The stabilizing osmolyte trimethylamine N-oxide (TMAO) is shown to be an efficient primary precipitant for protein crystal growth. In addition to TMAO, two other methylamine osmolytes, sarcosine and betaine, are shown to be effective cryoprotective agents for protein crystal cooling.
Keywords: crystallization, cryoprotection, osmolytes, trimethylamine N-oxide, sarcosine, betaine
Abstract
Both crystallization and cryoprotection are often bottlenecks for high-resolution X-ray structure determination of macromolecules. Methylamine osmolytes are known stabilizers of protein structure. One such osmolyte, trimethylamine N-oxide (TMAO), has seen occasional use as an additive to improve macromolecular crystal quality and has recently been shown to be an effective cryoprotective agent for low-temperature data collection. Here, TMAO and the related osmolytes sarcosine and betaine are investigated as primary precipitating agents for protein crystal growth. Crystallization experiments were undertaken with 14 proteins. Using TMAO, seven proteins crystallized in a total of 13 crystal forms, including a new tetragonal crystal form of trypsin. The crystals diffracted well, and eight of the 13 crystal forms could be effectively cryocooled as grown with TMAO as an in situ cryoprotective agent. Sarcosine and betaine produced crystals of four and two of the 14 proteins, respectively. In addition to TMAO, sarcosine and betaine were effective post-crystallization cryoprotective agents for two different crystal forms of thermolysin. Precipitation reactions of TMAO with several transition-metal ions (Fe3+, Co2+, Cu2+ and Zn2+) did not occur with sarcosine or betaine and were inhibited for TMAO at lower pH. Structures of proteins from TMAO-grown crystals and from crystals soaked in TMAO, sarcosine or betaine were determined, showing osmolyte binding in five of the 12 crystals tested. When an osmolyte was shown to bind, it did so near the protein surface, interacting with water molecules, side chains and backbone atoms, often at crystal contacts.
1. Introduction
Macromolecular crystallography is dependent upon effective methods for crystallizing macromolecules of interest as well as cryogenically cooling these crystals for exposure to high-intensity X-ray sources. Both techniques, crystallization and cryocooling, are grounded in physical chemistry, but are most effectively implemented via shotgun approaches in which many different solution conditions are tested. Crystallization methods have been classically based on reducing the solubility of the macromolecule via salting out (e.g. ammonium sulfate), excluded volume depletion forces (e.g. polyethylene glycol) or organic solvents (e.g. propanol and MPD, which operate via a combination of exclusion and modulation of the dielectric constant of the solution) (McPherson, 1999 ▶; Dumetz et al., 2008 ▶). More recently, other approaches have been discussed, including the use of organic molecules to bind at crystal contacts (e.g. silver bullets; McPherson & Cudney, 2006 ▶). Despite a long history with recent advances, both crystallization and cryoprotection can be bottlenecks for structure determinations and additional crystallization and cryoprotective agents would be beneficial for the field.
Osmolytes are naturally occurring solutes that are used for regulating cell volume in response to osmotic stress (Yancey et al., 1982 ▶; Hochachka & Somero, 2002 ▶). Inorganic ions are sometimes used for such regulation (e.g. in some archaea), but at high concentrations they can perturb cellular proteins, requiring molecular adaptations to maintain protein function. Organic osmolytes, on the other hand, help cells adapt to osmotic stress while maintaining normal protein structure and function, and are commonly used by eubacteria and eukaryotic organisms.
Five types of organic osmolytes have been identified: polyhydric alcohols and sugars, free amino acids and amino-acid derivatives, urea, methylsulfonium solutes and methylamines (Yancey et al., 1982 ▶; Yancey, 2005 ▶). Although the osmolytes in each of these categories differ in general structure and characteristics, they all share a few key qualities: they are highly soluble in water, typically have no net charge at physiological pH and are typically polar molecules (Hochachka & Somero, 2002 ▶).
As tools in macromolecular crystallography, alcohols and sugars have seen extensive use as cryoprotective agents and additives in crystallization screens (e.g. glycerol, glucose, sucrose and sorbitol). Amino acids (e.g. proline) and urea appear occasionally as additives in crystallization experiments. Although not currently extensively used for crystallography, the methylamines are of particular interest because they are known to increase protein stability (Yancey & Somero, 1979 ▶; Santoro et al., 1992 ▶; Bolen, 2004 ▶). In the marine cartilaginous fishes (e.g. sharks) methylamines appear in combination with urea, with the kosmotropic methylamines offsetting the chaotropic effects of urea and the combination of the two providing the correct osmotic balance (Yancey et al., 1982 ▶).
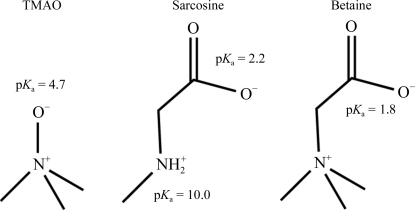
The methylamine osmolytes include trimethylamine N-oxide (TMAO), sarcosine and glycine betaine (or trimethylglycine, hereafter referred to as betaine) (Fig. 1 ▶). TMAO has been used in a few cases to improve crystal quality (Hill et al., 2002 ▶; Jiang et al., 2006 ▶) and appears as an additive in at least one commercial crystallization screen (Index Screen, Hampton Research, Aliso Viejo, California, USA). TMAO has also been shown to be an effective cryoprotective agent for low-temperature X-ray data collection (Mueller-Dieckmann et al., 2007 ▶, 2011 ▶; Pechkova et al., 2009 ▶).
Figure 1.
The three osmolytes investigated in this study: TMAO (pK a = 4.6–4.7; Lin & Timasheff, 1994 ▶; Qu, 2003 ▶), sarcosine (pK a = 2.2, 10.0; Dawson et al., 1959 ▶) and betaine (pK a = 1.8; Dawson et al., 1959 ▶). All are zwitterionic over nearly the whole pH range studied.
Here, we investigate TMAO, sarcosine and betaine for their ability to act as a primary precipitant for protein crystal growth. In concert, we further characterize TMAO, and test sarcosine and betaine, as cryoprotective agents for protein-crystal cryocooling.
2. Materials and methods
All chemicals were purchased from Sigma–Aldrich (St Louis, Missouri, USA). The TMAO (T0514), sarcosine (131776) and betaine (61692) were 98, 98 and 99% pure, respectively. Proteins were purchased from Sigma–Aldrich or Hampton Research or expressed and purified as previously reported (β-galactosidase, Juers et al., 2000 ▶; MntR, Glasfeld et al., 2003 ▶; Table 1 ▶). The following buffers were used: sodium acetate (pH 4.7), citric acid (pH 5.0), malic acid (pH 5.5), 2-(N-morpholino)ethanesulfonic acid (MES; pH 6.0), bis(2-hydroxyethyl)amino-tris(hydroxymethyl)methane (bis-tris; pH 7.0), tris(hydroxymethyl)aminomethane (Tris; pH 8.0, 8.5), boric acid (pH 9.0, 9.5, 10.0) and 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS; pH 10.5, 11.0). The pH of each buffer was adjusted with HCl or NaOH.
Table 1. Proteins used in crystallization screening.
The proteins shown in bold were successfully crystallized with TMAO as precipitating agent.
| Protein | pI† | Catalog No.‡ | Concentration§ (mg ml−1) | Buffer/cosolvents |
|---|---|---|---|---|
| HSA | 4.7 | A1653 | 40–400 | Water |
| Insulin | 5.3 | I5523 | 10 | 0.05 M CAPS pH 11 |
| β-Galactosidase | 5.3 | — | 12 | β-Gal buffer¶ |
| Catalase | 5.4 | C40 | 30 | Water |
| Thermolysin | 5.4 | P1512 | 25 | 0.05 M NaOH |
| MntR | 5.7 | — | 6 | MntR buffer¶ |
| Lipase B | 6.0 | HR7-099 | 40 | Water or 0.7% octylglucoside |
| Hemoglobin | 6.8 | H4131 | 5–30 | Water |
| Myoglobin | 7.3 | M0630 | 20–70 | Water |
| Thaumatin | 8.5 | T7638 | 12–40 | 0.1 M potassium tartrate |
| Chymotrypsinogen | 9.0 | C4879 | 15 | Water |
| Xylanase | 9.0 | HR7-106 | 11 | 14% glycerol, 60 mM Na/K phosphate pH 7.0 |
| HEWL | 9.3 | L6876 | 80 | Water |
| Trypsin | 10.3 | T8003 | 40 | 5–100 mM benzamidine |
pI is either provided by the manufacturer or predicted from the amino-acid sequence using ExPASy (http://expasy.org/).
Catalog Nos. are from Sigma–Aldrich, except for lipase B and xylanase, which were from Hampton Research.
Concentrations were based on searches of the Protein Data Bank and the literature and some optimization with small screens.
β-Gal buffer, 100 mM bis-tris pH 6.0, 200 mM MgCl2, 100 mM NaCl, 10 mM DTT. MntR buffer, 25 mM HEPES pH 7.5, 200 mM NaCl, 10%(v/v) glycerol.
2.1. Crystal growth
2.1.1. Crystallization screens
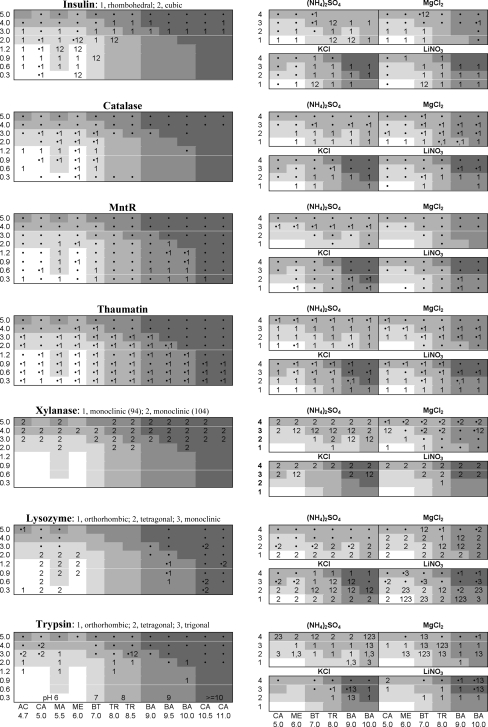
Crystals were grown using sitting-drop vapor diffusion in two crystallization screens designed to test the efficacy of TMAO as a primary precipitating agent. Screen 1 (TMAO–pH) was composed of TMAO (0.3–5.0 M) and buffer (0.1 M), while Screen 2 (TMAO–pH–salt) was composed of TMAO (1–4 M), buffer (0.1 M) and salt additives [0.2 M (NH4)2SO4, MgCl2, KCl or LiNO3], including ions of varying charge (monovalent versus divalent), size (Li+ versus K+) and geometry (planar NO3 − versus tetrahedral SO4 2−). All well solutions were made from stock solutions of 6 M TMAO, 1 M buffer at the pH stated above and 2 M salt, if present. To investigate TMAO as the primary precipitant without salting-out effects from other substituents, the pH of the solutions was not adjusted when mixing the well solutions, because pH adjustment would require high concentrations of counter-anions from strong acids. For example, the pH of 4.0 M TMAO dihydrate is 9.8 and to reach pH 5.0 using HCl requires 2.6 M Cl− to be present. As a consequence, the screens were biased towards basic pH, with the final pH in the well ranging from 6.0 to 11.0 (Fig. 2 ▶).
Figure 2.
Phase diagrams for the seven proteins that crystallized with TMAO, listed in order of increasing isoelectric point. Vertical axis, TMAO concentration in M. Horizontal axis (see trypsin diagram), buffer used with pH of the 1 M buffer stock solution below the two-letter code (AC, sodium acetate; CA, citric acid; MA, malic acid; ME, MES; BT, bis-tris; TR, Tris; BA, boric acid; CA, CAPS). Left, Screen 1: TMAO–pH. Right, Screen 2: TMAO–pH with 0.2 M salt additives. The crystal form is indicated numerically, precipitate is indicated by a bullet (•) and blank cells indicate neither precipitate nor crystals. The pH of the well solution is indicated by shading (see trypsin diagram; 6, pH 5.9–6.9; 7, pH 7.0–7.9; 8, pH 8.0–8.9; 9, pH 9.0–9.9; >=10, pH 10.0–11.9). The phase diagrams were based on observations made 6–8 weeks after tray setup.
Screens were also created using sarcosine (2.4–7.2 M; buffer stock solution pH 5–10; final well pH 5.9–8.5) and betaine (1.0–4.3 M; buffer stock solution pH 5–10; final well pH 5.9–12.6) both with and without the presence of 0.2 M (NH4)2SO4.. Higher concentrations of sarcosine were used than of TMAO and betaine because sarcosine is more soluble than the other two osmolytes (Table 2 ▶).
Table 2. Physical properties of the osmolytes related to their action as cryoprotective agents.
| Cryoprotective agent (MW in g mol−1) | ρRT† (σ) (g ml−1) | ρLT† (σ) (g ml−1) | Specific volume change‡ (σ) | Vitrification concentration§ (M) | Solubility (RT)† (M) |
|---|---|---|---|---|---|
| Betaine (117.2) | 1.095 (2) | 1.148 (1) | −0.048 (2) | 3.5 | 5.5 |
| Ethylene glycol (62.1) | 1.064 (3) | 1.139 (2) | −0.070 (3) | 5.4 | |
| Glycerol (92.1) | 1.130 (3) | 1.181 (2) | −0.046 (3) | 4.1 | |
| Sarcosine (89.2) | 1.151 (2) | 1.204 (7) | −0.046 (7) | 4.5 | 8.0 |
| TMAO (75.1) | 1.031 (2) | 1.098 (2) | −0.065 (3) | 3.0 | 6.0 |
RT, room temperature (294 K); LT, low temperature (liquid nitrogen at ∼72 K). Several measurements were made for each liquid (5–8 at each temperature), with σ representing the statistical variation in the measurements. All solutions were measured at 50%(w/w), except TMAO, which was measured at 44%(w/w). Values for glycerol and ethylene glycol, for comparison, are from Alcorn & Juers (2010 ▶).
The fractional change in the specific volume with cooling, (Δν/νRT) = (−Δρ/ρLT); σ is the propagated uncertainty. See Alcorn & Juers (2010 ▶).
Vitrification concentrations for glycerol and ethylene glycol were both 30%(v/v).
Two control screens were created: a negative control in which the TMAO was omitted from the TMAO–pH–salt screen and a positive control in which (NH4)2SO4 (0.8–3.6 M) replaced TMAO in the TMAO–pH screen.
Most of the screening was performed in 96-well plates (Intelli-Plates; Art Robbins Instruments, Sunnyvale, California), for which the well solution volume was 200 µl. For the TMAO screens, two drops were set up for each well (total volumes of 3 and 8 µl with equal volumes of protein solution and well solution). For the other screens, one 6 µl drop was used. The proteins used were hen egg-white lysozyme (HEWL), bovine pancreatic trypsin, equine skeletal muscle myoglobin, porcine hemoglobin, human serum albumin (HSA), bovine liver catalase, porcine insulin, Thaumatococcus danielli thaumatin, bovine α-chymotrypsinogen, Bacillus thermoproteolyticus Rokko thermolysin, B. subtilis manganese transport regulator (MntR), Trichoderma xylanase II, Escherichia coli β-galactosidase and Candida antarctica lipase B. See Table 1 ▶ for more information. The screens were created at room temperature (295 K) and were examined the day after setup, daily during the first week and then every few weeks after that for 3–8 weeks.
The crystals used for the structure determinations discussed below were grown using sitting-drop or hanging-drop vapor diffusion with the following conditions. Trypsin (P41212): 3.75 M TMAO, 0.1 M MES, 0.2 M (NH4)2SO4. Trypsin (P3121): 3 M TMAO, 0.1 M boric acid pH 10.0, 0.2 M LiNO3. Trypsin (P212121): 2 M TMAO, 0.1 M MES, 0.2 M (NH4)2SO4. Insulin (I213): 1.2 M TMAO, 0.1 M malic acid.
2.1.2. Crystals for cryoprotection and soaking experiments
For post-crystallization cryoprotection experiments, two crystal forms of thermolysin were used, both of which were grown by hanging-drop vapor diffusion in 24-well VDX plates (Hampton Research). The hexagonal crystal form (P6122) was grown using a protein solution consisting of 50 mg ml−1 protein in 45%(v/v) dimethyl sulfoxide, 1 M NaCl and 0.1 M MES pH 6.0 set up in 8 µl drops over 0–30% saturated ammonium sulfate. The tetragonal crystal form (P41212) was grown using identical conditions with the addition of 1 M ZnCl2 to the protein solution (Hausrath & Matthews, 2002 ▶). In both cases crystals appeared in a few days and grew to full size within two weeks. For sarcosine and betaine binding, trypsin crystals were grown with protein at 40 mg ml−1 using a well solution consisting of 20% PEG 8000, 0.1 M Tris pH 8.0, 0.2 M (NH4)2SO4, 0.1 M benzamidine (Leiros et al., 2001 ▶).
2.2. Cryoprotection experiments
Two types of cryoprotection experiment were carried out. In the first type of experiment, the osmolytes were used as post-crystallization treatments for crystals grown using previously published conditions. For hexagonal thermolysin, crystals were transferred using a cryoloop to sitting drops of pure cryoprotective agent (about 2 M) and then subsequently moved to other concentrations (i.e. 1–6 M) of cryoprotective agent in steps of 1 M, letting the crystal equilibrate for a few minutes at each concentration. An initial transfer to 2 M was necessary to prevent crystal cracking. These soaks were typically performed for at total of at least 30 min. Tetragonal thermolysin was marginally stable in pure osmolyte solutions, so short soaks (30–60 s) were performed using a range of osmolyte concentrations in the presence of 0.1 M MES pH 6.5 and 1 M NaCl. Although required for tetragonal crystal growth, ZnCl2 was not used in the cryoprotection solutions because of precipitation reactions with TMAO.
In the second type of experiment, crystals grown using TMAO as precipitant were cryocooled as grown and tested for diffraction. In these cases, the crystals were mounted directly from the crystallization drop by scooping with a cryoloop (Hampton Research), blotting the excess solution off the cryoloop and flash-cooling directly in the cryostream.
2.3. X-ray diffraction, data processing and structure determination
X-ray diffraction experiments were carried out on an Oxford Diffraction Xcalibur X-ray diffractometer with a Nova X-ray source and Onyx detector (Agilent Technologies, Santa Clara, California, USA) and an Oxford Cryojet temperature controller (Oxford Instruments, Oxford, England). Low-temperature data collection was performed by cryocooling the crystals directly in the cold stream at 100 K using 20 µm diameter nylon cryoloops (Hampton Research). Room-temperature (294 K) data collection was performed using MicroRT tubes (MiTeGen, Ithaca, New York, USA). Data-collection strategies were created with CrysAlisPro (Agilent Technologies) subject to contraints of desired overall redundancy (>3) and available beamtime, resulting in exposure times of 20–60 s, oscillation ranges of 0.3–1.0° and data sets of 69–454 images (Table 3 ▶). Data were integrated with CrysAlisPro and reduced with SCALA (Evans, 2006 ▶). Most structures were determined by starting with rigid-body refinement with REFMAC (Murshudov et al., 2011 ▶) using coordinates in the Protein Data Bank from a similar unit cell. The starting coordinates were as follows: for HEWL, 1bvx (tetragonal; Dong et al., 1999 ▶), 1f0w (orthorhombic; Biswal et al., 2000 ▶) or 5lym (monoclinic; Rao & Sundaralingam, 1996 ▶); for trypsin, 1tpo (orthorhombic; Marquart et al., 1983 ▶) or 1c1n (trigonal; Katz et al., 1998 ▶); for insulin, 1iza (hexagonal; Bentley et al., 1992 ▶) or 9ins (cubic; Gursky et al., 1992 ▶); for MntR, 2f5e (Kliegman et al., 2006 ▶); for xylanase, 1xyo (Törrönen & Rouvinen, 1995 ▶) or 2jic (Moukhametzianov et al., 2008 ▶); for thaumatin, 1kwn (Sauter et al., 2002 ▶); for thermolysin, 1l3f (tetragonal; Hausrath & Matthews, 2002 ▶) or 8tln (hexagonal; Holland et al., 1992 ▶). Refinements then continued with a few rounds of restrained refinement alternating with model building in Coot (Emsley et al., 2010 ▶). The tetragonal trypsin structure required molecular replacement, which was carried out using MOLREP (Vagin & Teplyakov, 2010 ▶) from the CCP4 package (Winn et al., 2011 ▶) with 1tpo as a search model prior to rigid-body refinement.
Table 3. Data-collection and refinement statistics.
Statistics are given for the whole data set, with values for the high-resolution bin given in parentheses. N ref, multiplicity, completeness, 〈I/σ(I)〉 and R meas are taken from the output from SCALA. Wilson B is from TRUNCATE. Mosaicity is the average of e 1, e 2 and e 3 from CrysAlisPro, which are the mosaicities in three directions defined in a coordinate system local to each reflection. e 1 and e 2 are the mosaicities in two orthogonal directions tangential to the Ewald sphere (on the image, e 2 is the mosaicity along the direction radial from the beam center), while e 3 is the mosaicity in a direction perpendicular to e 1 and p* = S − S 0, which is roughly the mosaicity in the scanning direction. Here, S is the scattered X-ray vector and S 0 is the incident X-ray vector (Schutt & Winkler, 1977 ▶; Kabsch, 2001 ▶). R and R free are refined crystallographic R values from REFMAC for all data (working and test set) and the test set, respectively. R.m.s. bond length and angle are root-mean-square deviations of bond lengths and angles from the average values in the geometry library of REFMAC. For structures listed as ‘precip’, TMAO was used as the precipitant to grow the crystal. For structures listed as ‘soak’, the crystal was grown using previously published conditions and then soaked in osmolyte (see §2). TLN, thermolysin.
| Protein/osmolyte | Trypsin/TMAO | Trypsin/TMAO | Trypsin/TMAO | Trypsin/sarcosine | Trypsin/betaine | Insulin/TMAO | TLN/TMAO | TLN/sarcosine | TLN/betaine |
|---|---|---|---|---|---|---|---|---|---|
| Space group | P41212 | P3121 | P212121 | P212121 | P212121 | I213 | P41212 | P41212 | P41212 |
| Unit-cell parameters (Å) | a = b = 53.9, c = 181.9 | a = b = 54.37, c = 105.36 | a = 54.9, b = 58.6, c = 67.9 | a = 54.9, b = 58.6, c = 67.7 | a = 55.1, b = 58.2, c = 68.9 | a = b = c = 79.1 | a = b = 96.9, c = 105.6 | a = b = 96.4, c = 105.9 | a = b = 96.2, c = 105.10 |
| [Osmolyte] (M) | 3.75, precip | 3.0, precip | 1.0, precip | 4.0, soak | 4.0, soak | 1.2, precip | 3.0, soak | 4.0, soak | 3.0, soak |
| Temperature (K) | 100 | 100 | 296 | 296 | 296 | 296 | 100 | 100 | 100 |
| Nref | 7217 (1017) | 18745 (2629) | 24788 (3555) | 24708 (3548) | 16718 (2383) | 4954 (721) | 37207 (5303) | 29727 (4252) | 33919 (4855) |
| No. of images | 454 | 213 | 215 | 273 | 182 | 69 | 162 | 172 | 90 |
| Detector distance (mm) | 90 | 90 | 65 | 65 | 65 | 65 | 74 | 90 | 280 |
| Scan width (°)/time (s) | 0.3/60 | 0.75/20 | 1.0/45 | 1.0/26.2 | 1.0/60 | 1.0/60 | 0.75/45 | 0.75/20 | 0.75/20 |
| Multiplicity | 10.0 (10.3) | 3.7 (2.7) | 5.3 (3.2) | 5.6 (4.0) | 5.8 (3.7) | 6.8 (5.0) | 6.9 (3.6) | 6.0 (4.0) | 8.6 (5.6) |
| Completeness (%) | 99.7 (100.0) | 99.5 (97.8) | 99.9 (100.0) | 99.9 (99.9) | 99.8 (99.3) | 99.9 (100.0) | 99.8 (99.3) | 99.9 (100.0) | 99.9 (100.0) |
| 〈I/σ(I)〉 | 7.0 (2.6) | 6.2 (3.1) | 9.1 (1.5) | 6.6 (1.3) | 7.6 (1.3) | 3.0 (1.8) | 5.4 (3.3) | 4.9 (1.3) | 14.0 (2.9) |
| Rmeas (%) | 10.5 (30.2) | 10.3 (30.3) | 7.4 (52.5) | 9.5 (59.4) | 9.7 (60.2) | 14.2 (46.4) | 9.4 (25.5) | 14.2 (68.4) | 12.5 (62.2) |
| Resolution limits (Å) | 51.7–2.80 (2.95–2.80) | 47.1–1.75 (1.84–1.75) | 44.4–1.70 (1.79–1.70) | 44.3–1.70 (1.79–1.70) | 44.5–1.95 (2.06–1.95) | 18.6–2.10 (2.21–2.10) | 71.4–1.95 (2.06–1.95) | 71.2–2.10 (2.21–2.10) | 70.9–2.00 (2.11–2.00) |
| Wilson B (Å2) | 40.6 | 13.0 | 13.4 | 14.4 | 18.5 | 27.2 | 13.9 | 19.4 | 18.1 |
| Mosaicity (°) | 0.57 | 0.76 | 0.71 | 0.67 | 0.71 | 0.72 | 0.83 | 0.73 | 0.71 |
| R (%) | 18.5 | 14.8 | 15.8 | 15.8 | 14.9 | 17.6 | 15.6 | 17.3 | 16.2 |
| Rfree (%) | 24.8 | 19.0 | 18.8 | 18.7 | 19.1 | 22.3 | 18.3 | 21.6 | 18.8 |
| 〈B〉prot (Å2) | 16.2 | 12.8 | 13.3 | 14.2 | 19.7 | 29.9 | 11.7 | 17.4 | 17.2 |
| 〈B〉wat (Å2) | 9.1 | 25.3 | 28.3 | 29.3 | 33.9 | 44.0 | 28.6 | 24.0 | 32.2 |
| No. of waters | 34 | 241 | 160 | 168 | 132 | 35 | 449 | 194 | 350 |
| R.m.s. bond length (Å) | 0.013 | 0.014 | 0.014 | 0.015 | 0.019 | 0.017 | 0.017 | 0.022 | 0.019 |
| R.m.s. bond angle (°) | 1.7 | 1.5 | 1.4 | 1.5 | 1.7 | 1.5 | 1.4 | 1.7 | 1.6 |
| PDB code | 3t28 | 3t29 | 3t25 | 3t26 | 3t27 | 3t2a | 3t2h | 3t2i | 3t2j |
In some cases, additional steps were taken to identify ligands in the crystals. For orthorhombic trypsin, a data set was collected from a crystal grown using control conditions (see above) and F o − F o difference Fourier maps (i.e. F o osmolyte − F o control phased using a model refined without bound ligands against the osmolyte data set) were used to identify TMAO, betaine and sarcosine binding locations. For the tetragonal thermolysin crystals soaked in the three osmolytes, anomalous difference Fourier maps were used to distinguish between water molecules and heavier ions (i.e. Ca2+, Zn2+ and Cl−). The latter were distinguished from each other based on known binding sites and ligand geometry.
2.4. Cryoprotection physical parameters
The physical parameters of the osmolytes were determined and compared with those of two of the more commonly used cryoprotective agents: glycerol and ethylene glycol. Thermal contractions of cryosolutions between 294 and 72 K were determined as previously reported (Alcorn & Juers, 2010 ▶). The minimum concentration of cryoprotective agent for vitrification was determined by using a cryoloop of 20 µm diameter nylon with dimensions of 0.4 × 0.6 mm (Hampton Research). The loop was passed through the cryosolution of interest and flash-cooled in the cryostream (100 K) by first blocking the cryostream, placing the sample on the goniometer and then removing the cryostream block. X-ray diffraction measurements were taken (30 s exposure time) and integrated azimuthally to yield a one-dimensional powder diffraction profile using CrysAlisPro. This was repeated in increments of 0.5 M TMAO, sarcosine or betaine or 2.5%(v/v) glycerol or ethylene glycol. The lowest concentration of cryoprotective agent that prevented the formation of cubic ice (Ic) was recorded as the vitrification concentration.
2.5. Precipitation reactions
Precipitation of the osmolytes with various metal-salt solutions was tested. Solutions of osmolyte in water were prepared and the pH was left as-made or adjusted to 7.0 or 4.0 using HCl (final concentration 4.0 M osmolyte). The osmolyte solutions were mixed with solutions of metal salts (1.0 M MgCl2, CaCl2, MnCl2, FeCl3, NiCl2, CoCl2, CuCl2 or ZnCl2) by adding 100 µl osmolyte solution to 10 µl salt solution in a microbatch crystallization tray (Nunc, Rochester, New York, USA). The drops were observed immediately after mixing in a stereomicroscope to assess for precipitate formation. For those drops that precipitated, the test was repeated with sulfate and/or acetate salts: Fe2(SO4)3, NiSO4, Ni(CH3CO2)2, Co(CH3CO2)2, CuSO4 and Zn(CH3CO2)2.
3. Results
3.1. Crystal growth
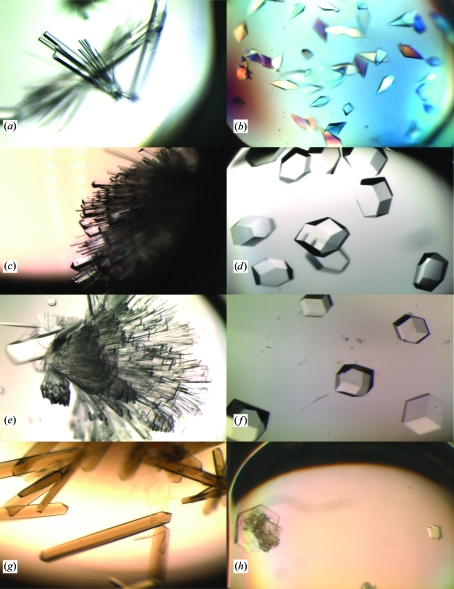
Fig. 3 ▶ shows example crystals produced with TMAO as precipitant. Of the 14 proteins tested, seven crystallized, some in multiple crystal forms. The TMAO–pH and TMAO–pH–salt screens produced crystals of lysozyme (three crystal forms), trypsin (three crystal forms), insulin (two crystal forms), catalase, xylanase (two crystal forms) and MntR (Fig. 2 ▶). Subsequent experiments, in which 0.1 M sodium/potassium tartrate was used as an additional additive, produced thaumatin crystals. A control screen lacking TMAO but including the salts and buffers failed to crystallize all proteins except for monoclinic lysozyme, which crystallized with 0.2 M nitrate. Diffraction experiments showed that all of the crystal forms observed have been reported before, with the exception of the tetragonal form of trypsin (P41212).
Figure 3.
Example crystals grown with TMAO as precipitant. (a) Orthorhombic trypsin; 3 M TMAO, 0.1 M pH 5.5 buffer. (b) Tetragonal trypsin; 4 M TMAO, 0.1 M pH 8.0 buffer, 0.2 M (NH4)2SO4. (c) Orthorhombic lysozyme; 2 M TMAO, 0.1 M pH 10.0 buffer, 0.2 M KCl. (d) Tetragonal lysozyme; 1 M TMAO, 0.1 M pH 9.0 buffer, 0.2 M MgCl2. (e) Monoclinic lysozyme; 1 M TMAO, 0.1 M pH 10.0 buffer, 0.2 M LiNO3. (f) Rhombohedral insulin; 1.2 M TMAO, 0.1 M pH 5.5 buffer. (g) Orthorhombic catalase; 0.9 M TMAO, 0.1 M pH 7.0 buffer. (h) Monoclinic xylanase (left, form 2; right, form 1); 3 M TMAO, 0.1 M pH 6.0 buffer, 0.2 M KCl.
Most of the protein crystal growth occurred in the first week or two following screen setup. In many cases, the drop size has an impact on whether crystals formed or which crystal form appeared. For example, in some cases smaller drops produced tetragonal lysozyme crystals while larger drops produced orthorhombic crystals.
Sarcosine produced crystals of four proteins: lysozyme, insulin, xylanase and thaumatin. Betaine produced crystals of insulin and lysozyme. The ammonium sulfate–pH screen produced crystals of nine of the 14 proteins: α-chymotrypsin, catalase, hemoglobin, insulin, myoglobin, thaumatin, trypsin and xylanase.
Structures of TMAO-grown crystals were determined for HEWL (all three forms), trypsin (all three forms), insulin (both forms), xylanase (form 2), thaumatin and MntR. Bound TMAO molecules were found in all trypsin structures and cubic insulin (see below), but not in the other TMAO-grown crystals.
3.2. Cryoprotection
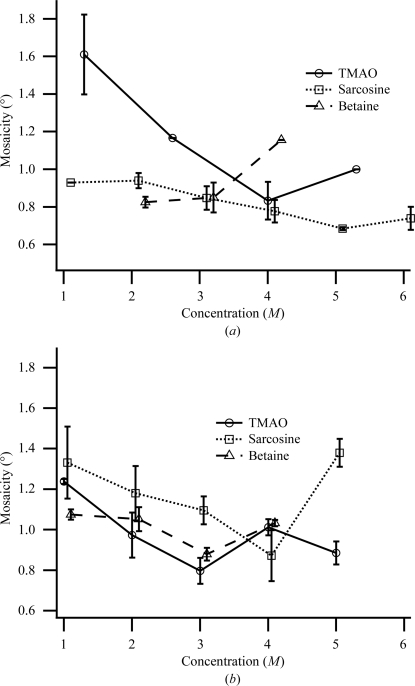
All three osmolytes can act as effective cryoprotective agents in post-crystallization treatment of thermolysin crystals. Figs. 4 ▶ and 5 ▶ show the diffraction properties of the two thermolysin crystal forms with the methylamine osmolytes as cryoprotective agents. In all cases the diffraction improves to a higher resolution limit and lower mosaicity with increasing concentrations of cryoprotective agent up to a certain concentration, at which point the diffraction degrades. In most cases, crystals grown from TMAO could be cryocooled with low mosaicity as grown without further treatment (Tables 3 ▶ and 4 ▶). Physical properties of the methylamine cryoprotective solutions are shown in Table 2 ▶ and are similar to those of solutions of glycerol and ethylene glycol.
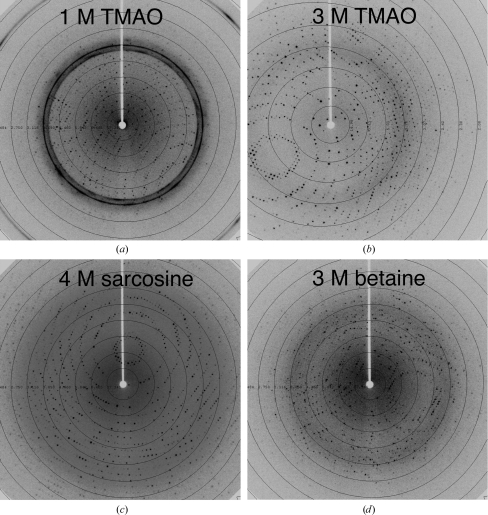
Figure 4.
Cryoprotective efficacy of methylamine osmolytes as post-growth treatments. Shown are diffraction images of cryocooled tetragonal thermolysin after 30–60 s soaks as described in §2. (a) Cryosolution composed of 1 M TMAO, 1 M NaCl and 0.1 M MES (see §2). Powder rings for ice Ic are visible and the protein diffraction limit is 3.3 Å. (b) Cryosolution using 3 M TMAO. The diffraction limit is now approximately 2.1 Å. (c) Cryosolution using 4 M sarcosine, giving a diffraction limit of ∼2.1 Å (1 M sarcosine yielded ∼3.5 Å diffraction). (d) Cryosolution using 3 M betaine, giving a diffraction limit of ∼2.4 Å (1 M betaine yielded ∼3.6 Å diffraction).
Figure 5.
Plots of mosaicity versus concentration of cryoprotective agent for hexagonal (a) and tetragonal (b) thermolysin at 100 K. The plots for sarcosine (squares) and betaine (triangles) are offset horizontally by 0.1 and 0.2 M, respectively. The room-temperature (294 K) mosaicities in the absence of cryoprotective agent are 0.6–0.7°. The error is the average of 2–6 measurements and the error bar is the standard error of the mean.
Table 4. Mosaicity of TMAO-grown crystals upon cryocooling.
| Protein | Crystal form | [TMAO] (M) | η† (°) |
|---|---|---|---|
| Catalase | Orthorhombic | 2.0 | No diffraction |
| Insulin | Hexagonal | 1.0 | 0.81 |
| Cubic | 1.0 | 0.57 | |
| Lysozyme | Tetragonal | 1.0 | 0.82 |
| Orthorhombic | 2.0, 4.0‡ | 0.67 | |
| Monoclinic | 1.0 | 2.03 | |
| Thaumatin | Tetragonal | 3.0 | 0.72 |
| Trypsin | Orthorhombic | 2.0 | 0.87 |
| Tetragonal | 3.8 | 0.63 | |
| Trigonal | 3.0 | 0.78 | |
| Xylanase | Monoclinic (1) | 2.0 | 1.10 |
Average mosaicity from CrysAlisPro (i.e. average of e 1, e 2 and e 3; see headnote to Table 3 ▶).
For orthorhombic lysozyme, two crystals were tested (one at 2 M TMAO and one at 4 M TMAO) and the average η is given. For the other proteins, one crystal was tested.
Monoclinic xylanase with β ≃ 94°.
3.3. Precipitation
Precipitation on adding 100 µl 4.0 M TMAO to 10 µl 1.0 M chloride salt (final concentrations 3.6 M osmolyte/90 mM salt) occurred with FeCl3, CoCl2, CuCl2 and ZnCl2 at pH 9.8, the pH of 4.0 M TMAO dihydrate. At pH 7.0 FeCl3 and CuCl2 precipitated and at pH 4.0 only FeCl3 precipitated. Precipitation also occurred with sulfate and/or acetate salts of the same four cations (Fe3+, Co2+, Cu2+ and Zn2+). Sarcosine and betaine did not precipitate with the same salts under identical concentration and pH conditions, but did cause a color change, turning the solutions of FeCl3 and CuCl2 darker red/brown and blue, respectively.
4. Discussion
4.1. TMAO is an efficient primary precipitant for protein crystal growth
Previous investigations have shown that when used as an additive in crystallization experiments, TMAO can improve crystal order, a consequence of the well known protein structure-stabilizing aspect of this osmolyte (Hill et al., 2002 ▶; Jiang et al., 2006 ▶). The effects of TMAO on protein solubility, however, have been less well studied. Here, we have demonstrated that TMAO can function effectively as the primary precipitant for protein crystal growth, producing crystals of seven of the 14 proteins studied in a total of 13 different crystal forms (Figs. 2 ▶ and 3 ▶). In most cases, the presence of salt additives [(NH4)2SO4, MgCl2, KCl or LiNO3] seemed to increase the probability of crystals appearing rather than precipitate (Fig. 2 ▶). The control screen using ammonium sulfate in place of TMAO produced crystals of nine of the 14 proteins. Thus, the efficiency of TMAO as a crystallization agent is slightly lower than that of ammonium sulfate.
Sarcosine and betaine were less efficient, producing crystals of four and two of the 14 proteins studied, respectively. In most cases, betaine did not cause precipitation during tray setup or after equilibration. The exceptions were insulin and thermolysin. For thermolysin, which has a low solubility in pure water, there was less precipitation at higher betaine concentrations. Sarcosine precipitated about half of the proteins, including thermolysin, with more precipitate at higher sarcosine concentrations.
Methylamine osmolytes are depleted from the protein backbone by about 50% relative to the bulk solution concentration, which stabilizes the more compact folded form of the protein (Bolen, 2004 ▶; Street et al., 2006 ▶; Hu et al., 2010 ▶). This osmolyte exclusion from the protein backbone should also reduce protein solubility, since crystallization involves burying at least some backbone groups at crystal contacts. However, side-chain interactions must also be considered (Auton et al., 2011 ▶). For example, proline is also depleted from backbone atoms, but makes favorable side-chain interactions which dominate, thus increasing solubility (Bolen, 2004 ▶). TMAO, on the other hand, has only marginally favorable side-chain interactions. The net result is overall unfavorable interactions between TMAO and the folded protein, which decreases the solubility of the protein relative to pure water.
Betaine behaves similarly to proline, being strongly excluded from the protein backbone, but with more favorable side-chain interactions than TMAO, especially with basic side chains. Betaine is therefore a protein solubilizer, as has been observed experimentally (Paleg et al., 1984 ▶). Sarcosine, while being similar in structure to betaine, is expected to reduce protein solubility nearly as effectively as TMAO and should be a protein precipitant (Auton et al., 2011 ▶). Sarcosine is strongly excluded from the backbone but interacts less favourably with side chains than betaine. As a crystallization agent, sarcosine was intermediate in effectiveness between betaine and TMAO. Below, we discuss some specific aspects of the proteins studied.
4.1.1. Insulin
The TMAO screens produced two crystal forms of porcine insulin, rhombohedral (R3; unit-cell parameters a = 82.3, c = 34.1 Å) and cubic (I213; a = 78.9 Å), both of which diffracted beyond 2.0 Å resolution. Insulin crystals have been grown from a variety of conditions, generally using salts including phosphate, citrate and sodium chloride (Harding et al., 1966 ▶; Gursky et al., 1992 ▶; Mueller-Dieckmann et al., 2007 ▶). Here, the rhombohedral form appeared in a large range of conditions, more readily at lower pH values closer to the pI of 5.3. Structure determination showed the rhombohedral form to be the 2Zn variety (Harding et al., 1966 ▶) with two bound zinc ions along a threefold crystal axis, although the crystallization buffer included no supplemental zinc. Cubic insulin crystals are commonly grown in the presence of EDTA to chelate zinc ions, which are absent from the cubic lattice. Here, the cubic form appeared sporadically throughout the screens. No TMAO molecules were observed in the rhombohedral form (based on a 294 K 2.0 Å resolution X-ray data set from a 1 M TMAO-grown crystal), but in the cubic form (294 K 1.8 Å resolution data set from a 1.2 M TMAO-grown crystal) a TMAO molecule was found at a crystal contact on a threefold axis in a location normally occupied by five water molecules.
4.1.2. Catalase
Bovine liver catalase crystals have been grown using ammonium sulfate and polyethylene glycol as precipitants (Ko et al., 1999 ▶; Mueller-Dieckmann et al., 2011 ▶; Purwar et al., 2011 ▶). As with insulin, the TMAO-grown crystals appeared more readily at lower pH values closer to the pI of 5.4. The crystals were orthorhombic, diffracting to 3.5 Å resolution, with unit-cell parameters a = 88, b = 141, c = 231 Å, similar to previously reported catalase crystals. A full X-ray diffraction data set was not collected from the catalase crystals owing to the relatively poor diffraction.
4.1.3. MntR
Crystals of the manganese transport regulator (MntR) are typically grown using PEG 4000 or PEG 400 as the precipitating agent (Kliegman et al., 2006 ▶). Here, MntR crystals appeared in both TMAO screens, principally at lower concentrations of TMAO (space group P21; a = 50.5, b = 46.5, c = 75.7 Å; diffraction to 2.5 Å resolution). Two manganese coordination geometries called ‘AB’ and ‘AC’ have previously been reported for MntR and seem to be dependent on the temperature of X-ray data collection. Structure determination to 2.8 Å resolution at 294 K of a crystal grown in 0.6 M TMAO confirmed the expected AC coordination geometry, while no TMAO molecules were observed bound to the crystal.
4.1.4. Thaumatin
Tetragonal thaumatin is normally grown using a variety of precipitating agents in the presence of tartrate (McPherson, 2001 ▶). This was also true for TMAO. With 0.1 M tartrate present, TMAO was necessary and sufficient to produce thaumatin crystals (space group P41212; a = 58.5, c = 151.8 Å; diffraction beyond 2.0 Å resolution), but without tartrate no crystals were produced. A 1.8 Å resolution structure was determined from a crystal grown in 2 M TMAO and flash-cooled directly from the crystallization drop into the cryostream; no bound TMAO molecules were observed.
4.1.5. Xylanase II
Xylanase II crystals are typically grown using ammonium sulfate as the precipitant (Törrönen & Rouvinen, 1995 ▶; Moukhametzianov et al., 2008 ▶). Two monoclinic crystal forms of xylanase appeared in the TMAO screens under a variety of conditions. The first form (space group P21; a = 56.3, b = 40.1, c = 82.3 Å, β = 94.7°), resembling the published coordinates of PDB entry 1xyo, were thick plates (Fig. 3 ▶) that appeared only in the presence of the salt additives. They diffracted to maximum resolution of 2.0 Å. The second form, with larger β angle (space group P21; a = 40.9, b = 39.3, c = 57.4 Å, β = 110.8), resembled the published coordinates of PDB entry 2jic, and were thin plates (Fig. 3 ▶) that appeared with or without salt additives. They diffracted to about 2.5 Å resolution. A structure of the first form was determined to 2.2 Å resolution at 294 K from a 1 M TMAO-grown crystal and no TMAO molecules were found bound to the crystal.
4.1.6. Lysozyme
Tetragonal HEWL crystals have previously been obtained using chloride, bromide, nitrate and sulfate anions as precipitating agents (Forsythe et al., 1999 ▶; Pusey & Nadarajah, 2002 ▶). Fig. 2 ▶ shows that tetragonal crystals appear broadly across Screen 2 with chloride, nitrate and sulfate, but interestingly they also appear in the TMAO–pH screen. Under these conditions, the crystallization solution contains only TMAO and 0.1 M buffer, so the only anions present should be those from the buffer.
Orthorhombic lysozyme crystals usually appear under similar conditions to tetragonal lysozyme crystals, but at higher temperature and higher protein concentrations (Forsythe et al., 1999 ▶; Pusey & Nadarajah, 2002 ▶). Here, higher concentrations of TMAO as well as a larger drop size favored orthorhombic crystals.
Monoclinic lysozyme crystals have been obtained using nitrate, iodide and thiocyanate anions (Pusey & Nadarajah, 2002 ▶). Here, the monoclinic form appeared in control screens without TMAO. The monoclinic crystals could be grown in a few hours to 1 d using 0.2–0.4 M LiNO3 at pH 9 with protein at 40–80 mg ml−1. With 1 M TMAO present under otherwise identical conditions, the crystals appeared at the same time to 1 d sooner than without TMAO.
Structures were determined of all three lysozyme crystal forms at 294 K (tetragonal, 1.8 Å resolution, 1 M TMAO; orthorhombic, 2.1 Å resolution, 2 M TMAO; monoclinic, 1.7 Å resolution, 1 M TMAO) and no bound TMAO molecules were observed.
4.1.7. Trypsin
Bovine trypsin appeared in three crystal forms. The orthorhombic form is commonly grown using ammonium sulfate or PEG (Kurinov & Harrison, 1994 ▶; Leiros et al., 2001 ▶). Here, the orthorhombic form occurred under a variety of conditions. The trigonal form of trypsin is normally grown with ammonium sulfate and often appears in the same drop as the orthorhombic form, which was also true with TMAO as precipitant. The refined structures of the orthorhombic and trigonal TMAO-grown crystals showed several TMAO molecules bound to the protein. In contrast, in a recent report trigonal trypsin crystals showed no bound TMAO molecules after a 10 s soak in 4 M TMAO (Mueller-Dieckmann et al., 2011 ▶).
The third crystal form of trypsin produced with TMAO was a tetragonal form (space group P41212; a = 53.9, c = 181.9 Å; 57% solvent content), which took longer to appear than the orthorhombic form, growing to maximum dimensions of 0.5 × 0.2 × 0.2 mm in about two weeks and typically diffracting to about 2.8 Å resolution at both 294 K and when directly cooled to 100 K as-grown. Another tetragonal form of trypsin has previously been reported with similar unit-cell parameters (Tsunogae et al., 1986 ▶; Koepke et al., 2000 ▶; PDB entry 1tab; space group P41212; a = 55.4, c = 181.7 Å; 48% solvent content) but with trypsin in complex with a 97-amino-acid inhibitor (the Bowman–Birk inhibitor, BBI) and crystallized using ammonium sulfate and magnesium sulfate. The crystal packing in the TMAO-grown tetragonal trypsin crystals is quite different from that of the trypsin–BBI complex. About 40 and 50 residues make 4.0 Å trypsin–trypsin crystal contacts in the TMAO-grown trypsin and the trypsin–BBI crystals, respectively, but only about 20 of these residues are in common. It is therefore coincidental that the two crystal forms occur in the same space group with similar unit-cell parameters. TMAO thus produced a new form of the commonly crystallized protein trypsin.
The three trypsin crystal forms produced with TMAO, orthorhombic, trigonal and tetragonal, have four, six and eight molecules per unit cell, respectively. There is one crystal contact in common amongst the three crystal forms, creating a crystallographic dimer that appears in all three forms and binds a TMAO molecule. There are TMAO molecules involved in other crystal contacts, but none offer a ready explanation for the growth of the tetragonal crystal form with TMAO.
4.2. Methylamines as cryoprotective agents
Figs. 4 ▶ and 5 ▶ show that the three methylamine osmolytes TMAO, sarcosine and betaine are all effective post-crystallization cryoprotective agents. The cryocooled crystals diffracted poorly at 1 M osmolyte but to higher resolution and lower mosaicity at higher osmolyte concentrations, indicating that (i) the crystals require cryoprotective agents for successful cooling and (ii) the osmolytes are effective in the role of cryoprotection. It has recently been reported that TMAO can act as an effective cryoprotective agent (Mueller-Dieckmann et al., 2011 ▶). Here, we have demonstrated the effectiveness of TMAO for two additional crystals and that the related osmolytes sarcosine and betaine are also effective cryoprotective agents.
In post-crystallization soaks of tetragonal thermolysin in TMAO, a precipitation reaction occurred which was traced to TMAO and zinc chloride. Subsequent experiments showed that precipitation occurs with TMAO in the presence of salts of Fe3+, Co2+, Cu2+ or Zn2+ and that the precipitation reactions diminish with lower pH. TMAO-induced precipitation of iron is thought to be involved in iron deficiency in mink fed certain marine fish high in TMAO (Cheeke & Dierenfeld, 2010 ▶). Color changes of Cu2+ and Fe3+ solutions when mixed with sarcosine or betaine suggest changes in the metal ligands or a change of the oxidation state of the metal. The structure of tetragonal thermolysin in the presence of betaine showed direct interactions between zinc ions and betaine carboxylate groups (see below). Because they did not precipitate with the metal-salt solutions, sarcosine and betaine are good alternative cryoprotective agents to TMAO if metal ions are present.
TMAO-grown crystals could also be cryocooled in situ without further treatment (Tables 3 ▶ and 4 ▶; P41212 trypsin and P3121 trypsin). Since in some cases crystals appear at relatively low TMAO concentrations (i.e. < 2 M; Fig. 2 ▶) cryocooling may be improved by equilibrating the TMAO-grown crystal with higher TMAO concentrations prior to cooling or by using an external oil to eliminate ice formation on the outside of the crystal (Kwong & Liu, 1999 ▶).
TMAO prevented the formation of cubic ice at the lowest concentration (3.0 M) of the cryoprotective agents tested (Table 2 ▶). With respect to cryosolution thermal contraction, TMAO was similar to ethylene glycol, while sarcosine and betaine were closer to glycerol.
4.3. Osmolyte binding
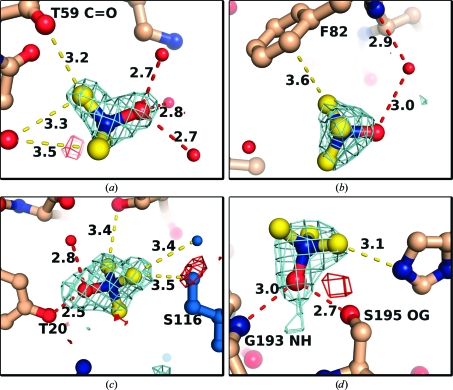
Several studies have shown that TMAO and other stabilizing osmolytes are excluded from the protein surface (Lee & Timasheff, 1981 ▶; Lin & Timasheff, 1994 ▶; Bolen, 2004 ▶). Since this exclusion is relative to bulk solvent, we may still expect to find TMAO molecules in the vicinity of the protein. For example, a simple model predicts that in 1 M TMAO the local TMAO concentration near backbone groups should be about 0.5 M (Street et al., 2006 ▶). There were 12 unique places that TMAO molecules were bound in the five crystals (the three crystal forms of trypsin, cubic insulin and thermolysin), including seven at crystal contacts. The pseudotetrahedral symmetry of the TMAO molecule made it necessary to deduce the binding orientation from interaction partners, which was possible in most cases. The oxide group forms polar contacts with water molecules (Figs. 6 ▶ a, 6 ▶ b and 6 ▶ c), protein side chains (Figs. 6 ▶ c and 6 ▶ d) and backbone amides (Fig. 6 ▶ d). In about half of the cases the TMAO molecule appears to interact directly with only solvent molecules. The methyl groups, which each carry a partial positive charge (Zou et al., 2002 ▶), sometimes make van der Waals contacts with a variety of protein atoms, including backbone carbonyl groups, which bear a partial negative charge (Fig. 6 ▶ a). In other cases, the trimethyl group projects into bulk solvent (Figs. 6 ▶ b and 6 ▶ d).
Figure 6.
Examples of bound TMAO molecules. Red, oxygen; blue, nitrogen; yellow or wheat, carbon; light blue, symmetry molecule. Numbers indicate contact distances in Å. Red dashed lines, contacts of 3.0 Å or less. Yellow dashed lines, contacts of >3.0 Å. The maps show OMIT DELFWT/PHDELFWT electron density from REFMAC contoured at +3σ/−3σ (cyan/red) (Murshudov et al., 2011 ▶). (a–c) Examples from trigonal trypsin at 1.75 Å resolution. The maps were calculated after ten cycles of restrained refinement of the final model with the TMAO molecules removed. (d) Example from orthorhombic trypsin at 1.70 Å resolution. The map was calculated after restrained refinement of the initial model with waters in the vicinity of the TMAO molecules removed but before building TMAO molecules into the model. This figure was prepared with PyMOL (Schrödinger LLC).
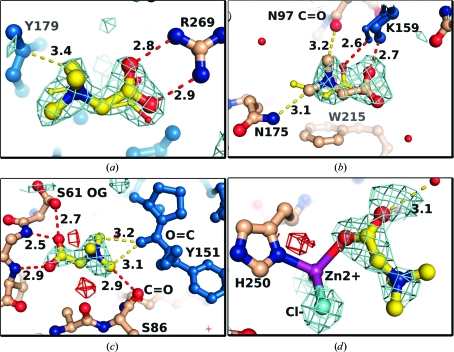
Structures were determined of proteins in the presence of sarcosine and betaine for comparison with TMAO binding. Orthorhombic trypsin and tetragonal thermolysin crystals were grown using previously published conditions and soaked in solutions of sarcosine and betaine prior to X-ray data collection (see §2). Trypsin and thermolysin bound four and six betaine molecules, respectively. In each case, the betaine carboxylate group contacts polar atoms on the protein, including arginine (Fig. 7 ▶ a), lysine (Fig. 7 ▶ b) and histidine side chains and backbone amide groups (Fig. 7 ▶ c) as well as water molecules (Fig. 7 ▶ d). In one case the betaine molecule interacts directly with primarily water molecules. Interactions between carboxylate groups of betaine and basic side chains are consistent with a free-energy transfer model in which the increase in protein solubility in the presence of betaine is largely a consequence of interactions between betaine and charged side chains (Auton et al., 2011 ▶). As with TMAO, the betaine methyl groups make van der Waals contacts with some carbonyl atoms (Figs. 7 ▶ b and 7 ▶ c). Two of the betaine molecules bound to thermolysin interact with bound zinc ions directly via their carboxylate groups: one in the active site and another in a different region of the protein (Fig. 7 ▶ d). In the sarcosine-soaked crystals, five sarcosine molecules were observed, all of which were bound at betaine sites with similar osmolyte–protein interactions (Figs. 7 ▶ a and 7 ▶ c).
Figure 7.
Examples of bound betaine and sarcosine molecules. The color scheme is identical to that in Fig. 6 ▶. Cyan spheres, Cl− ions. Large sticks, betaine. Small sticks, sarcosine. The maps show OMIT DELFWT/PHDELFWT electron density from REFMAC contoured at +3σ/−3σ (cyan/red) (Murshudov et al., 2011 ▶). The maps were calculated after restrained refinement of the initial model with water molecules in the vicinity of the ligands removed but before building betaine molecules into the model. (a, c) Two examples from tetragonal thermolysin at 2.00 Å resolution (betaine) and 2.10 Å resolution (sarcosine). (b, d) Two examples from orthorhombic trypsin at 1.95 Å resolution (betaine) and 1.70 Å resolution (sarcosine). This figure was prepared with PyMOL (Schrödinger LLC).
4.4. Stabilizing methylamine osmolytes for protein crystal growth and cryoprotection
TMAO is a crystallization agent with unique chemical and physical properties. Like many precipitants, it reduces protein solubility via an exclusion mechanism, which for TMAO is based on the preference of backbone atoms to interact with water rather than osmolyte (Bolen & Rose, 2008 ▶). In contrast, the exclusion mechanism for polyethylene glycol is based on excluded volume (Arakawa & Timasheff, 1985 ▶). Unlike some precipitants (e.g. MPD and PEG), TMAO increases protein stability (Yancey & Somero, 1979 ▶; Bolen, 2004 ▶). Above pH ∼4.6–4.8 TMAO is zwitterionic, a feature shared with none of the commonly used precipitating agents. TMAO produced diffraction-quality crystals of proteins normally grown using either salt or polyethylene glycol as precipitants. Furthermore, TMAO was able to produce a heretofore unreported crystal form of trypsin. The binding of TMAO at crystal contacts suggests that this osmolyte may not only be able to provide the overall thermodynamic driving force for crystallization, but may also be able to provide specificity in mediating crystal contacts to yield new crystal forms. Sarcosine and betaine also produced crystals, but less effectively than TMAO.
For cryoprotection, both protein stability and solubility are important. Ideally, the cryoprotective agent will increase stability and decrease solubility, but both criteria are not always met. Some cryoprotective agents destabilize proteins [e.g. low-molecular-weight PEGs (Arakawa & Timasheff, 1985 ▶) and DMSO (Arakawa et al., 2007 ▶)]. Glycerol, while being a known protein stabilizer (Vagenende et al., 2009 ▶), increases solubility (Zukoski & Farnum, 1999 ▶; Schall et al., 2009 ▶; Auton et al., 2011 ▶) and sometimes dissolves crystals when used as a post-crystallization cryoprotective agent. Other cryoprotective agents that solubilize proteins include sucrose (Henry et al., 2005 ▶) and ethylene glycol (Zukoski & Kulkarni, 2002 ▶). Here, all three methylamine osmolytes were effective cryoprotective agents and all three increase protein stability. Betaine, however, increases solubility (Paleg et al., 1984 ▶) and should therefore tend to dissolve crystals, similar to the effect of glycerol. Here, the betaine concentrations (3–4 M) tolerated by thermolysin crystals were lower than for sarcosine or TMAO (5–6 M), but were adequate for successful cryocooling. TMAO and sarcosine reduce solubility and should therefore stabilize both the crystal and the protein when used as post-crystallization cryoprotective agents.
5. Conclusions
The stabilizing osmolyte TMAO is an efficient primary precipitant for protein crystal growth, providing new routes for crystallographers to find crystallization conditions. All three methylamine osmolytes explored (TMAO, sarcosine and betaine) can function as effective cryoprotective agents. In cases where transition-metal ions are present, the use of TMAO as cryoprotective agent may produce precipitation reactions, which can avoided by using sarcosine or betaine. The combined properties of protein stabilization and cryoprotection give the methylamine osmolytes in general, and owing to its efficient crystallization capabilities TMAO in particular, great potential as tools for macromolecular crystallography.
Supplementary Material
PDB reference: trypsin, TMAO-grown, tetragonal, 3t28
PDB reference: TMAO-grown, trigonal, 3t29
PDB reference: TMAO-grown, orthorhombic, 3t25
PDB reference: in the presence of sarcosine, 3t26
PDB reference: in the presence of betaine, 3t27
PDB reference: insulin, TMAO-grown, 3t2a
PDB reference: thermolysin, in the presence of TMAO, 3t2h
PDB reference: in the presence of sarcosine, 3t2i
PDB reference: in the presence of betaine, 3t2j
Acknowledgments
This work was supported in part by NIH Grant GM090248 (DHJ) and in part by a grant from HHMI to Whitman College. We would like to thank Paul Yancey for helpful discussions.
References
- Alcorn, T. & Juers, D. H. (2010). Acta Cryst. D66, 366–373. [DOI] [PMC free article] [PubMed]
- Arakawa, T., Kita, Y. & Timasheff, S. N. (2007). Biophys. Chem. 131, 62–70. [DOI] [PubMed]
- Arakawa, T. & Timasheff, S. N. (1985). Biochemistry, 24, 6756–6762. [DOI] [PubMed]
- Auton, M., Rösgen, J., Sinev, M., Holthauzen, L. M. F. & Bolen, D. W. (2011). Biophys. Chem. 159, 90–99. [DOI] [PMC free article] [PubMed]
- Bentley, G. A., Brange, J., Derewenda, Z., Dodson, E. J., Dodson, G. G., Markussen, J., Wilkinson, A. J., Wollmer, A. & Xiao, B. (1992). J. Mol. Biol. 228, 1163–1176. [DOI] [PubMed]
- Biswal, B. K., Sukumar, N. & Vijayan, M. (2000). Acta Cryst. D56, 1110–1119. [DOI] [PubMed]
- Bolen, D. W. (2004). Methods, 34, 312–322. [DOI] [PubMed]
- Bolen, D. W. & Rose, G. D. (2008). Annu. Rev. Biochem. 77, 339–362. [DOI] [PubMed]
- Cheeke, P. R. & Dierenfeld, E. S. (2010). Comparative Animal Nutrition and Metabolism Cambridge: CABI.
- Dawson, R. M. C., Elliott, D. C., Elliott, W. H. & Jones, K. M. (1959). Editors. Data for Biochemical Research Oxford University Press.
- Dong, J., Boggon, T. J., Chayen, N. E., Raftery, J., Bi, R.-C. & Helliwell, J. R. (1999). Acta Cryst. D55, 745–752. [DOI] [PubMed]
- Dumetz, A. C., Chockla, A. M., Kaler, E. W. & Lenhoff, A. M. (2008). Cryst. Growth Des. 9, 682–691.
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Forsythe, E. L., Snell, E. H., Malone, C. C. & Pusey, M. L. (1999). J. Cryst. Growth, 196, 332–343.
- Glasfeld, A., Guedon, E., Helmann, J. D. & Brennan, R. G. (2003). Nature Struct. Biol. 10, 652–657. [DOI] [PubMed]
- Gursky, O., Li, Y., Badger, J. & Caspar, D. L. (1992). Biophys. J. 61, 604–611. [DOI] [PMC free article] [PubMed]
- Harding, M. M., Hodgkin, D. C., Kennedy, A. F., O’Conor, A. & Weitzmann, P. D. (1966). J. Mol. Biol. 16, 212–226. [DOI] [PubMed]
- Hausrath, A. C. & Matthews, B. W. (2002). Acta Cryst. D58, 1002–1007. [DOI] [PubMed]
- Henry, C. S., Valente, J. J., Verma, K. S., Manning, M. C. & Wilson, W. W. (2005). Biophys. J. 89, 4211–4218. [DOI] [PMC free article] [PubMed]
- Hill, C. M., Bates, I. R., White, G. F., Hallett, F. R. & Harauz, G. (2002). J. Struct. Biol. 139, 13–26. [DOI] [PubMed]
- Hochachka, P. W. & Somero, G. N. (2002). Biochemical Adaptation. Oxford University Press.
- Holland, D. R., Tronrud, D. E., Pley, H. W., Flaherty, K. M., Stark, W., Jansonius, J. N., McKay, D. B. & Matthews, B. W. (1992). Biochemistry, 31, 11310–11316. [DOI] [PubMed]
- Hu, C. Y., Lynch, G. C., Kokubo, H. & Pettitt, B. M. (2010). Proteins, 78, 695–704. [DOI] [PMC free article] [PubMed]
- Jiang, J., Lafer, E. M. & Sousa, R. (2006). Acta Cryst. F62, 39–43. [DOI] [PMC free article] [PubMed]
- Juers, D. H., Jacobson, R. H., Wigley, D., Zhang, X.-J., Huber, R. E., Tronrud, D. E. & Matthews, B. W. (2000). Protein Sci. 9, 1685–1699. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2001). International Tables for Crystallography, Vol. F, edited by M. G. Rossmann & E. Arnold, pp. 218–225. Dordrecht: Kluwer Academic Publishers.
- Katz, B. A., Clark, J. M., Finer-Moore, J. S., Jenkins, T. E., Johnson, C. R., Ross, M. J., Luong, C., Moore, W. R. & Stroud, R. M. (1998). Nature (London), 391, 608–612. [DOI] [PubMed]
- Kliegman, J. I., Griner, S. L., Helmann, J. D., Brennan, R. G. & Glasfeld, A. (2006). Biochemistry, 45, 3493–3505. [DOI] [PMC free article] [PubMed]
- Ko, T.-P., Day, J., Malkin, A. J. & McPherson, A. (1999). Acta Cryst. D55, 1383–1394. [DOI] [PubMed]
- Koepke, J., Ermler, U., Warkentin, E., Wenzl, G. & Flecker, P. (2000). J. Mol. Biol. 298, 477–491. [DOI] [PubMed]
- Kurinov, I. V. & Harrison, R. W. (1994). Nature Struct. Biol. 1, 735–743. [DOI] [PubMed]
- Kwong, P. D. & Liu, Y. (1999). J. Appl. Cryst. 32, 102–105.
- Lee, J. C. & Timasheff, S. N. (1981). J. Biol. Chem. 256, 7193–7201. [PubMed]
- Leiros, H.-K. S., McSweeney, S. M. & Smalås, A. O. (2001). Acta Cryst. D57, 488–497. [DOI] [PubMed]
- Lin, T. Y. & Timasheff, S. N. (1994). Biochemistry, 33, 12695–12701. [DOI] [PubMed]
- Marquart, M., Walter, J., Deisenhofer, J., Bode, W. & Huber, R. (1983). Acta Cryst. B39, 480–490.
- McPherson, A. (1999). Cryst. Growth Des. 9, 682–691.
- McPherson, A. (2001). Protein Sci. 10, 418–422. [DOI] [PMC free article] [PubMed]
- McPherson, A. & Cudney, B. (2006). J. Struct. Biol. 156, 387–406. [DOI] [PubMed]
- Moukhametzianov, R., Burghammer, M., Edwards, P. C., Petitdemange, S., Popov, D., Fransen, M., McMullan, G., Schertler, G. F. X. & Riekel, C. (2008). Acta Cryst. D64, 158–166. [DOI] [PMC free article] [PubMed]
- Mueller-Dieckmann, C., Kauffmann, B. & Weiss, M. S. (2011). J. Appl. Cryst. 44, 433–436.
- Mueller-Dieckmann, C., Panjikar, S., Schmidt, A., Mueller, S., Kuper, J., Geerlof, A., Wilmanns, M., Singh, R. K., Tucker, P. A. & Weiss, M. S. (2007). Acta Cryst. D63, 366–380. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Paleg, L. G., Stewart, G. R. & Bradbeer, J. W. (1984). Plant Physiol. 75, 974–978. [DOI] [PMC free article] [PubMed]
- Pechkova, E., Tripathi, S., Ravelli, R. B., McSweeney, S. & Nicolini, C. (2009). J. Struct. Biol. 168, 409–418. [DOI] [PubMed]
- Purwar, N., McGarry, J. M., Kostera, J., Pacheco, A. A. & Schmidt, M. (2011). Biochemistry, 50, 4491–4503. [DOI] [PMC free article] [PubMed]
- Pusey, M. L. & Nadarajah, A. (2002). Cryst. Growth Des. 2, 475–483.
- Qu, Y. & Bolen, D. W. (2003). Biochemistry, 42, 5837–5849. [DOI] [PubMed]
- Rao, S. T. & Sundaralingam, M. (1996). Acta Cryst. D52, 170–175. [DOI] [PubMed]
- Santoro, M. M., Liu, Y., Khan, S. M., Hou, L. X. & Bolen, D. W. (1992). Biochemistry, 31, 5278–5283. [DOI] [PubMed]
- Sauter, C., Lorber, B. & Giegé, R. (2002). Proteins, 48, 146–150. [DOI] [PubMed]
- Schall, C. A., Gosavi, R. A., Bhamidi, V. & Varanasi, S. (2009). Langmuir, 25, 4579–4587. [DOI] [PubMed]
- Schutt, C. E. & Winkler, F. K. (1977). The Rotation Method in Crystallography, edited by U. W. Arndt & A. J. Wonacott, pp. 173–186. Amsterdam: North-Holland Publishing Company.
- Street, T. O., Bolen, D. W. & Rose, G. D. (2006). Proc. Natl Acad. Sci. USA, 103, 13997–14002. [DOI] [PMC free article] [PubMed]
- Törrönen, A. & Rouvinen, J. (1995). Biochemistry, 34, 847–856. [DOI] [PubMed]
- Tsunogae, Y., Tanaka, I., Yamane, T., Kikkawa, J., Ashida, T., Ishikawa, C., Watanabe, K., Nakamura, S. & Takahashi, K. (1986). J. Biochem. 100, 1637–1646. [DOI] [PubMed]
- Vagenende, V., Yap, M. G. & Trout, B. L. (2009). Biochemistry, 48, 11084–11096. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yancey, P. H. (2005). J. Exp. Biol. 208, 2819–2830. [DOI] [PubMed]
- Yancey, P. H., Clark, M. E., Hand, S. C., Bowlus, R. D. & Somero, G. N. (1982). Science, 217, 1214–1222. [DOI] [PubMed]
- Yancey, P. H. & Somero, G. N. (1979). Biochem. J. 183, 317–323. [DOI] [PMC free article] [PubMed]
- Zou, Q., Bennion, B. J., Daggett, V. & Murphy, K. P. (2002). J. Am. Chem. Soc. 124, 1192–1202. [DOI] [PubMed]
- Zukoski, C. & Farnum, M. (1999). Biophys. J. 76, 2716–2726. [DOI] [PMC free article] [PubMed]
- Zukoski, C. F. & Kulkarni, A. M. (2002). Langmuir, 18, 3090–3099.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: trypsin, TMAO-grown, tetragonal, 3t28
PDB reference: TMAO-grown, trigonal, 3t29
PDB reference: TMAO-grown, orthorhombic, 3t25
PDB reference: in the presence of sarcosine, 3t26
PDB reference: in the presence of betaine, 3t27
PDB reference: insulin, TMAO-grown, 3t2a
PDB reference: thermolysin, in the presence of TMAO, 3t2h
PDB reference: in the presence of sarcosine, 3t2i
PDB reference: in the presence of betaine, 3t2j