio was significantly increased in the hippocampus. Mitochondria purified from the hippocampus of ketogenic diet–fed rats also showed a reduction in H2O2 production and were resistant to the H2O2-induced mtDNA damage (304).
Rat neocortical brain slices and neurons exposed to H2O2 showed protection against electrophysiological changes, loss of cell viability, and quenching of DCF (2′,7′-dichlorofluorescin) fluorescence when incubated with beta-hydroxybutyrate and acetoacetate, which were also protective against diamide thiol oxidation–induced electrophysiological changes and neuronal cell death. Ketones also directly increased the Ca2+ threshold of the mtPTP. The protective effects of ketones on electrophysiological effects were mimicked by the mtPTP inhibitors, cyclosporin A acting on cyclophilin D, and bongkrekic acid acting on ANT (305). Therefore, ketone bodies reduce mitochondrial ROS production, increase antioxidant defenses, and stabilize the mtPTP.
The reduction in mitochondrial ROS production and the stabilization of the mtPTP could also arise from increased mitochondrial OXPHOS. The resulting increased ΔP would also stabilize the mtPTP. The increased GSH/GSSG ratio could be the result of increased mitochondrial NADPH levels generated by the mitochondrial NNT, which in turn is driven by the increased mitochondrial ΔP. Increased mitochondrial NADPH levels would increase the mitochondrial GSH/GSSG ratio. Similarly, the antioxidant defenses in the cytosol could be enhanced by the export of the excess mitochondrial citrate into the cytosol, where it can be cleaved by ATP-citrate lyase into oxaloacetate and acetyl-CoA. The oxaloacetate could then be reduced by malate dehydrogenase, and the malate could be oxidized to pyruvate by malic enzyme, thereby reducing NADP+ to NADPH. The increased cytosolic NADPH could then reduce the cytosolicGSSG to GSH (Figure 3).
The induction of mitochondrial bioenergetic genes could be explained by the increased generation of cytosolic acetyl-CoA produced from the mitochondrial citrate cleaved by ATP-citrate lyase. Elevated acetyl-CoA could then drive nuclear histone acetyltransferases to acetylate histones. This process would open the chromatin and increase the expression of the nDNA-encoded mitochondrial genes (Figure 1) (121). This speculation is supported by the observation that the antiepileptic drug valproic acid is a histone deacetylase inhibitor. Valproate treatment would increase histone acetylation and perhaps increase mitochondrial gene transcription (Figure 3).
Furthermore, during the glycolytic conversion of glucose to pyruvate, cytosolic NAD+ is reduced to NADH. The high-cytosolic acetyl-CoA generated from the pyruvate via mitochondrial citrate would drive not only histone acetylation but also FOXO and PGC-1α acetylation, inactivating these proteins. Because the Sirt1 deacetylase cannot use NADH as a substrate, the acetylated FOXOs and PGC-1α would remain inactive, thus downregulating mitochondrial biogenesis and OXPHOS and shifting metabolism toward glycolysis. By contrast, oxidation of fatty acids or ketones occurs entirely within the mitochondrion. The mitochondrial acetyl-CoA would still be exported to the cytosol via citrate to acetylate histones and stimulate transcription. However, only the mitochondrial NAD+ would be actively reduced to NADH, leaving the cytosolic NAD+ more oxidized. The high-cytosolic NAD+ levels would then stimulate the Sirt1 deacetylation of the FOXOs and PGC-1α, thus causing the induction of mitochondrial biogenesis and shifting metabolism toward OXPHOS and away from glycolysis (Figure 3). These speculations suggest that the ketogenic diet may act at multiple levels: It may decrease excitatory neuronal activity, increase the expression of bioenergetic genes, increase mitochondrial biogenesis and oxidative energy production, and increase mitochondrial NADPH production, thus decreasing mitochondrial oxidative stress.
CONCLUSION
Although it is becoming increasingly clear that mitochondrial dysfunction is a common factor in a wide range of complex diseases, effective therapeutic interventions are still not readily available. New approaches for genetic therapies for nDNA-encoded mitochondrial diseases as well as for mtDNA diseases are beginning to offer alternatives for individuals suffering from these devastating disorders. However, these approaches are not as likely to relieve the devastating symptoms suffered by individuals with bioenergetic diseases. Current metabolic interventions have been consistently disappointing, but new insights into the integrated energetic network of the cell are providing fresh insight into the mode of action of the older therapeutic approaches, such as fasting and the ketogenic diet, and suggesting more integrated approaches for restoring cellular energetics to homeostasis. Therefore, there is hope that more sophisticated and potentially more efficacious therapeutic approaches will evolve from our current knowledge.
SUMMARY POINTS.
Mitochondrial disorders are a heterogeneous group of multisystem diseases. They can result from mutations in hundreds of genes distributed across all of the chromosomes as well as the mtDNA.
mtDNA genetics follows the specific rules of maternal inheritance, heteroplasmy, mitotic and meiotic segregation, and threshold effect.
Mitochondrial dysfunction is a common factor in a wide spectrum of metabolic and generative diseases, cancers, and aging.
There are two main approaches to mitochondrial disease therapy: genetic and metabolic pharmacological.
Progress is being made in developing genetic therapies for mitochondrial disease, although efforts to develop effective metabolic and pharmacological therapies have been surprisingly disappointing. A primary reason for difficulties in developing metabolic and pharmacological therapies is our lack of understanding of the complex bioenergetic and redox networks of the mitochondrion and the cell.
Consideration must be given to how the genetic defect and the therapeutic approach perturb the cellular bioenergetic and redox network and how these networks respond to the perturbation. Hence, extensive research on cellular and mitochondrial bioenergetics, redox systems, and mitochondrial-nuclear interactions should be performed.
As metabolic and pharmacological therapies are developed for mitochondrial dysfunction, randomized clinical trials of homogeneous patient cohorts must be conducted to ensure that the treatments are not harmful and to affirm their therapeutic benefit.
The development of effective metabolic and pharmacological treatments for defined mitochondrial diseases holds promise for future therapies that could ameliorate the symptoms of a broad range of common metabolic and degenerative disorders, cancers, and even aging.
ACKNOWLEDGMENTS
This research was supported in part by National Institutes of Health grants R01 NS21328, AG24373, DK73691, AG16573, and NS41850; a Doris Duke Translational Research Grant; CIRM Comprehensive Grant RC1-00353-1; an Autism Speaks High-Impact Grant awarded to D.C.W.; and a CIRM Predoctoral Fellowship awarded to W.F.
Glossary
- Mitochondrial DNA (mtDNA)
maternally inherited circular DNA located in the mitochondrial matrix; contains 13 critical oxidative phosphorylation subunits, 2 ribosomal RNA genes, and 22 transfer RNA genes
- Mitochondrion
cytosolic organelle of endosymbiotic origin that produces energy by oxidizing substrates with oxygen by oxidative phosphorylation
- nDNA
nuclear DNA
- Reactive oxygen species (ROS)
oxidative agents, generated as by-products of oxidative phosphorylation, that are central to the signal transduction systems of the cell but that, when produced in excess, can damage cellular mitochondrial, cytosolic, and nuclear components (DNA, proteins, and lipids)
- Apoptosis
programmed cell death resulting in the activation of caspase enzymes, leading to cellular degradation. The intrinsic apoptosis pathway can be initiated via the mitochondrial permeability transition pore
- mtPTP
mitochondrial permeability transition pore
- Oxidative phosphorylation (OXPHOS)
process by which the mitochondrion oxidizes reduced substrates with oxygen to generate energy in a variety of forms, including ATP, heat, membrane potentials, and redox reactions
- NADH
reduced nicotinamide adenine nucleotide
- CoQ
coenzyme Q10
- Heteroplasmy
situation in which two different mtDNAs, mutant and normal, coexist in a cell
- Homoplasmy
situation in which the mtDNAs in a cell are either 100% normal or 100% mutant
- Threshold expression
percentage of mutant mtDNAs and/or severity of a biochemical defect that is sufficient to cause a specific clinical phenotype
- Transmitochondrial cybrid cells
cells formed by the transfer of the mitochondria and mtDNAs from a donor cell to a recipient cell of a different nDNA origin
- MnSOD
manganese superoxide dismutase (Sod2)
- PGC-1
peroxisome proliferator–activated receptor gamma coactivator 1
- Cu/ZnSOD
copper/zinc superoxide dismutase (Sod1 or Sod3)
- NOS
nitric oxide synthase
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallace DC. Mitochondria as chi. Genetics. 2008;179:727–735. doi: 10.1534/genetics.104.91769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallace DC, Lott MT, Procaccio V. Mitochondrial genes in degenerative diseases, cancer and aging. In: Rimoin DL, Connor JM, Pyeritz RE, Korf BR, editors. Emery and Rimoin’s Principles and Practice of Medical Genetics. 5th ed. Vol. 1. Philadelphia: Churchill Livingstone Elsevier; 2007. pp. 194–298. [Google Scholar]
- 4.Holt IJ, Harding AE, Morgan-Hughes JA. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature. 1988;331:717–719. doi: 10.1038/331717a0. [DOI] [PubMed] [Google Scholar]
- 5.Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, et al. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science. 1988;242:1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- 6.Wallace DC, Zheng X, Lott MT, Shoffner JM, Hodge JA, et al. Familial mitochondrial encephalomyopathy (MERRF): genetic, pathophysiological, and biochemical characterization of a mitochondrial DNA disease. Cell. 1988;55:601–610. doi: 10.1016/0092-8674(88)90218-8. [DOI] [PubMed] [Google Scholar]
- 7.Shoffner JM, Lott MT, Lezza AM, Seibel P, Ballinger SW, Wallace DC. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNALys mutation. Cell. 1990;61:931–937. doi: 10.1016/0092-8674(90)90059-n. [DOI] [PubMed] [Google Scholar]
- 8.Schaefer AM, Taylor RW, Turnbull DM, Chinnery PF. The epidemiology of mitochondrial disorders—past, present and future. Biochim. Biophys. Acta. 2004;1659:115–120. doi: 10.1016/j.bbabio.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 9.Schaefer AM, McFarland R, Blakely EL, He L, Whittaker RG, et al. Prevalence of mitochondrial DNA disease in adults. Ann. Neurol. 2007;63:35–39. doi: 10.1002/ana.21217. [DOI] [PubMed] [Google Scholar]
- 10.Elliott HR, Samuels DC, Eden JA, Relton CL, Chinnery PF. Pathogenic mitochondrial DNA mutations are common in the general population. Am. J. Hum. Genet. 2008;83:254–260. doi: 10.1016/j.ajhg.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wallace DC. Why do we have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu. Rev. Biochem. 2007;76:781–821. doi: 10.1146/annurev.biochem.76.081205.150955. [DOI] [PubMed] [Google Scholar]
- 12.Wallace DC, Ruiz-Pesini E, Mishmar D. mtDNA variation, climatic adaptation, degenerative diseases, and longevity. Cold Spring Harb. Symp. Quant. Biol. 2003;68:479–486. doi: 10.1101/sqb.2003.68.471. [DOI] [PubMed] [Google Scholar]
- 13.Mishmar D, Ruiz-Pesini EE, Golik P, Macaulay V, Clark AG, et al. Natural selection shaped regional mtDNA variation in humans. Proc. Natl. Acad. Sci. USA. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruiz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace DC. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science. 2004;303:223–226. doi: 10.1126/science.1088434. [DOI] [PubMed] [Google Scholar]
- 15.Ruiz-Pesini E, Wallace DC. Evidence for adaptive selection acting on the tRNA and rRNA genes of the human mitochondrial DNA. Hum. Mutat. 2006;27:1072–1081. doi: 10.1002/humu.20378. [DOI] [PubMed] [Google Scholar]
- 16.Wallace DC. Mitochondrial genetics: a paradigm for aging and degenerative diseases? Science. 1992;256:628–632. doi: 10.1126/science.1533953. [DOI] [PubMed] [Google Scholar]
- 17.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 18.Montoya J, Ojala D, Attardi G. Distinctive features of the 5′-terminal sequences of the human mitochondrial mRNAs. Nature. 1981;290:465–470. doi: 10.1038/290465a0. [DOI] [PubMed] [Google Scholar]
- 19.Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 20.Wallace DC, Lott MT, Brown MD, Kerstann K. Mitochondria and neuro-ophthalmological diseases. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic and Molecular Basis of Inherited Disease. Vol. 2. New York: McGraw-Hill; 2001. pp. 2425–2512. [Google Scholar]
- 21.Nachman MW, Brown WM, Stoneking M, Aquadro CF. Nonneutral mitochondrial DNA variation in humans and chimpanzees. Genetics. 1996;142:953–963. doi: 10.1093/genetics/142.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schriner SE, Ogburn CE, Smith AC, Newcomb TG, Ladiges WC, et al. Levels of DNA damage are unaltered in mice overexpressing human catalase in nuclei. Free Radic. Biol. Med. 2000;29:664–673. doi: 10.1016/s0891-5849(00)00352-x. [DOI] [PubMed] [Google Scholar]
- 23.Wallace DC. The mitochondrial genome in human adaptive radiation and disease: on the road to therapeutics and performance enhancement. Gene. 2005;354:169–180. doi: 10.1016/j.gene.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Giles RE, Blanc H, Cann HM, Wallace DC. Maternal inheritance of human mitochondrial DNA. Proc. Natl. Acad. Sci. USA. 1980;77:6715–6719. doi: 10.1073/pnas.77.11.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz M, Vissing J. Paternal inheritance of mitochondrial DNA. N. Engl. J. Med. 2002;347:576–580. doi: 10.1056/NEJMoa020350. [DOI] [PubMed] [Google Scholar]
- 26.Mita S, Schmidt B, Schon EA, DiMauro S, Bonilla E. Detection of ‘deleted’ mitochondrial genomes in cytochrome c oxidase–deficient muscle fibers of a patient with Kearns-Sayre syndrome. Proc. Natl. Acad. Sci. USA. 1989;86:9509–9513. doi: 10.1073/pnas.86.23.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wallace DC, Lott MT. Mitochondrial genes in degenerative diseases, cancer and aging. In: Rimoin DL, Connor JM, Pyeritz RE, Korf BR, editors. Emery and Rimoin’s Principles and Practice of Medical Genetics. 4th ed. Vol. 1. London: Churchill Livingstone Elsevier; 2002. pp. 299–409. [Google Scholar]
- 28.Kapsa R, Thompson GN, Thorburn DR, Dahl HH, Marzuki S, et al. A novel mtDNA deletion in an infant with Pearson syndrome. J. Inherit. Metab. Dis. 1994;17:521–526. doi: 10.1007/BF00711584. [DOI] [PubMed] [Google Scholar]
- 29.Rotig A, Bourgeron T, Chretien D, Rustin P, Munnich A. Spectrum of mitochondrial DNA rearrangements in the Pearson marrow-pancreas syndrome. Hum. Mol. Genet. 1995;4:1327–1330. doi: 10.1093/hmg/4.8.1327. [DOI] [PubMed] [Google Scholar]
- 30.Holt IJ, Harding AE, Petty RK, Morgan-Hughes JA. A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am. J. Hum. Genet. 1990;46:428–433. [PMC free article] [PubMed] [Google Scholar]
- 31.Andreu AL, Bruno C, Shanske S, Shtilbans A, Hirano M, et al. Missense mutation in the mtDNA cytochrome b gene in a patient with myopathy. Neurology. 1998;51:1444–1447. doi: 10.1212/wnl.51.5.1444. [DOI] [PubMed] [Google Scholar]
- 32.Andreu AL, Hanna MG, Reichmann H, Bruno C, Penn AS, et al. Exercise intolerance due to mutations in the cytochrome b gene of mitochondrial DNA. N. Engl. J. Med. 1999;341:1037–1044. doi: 10.1056/NEJM199909303411404. [DOI] [PubMed] [Google Scholar]
- 33.Andreu AL, Bruno C, Dunne TC, Tanji K, Shanske S, et al. A nonsense mutation (G15059A) in the cytochrome b gene in a patient with exercise intolerance and myoglobinuria. Ann. Neurol. 1999;45:127–130. doi: 10.1002/1531-8249(199901)45:1<127::aid-art20>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 34.Goto Y, Nonaka I, Horai S. A mutation in the tRNALeu(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1990;348:651–653. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- 35.Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- 36.Wallace DC. Mitochondria and cancer: Warburg address. Cold Spring Harb. Symp. Quant. Biol. 2005;70:363–374. doi: 10.1101/sqb.2005.70.035. [DOI] [PubMed] [Google Scholar]
- 37.Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc. Natl. Acad. Sci. USA. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, et al. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 39.Wallace DC, Brown MD, Lott MT. Mitochondrial DNA variation in human evolution and disease. Gene. 1999;238:211–230. doi: 10.1016/s0378-1119(99)00295-4. [DOI] [PubMed] [Google Scholar]
- 40.Brown MD, Torroni A, Reckord CL, Wallace DC. Phylogenetic analysis of Leber’s hereditary optic neuropathy mitochondrial DNA’s indicates multiple independent occurrences of the common mutations. Hum. Mutat. 1995;6:311–325. doi: 10.1002/humu.1380060405. [DOI] [PubMed] [Google Scholar]
- 41.Brown MD, Sun F, Wallace DC. Clustering of Caucasian Leber hereditary optic neuropathy patients containing the 11778 or 14484 mutations on an mtDNA lineage. Am. J. Hum. Genet. 1997;60:381–387. [PMC free article] [PubMed] [Google Scholar]
- 42.Brown MD, Starikovskaya E, Derbeneva O, Hosseini S, Allen JC, et al. The role of mtDNA background in disease expression: a new primary LHON mutation associated with Western Eurasian haplogroup J. Hum. Genet. 2002;110:130–138. doi: 10.1007/s00439-001-0660-8. [DOI] [PubMed] [Google Scholar]
- 43.Torroni A, Petrozzi M, D’Urbano L, Sellitto D, Zeviani M, et al. Haplotype and phylogenetic analyses suggest that one European-specific mtDNA background plays a role in the expression of Leber hereditary optic neuropathy by increasing the penetrance of the primary mutations 11778 and 14484. Am. J. Hum. Genet. 1997;60:1107–1121. [PMC free article] [PubMed] [Google Scholar]
- 44.Jones MM, Manwaring N, Wang JJ, Rochtchina E, Mitchell P, Sue CM. Mitochondrial DNA haplogroups and age-related maculopathy. Arch. Ophthalmol. 2007;125:1235–1240. doi: 10.1001/archopht.125.9.1235. [DOI] [PubMed] [Google Scholar]
- 45.Udar N, Atilano SR, Memarzadeh M, Boyer DS, Chwa M, et al. Mitochondrial DNA haplogroups associated with age-related macular degeneration. Investig. Ophthalmol. Vis. Sci. 2009;50:2966–2974. doi: 10.1167/iovs.08-2646. [DOI] [PubMed] [Google Scholar]
- 46.Van Der Walt JM, Nicodemus KK, Martin ER, Scott WK, Nance MA, et al. Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. Am. J. Hum. Genet. 2003;72:804–811. doi: 10.1086/373937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghezzi D, Marelli C, Achilli A, Goldwurm S, Pezzoli G, et al. Mitochondrial DNA haplogroup K is associated with a lower risk of Parkinson’s disease in Italians. Eur. J. Hum. Genet. 2005;13:748–752. doi: 10.1038/sj.ejhg.5201425. [DOI] [PubMed] [Google Scholar]
- 48.Khusnutdinova E, Gilyazova I, Ruiz-Pesini E, Derbeneva O, Khusainova R, et al. A mitochondrial etiology of neurodegenerative diseases: evidence from Parkinson’s disease. Ann. N.Y. Acad. Sci. 2008;1147:1–20. doi: 10.1196/annals.1427.001. [DOI] [PubMed] [Google Scholar]
- 49.Shoffner JM, Brown MD, Torroni A, Lott MT, Cabell MR, et al. Mitochondrial DNA variants observed in Alzheimer disease and Parkinson disease patients. Genomics. 1993;17:171–184. doi: 10.1006/geno.1993.1299. [DOI] [PubMed] [Google Scholar]
- 50.Chagnon P, Gee M, Filion M, Robitaille Y, Belouchi M, Gauvreau D. Phylogenetic analysis of the mitochondrial genome indicates significant differences between patients with Alzheimer disease and controls in a French-Canadian founder population. Am. J. Med. Genet. 1999;85:20–30. doi: 10.1002/(sici)1096-8628(19990702)85:1<20::aid-ajmg6>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 51.Van Der Walt JM, Dementieva YA, Martin ER, Scott WK, Nicodemus KK, et al. Analysis of European mitochondrial haplogroups with Alzheimer disease risk. Neurosci. Lett. 2004;365:28–32. doi: 10.1016/j.neulet.2004.04.051. [DOI] [PubMed] [Google Scholar]
- 52.Carrieri G, Bonafe M, De Luca M, Rose G, Varcasia O, et al. Mitochondrial DNA haplogroups and APOE4 allele are non-independent variables in sporadic Alzheimer’s disease. Hum. Genet. 2001;108:194–198. doi: 10.1007/s004390100463. [DOI] [PubMed] [Google Scholar]
- 53.De Benedictis G, Rose G, Carrieri G, De Luca M, Falcone E, et al. Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. FASEB J. 1999;13:1532–1536. doi: 10.1096/fasebj.13.12.1532. [DOI] [PubMed] [Google Scholar]
- 54.Rose G, Passarino G, Carrieri G, Altomare K, Greco V, et al. Paradoxes in longevity: sequence analysis of mtDNA haplogroup J in centenarians. Eur. J. Hum. Genet. 2001;9:701–707. doi: 10.1038/sj.ejhg.5200703. [DOI] [PubMed] [Google Scholar]
- 55.Niemi AK, Hervonen A, Hurme M, Karhunen PJ, Jylha M, Majamaa K. Mitochondrial DNA polymorphisms associated with longevity in a Finnish population. Hum. Genet. 2003;112:29–33. doi: 10.1007/s00439-002-0843-y. [DOI] [PubMed] [Google Scholar]
- 56.Ivanova R, Lepage V, Charron D, Schachter F. Mitochondrial genotype associated with French Caucasian centenarians. Gerontology. 1998;44:349. doi: 10.1159/000022041. [DOI] [PubMed] [Google Scholar]
- 57.Tanaka M, Gong JS, Zhang J, Yoneda M, Yagi K. Mitochondrial genotype associated with longevity. Lancet. 1998;351:185–186. doi: 10.1016/S0140-6736(05)78211-8. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka M, Gong J, Zhang J, Yamada Y, Borgeld HJ, Yagi K. Mitochondrial genotype associated with longevity and its inhibitory effect on mutagenesis. Mech. Ageing Dev. 2000;116:65–76. doi: 10.1016/s0047-6374(00)00149-4. [DOI] [PubMed] [Google Scholar]
- 59.Crispim D, Canani LH, Gross JL, Tschiedel B, Souto KE, Roisenberg I. The European-specific mitochondrial cluster J/T could confer an increased risk of insulin-resistance and type 2 diabetes: an analysis of the m.4216T > C and m.4917A > G variants. Ann. Hum. Genet. 2006;70:488–495. doi: 10.1111/j.1469-1809.2005.00249.x. [DOI] [PubMed] [Google Scholar]
- 60.Mohlke KL, Jackson AU, Scott LJ, Peck EC, Suh YD, et al. Mitochondrial polymorphisms and susceptibility to type 2 diabetes–related traits in Finns. Hum. Genet. 2005;118:245–254. doi: 10.1007/s00439-005-0046-4. [DOI] [PubMed] [Google Scholar]
- 61.Saxena R, de Bakker PI, Singer K, Mootha V, Burtt N, et al. Comprehensive association testing of common mitochondrial DNA variation in metabolic disease. Am. J. Hum. Genet. 2006;79:54–61. doi: 10.1086/504926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fuku N, Park KS, Yamada Y, Nishigaki Y, Cho YM, et al. Mitochondrial haplogroup N9a confers resistance against type 2 diabetes in Asians. Am. J. Hum. Genet. 2007;80:407–415. doi: 10.1086/512202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nishigaki Y, Yamada Y, Fuku N, Matsuo H, Segawa T, et al. Mitochondrial haplogroup N9b is protective against myocardial infarction in Japanese males. Hum. Genet. 2007;120:827–836. doi: 10.1007/s00439-006-0269-z. [DOI] [PubMed] [Google Scholar]
- 64.Wallace DC, Chuang L-M, Wang PH, Chang Y-C, Derbeneva O, et al. Asian mitochondrial DNA (mtDNA) lineages are associated with altered risk of developing type 2 diabetes and metabolic syndrome. Presented at Annu. Meet. Am. Soc. Hum. Genet., 57th; San Diego. 2007. Abstr. 1549. [Google Scholar]
- 65.Raby BA, Klanderman B, Murphy A, Mazza S, Camargo CA, Jr, et al. A common mitochondrial haplogroup is associated with elevated total serum IgE levels. J. Allergy Clin. Immunol. 2007;120:351–358. doi: 10.1016/j.jaci.2007.05.029. [DOI] [PubMed] [Google Scholar]
- 66.Baudouin SV, Saunders D, Tiangyou W, Elson JL, Poynter J, et al. Mitochondrial DNA and survival after sepsis: a prospective study. Lancet. 2005;366:2118–2121. doi: 10.1016/S0140-6736(05)67890-7. [DOI] [PubMed] [Google Scholar]
- 67.Hendrickson SL, Hutcheson HB, Ruiz-Pesini E, Poole JC, Lautenberger J, et al. Mitochondrial DNA haplogroups influence AIDS progression. AIDS. 2008;22:2429–2439. doi: 10.1097/QAD.0b013e32831940bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hendrickson SL, Kingsley LA, Ruiz-Pesini E, Poole JC, Jacobson LP, et al. Mitochondrial DNA haplogroups influence lipoatrophy after highly active anti-retroviral therapy. J. Acquir. Immune Defic. Syndr. 2009;51:111–116. doi: 10.1097/QAI.0b013e3181a324d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Booker LM, Habermacher GM, Jessie BC, Sun QC, Baumann AK, et al. North American white mitochondrial haplogroups in prostate and renal cancer. J. Urol. 2006;175:468–472. doi: 10.1016/S0022-5347(05)00163-1. [DOI] [PubMed] [Google Scholar]
- 70.Bai RK, Leal SM, Covarrubias D, Liu A, Wong LJ. Mitochondrial genetic background modifies breast cancer risk. Cancer Res. 2007;67:4687–4694. doi: 10.1158/0008-5472.CAN-06-3554. [DOI] [PubMed] [Google Scholar]
- 71.Darvishi K, Sharma S, Bhat AK, Rai E, Bamezai RN. Mitochondrial DNA G10398A polymorphism imparts maternal haplogroup N a risk for breast and esophageal cancer. Cancer Lett. 2007;249:249–255. doi: 10.1016/j.canlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 72.Gottlieb E, Tomlinson IP. Mitochondrial tumour suppressors: a genetic and biochemical update. Nat. Rev. Cancer. 2005;5:857–866. doi: 10.1038/nrc1737. [DOI] [PubMed] [Google Scholar]
- 73.Procaccio V, Wallace DC. Late-onset Leigh syndrome in a patient with mitochondrial complex I NDUFS8 mutations. Neurology. 2004;62:1899–1901. doi: 10.1212/01.wnl.0000125251.56131.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu Z, Yao J, Johns T, Fu F, De Bie I, et al. SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat. Genet. 1998;20:337–343. doi: 10.1038/3804. [DOI] [PubMed] [Google Scholar]
- 75.Spelbrink JN, Li FY, Tiranti V, Nikali K, Yuan QP, et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4–like protein localized in mitochondria. Nat. Genet. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 76.Van Goethem G, Dermaut B, Lofgren A, Martin JJ, Van Broeckhoven C. Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions. Nat. Genet. 2001;28:211–212. doi: 10.1038/90034. [DOI] [PubMed] [Google Scholar]
- 77.Palmieri L, Alberio S, Pisano I, Lodi T, Meznaric-Petrusa M, et al. Complete loss-of-function of the heart/muscle-specific adenine nucleotide translocator is associated with mitochondrial myopathy and cardiomyopathy. Hum. Mol. Genet. 2005;14:3079–3088. doi: 10.1093/hmg/ddi341. [DOI] [PubMed] [Google Scholar]
- 78.Kaukonen J, Juselius JK, Tiranti V, Kyttala A, Zeviani M, et al. Role of adenine nucleotide translocator 1 in mtDNA maintenance. Science. 2000;289:782–785. doi: 10.1126/science.289.5480.782. [DOI] [PubMed] [Google Scholar]
- 79.Mandel H, Szargel R, Labay V, Elpeleg O, Saada A, et al. The deoxyguanosine kinase gene is mutated in individuals with depleted hepatocerebral mitochondrial DNA. Nat. Genet. 2001;29:337–341. doi: 10.1038/ng746. [DOI] [PubMed] [Google Scholar]
- 80.Saada A, Shaag A, Mandel H, Nevo Y, Eriksson S, Elpeleg O. Mutant mitochondrial thymidine kinase in mitochondrial DNA depletion myopathy. Nat. Genet. 2001;29:342–344. doi: 10.1038/ng751. [DOI] [PubMed] [Google Scholar]
- 81.Carrozzo R, Dionisi-Vici C, Steuerwald U, Lucioli S, Deodato F, et al. SUCLA2 mutations are associated with mild methylmalonic aciduria, Leigh-like encephalomyopathy, dystonia and deafness. Brain. 2007;130:862–874. doi: 10.1093/brain/awl389. [DOI] [PubMed] [Google Scholar]
- 82.Nishino I, Spinazzola A, Hirano M. Thymidine phosphorylase gene mutations in MNGIE, a human mitochondrial disorder. Science. 1999;283:689–692. doi: 10.1126/science.283.5402.689. [DOI] [PubMed] [Google Scholar]
- 83.Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, et al. Nuclear gene OPA1, encoding a mitochondrial dynamin–related protein, is mutated in dominant optic atrophy. Nat. Genet. 2000;26:207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- 84.Zuchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat. Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- 85.Graham BH, Waymire KG, Cottrell B, Trounce IA, MacGregor GR, Wallace DC. A mouse model for mitochondrial myopathy and cardiomyopathy resulting from a deficiency in the heart/skeletal muscle isoform of the adenine nucleotide translocator. Nat. Genet. 1997;16:226–234. doi: 10.1038/ng0797-226. [DOI] [PubMed] [Google Scholar]
- 86.Flierl A, Chen Y, Coskun PE, Samulski RJ, Wallace DC. Adeno-associated virus-mediated gene transfer of the heart/muscle adenine nucleotide translocator (ANT) in mouse. Gene Ther. 2005;12:570–578. doi: 10.1038/sj.gt.3302443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ruiz-Pesini E, Lott MT, Procaccio V, Poole J, Brandon MC, et al. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007;35:D823–D828. doi: 10.1093/nar/gkl927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Oca-Cossio J, Kenyon L, Hao H, Moraes CT. Limitations of allotopic expression of mitochondrial genes in mammalian cells. Genetics. 2003;165:707–720. doi: 10.1093/genetics/165.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Manfredi G, Fu J, Ojaimi J, Sadlock JE, Kwong JQ, et al. Rescue of a deficiency in ATP synthesis by transfer of MTATP6, a mitochondrial DNA–encoded gene, to the nucleus. Nat. Genet. 2002;30:394–399. doi: 10.1038/ng851. [DOI] [PubMed] [Google Scholar]
- 90.Guy J, Qi X, Pallotti F, Schon EA, Manfredi G, et al. Rescue of a mitochondrial deficiency causing Leber hereditary optic neuropathy. Ann. Neurol. 2002;52:534–542. doi: 10.1002/ana.10354. [DOI] [PubMed] [Google Scholar]
- 91.Kolesnikova OA, Entelis NS, Jacquin-Becker C, Goltzene F, Chrzanowska-Lightowlers ZM, et al. Nuclear DNA–encoded tRNAs targeted into mitochondria can rescue a mitochondrial DNA mutation associated with the MERRF syndrome in cultured human cells. Hum. Mol. Genet. 2004;13:2519–2534. doi: 10.1093/hmg/ddh267. [DOI] [PubMed] [Google Scholar]
- 92.Mahata B, Mukherjee S, Mishra S, Bandyopadhyay A, Adhya S. Functional delivery of a cytosolic tRNA into mutant mitochondria of human cells. Science. 2006;314:471–474. doi: 10.1126/science.1129754. [DOI] [PubMed] [Google Scholar]
- 93.Seo BB, Nakamaru-Ogiso E, Cruz P, Flotte TR, Yagi T, Matsuno-Yagi A. Functional expression of the single subunit NADH dehydrogenase in mitochondria in vivo: a potential therapy for complex I deficiencies. Hum. Gene Ther. 2004;15:887–895. doi: 10.1089/hum.2004.15.887. [DOI] [PubMed] [Google Scholar]
- 94.Marella M, Seo BB, Nakamaru-Ogiso E, Greenamyre JT, Matsuno-Yagi A, Yagi T. Protection by the NDI1 gene against neurodegeneration in a rotenone rat model of Parkinson’s disease. PLoS ONE. 2008;3:e1433. doi: 10.1371/journal.pone.0001433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Qi X, Lewin AS, Hauswirth WW, Guy J. Optic neuropathy induced by reductions in mitochondrial superoxide dismutase. Investig. Ophthalmol. Vis. Sci. 2003;44:1088–1096. doi: 10.1167/iovs.02-0864. [DOI] [PubMed] [Google Scholar]
- 96.Qi X, Lewin AS, Hauswirth WW, Guy J. Suppression of complex I gene expression induces optic neuropathy. Ann. Neurol. 2003;53:198–205. doi: 10.1002/ana.10426. [DOI] [PubMed] [Google Scholar]
- 97.Qi X, Lewin AS, Sun L, Hauswirth WW, Guy J. SOD2 gene transfer protects against optic neuropathy induced by deficiency of complex I. Ann. Neurol. 2004;56:182–191. doi: 10.1002/ana.20175. [DOI] [PubMed] [Google Scholar]
- 98.Hakkaart GA, Dassa EP, Jacobs HT, Rustin P. Allotopic expression of a mitochondrial alternative oxidase confers cyanide resistance to human cell respiration. EMBO Rep. 2006;7:341–345. doi: 10.1038/sj.embor.7400601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Perales-Clemente E, Bayona-Bafaluy MP, Perez-Martos A, Barrientos A, Fernandez-Silva P, Enriquez JA. Restoration of electron transport without proton pumping in mammalian mitochondria. Proc. Natl. Acad. Sci. USA. 2008;105:18735–18739. doi: 10.1073/pnas.0810518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Santra S, Gilkerson RW, Davidson M, Schon EA. Ketogenic treatment reduces deleted mitochondrial DNAs in cultured human cells. Ann. Neurol. 2004;56:662–669. doi: 10.1002/ana.20240. [DOI] [PubMed] [Google Scholar]
- 101.Manfredi G, Gupta N, Vazquez-Memije ME, Sadlock JE, Spinazzola A, et al. Oligomycin induces a decrease in the cellular content of a pathogenic mutation in the human mitochondrial ATPase 6 gene. J. Biol. Chem. 1999;274:9386–9391. doi: 10.1074/jbc.274.14.9386. [DOI] [PubMed] [Google Scholar]
- 102.Tanaka M, Borgeld HJ, Zhang J, Muramatsu S, Gong JS, et al. Gene therapy for mitochondrial disease by delivering restriction endonuclease SmaI into mitochondria. J. Biomed. Sci. 2002;9:534–541. doi: 10.1159/000064726. [DOI] [PubMed] [Google Scholar]
- 103.Bayona-Bafaluy MP, Blits B, Battersby BJ, Shoubridge EA, Moraes CT. Rapid directional shift of mitochondrial DNA heteroplasmy in animal tissues by a mitochondrially targeted restriction endonuclease. Proc. Natl. Acad. Sci. USA. 2005;102:14392–14397. doi: 10.1073/pnas.0502896102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Taylor RW, Chinnery PF, Turnbull DM, Lightowlers RN. Selective inhibition of mutant human mitochondrial DNA replication in vitro by peptide nucleic acids. Nat. Genet. 1997;15:212–215. doi: 10.1038/ng0297-212. [DOI] [PubMed] [Google Scholar]
- 105.Chinnery PF, Taylor RW, Diekert K, Lill R, Turnbull DM, Lightowlers RN. Peptide nucleic acid delivery to human mitochondria. Gene Ther. 1999;6:1919–1928. doi: 10.1038/sj.gt.3301061. [DOI] [PubMed] [Google Scholar]
- 106.Flierl A, Jackson C, Cottrell B, Murdock D, Seibel P, Wallace DC. Targeted delivery of DNA to the mitochondrial compartment via import sequence–conjugated peptide nucleic acid. Mol. Ther. 2003;7:550–557. doi: 10.1016/s1525-0016(03)00037-6. [DOI] [PubMed] [Google Scholar]
- 107.Hirano M, Marti R, Casali C, Tadesse S, Uldrick T, et al. Allogeneic stem cell transplantation corrects biochemical derangements in MNGIE. Neurology. 2006;67:1458–1460. doi: 10.1212/01.wnl.0000240853.97716.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bonnet D, Rustin P, Rotig A, Le Bidois J, Munnich A, et al. Heart transplantation in children with mitochondrial cardiomyopathy. Heart. 2001;86:570–573. doi: 10.1136/heart.86.5.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dimmock DP, Dunn JK, Feigenbaum A, Rupar A, Horvath R, et al. Abnormal neurological features predict poor survival and should preclude liver transplantation in patients with deoxyguanosine kinase deficiency. Liver Transpl. 2008;14:1480–1485. doi: 10.1002/lt.21556. [DOI] [PubMed] [Google Scholar]
- 110.Chinnery PF, DiMauro S, Shanske S, Schon EA, Zeviani M, et al. Risk of developing a mitochondrial DNA deletion disorder. Lancet. 2004;364:592–596. doi: 10.1016/S0140-6736(04)16851-7. [DOI] [PubMed] [Google Scholar]
- 111.Jacobs LJ, deWert G, Geraedts JP, de Coo IF, Smeets HJ. The transmission of OXPHOS disease and methods to prevent this. Hum. Reprod. Update. 2006;12:119–136. doi: 10.1093/humupd/dmi042. [DOI] [PubMed] [Google Scholar]
- 112.Bouchet C, Steffann J, Corcos J, Monnot S, Paquis V, et al. Prenatal diagnosis of myopathy, encephalopathy, lactic acidosis, and stroke-like syndrome: contribution to understanding mitochondrial DNA segregation during human embryofetal development. J. Med. Genet. 2006;43:788–792. doi: 10.1136/jmg.2005.034140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Steffann J, Gigarel N, Corcos J, Bonniere M, Encha-Razavi F, et al. Stability of the m.8993T→G mtDNA mutation load during human embryofetal development has implications for the feasibility of prenatal diagnosis in NARP syndrome. J. Med. Genet. 2007;44:664–669. doi: 10.1136/jmg.2006.048553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tachibana M, Sparman M, Sritanaudomchai H, Ma H, Clepper L, et al. Mitochondrial gene replacement in primate offspring and embryonic stem cells. Nature. 2009:08368. doi: 10.1038/nature08368. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bredenoord A, Dondorp W, Pennings G, de Die–Smulders C, Smeets B, deWert G. Preimplantation genetic diagnosis for mitochondrial DNA disorders: ethical guidance for clinical practice. Eur. J. Hum. Genet. 2009 doi: 10.1038/ejhg.2009.88. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bredenoord AL, Pennings G, Smeets HJ, de Wert G. Dealing with uncertainties: ethics of prenatal diagnosis and preimplantation genetic diagnosis to prevent mitochondrial disorders. Hum. Reprod. Update. 2008;14:83–94. doi: 10.1093/humupd/dmm037. [DOI] [PubMed] [Google Scholar]
- 117.Wallace DC. Maternal genes: mitochondrial diseases. In: McKusick VA, Roderick TH, Mori J, Paul MW, editors. Medical and Experimental Mammalian Genetics: A Perspective. Vol. 23. New York: Liss; 1987. pp. 137–190. [Google Scholar]
- 118.Trounce I, Wallace DC. Production of transmitochondrial mouse cell lines by cybrid rescue of rhodamine-6G pre-treated L-cells. Somat. Cell Mol. Genet. 1996;22:81–85. doi: 10.1007/BF02374379. [DOI] [PubMed] [Google Scholar]
- 119.Chinnery P, Majamaa K, Turnbull D, Thorburn D. Treatment for mitochondrial disorders. Cochrane Database Syst. Rev. 2006;1 doi: 10.1002/14651858.CD004426.pub2. CD004426. [DOI] [PubMed] [Google Scholar]
- 120.Kaufmann P, Engelstad K, Wei Y, Jhung S, Sano MC, et al. Dichloroacetate causes toxic neuropathy in MELAS: a randomized, controlled clinical trial. Neurology. 2006;66:324–330. doi: 10.1212/01.wnl.0000196641.05913.27. [DOI] [PubMed] [Google Scholar]
- 121.Wallace DC, Fan W. The pathophysiology of mitochondrial diseases as modeled in the mouse. Genes Dev. 2009;23:1714–1736. doi: 10.1101/gad.1784909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Burdon RH. Superoxide and hydrogen peroxide in relation to mammalian cell proliferation. Free Radic. Biol. Med. 1995;18:775–794. doi: 10.1016/0891-5849(94)00198-s. [DOI] [PubMed] [Google Scholar]
- 123.McCord JM. The evolution of free radicals and oxidative stress. Am. J. Med. Genet. 2000;108:652–659. doi: 10.1016/s0002-9343(00)00412-5. [DOI] [PubMed] [Google Scholar]
- 124.Evans AR, Limp-Foster M, Kelley MR. Going APE over ref-1. Mutat. Res. 2000;461:83–108. doi: 10.1016/s0921-8777(00)00046-x. [DOI] [PubMed] [Google Scholar]
- 125.Kelley MR, Parsons SH. Redox regulation of the DNA repair function of the human AP endonuclease Ape1/ref-1. Antioxid. Redox Signal. 2001;3:671–683. doi: 10.1089/15230860152543014. [DOI] [PubMed] [Google Scholar]
- 126.Jones DP. Disruption of mitochondrial redox circuitry in oxidative stress. Chem. Biol. Interact. 2006;163:38–53. doi: 10.1016/j.cbi.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 127.Hansen JM, Go YM, Jones DP. Nuclear and mitochondrial compartmentation of oxidative stress and redox signaling. Annu. Rev. Pharmacol. Toxicol. 2006;46:215–234. doi: 10.1146/annurev.pharmtox.46.120604.141122. [DOI] [PubMed] [Google Scholar]
- 128.Cassarino DS, Fall CP, Swerdlow RH, Smith TS, Halvorsen EM, et al. Elevated reactive oxygen species and antioxidant enzyme activities in animal and cellular models of Parkinson’s disease. Biochim. Biophys. Acta. 1997;1362:77–86. doi: 10.1016/s0925-4439(97)00070-7. [DOI] [PubMed] [Google Scholar]
- 129.Cassarino DS, Swerdlow RH, Parks JK, Parker WD, Jr, Bennett JP., Jr Cyclosporin A increases resting mitochondrial membrane potential in SY5Y cells and reverses the depressed mitochondrial membrane potential of Alzheimer’s disease cybrids. Biochem. Biophys. Res. Commun. 1998;248:168–173. doi: 10.1006/bbrc.1998.8866. [DOI] [PubMed] [Google Scholar]
- 130.Bredt DS, Hwang PM, Glatt CE, Lowenstein C, Reed RR, Snyder SH. Cloned and expressed nitric oxide synthase structurally resembles cytochrome P-450 reductase. Nature. 1991;351:714–718. doi: 10.1038/351714a0. [DOI] [PubMed] [Google Scholar]
- 131.Lowenstein CJ, Glatt CS, Bredt DS, Snyder SH. Cloned and expressed macrophage nitric oxide synthase contrasts with the brain enzyme. Proc. Natl. Acad. Sci. USA. 1992;89:6711–6715. doi: 10.1073/pnas.89.15.6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Marsden PA, Schappert KT, Chen HS, Flowers M, Sundell CL, et al. Molecular cloning and characterization of human endothelial nitric oxide synthase. FEBS Lett. 1992;307:287–293. doi: 10.1016/0014-5793(92)80697-f. [DOI] [PubMed] [Google Scholar]
- 133.Rajfer J, Aronson WJ, Bush PA, Dorey FJ, Ignarro LJ. Nitric oxide as a mediator of relaxation of the corpus cavernosum in response to nonadrenergic, noncholinergic neurotransmission. N. Engl. J. Med. 1992;326:90–94. doi: 10.1056/NEJM199201093260203. [DOI] [PubMed] [Google Scholar]
- 134.Koga Y, Ishibashi M, Ueki I, Yatsuga S, Fukiyama R, et al. Effects of L-arginine on the acute phase of strokes in three patients with MELAS. Neurology. 2002;58:827–828. doi: 10.1212/wnl.58.5.827. [DOI] [PubMed] [Google Scholar]
- 135.Pacher P, Beckman JS, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol. Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lane N. Oxygen: The Molecule That Made the World. Oxford: Oxford Univ. Press; 2002. p. 374. [Google Scholar]
- 137.Lane N. Power, Sex, Suicide: Mitochondria and the Meaning of Life. Oxford: Oxford Univ. Press; 2005. p. 354. [Google Scholar]
- 138.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc. Natl. Acad. Sci. USA. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ahn BH, Kim HS, Song S, Lee IH, Liu J, et al. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc. Natl. Acad. Sci. USA. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jones DP. Radical-free biology of oxidative stress. Am. J. Physiol. Cell Physiol. 2008;295:C849–C868. doi: 10.1152/ajpcell.00283.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kemp M, Go YM, Jones DP. Nonequilibrium thermodynamics of thiol/disulfide redox systems: a perspective on redox systems biology. Free Radic. Biol. Med. 2008;44:921–937. doi: 10.1016/j.freeradbiomed.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jones DP. Redefining oxidative stress. Antioxid. Redox Signal. 2006;8:1865–1879. doi: 10.1089/ars.2006.8.1865. [DOI] [PubMed] [Google Scholar]
- 143.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic. Biol. Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 144.Dalton TP, Dieter MZ, Yang Y, Shertzer HG, Nebert DW. Knockout of the mouse glutamate cysteine ligase catalytic subunit (Gclc) gene: embryonic lethal when homozygous, and proposed model for moderate glutathione deficiency when heterozygous. Biochem. Biophys. Res. Commun. 2000;279:324–329. doi: 10.1006/bbrc.2000.3930. [DOI] [PubMed] [Google Scholar]
- 145.Shi ZZ, Osei-Frimpong J, Kala G, Kala SV, Barrios RJ, et al. Glutathione synthesis is essential for mouse development but not for cell growth in culture. Proc. Natl. Acad. Sci. USA. 2000;97:5101–5106. doi: 10.1073/pnas.97.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chen Y, Yang Y, Miller ML, Shen D, Shertzer HG, et al. Hepatocyte-specific Gclc deletion leads to rapid onset of steatosis with mitochondrial injury and liver failure. Hepatology. 2007;45:1118–1128. doi: 10.1002/hep.21635. [DOI] [PubMed] [Google Scholar]
- 147.Yang Y, Dieter MZ, Chen Y, Shertzer HG, Nebert DW, Dalton TP. Initial characterization of the glutamate-cysteine ligase modifier subunit Gclm−/− knockout mouse. Novel model system for a severely compromised oxidative stress response. J. Biol. Chem. 2002;277:49446–49452. doi: 10.1074/jbc.M209372200. [DOI] [PubMed] [Google Scholar]
- 148.Matsui M, Oshima M, Oshima H, Takaku K, Maruyama T, et al. Early embryonic lethality caused by targeted disruption of the mouse thioredoxin gene. Dev. Biol. 1996;178:179–185. doi: 10.1006/dbio.1996.0208. [DOI] [PubMed] [Google Scholar]
- 149.Nonn L, Williams RR, Erickson RP, Powis G. The absence of mitochondrial thioredoxin 2 causes massive apoptosis, exencephaly, and early embryonic lethality in homozygous mice. Mol. Cell. Biol. 2003;23:916–922. doi: 10.1128/MCB.23.3.916-922.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jakupoglu C, Przemeck GK, Schneider M, Moreno SG, Mayr N, et al. Cytoplasmic thioredoxin reductase is essential for embryogenesis but dispensable for cardiac development. Mol. Cell. Biol. 2005;25:1980–1988. doi: 10.1128/MCB.25.5.1980-1988.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Conrad M, Jakupoglu C, Moreno SG, Lippl S, Banjac A, et al. Essential role for mitochondrial thioredoxin reductase in hematopoiesis, heart development, and heart function. Mol. Cell. Biol. 2004;24:9414–9423. doi: 10.1128/MCB.24.21.9414-9423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Go YM, Jones DP. Redox compartmentalization in eukaryotic cells. Biochim. Biophys. Acta. 2008;1780:1273–1290. doi: 10.1016/j.bbagen.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jones DP, Go YM, Anderson CL, Ziegler TR, Kinkade JM, Jr, Kirlin WG. Cysteine/cystine couple is a newly recognized node in the circuitry for biologic redox signaling and control. FASEB J. 2004;18:1246–1248. doi: 10.1096/fj.03-0971fje. [DOI] [PubMed] [Google Scholar]
- 154.Li L, Shoji W, Takano H, Nishimura N, Aoki Y, et al. Increased susceptibility of MER5 (peroxiredoxin III) knockout mice to LPS-induced oxidative stress. Biochem. Biophys. Res. Commun. 2007;355:715–721. doi: 10.1016/j.bbrc.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 155.He M, Cai J, Go YM, Johnson JM, Martin WD, et al. Identification of thioredoxin-2 as a regulator of the mitochondrial permeability transition. Toxicol. Sci. 2008;105:44–50. doi: 10.1093/toxsci/kfn116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhang H, Go YM, Jones DP. Mitochondrial thioredoxin-2/peroxiredoxin-3 system functions in parallel with mitochondrial GSH system in protection against oxidative stress. Arch. Biochem. Biophys. 2007;465:119–126. doi: 10.1016/j.abb.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 157.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, et al. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. USA. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metabol. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 160.Ho YS, Xiong Y, Ho DS, Gao J, Chua BH, et al. Targeted disruption of the glutaredoxin 1 gene does not sensitize adult mice to tissue injury induced by ischemia/reperfusion and hyperoxia. Free Radic. Biol. Med. 2007;43:1299–1312. doi: 10.1016/j.freeradbiomed.2007.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Neumann CA, Krause DS, Carman CV, Das S, Dubey DP, et al. Essential role for the peroxiredoxin Prdx1 in erythrocyte antioxidant defence and tumour suppression. Nature. 2003;424:561–565. doi: 10.1038/nature01819. [DOI] [PubMed] [Google Scholar]
- 162.Nagy N, Malik G, Fisher AB, Das DK. Targeted disruption of peroxiredoxin 6 gene renders the heart vulnerable to ischemia-reperfusion injury. Am. J. Physiol. Heart Circ. Physiol. 2006;291:H2636–H2640. doi: 10.1152/ajpheart.00399.2006. [DOI] [PubMed] [Google Scholar]
- 163.Wang X, Phelan SA, Forsman-Semb K, Taylor EF, Petros C, et al. Mice with targeted mutation of peroxiredoxin 6 develop normally but are susceptible to oxidative stress. J. Biol. Chem. 2003;278:25179–25190. doi: 10.1074/jbc.M302706200. [DOI] [PubMed] [Google Scholar]
- 164.Lee TH, Kim SU, Yu SL, Kim SH, Park DS, et al. Peroxiredoxin II is essential for sustaining life span of erythrocytes in mice. Blood. 2003;101:5033–5038. doi: 10.1182/blood-2002-08-2548. [DOI] [PubMed] [Google Scholar]
- 165.Saitoh M, Nishitoh H, Fujii M, Takeda K, Tobiume K, et al. Mammalian thioredoxin is a direct inhibitor of apoptosis signal–regulating kinase (ASK) 1. EMBO J. 1998;17:2596–2606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Song JJ, Lee YJ. Differential role of glutaredoxin and thioredoxin in metabolic oxidative stress-induced activation of apoptosis signal–regulating kinase 1. Biochem. J. 2003;373:845–853. doi: 10.1042/BJ20030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hansen JM, Zhang H, Jones DP. Mitochondrial thioredoxin-2 has a key role in determining tumor necrosis factor-α–induced reactive oxygen species generation, NF-κB activation, and apoptosis. Toxicol. Sci. 2006;91:643–650. doi: 10.1093/toxsci/kfj175. [DOI] [PubMed] [Google Scholar]
- 168.Hansen JM, Watson WH, Jones DP. Compartmentation of Nrf-2 redox control: regulation of cytoplasmic activation by glutathione and DNA binding by thioredoxin-1. Toxicol. Sci. 2004;82:308–317. doi: 10.1093/toxsci/kfh231. [DOI] [PubMed] [Google Scholar]
- 169.Guo Y, Einhorn L, Kelley M, Hirota K, Yodoi J, et al. Redox regulation of the embryonic stem cell transcription factor oct-4 by thioredoxin. Stem Cells. 2004;22:259–264. doi: 10.1634/stemcells.22-3-259. [DOI] [PubMed] [Google Scholar]
- 170.Kirlin WG, Cai J, Thompson SA, Diaz D, Kavanagh TJ, Jones DP. Glutathione redox potential in response to differentiation and enzyme inducers. Free Radic. Biol. Med. 1999;27:1208–1218. doi: 10.1016/s0891-5849(99)00145-8. [DOI] [PubMed] [Google Scholar]
- 171.Ramirez A, Ramadan B, Ritzenthaler JD, Rivera HN, Jones DP, Roman J. Extracellular cysteine/cystine redox potential controls lung fibroblast proliferation and matrix expression through upregulation of transforming growth factor-β. Am. J. Physiol. Lung Cell. Mol. Physiol. 2007;293:L972–L981. doi: 10.1152/ajplung.00010.2007. [DOI] [PubMed] [Google Scholar]
- 172.Pisoni RL, Acker TL, Lisowski KM, Lemons RM, Thoene JG. A cysteine-specific lysosomal transport system provides a major route for the delivery of thiol to human fibroblast lysosomes: possible role in supporting lysosomal proteolysis. J. Cell Biol. 1990;110:327–335. doi: 10.1083/jcb.110.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Thoene J, Lemons R, Anikster Y, Mullet J, Paelicke K, et al. Mutations of CTNS causing intermediate cystinosis. Mol. Genet. Metab. 1999;67:283–293. doi: 10.1006/mgme.1999.2876. [DOI] [PubMed] [Google Scholar]
- 174.Laube GF, Shah V, Stewart VC, Hargreaves IP, Haq MR, et al. Glutathione depletion and increased apoptosis rate in human cystinotic proximal tubular cells. Pediatr. Nephrol. 2006;21:503–509. doi: 10.1007/s00467-006-0005-x. [DOI] [PubMed] [Google Scholar]
- 175.Chol M, Nevo N, Cherqui S, Antignac C, Rustin P. Glutathione precursors replenish decreased glutathione pool in cystinotic cell lines. Biochem. Biophys. Res. Commun. 2004;324:231–235. doi: 10.1016/j.bbrc.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 176.Levtchenko E, de Graaf–Hess A, Wilmer M, Van Den Heuvel L, Monnens L, Blom H. Altered status of glutathione and its metabolites in cystinotic cells. Nephrol. Dial. Transplant. 2005;20:1828–1832. doi: 10.1093/ndt/gfh932. [DOI] [PubMed] [Google Scholar]
- 177.Levtchenko EN, Wilmer MJ, Janssen AJ, Koenderink JB, Visch HJ, et al. Decreased intracellular ATP content and intact mitochondrial energy generating capacity in human cystinotic fibroblasts. Pediatr. Res. 2006;59:287–292. doi: 10.1203/01.pdr.0000196334.46940.54. [DOI] [PubMed] [Google Scholar]
- 178.Wilmer MJ, de Graaf–Hess A, Blom HJ, Dijkman HB, Monnens LA, et al. Elevated oxidized glutathione in cystinotic proximal tubular epithelial cells. Biochem. Biophys. Res. Commun. 2005;337:610–614. doi: 10.1016/j.bbrc.2005.09.094. [DOI] [PubMed] [Google Scholar]
- 179.Armstrong JS, Whiteman M, Yang H, Jones DP, Sternberg P., Jr Cysteine starvation activates the redox-dependent mitochondrial permeability transition in retinal pigment epithelial cells. Investig. Ophthalmol. Vis. Sci. 2004;45:4183–4189. doi: 10.1167/iovs.04-0570. [DOI] [PubMed] [Google Scholar]
- 180.Park MA, Pejovic V, Kerisit KG, Junius S, Thoene JG. Increased apoptosis in cystinotic fibroblasts and renal proximal tubule epithelial cells results from cysteinylation of protein kinase Cδ. J. Am. Soc. Nephrol. 2006;17:3167–3175. doi: 10.1681/ASN.2006050474. [DOI] [PubMed] [Google Scholar]
- 181.Thoene JG. A review of the role of enhanced apoptosis in the pathophysiology of cystinosis. Mol. Genet. Metab. 2007;92:292–298. doi: 10.1016/j.ymgme.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 182.Cherqui S, Sevin C, Hamard G, Kalatzis V, Sich M, et al. Intralysosomal cystine accumulation in mice lacking cystinosin, the protein defective in cystinosis. Mol. Cell. Biol. 2002;22:7622–7632. doi: 10.1128/MCB.22.21.7622-7632.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kimonis VE, Troendle J, Rose SR, Yang ML, Markello TC, Gahl WA. Effects of early cysteamine therapy on thyroid function and growth in nephropathic cystinosis. J. Clin. Endocrinol. Metab. 1995;80:3257–3261. doi: 10.1210/jcem.80.11.7593434. [DOI] [PubMed] [Google Scholar]
- 184.Jones AC, Austin J, Hansen N, Hoogendoorn B, Oefner PJ, et al. Optimal temperature selection for mutation detection by denaturing HPLC and comparison to single-stranded conformation polymorphism and heteroduplex analysis. Clin. Chem. 1999;45:1133–1140. [PubMed] [Google Scholar]
- 185.Sumimoto H, Miyano K, Takeya R. Molecular composition and regulation of the Nox family NAD(P)H oxidases. Biochem. Biophys. Res. Commun. 2005;338:677–686. doi: 10.1016/j.bbrc.2005.08.210. [DOI] [PubMed] [Google Scholar]
- 186.Arnold RS, Shi J, Murad E, Sun CQ, Polavarapu R, et al. Hydrogen peroxide mediates the cell growth and transformation caused by the mitogenic oxidase NOX1. Proc. Natl. Acad. Sci. USA. 2001;98:5550–5555. doi: 10.1073/pnas.101505898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Finkel T. Intracellular redox regulation by the family of small GTPases. Antioxid. Redox Signal. 2006;8:1857–1863. doi: 10.1089/ars.2006.8.1857. [DOI] [PubMed] [Google Scholar]
- 188.Selvakumar B, Hess DT, Goldschmidt-Clermont PJ, Stamler JS. Co-regulation of constitutive nitric oxide synthases and NADPH oxidase by the small GTPase Rac. FEBS Lett. 2008;582:2195–2202. doi: 10.1016/j.febslet.2008.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hsu HH, Hoffmann S, Endlich N, Velic A, Schwab A, et al. Mechanisms of angiotensin II signaling on cytoskeleton of podocytes. J. Mol. Med. 2008;86:1379–1394. doi: 10.1007/s00109-008-0399-y. [DOI] [PubMed] [Google Scholar]
- 190.Kim YS, Morgan MJ, Choksi S, Liu ZG. TNF-induced activation of the Nox1 NADPH oxidase and its role in the induction of necrotic cell death. Mol. Cell. 2007;26:675–687. doi: 10.1016/j.molcel.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 191.Vanden Berghe T, Declercq W, Vandenabeele P. NADPH oxidases: new players in TNF-induced necrotic cell death. Mol. Cell. 2007;26:769–771. doi: 10.1016/j.molcel.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 192.Gianni D, Bohl B, Courtneidge SA, Bokoch GM. The involvement of the tyrosine kinase c-Src in the regulation of reactive oxygen species generation mediated by NADPH oxidase-1. Mol. Biol. Cell. 2008;19:2984–2994. doi: 10.1091/mbc.E08-02-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Lambeth JD. Nox enzymes, ROS, and chronic disease: an example of antagonistic pleiotropy. Free Radic. Biol. Med. 2007;43:332–347. doi: 10.1016/j.freeradbiomed.2007.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Li Y, Huang TT, Carlson EJ, Melov S, Ursell PC, et al. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat. Genet. 1995;11:376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- 195.Lebovitz RM, Zhang H, Vogel H, Cartwright J, Jr, Dionne L, et al. Neurodegeneration, myocardial injury, and perinatal death in mitochondrial superoxide dismutase–deficient mice. Proc. Natl. Acad. Sci. USA. 1996;93:9782–9787. doi: 10.1073/pnas.93.18.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Reaume AG, Elliott JL, Hoffman EK, Kowall NW, Ferrante RJ, et al. Motor neurons in Cu/Zn superoxide dismutase–deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat. Genet. 1996;13:43–47. doi: 10.1038/ng0596-43. [DOI] [PubMed] [Google Scholar]
- 197.Carlsson LM, Jonsson J, Edlund T, Marklund SL. Mice lacking extracellular superoxide dismutase are more sensitive to hyperoxia. Proc. Natl. Acad. Sci. USA. 1995;92:6264–6268. doi: 10.1073/pnas.92.14.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Semenza GL. Hypoxia-inducible factor 1 (HIF-1) pathway. Sci. Signal Transduct. Knowl. Environ. 2007;407:cm8. doi: 10.1126/stke.4072007cm8. [DOI] [PubMed] [Google Scholar]
- 199.Semenza GL. Life with oxygen. Science. 2007;318:62–64. doi: 10.1126/science.1147949. [DOI] [PubMed] [Google Scholar]
- 200.Semenza GL. Oxygen-dependent regulation of mitochondrial respiration by hypoxia-inducible factor 1. Biochem. J. 2007;405:1–9. doi: 10.1042/BJ20070389. [DOI] [PubMed] [Google Scholar]
- 201.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 202.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 203.Simon MC. Coming up for air: HIF-1 and mitochondrial oxygen consumption. Cell Metab. 2006;3:150–151. doi: 10.1016/j.cmet.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 204.Fukuda R, Zhang H, Kim JW, Shimoda L, Dang CV, Semenza GL. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129:111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 205.Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, et al. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 206.Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J. Biol. Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 207.Semenza GL. Mitochondrial autophagy: life and breath of the cell. Autophagy. 2008;4:534–536. doi: 10.4161/auto.5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Hansen JM, Zhang H, Jones DP. Differential oxidation of thioredoxin-1, thioredoxin-2, and glutathione by metal ions. Free Radic. Biol. Med. 2006;40:138–145. doi: 10.1016/j.freeradbiomed.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 209.Iyer SS, Jones DP, Brigham KL, Rojas M. Oxidation of plasma cysteine/cystine redox state in endotoxin-induced lung injury. Am. J. Respir. Cell Mol. Biol. 2009;40:90–98. doi: 10.1165/rcmb.2007-0447OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Nishi K, Oda T, Takabuchi S, Oda S, Fukuda K, et al. LPS induces hypoxia-inducible factor 1 activation in macrophage-differentiated cells in a reactive oxygen species–dependent manner. Antioxid. Redox Signal. 2008;10:983–995. doi: 10.1089/ars.2007.1825. [DOI] [PubMed] [Google Scholar]
- 211.Csete M. Oxygen in the cultivation of stem cells. Ann. N.Y. Acad. Sci. 2005;1049:1–8. doi: 10.1196/annals.1334.001. [DOI] [PubMed] [Google Scholar]
- 212.Studer L, Csete M, Lee SH, Kabbani N, Walikonis J, et al. Enhanced proliferation, survival, and dopaminergic differentiation of CNS precursors in lowered oxygen. J. Neurosci. 2000;20:7377–7383. doi: 10.1523/JNEUROSCI.20-19-07377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Morrison SJ, Csete M, Groves AK, Melega W, Wold B, Anderson DJ. Culture in reduced levels of oxygen promotes clonogenic sympathoadrenal differentiation by isolated neural crest stem cells. J. Neurosci. 2000;20:7370–7376. doi: 10.1523/JNEUROSCI.20-19-07370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Csete M, Walikonis J, Slawny N, Wei Y, Korsnes S, et al. Oxygen-mediated regulation of skeletal muscle satellite cell proliferation and adipogenesis in culture. J. Cell. Physiol. 2001;189:189–196. doi: 10.1002/jcp.10016. [DOI] [PubMed] [Google Scholar]
- 215.Lee S, Shin HS, Shireman PK, Vasilaki A, Van Remmen H, Csete ME. Glutathione-peroxidase-1 null muscle progenitor cells are globally defective. Free Radic. Biol. Med. 2006;41:1174–1184. doi: 10.1016/j.freeradbiomed.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 216.Ku HT, Zhang N, Kubo A, O’Connor R, Mao M, et al. Committing embryonic stem cells to early endocrine pancreas in vitro. Stem Cells. 2004;22:1205–1217. doi: 10.1634/stemcells.2004-0027. [DOI] [PubMed] [Google Scholar]
- 217.Garcia CK, Wright WE, Shay JW. Human diseases of telomerase dysfunction: insights into tissue aging. Nucleic Acids Res. 2007;35:7406–7416. doi: 10.1093/nar/gkm644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 219.Shay JW, Wright WE. Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis. 2005;26:867–874. doi: 10.1093/carcin/bgh296. [DOI] [PubMed] [Google Scholar]
- 220.Shay JW, Wright WE. Telomeres and telomerase in aging and cancer. In: Guarente LP, Partridge L, Wallace DC, editors. Molecular Biology of Aging. Cold Spring Harbor, NY: Cold Spring Harb. Lab.; 2008. pp. 575–597. [Google Scholar]
- 221.Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat. Cell Biol. 2003;5:741–747. doi: 10.1038/ncb1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Saretzki G, Walter T, Atkinson S, Passos JF, Bareth B, et al. Downregulation of multiple stress defense mechanisms during differentiation of human embryonic stem cells. Stem Cells. 2008;26:455–464. doi: 10.1634/stemcells.2007-0628. [DOI] [PubMed] [Google Scholar]
- 223.Yang C, Przyborski S, Cooke MJ, Zhang X, Stewart R, et al. A key role for telomerase reverse transcriptase unit in modulating human embryonic stem cell proliferation, cell cycle dynamics, and in vitro differentiation. Stem Cells. 2008;26:850–863. doi: 10.1634/stemcells.2007-0677. [DOI] [PubMed] [Google Scholar]
- 224.von Zglinicki T. Oxidative stress shortens telomeres. Trends Biochem. Sci. 2002;27:339–344. doi: 10.1016/s0968-0004(02)02110-2. [DOI] [PubMed] [Google Scholar]
- 225.Passos JF, von Zglinicki T. Mitochondria, telomeres and cell senescence. Exp. Gerontol. 2005;40:466–472. doi: 10.1016/j.exger.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 226.Saretzki G, Murphy MP, von Zglinicki T. MitoQ counteracts telomere shortening and elongates lifespan of fibroblasts under mild oxidative stress. Aging Cell. 2003;2:141–143. doi: 10.1046/j.1474-9728.2003.00040.x. [DOI] [PubMed] [Google Scholar]
- 227.Richter T, von Zglinicki T. A continuous correlation between oxidative stress and telomere shortening in fibroblasts. Exp. Gerontol. 2007;42:1039–1042. doi: 10.1016/j.exger.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 228.Richter T, Saretzki G, Nelson G, Melcher M, Olijslagers S, von Zglinicki T. TRF2 overexpression diminishes repair of telomeric single-strand breaks and accelerates telomere shortening in human fibroblasts. Mech. Ageing Dev. 2007;128:340–345. doi: 10.1016/j.mad.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 229.Passos JF, Saretzki G, Ahmed S, Nelson G, Richter T, et al. Mitochondrial dysfunction accounts for the stochastic heterogeneity in telomere-dependent senescence. PLoS Biol. 2007;5:e110. doi: 10.1371/journal.pbio.0050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Santos JH, Meyer JN, Skorvaga M, Annab LA, Van Houten B. Mitochondrial hTERT exacerbates free-radical-mediated mtDNA damage. Aging Cell. 2004;3:399–411. doi: 10.1111/j.1474-9728.2004.00124.x. [DOI] [PubMed] [Google Scholar]
- 231.Haendeler J, Hoffmann J, Brandes RP, Zeiher AM, Dimmeler S. Hydrogen peroxide triggers nuclear export of telomerase reverse transcriptase via Src kinase family–dependent phosphorylation of tyrosine 707. Mol. Cell. Biol. 2003;23:4598–4610. doi: 10.1128/MCB.23.13.4598-4610.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Haendeler J, Hoffmann J, Diehl JF, Vasa M, Spyridopoulos I, et al. Antioxidants inhibit nuclear export of telomerase reverse transcriptase and delay replicative senescence of endothelial cells. Circ. Res. 2004;94:768–775. doi: 10.1161/01.RES.0000121104.05977.F3. [DOI] [PubMed] [Google Scholar]
- 233.Del Bufalo D, Rizzo A, Trisciuoglio D, Cardinali G, Torrisi MR, et al. Involvement of hTERT in apoptosis induced by interference with Bcl-2 expression and function. Cell Death Differ. 2005;12:1429–1438. doi: 10.1038/sj.cdd.4401670. [DOI] [PubMed] [Google Scholar]
- 234.Ahmed S, Passos JF, Birket MJ, Beckmann T, Brings S, et al. Telomerase does not counteract telomere shortening but protects mitochondrial function under oxidative stress. J. Cell Sci. 2008;121:1046–1053. doi: 10.1242/jcs.019372. [DOI] [PubMed] [Google Scholar]
- 235.Nkabyo YS, Go YM, Ziegler TR, Jones DP. Extracellular cysteine/cystine redox regulates the p44/p42 MAPK pathway by metalloproteinase-dependent epidermal growth factor receptor signaling. Am. J. Physiol. Gastrointest. Liver Physiol. 2005;289:G70–G88. doi: 10.1152/ajpgi.00280.2004. [DOI] [PubMed] [Google Scholar]
- 236.Jonas CR, Gu LH, Nkabyo YS, Mannery YO, Avissar NE, et al. Glutamine and KGF each regulate extracellular thiol/disulfide redox and enhance proliferation in Caco-2 cells. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2003;285:R1421–R1429. doi: 10.1152/ajpregu.00702.2002. [DOI] [PubMed] [Google Scholar]
- 237.Rotig A, Appelkvist EL, Geromel V, Chretien D, Kadhom N, et al. Quinone-responsive multiple respiratory-chain dysfunction due to widespread coenzyme Q10 deficiency. Lancet. 2000;356:391–395. doi: 10.1016/S0140-6736(00)02531-9. [DOI] [PubMed] [Google Scholar]
- 238.Quinzii CM, DiMauro S, Hirano M. Human coenzyme Q10 deficiency. Neurochem. Res. 2007;32:723–727. doi: 10.1007/s11064-006-9190-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Geromel V, Darin N, Chretien D, Benit P, DeLonlay P, et al. Coenzyme Q(10) and idebenone in the therapy of respiratory chain diseases: rationale and comparative benefits. Mol. Genet. Metab. 2002;77:21–30. doi: 10.1016/s1096-7192(02)00145-2. [DOI] [PubMed] [Google Scholar]
- 240.Shoffner JM, Wallace DC. Oxidative phosphorylation diseases and mitochondrial DNA mutations: diagnosis and treatment. Annu. Rev. Nutr. 1994;14:535–568. doi: 10.1146/annurev.nu.14.070194.002535. [DOI] [PubMed] [Google Scholar]
- 241.Rustin P, von Kleist–Retzow JC, Chantrel-Groussard K, Sidi D, Munnich A, Rotig A. Effect of idebenone on cardiomyopathy in Friedreich’s ataxia: a preliminary study. Lancet. 1999;354:477–479. doi: 10.1016/S0140-6736(99)01341-0. [DOI] [PubMed] [Google Scholar]
- 242.Hart PE, Lodi R, Rajagopalan B, Bradley JL, Crilley JG, et al. Antioxidant treatment of patients with Friedreich ataxia: four-year follow-up. Arch. Neurol. 2005;62:621–626. doi: 10.1001/archneur.62.4.621. [DOI] [PubMed] [Google Scholar]
- 243.Di Prospero NA, Baker A, Jeffries N, Fischbeck KH. Neurological effects of high-dose idebenone in patients with Friedreich’s ataxia: a randomised, placebo-controlled trial. Lancet Neurol. 2007;6:878–886. doi: 10.1016/S1474-4422(07)70220-X. [DOI] [PubMed] [Google Scholar]
- 244.Shoffner JM, Lott MT, Voljavec AS, Soueidan SA, Costigan DA, Wallace DC. Spontaneous Kearns-Sayre/chronic external ophthalmoplegia plus syndrome associated with a mitochondrial DNA deletion: a slip-replication model and metabolic therapy. Proc. Natl. Acad. Sci. USA. 1989;86:7952–7956. doi: 10.1073/pnas.86.20.7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Keightley JA, Anitori R, Burton MD, Quan F, Buist NR, Kennaway NG. Mitochondrial encephalomyopathy and complex III deficiency associated with a stop-codon mutation in the cytochrome b gene. Am. J. Hum. Genet. 2000;67:1400–1410. doi: 10.1086/316900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Mowat D, Kirby DM, Kamath KR, Kan A, Thorburn DR, Christodoulou J. Respiratory chain complex III [correction of complex] in deficiency with pruritus: a novel vitamin responsive clinical feature. J. Pediatr. 1999;134:352–354. doi: 10.1016/s0022-3476(99)70463-4. [DOI] [PubMed] [Google Scholar]
- 247.Argov Z, Bank WJ, Maris J, Eleff S, Kennaway NG, et al. Treatment of mitochondrial myopathy due to complex III deficiency with vitamins K3 and C: a 31P-NMR follow-up study. Ann. Neurol. 1986;19:598–602. doi: 10.1002/ana.410190615. [DOI] [PubMed] [Google Scholar]
- 248.Marriage BJ, Clandinin MT, Macdonald IM, Glerum DM. Cofactor treatment improves ATP synthetic capacity in patients with oxidative phosphorylation disorders. Mol. Genet. Metab. 2004;81:263–272. doi: 10.1016/j.ymgme.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 249.Naito E, Ito M, Yokota I, Saijo T, Matsuda J, et al. Thiamine-responsive pyruvate dehydrogenase deficiency in two patients caused by a point mutation (F205L and L216F) within the thiamine pyrophosphate binding region. Biochim. Biophys. Acta. 2002;1588:79–84. doi: 10.1016/s0925-4439(02)00142-4. [DOI] [PubMed] [Google Scholar]
- 250.Sedel F, Challe G, Mayer JM, Boutron A, Fontaine B, et al. Thiamine responsive pyruvate dehydrogenase deficiency in an adult with peripheral neuropathy and optic neuropathy. J. Neurol. Neurosurg. Psychiatry. 2008;79:846–847. doi: 10.1136/jnnp.2007.136630. [DOI] [PubMed] [Google Scholar]
- 251.Gibson GE, Blass JP. Thiamine-dependent processes and treatment strategies in neurodegeneration. Antioxid. Redox Signal. 2007;9:1605–1619. doi: 10.1089/ars.2007.1766. [DOI] [PubMed] [Google Scholar]
- 252.Longo N, Amat di San Filippo C, Pasquali M. Disorders of carnitine transport and the carnitine cycle. Am. J. Med. Genet. Part C, Semin. Med. Genet. 2006;142C:77–85. doi: 10.1002/ajmg.c.30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Hsu CC, Chuang YH, Tsai JL, Jong HJ, Shen YY, et al. CPEO and carnitine deficiency overlapping in MELAS syndrome. Acta Neurol. Scand. 1995;92:252–255. doi: 10.1111/j.1600-0404.1995.tb01697.x. [DOI] [PubMed] [Google Scholar]
- 254.King MP, Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 255.Klivenyi P, Gardian G, Calingasan NY, Yang L, von Borstel R, et al. Neuroprotective effects of oral administration of triacetyluridine against MPTP neurotoxicity. Neuromol. Med. 2004;6:87–92. doi: 10.1385/NMM:6:2-3:087. [DOI] [PubMed] [Google Scholar]
- 256.Stacpoole PW, Kerr DS, Barnes C, Bunch ST, Carney PR, et al. Controlled clinical trial of dichloroacetate for treatment of congenital lactic acidosis in children. Pediatrics. 2006;117:1519–1531. doi: 10.1542/peds.2005-1226. [DOI] [PubMed] [Google Scholar]
- 257.Barshop BA. Metabolomic approaches to mitochondrial disease: correlation of urine organic acids. Mitochondrion. 2004;4:521–527. doi: 10.1016/j.mito.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 258.Koga Y, Akita Y, Nishioka J, Yatsuga S, Povalko N, et al. L-arginine improves the symptoms of strokelike episodes in MELAS. Neurology. 2005;64:710–712. doi: 10.1212/01.WNL.0000151976.60624.01. [DOI] [PubMed] [Google Scholar]
- 259.Murphy MP, Smith RA. Targeting antioxidants to mitochondria by conjugation to lipophilic cations. Annu. Rev. Pharmacol. Toxicol. 2007;47:629–656. doi: 10.1146/annurev.pharmtox.47.120505.105110. [DOI] [PubMed] [Google Scholar]
- 260.Murphy MP, Smith RA. Drug delivery to mitochondria: the key to mitochondrial medicine. Adv. Drug Deliv. Rev. 2000;41:235–250. doi: 10.1016/s0169-409x(99)00069-1. [DOI] [PubMed] [Google Scholar]
- 261.Marangon K, Devaraj S, Tirosh O, Packer L, Jialal I. Comparison of the effect of α-lipoic acid and α-tocopherol supplementation on measures of oxidative stress. Free Radic. Biol. Med. 1999;27:1114–1121. doi: 10.1016/s0891-5849(99)00155-0. [DOI] [PubMed] [Google Scholar]
- 262.Schriner SE, Linford NJ, Martin GM, Treuting P, Ogburn CE, et al. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–1911. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- 263.Melov S, Schneider JA, Day BJ, Hinerfeld D, Coskun P, et al. A novel neurological phenotype in mice lacking mitochondrial manganese superoxide dismutase. Nat. Genet. 1998;18:159–163. doi: 10.1038/ng0298-159. [DOI] [PubMed] [Google Scholar]
- 264.Melov S, Doctrow SR, Schneider JA, Haberson J, Patel M, et al. Lifespan extension and rescue of spongiform encephalopathy in superoxide dismutase 2 nullizygous mice treated with superoxide dismutase-catalase mimetics. J. Neurosci. 2001;21:8348–8353. doi: 10.1523/JNEUROSCI.21-21-08348.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Rasola A, Bernardi P. The mitochondrial permeability transition pore and its involvement in cell death and in disease pathogenesis. Apoptosis. 2007;12:815–833. doi: 10.1007/s10495-007-0723-y. [DOI] [PubMed] [Google Scholar]
- 266.Irwin WA, Bergamin N, Sabatelli P, Reggiani C, Megighian A, et al. Mitochondrial dysfunction and apoptosis in myopathic mice with collagen VI deficiency. Nat. Genet. 2003;35:367–371. doi: 10.1038/ng1270. [DOI] [PubMed] [Google Scholar]
- 267.Angelin A, Tiepolo T, Sabatelli P, Grumati P, Bergamin N, et al. Mitochondrial dysfunction in the pathogenesis of Ullrich congenital muscular dystrophy and prospective therapy with cyclosporins. Proc. Natl. Acad. Sci. USA. 2007;104:991–996. doi: 10.1073/pnas.0610270104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Porcelli AM, Angelin A, Ghelli A, Mariani E, Martinuzzi A, et al. Respiratory complex I dysfunction due to mitochondrial DNA mutations shifts the voltage threshold for opening of the permeability transition pore toward resting levels. J. Biol. Chem. 2009;284:2045–2052. doi: 10.1074/jbc.M807321200. [DOI] [PubMed] [Google Scholar]
- 269.Wang H, Guan Y, Wang X, Smith K, Cormier K, et al. Nortriptyline delays disease onset in models of chronic neurodegeneration. Eur. J. Neurosci. 2007;26:633–641. doi: 10.1111/j.1460-9568.2007.05663.x. [DOI] [PubMed] [Google Scholar]
- 270.Jouaville LS, Pinton P, Bastianutto C, Rutter GA, Rizzuto R. Regulation of mitochondrial ATP synthesis by calcium: evidence for a long-term metabolic priming. Proc. Natl. Acad. Sci. USA. 1999;96:13807–13812. doi: 10.1073/pnas.96.24.13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Brini M, Pinton P, King MP, Davidson M, Schon EA, Rizzuto R. A calcium signaling defect in the pathogenesis of a mitochondrial DNA inherited oxidative phosphorylation deficiency. Nat. Med. 1999;5:951–954. doi: 10.1038/11396. [DOI] [PubMed] [Google Scholar]
- 272.Haigis MC, Guarente LP. Mammalian sirtuins—emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 273.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 274.Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Srivastava S, Barrett JN, Moraes CT. PGC-1α/β upregulation is associated with improved oxidative phosphorylation in cells harboring nonsense mtDNA mutations. Hum. Mol. Genet. 2007;16:993–1005. doi: 10.1093/hmg/ddm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Wenz T, Diaz F, Spiegelman BM, Moraes CT. Activation of the PPAR/PGC-1α pathway prevents a bioenergetic deficit and effectively improves a mitochondrial myopathy phenotype. Cell Metab. 2008;8:249–256. doi: 10.1016/j.cmet.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 277.Bastin J, Aubey F, Rotig A, Munnich A, Djouadi F. Activation of peroxisome proliferator–activated receptor pathway stimulates the mitochondrial respiratory chain and can correct deficiencies in patients’ cells lacking its components. J. Clin. Endocrinol. Metab. 2008;93:1433–1441. doi: 10.1210/jc.2007-1701. [DOI] [PubMed] [Google Scholar]
- 278.Djouadi F, Bastin J. PPARs as therapeutic targets for correction of inborn mitochondrial fatty acid oxidation disorders. J. Inherit. Metab. Dis. 2008;31:217–225. doi: 10.1007/s10545-008-0844-7. [DOI] [PubMed] [Google Scholar]
- 279.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Taivassalo T, Gardner JL, Taylor RW, Schaefer AM, Newman J, et al. Endurance training and detraining in mitochondrial myopathies due to single large-scale mtDNA deletions. Brain. 2006;129:3391–3401. doi: 10.1093/brain/awl282. [DOI] [PubMed] [Google Scholar]
- 281.Jeppesen TD, Schwartz M, Olsen DB, Wibrand F, Krag T, et al. Aerobic training is safe and improves exercise capacity in patients with mitochondrial myopathy. Brain. 2006;129:3402–3412. doi: 10.1093/brain/awl149. [DOI] [PubMed] [Google Scholar]
- 282.Taivassalo T, Fu K, Johns T, Arnold D, Karpati G, Shoubridge EA. Gene shifting: a novel therapy for mitochondrial myopathy. Hum. Mol. Genet. 1999;8:1047–1052. doi: 10.1093/hmg/8.6.1047. [DOI] [PubMed] [Google Scholar]
- 283.Murphy JL, Blakely EL, Schaefer AM, He L, Wyrick P, et al. Resistance training in patients with single, large-scale deletions of mitochondrial DNA. Brain. 2008;131:2832–2840. doi: 10.1093/brain/awn252. [DOI] [PubMed] [Google Scholar]
- 284.Clark KM, Bindoff LA, Lightowlers RN, Andrews RM, Griffiths PG, et al. Reversal of a mitochondrial DNA defect in human skeletal muscle. Nat. Genet. 1997;16:222–224. doi: 10.1038/ng0797-222. [DOI] [PubMed] [Google Scholar]
- 285.Veech RL. The therapeutic implications of ketone bodies: the effects of ketone bodies in pathological conditions: ketosis, ketogenic diet, redox states, insulin resistance, and mitochondrial metabolism. Prostaglandins Leukot. Essent. Fatty Acids. 2004;70:309–319. doi: 10.1016/j.plefa.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 286.Bentourkia M, Tremblay S, Pifferi F, Rousseau J, Lecomte R, Cunnane S. PET study of 11C-acetoacetate kinetics in rat brain during dietary treatments affecting ketosis. Am. J. Physiol. Endocrinol. Metab. 2009;296:E796–E801. doi: 10.1152/ajpendo.90644.2008. [DOI] [PubMed] [Google Scholar]
- 287.Kashiwaya Y, Takeshima T, Mori N, Nakashima K, Clarke K, Veech RL. D-β-hydroxybutyrate protects neurons in models of Alzheimer’s and Parkinson’s disease. Proc. Natl. Acad. Sci. USA. 2000;97:5440–5444. doi: 10.1073/pnas.97.10.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Bough K. Energy metabolism as part of the anticonvulsant mechanism of the ketogenic diet. Epilepsia. 2008;49 Suppl. 8:91–93. doi: 10.1111/j.1528-1167.2008.01846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Panetta J, Smith LJ, Boneh A. Effect of high-dose vitamins, coenzyme Q and high-fat diet in paediatric patients with mitochondrial diseases. J. Inherit. Metab. Dis. 2004;27:487–498. doi: 10.1023/B:BOLI.0000037354.66587.38. [DOI] [PubMed] [Google Scholar]
- 290.Kang HC, Lee YM, Kim HD, Lee JS, Slama A. Safe and effective use of the ketogenic diet in children with epilepsy and mitochondrial respiratory chain complex defects. Epilepsia. 2007;48:82–88. doi: 10.1111/j.1528-1167.2006.00906.x. [DOI] [PubMed] [Google Scholar]
- 291.Erecinska M, Nelson D, Daikhin Y, Yudkoff M. Regulation of GABA level in rat brain synaptosomes: fluxes through enzymes of the GABA shunt and effects of glutamate, calcium, and ketone bodies. J. Neurochem. 1996;67:2325–2334. doi: 10.1046/j.1471-4159.1996.67062325.x. [DOI] [PubMed] [Google Scholar]
- 292.Yudkoff M, Daikhin Y, Nissim I, Grunstein R, Nissim I. Effects of ketone bodies on astrocyte amino acid metabolism. J. Neurochem. 1997;69:682–692. doi: 10.1046/j.1471-4159.1997.69020682.x. [DOI] [PubMed] [Google Scholar]
- 293.Yudkoff M, Daikhin Y, Nissim I, Lazarow A, Nissim I. Brain amino acid metabolism and ketosis. J. Neurosci. Res. 2001;66:272–281. doi: 10.1002/jnr.1221. [DOI] [PubMed] [Google Scholar]
- 294.Yudkoff M, Daikhin Y, Nissim I, Lazarow A, Nissim I. Ketogenic diet, amino acid metabolism, and seizure control. J. Neurosci. Res. 2001;66:931–940. doi: 10.1002/jnr.10083. [DOI] [PubMed] [Google Scholar]
- 295.Yudkoff M, Daikhin Y, Nissim I, Lazarow A, Nissim I. Ketogenic diet, brain glutamate metabolism and seizure control. Prostaglandins Leukot. Essent. Fatty Acids. 2004;70:277–285. doi: 10.1016/j.plefa.2003.07.005. [DOI] [PubMed] [Google Scholar]
- 296.Yudkoff M, Daikhin Y, Nissim I, Horyn O, Lazarow A, et al. Response of brain amino acid metabolism to ketosis. Neurochem. Int. 2005;47:119–128. doi: 10.1016/j.neuint.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 297.Yudkoff M, Daikhin Y, Melo TM, Nissim I, Sonnewald U, Nissim I. The ketogenic diet and brain metabolism of amino acids: relationship to the anticonvulsant effect. Annu. Rev. Nutr. 2007;27:415–430. doi: 10.1146/annurev.nutr.27.061406.093722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Yudkoff M, Daikhin Y, Horyn O, Nissim I, Nissim I. Ketosis and brain handling of glutamate, glutamine, and GABA. Epilepsia. 2008;49 Suppl. 8:73–75. doi: 10.1111/j.1528-1167.2008.01841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Lund TM, Risa O, Sonnewald U, Schousboe A, Waagepetersen HS. Availability of neurotransmitter glutamate is diminished when β-hydroxybutyrate replaces glucose in cultured neurons. J. Neurochem. 2009;110:80–91. doi: 10.1111/j.1471-4159.2009.06115.x. [DOI] [PubMed] [Google Scholar]
- 300.Yudkoff M, Daikhin Y, Nissim I, Horyn O, Luhovyy B, et al. Short-term fasting, seizure control and brain amino acid metabolism. Neurochem. Int. 2006;48:650–656. doi: 10.1016/j.neuint.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 301.Sato K, Kashiwaya Y, Keon CA, Tsuchiya N, King MT, et al. Insulin, ketone bodies, and mitochondrial energy transduction. FASEB J. 1995;9:651–658. doi: 10.1096/fasebj.9.8.7768357. [DOI] [PubMed] [Google Scholar]
- 302.Nylen K, Velazquez JL, Sayed V, Gibson KM, Burnham WM, Snead OC., 3rd The effects of a ketogenic diet on ATP concentrations and the number of hippocampal mitochondria in Aldh5a1−/− mice. Biochim. Biophys. Acta. 2009;1790:208–212. doi: 10.1016/j.bbagen.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Maalouf M, Sullivan PG, Davis L, Kim DY, Rho JM. Ketones inhibit mitochondrial production of reactive oxygen species production following glutamate excitotoxicity by increasing NADH oxidation. Neuroscience. 2007;145:256–264. doi: 10.1016/j.neuroscience.2006.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Jarrett SG, Milder JB, Liang LP, Patel M. The ketogenic diet increases mitochondrial glutathione levels. J. Neurochem. 2008;106:1044–1051. doi: 10.1111/j.1471-4159.2008.05460.x. [DOI] [PubMed] [Google Scholar]
- 305.Kim do Y, Davis LM, Sullivan PG, Maalouf M, Simeone TA, et al. Ketone bodies are protective against oxidative stress in neocortical neurons. J. Neurochem. 2007;101:1316–1326. doi: 10.1111/j.1471-4159.2007.04483.x. [DOI] [PubMed] [Google Scholar]
- 306.Galimberti CA, Diegoli M, Sartori I, Uggetti C, Brega A, et al. Brain pseudoatrophy and mental regression on valproate and a mitochondrial DNA mutation. Neurology. 2006;67:1715–1717. doi: 10.1212/01.wnl.0000242882.58086.9a. [DOI] [PubMed] [Google Scholar]
- 307.Chen RS, Huang CC, Chu NS. Coenzyme Q10 treatment in mitochondrial encephalomyopathies. Short-term double-blind, crossover study. Eur. Neurol. 1997;37:212–218. doi: 10.1159/000117445. [DOI] [PubMed] [Google Scholar]
- 308.Muller W, Reimers CD, Berninger K-P, Frey A, et al. Coenzyme Q10 in ophthalmoplegia plus—a double blind, cross over therapeutic trial. J. Neurol. Sci. 1990;98 Suppl.:442. [Google Scholar]
- 309.Lerman-Sagie T, Rustin P, Lev D, Yanoov M, Leshinsky-Silver E, et al. Dramatic improvement in mitochondrial cardiomyopathy following treatment with idebenone. J. Inherit. Metab. Dis. 2001;24:28–34. doi: 10.1023/a:1005642302316. [DOI] [PubMed] [Google Scholar]
- 310.Liet JM, Pelletier V, Robinson BH, Laryea MD, Wendel U, et al. The effect of short-term dimethylglycine treatment on oxygen consumption in cytochrome oxidase deficiency: a double-blind randomized crossover clinical trial. J. Pediatr. 2003;142:62–66. doi: 10.1067/mpd.2003.mpd0333. [DOI] [PubMed] [Google Scholar]
- 311.Remes AM, Liimatta EV, Winqvist S, Tolonen U, Ranua JA, et al. Ubiquinone and nicotinamide treatment of patients with the 3243A→G mtDNA mutation. Neurology. 2002;59:1275–1277. doi: 10.1212/wnl.59.8.1275. [DOI] [PubMed] [Google Scholar]
- 312.Barshop BA, Naviaux RK, McGowan KA, Levine F, Nyhan WL, et al. Chronic treatment of mitochondrial disease patients with dichloroacetate. Mol. Genet. Metab. 2004;83:138–149. doi: 10.1016/j.ymgme.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 313.De Stefano N, Matthews PM, Ford B, Genge A, Karpati G, Arnold DL. Short-term dichloroacetate treatment improves indices of cerebral metabolism in patients with mitochondrial disorders. Neurology. 1995;45:1193–1198. doi: 10.1212/wnl.45.6.1193. [DOI] [PubMed] [Google Scholar]
- 314.Klopstock T, Querner V, Schmidt F, Gekeler F, Walter M, et al. A placebo-controlled crossover trial of creatine in mitochondrial diseases. Neurology. 2000;55:1748–1751. doi: 10.1212/wnl.55.11.1748. [DOI] [PubMed] [Google Scholar]
- 315.Tarnopolsky MA, Roy BD, MacDonald JR. A randomized, controlled trial of creatine monohydrate in patients with mitochondrial cytopathies. Muscle Nerve. 1997;20:1502–1509. doi: 10.1002/(sici)1097-4598(199712)20:12<1502::aid-mus4>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]