Interleukin-10 (IL-10) is an immunoregulatory cytokine that influences the clinical outcome of chronic viral infections. Results show that IL-10-promoter genetic variants influence human immunodeficiency virus-1 pathogenesis possibly via regulating IL-10 plasma levels and the breadth of CD8+ T-cell immune responses.
Abstract
Background. Interleukin-10 (IL-10) is a potent immunoregulatory cytokine. IL-10-promoter polymorphisms have been shown to affect human immunodeficiency virus type 1 (HIV-1) clinical outcomes but the underlying mechanisms are poorly understood.
Methods. We investigated the relationship between IL-10-promoter variants, plasma cytokine levels, immune responses and markers of disease outcome in antiretroviral-naïve HIV-1 chronically infected individuals from South Africa. Two IL-10-promoter single nucleotide polymorphisms (SNPs) were genotyped in 451 participants. Baseline plasma levels of select cytokines were measured for 112 individuals. Viral load, CD4+ T-cell counts and HIV-1-specific interferon-gamma CD8+ T-cell immune responses were measured at baseline. CD4+ T-cell counts were measured longitudinally and rates of CD4+ T-cell decline computed for 300 study subjects.
Results. The minor IL-10-1082G and -592A variants occurred at frequencies of 0.31 and 0.34, respectively. The -592AA genotype associated significantly with attenuated loss of CD4+ T cells (P = .0496). Individuals possessing -1082GG had significantly higher IL-10 levels compared to -1082AA/AG (P = .0006). The -592AA genotype was associated with greater breadth of virus-specific CD8+ T-cell responses compared to CC and CA (P = .002 and .004 respectively).
Conclusions. IL-10-promoter variants may influence the rate of HIV-1 disease progression by regulating IL-10 levels and the breadth of CD8+ T-cell immune responses.
It is now approximately 3 decades since the human immunodeficiency virus type 1 (HIV-1) was first described, and the virus has since spread to become a pandemic with high morbidity and mortality. Almost two-thirds of the world’s HIV-infected individuals are found in sub–Saharan Africa, including South Africa [1]. Although antiretroviral drugs are now widely available for the clinical management of HIV-1 infection, significant challenges in the roll-out and subsequent lifelong use of these drugs remain, and it is unlikely that the spread of HIV-1 will be substantially curtailed without a preventive vaccine or immunotherapy. However, vaccine development efforts have been significantly hampered by the limited knowledge of the biological factors that underlie HIV-1 pathogenesis, and so far, only modest success or failure has been achieved with candidate HIV-1 vaccines that have undergone large-scale human trials [2–4]. The considerable variation in the natural history of HIV/AIDS and disproportionate global distribution of HIV infection present an opportunity to study biological factors that may influence HIV/AIDS pathogenesis, and have the potential to be manipulated for effective vaccination or therapy [5, 6].
Geographical differences of host genetic factors that influence HIV/AIDS exposure or infection are well illustrated in the distribution of the CCR5 chemokine receptor mutation CCR5-Δ32, which has been associated with protection against HIV-1 infection by R5 HIV-1 variants [7, 8]. Studies have shown that the frequency of the CCR5-Δ32 alleles is higher among Northern Europeans, decreasing geographically further south [9]. Likewise, the HLA locus, which codes for HLA class I molecules (which function to present pathogen-derived peptides on the cell surface of infected cells for recognition by CD8+ T lymphocytes [5, 9]), displays significant natural variation among geographically diverse populations, and has been associated with differences in HIV/AIDS disease progression and natural viral control [9–11].
Interleukin-10 (IL-10) is a potent anti-inflammatory cytokine, which plays a key role in regulating the immune response [12, 13]. IL-10 has also been shown to downregulate the expression of proinflammatory cytokines as well as the expression of major histocompatibility complex class I and II molecules [14–17]. Previous studies have focused on the 3 classic single-nucleotide polymorphisms (SNPs) found in the proximal promoter region [18–21]. These polymorphisms are found at positions -1082 (rs1800896), an A to G transition; -819 (rs1800871), a C to T transition; and -592 (rs1800872), a C to A transversion. The 819 and -592 mutations are in complete linkage disequilibrium. IL-10 variants are associated with differential IL-10 production [19, 22–24]; -1082G with high IL-10 production and the -592A with low IL-10 production. Genetic association data suggests that IL-10 variant -592A, a low IL-10 producer variant, is linked with increased susceptibility to HIV-1 infection and an accelerated progression to AIDS, particularly in the late stages of the disease among European Americans [20, 21, 25]. Consistent with these data, it was demonstrated in an African cohort that survival was doubled in carriers of the IL-10-1082G allele, which is associated with increased IL-10 production [18]. Therefore, there is evidence that suggests that IL-10 polymorphisms associated with increased IL-10 production have a protective role against disease progression, possibly by decreasing the chronic immune activation that is a major factor in HIV pathogenesis.
On the other hand, we have recently shown in a cohort of high-risk black African women that although the IL-10 polymorphisms -1082AA and -592AA associated with decreased IL-10 production were overrepresented among seroconverters compared with those who remained HIV-1 negative in longitudinal follow-up, these polymorphisms also associated with high viral loads and low CD4 counts during the acute/early phases of infection, suggesting that the effects of IL-10 polymorphisms may be infection-phase dependent and that high IL-10 levels during the early phase may be detrimental [26]. Mechanistic studies of the lymphocytic choriomeningitis virus (LCMV) in mice showed that IL-10 gene knock-out or signaling blockade enhanced T-cell immune responses, resulting in rapid viral elimination and the development of antiviral memory T-cell responses [27, 28]. We have also shown in vitro that IL-10 blockade in peripheral blood mononuclear cells (PBMCs) from HIV-infected individuals resulted in restoration of proliferative and effector CD4 T-cell function [29]. IL-10 is also reported to enhance detrimental deletion of dendritic cells by natural killer cells, further exacerbating immune dysfunction in chronic HIV-1 infection [30]. Taken together, these studies suggest a complicated but significant role for IL-10 in viral pathogenesis, and considering that manipulation of the IL-10 pathway to boost antiviral immune responses and improve vaccine effectiveness has been suggested, there is a clear and urgent need to better decipher underlying mechanisms of pathogenesis for this immunoregulatory cytokine, particularly in geographical regions most severely affected by the HIV-1 epidemic, as this may have implications for immunotherapeutic strategies and vaccine design.
Therefore, the purpose of this study was to investigate the frequency of IL-10-promoter polymorphisms in a large cohort of antiretroviral-naive chronically HIV-1-infected predominantly Zulu/Xhosa individuals to determine whether these polymorphisms affect markers of HIV-1 disease progression; namely, viral load, CD4+ T-cell counts, and the rate of CD4+ T-cell decline. We also wanted to determine whether these polymorphisms are associated with differential levels of select pro- and anti-inflammatory plasma cytokines. Furthermore, we sought to explore the link between these polymorphisms and levels of cytotoxic T-cell immune responses, which have been shown to play a role in the control of HIV-1 replication. Our data suggest that IL-10-promoter polymorphisms may modulate HIV-1 pathogenesis, possibly through effects on plasma IL-10 levels and the breadth of immune responses, among other mechanisms.
MATERIALS AND METHODS
Study Population, Materials, and Methods
The HIV Pathogenesis Programme (HPP) Sinikithemba cohort is described in detail elsewhere [31]. This cohort is based at McCord Hospital in Durban, South Africa. The cohort was established in August 2003, with 451 antiretroviral-naive chronically HIV-1-infected adult study subjects enrolled until June 2006.
CD4 cell counts and plasma viral loads were performed routinely for study participants. CD4+ T-cell counts were enumerated by flow cytometry (Becton Dickinson, San Jose, CA), while plasma viral loads were measured using the Roche Amplicor version 1.5 assay (Roche, NJ). CD4+ T-cell counts were performed at 3-month and plasma viral loads at 6-month intervals. The number and magnitude of HIV peptides targeted by cytotoxic T lymphocytes (CTLs) were measured at baseline by interferon-γ (IFN- γ) enzyme-linked immunospot (ELISPOT) assay using a panel of 410 overlapping peptides spanning the HIV-1C proteome [31, 32].
IL-10 genotype data for SNPs -1082 and -592 were generated using predesigned TaqMan SNP Genotyping assays (Applied Biosystems, CA). Plasma IFN-γ, IL-2, IL-6, IL-10, and tumor necrosis factor (TNF)–α concentrations were determined by Luminex, using the Millipore Milliplex MAP High Sensitivity Human Cytokine Kit.
Different methods of statistical analysis were used to determine the association and correlation of IL-10 variants/levels with aforementioned biomarkers. We used the χ2 test to compare the allelic frequencies of the IL-10 variants, confirming their fit to Hardy–Weinberg equilibrium. Kruskal–Wallis tests were used to determine the association between IL-10 variants and viral load or CD4+ T-cell count. The CD4 decline over 24 months of follow-up was estimated using multivariate mixed effects models. The Kruskal–Wallis test was used to determine the association between IL-10 variants and magnitude or breadth of immune responses. To determine if there was an association between IL-10 variant and plasma IL-10 concentration, the Kruskal–Wallis test was used. The Pearson product moment correlation test was used to determine the association between IL-10 concentration and markers of HIV-1 pathogenesis or plasma cytokines.
RESULTS
IL-10-Promoter Genotyping
The 2 IL-10 promoter positions -1082 and -592 were polymorphic in this cohort of HIV-1C chronically infected individuals. The allele frequencies of the minor -1082G and -592A polymorphisms were 0.31 and 0.34, respectively. Allele frequencies were confirmed by χ2 tests to be in Hardy–Weinberg equilibrium for the entire cohort and for subgroups analyzed in this study. The baseline characteristics of the cohort are shown in Table 1. The median age of the study group was 31 years, 82% were female, 36% had a baseline viral load (VL) >100 000 copies/mL, 62% had a baseline CD4 count >350 cells/μL, and had a median follow-up of 24.7 months.
Table 1.
Baseline Characteristics of the Study Group Based on Genotype
| -1082 Genotype |
-592 Genotype |
||||||
| Characteristic | AA (n = 141) | AG (n = 122) | GG (n = 35) | CC (n = 142) | CA (n = 127) | AA (n = 30) | Total (n = 300) |
| Age, median years (IQR) | 32 (27–37) | 31 (27–38) | 30 (26–35) | 30 (26–36) | 31 (28–37) | 34 (29–37) | 31 (27–37) |
| Sex, female (%) | 109 (77) | 102 (84) | 32 (91) | 116 (82) | 104 (82) | 24 (80) | 245 (82) |
| Baseline pVL >100 000 copies/mL (%) | 52 (37) | 46 (38) | 9 (26) | 48 (34) | 47 (37) | 12 (40) | 107 (36) |
| Baseline CD4 >350 cells/μL (%) | 84 (60) | 76 (62) | 25 (71) | 92 (65) | 76 (60) | 17 (57) | 186 (62) |
| Median follow-up, months (IQR) | 24.2 (14.4–37.8) | 24.4 (12.9–41.0) | 28.9 (21.9–45.3) | 24.7 (14.5–41.0) | 24.2 (15.4–41.5) | 27.1 (15.9–42.6) | 24.7 (15.0–41.0) |
Abbreviations: IQR, interquartile range; pVL, plasma viral load.
Viral Load, CD4+ T-Cell Count, and CD4 Decline
For the 409 individuals with baseline data, we investigated the association between the IL-10 genotypes and baseline viral load and CD4+ T-cell counts. Table 2 shows the association between baseline viral load and CD4+ T cells. In this cross-sectional analysis, we found no association between any IL-10 variant and viral load or CD4+ T-cell count.
Table 2.
Baseline Viral Load and CD4+ T-Cell Count Based on IL-10 Genotype
| −1082 Genotype |
−592 Genotype |
|||||||
| AA | AG | GG | P value | CC | CA | AA | P value | |
| Baseline CD4 (cells/μL) | 341 | 375 | 431 | 0.23 | 397 | 339 | 341 | .22 |
| Log mean pVL (log copies/mL) | 4.8 | 4.8 | 4.7 | 0.40 | 4.8 | 4.8 | 4.8 | .41 |
Abbreviation: pVL, plasma viral load.
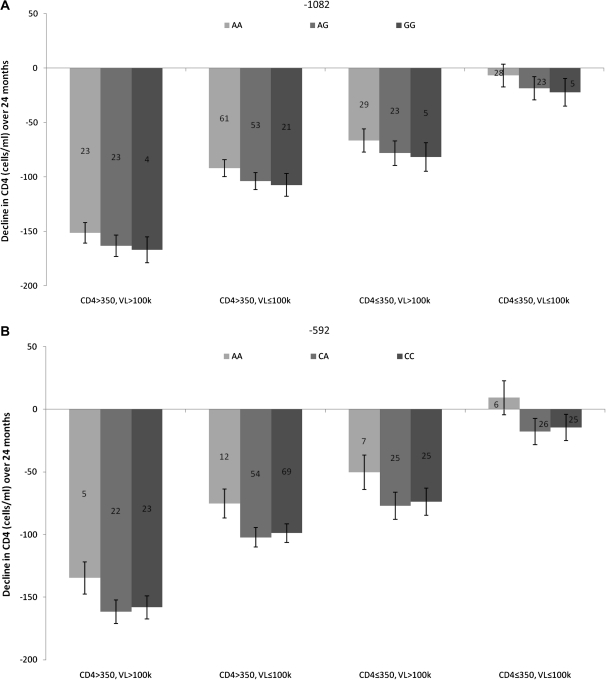
We investigated the rate of CD4+ T-cell decline in 300 individuals with follow-up data. Figure 1 shows the rate of CD4+ T-cell decline over 24 months based on genotype. CD4 decline was stratified according to CD4 and viral load (ie, CD4 >350, VL >100 000; CD4 >350, VL ≤100 000; CD4 ≤350, VL >100 000; CD4 ≤350, VL ≤100 000). The low-IL-10-producing -1082AA genotype had an attenuated CD4 cell loss compared with the -1082AG or -1082GG groups (Figure 1A). This trend was seen in each strata; however, there was no significant association (P = .15). The low-IL-10-producing -592AA genotype had an attenuated loss of CD4 cells over all strata (Figure 1B), with a borderline significant association between the -592 genotype and CD4 decline over 24 months (P = .0496). Overall, low-IL-10-producing variants had an attenuated loss of CD4 cells over all strata in this study cohort.
Figure 1.
The rate of CD4+ T-cell decline over 24 months based on IL-10 variants. Data was stratified into different groups based on viral load and CD4 count (ie, CD4 >350, VL >100 000; CD4 >350, VL ≤100 000; CD4 ≤350, VL >100 000; and CD4 ≤350, VL ≤100 000). The numbers in the bars indicate the number of people in each category. A, The rate of CD4+ T-cell decline over 24 months based on -1082 genotype. The low IL-10-producing -1082AA groups tended to have an attenuated loss of CD4 cells as compared to the other groups; however, this was not significant (P = .15). B, The rate of CD4+ T-cell decline over 24 months based on -592 genotype. The low IL-10-producing -592AA group had an attenuated loss of CD4 cells over 24 months. The was a significant association between -592 genotype and CD4 cell loss (P = .0496). Abbreviations: IL, interleukin; VL, viral load; k, thousands.
Cytokine Expression Analysis
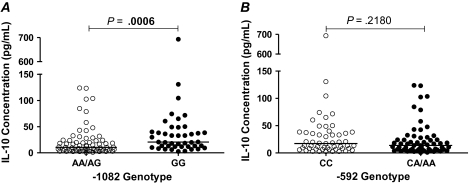
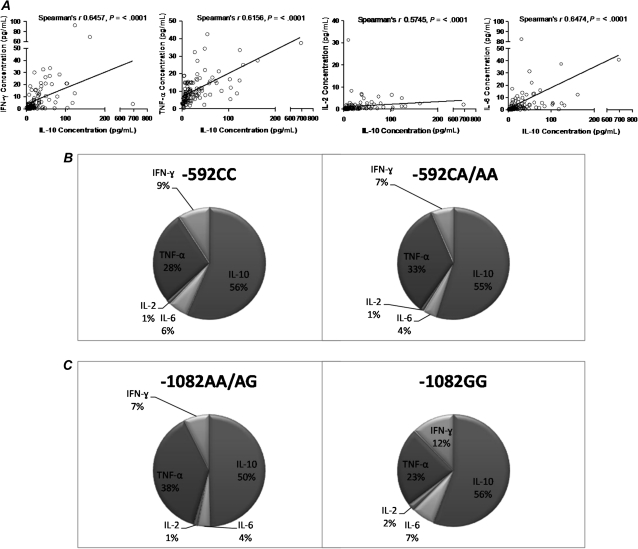
We next investigated the association between IL-10-promoter polymorphisms and plasma IL-10 and/or plasma proinflammatory cytokines IFN-γ, IL-2, IL-6, and TNF-α in a subset of 112 individuals. To determine if IL-10 levels associate with IL-10 polymorphisms in an African setting of chronic HIV-1C infection, we investigated if plasma IL-10 levels associate with IL-10 variants (Figure 2). We grouped the genotypes according to dominance patterns previously described by Shin et al. [21]. Figure 2A shows that based on a recessive model, the -1082GG group had significantly higher median plasma IL-10 concentration compared with the combined -1082AA/AG groups (P = .0006). Figure 2B shows that when considering a dominant model [33], the -592CC group had a higher median IL-10 concentration as compared with the combined -592CA/AA groups; however, this was not significant (P = .2180). In a cross-sectional analysis, and in contrast to what had been seen in other cohorts of different ethnicities located in other geographic areas [29], IL-10 plasma levels did not significantly correlate with viral load (Pearson correlation = 0.08465, P = .3838), CD4 count (Pearson correlation = −0.02797, P = .7749), the breadth of immune responses (Pearson correlation = −0.0613, P = .5304), or the magnitude of immune responses (Pearson correlation = 0.0597, P = .5412), data not shown. As IL-10 is a major inhibitory immunoregulator, we next examined whether the different IL-10-promoter polymorphisms were associated with distinct cytokine profiles in peripheral blood. Figure 3 represents the analysis of the 5 cytokines that were measured. Figure 3A shows that there was significant positive correlation between the levels of each of the proinflammatory cytokines (IFN-γ, IL-2, and IL-6 and TNF-α) and IL-10 levels (P < .0001, Spearman rank correlation [ρ]). Figure 3B and 3C shows that, overall, IL-10 dominated the measured plasma cytokine levels in this chronic HIV-1C setting irrespective of the IL-10 genotype.
Figure 2.
The association of IL-10 expression based on IL-10 genotype. A, the association of IL-10 expression based on -1082 genotype. Genotypic groups were grouped according to a recessive model of inheritance. We found a significant association between -1082 genotype and IL-10 expression (P = .0006). B, The association of IL-10 expression based on -592 genotype. Genotypic groups were grouped according to a dominant model of dominance. There was no significant association between -592 genotype and IL-10 expression (P = .2180). Abbreviations: IL, interleukin; VL, viral load; k, thousands.
Figure 3.
Cytokine expression during chronic HIV-1C infection. IL-10, IFN-γ, TNF-α, IL-2, and IL-6 were measured by Luminex methodology in 112 individuals. A, The correlation between IL-10 and IFN-γ, TNF-α, IL-2, and IL-6. Overall, all the cytokines had a significantly positive correlation with IL-10 expression (P < .0001 for each cytokine, Spearman ρ). B, Proportion of cytokine expression based on IL-10 -592 genotype. IL-10 seemed to dominate cytokine expression overall. C, Proportion of cytokine expression based on IL-10 -1082 genotype. IL-10 seemed to dominate cytokine expression overall. Abbreviations: HIV, human immunodeficiency virus; IL, interleukin; IFN, interferon; TNF, tumor necrosis factor.
Breadth and Magnitude of Immune Responses
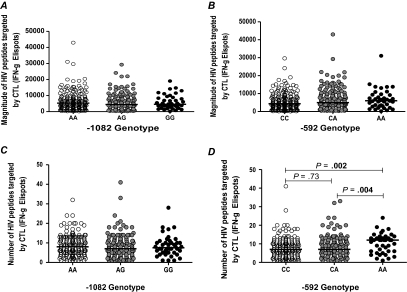
In the LCMV murine model of chronic viral infection, IL-10-deficient mice showed an increased frequency of tetramer-positive virus-specific CD8+ T cells, and IL-10 receptor blockade increased IFN-γ production by virus-specific CD8+ T cells [27, 28] We therefore reasoned that IL-10 variants that affect IL-10 production and influence disease progression may also be linked with the magnitude (number of IFN-γ-producing cells per million PBMCs) and breadth (number of HIV peptides targeted by CTLs) of HIV-1-specific immune response in vivo, as measured by IFN-γ ELISPOT. We thus investigated the association between IL-10 variants and the magnitude and breadth of CD8+ T-cell immune responses. Figure 4A and 4B shows the association between the magnitude of immune responses based on the 1082 and -592 genotypes, respectively. We found no significant association between the magnitude of HIV-1-specific immune responses and -1082 genotype (P = .44) or the -592 genotype (P = .17). We then assessed the breadth of immune responses in relation to IL-10 genotype. We found no significant association between -1082 genotype and the number of HIV peptides targeted by CTLs (P = .2316). However, there was a significant association between the number of HIV peptides targeted and the -592 genotype (P = .0069). The low-IL-10-producing -592AA group had a median of 12 HIV peptides versus 7 peptides targeted for the -592CC or -592CA genotypes (P = .002 and 0.004, respectively).
Figure 4.
Magnitude and breadth of immune responses based on genotype. A, the magnitude of HIV peptides targeted by CTLs (IFN-γ) based on -1082 genotype. There was no significant association between -1082 genotype and the magnitude of immune responses (P = .044). B, The magnitude of HIV peptides targeted by CTLs (IFN-γ) based on -592 genotype. There was no significant association between -592 genotype and the magnitude of immune responses (P = .17). C, The number of HIV peptides targeted by CTLs (IFN-γ) based on -1082 genotype. There was no significant association between the number of HIV peptides targeted by CTLs and -1082 genotype (P = .23). D, the number of HIV peptides targeted by CTLs (IFN-γ) based on -592 genotype. There was a significant association between the number of HIV peptides targeted by CTLs and -592 genotype (P = .007). The -592AA group targeted a significantly larger number of HIV peptides as compared to the -592CC or 592CA groups (P = .002 and .004, respectively). Abbreviations: HIV, human immunodeficiency virus; CTLs, cytotoxic T lymphocytes; IFN, interferon.
DISCUSSION
In this study, we sought to extend earlier observations on the modulation of HIV-1 infection by genetic polymorphisms of the IL-10 promoter to an investigation of possible underlying mechanisms in vivo. We therefore characterized IL-10-promoter SNPs in a large, predominantly black African cohort chronically infected with HIV-1 subtype C. IL-10 polymorphisms were investigated for association with biomarkers of disease progression; namely, plasma viral load, CD4+ T-cell counts, and the rate of CD4+ T-cell decline. These polymorphisms were then analyzed for association with plasma IL-10 and select proinflammatory cytokines levels (as this had not previously been investigated in an HIV setting), and the breadth and magnitude of CD8+ T-cell immune responses. Our data suggest an association between IL-10-promoter genotypes with the rate of CD4+ T-cell loss during chronic HIV-1 infection, an association with plasma IL-10 levels, a predominance of the anti-inflammatory IL-10 over proinflammatory cytokines in the plasma of HIV-1-infected individuals, and an effect of IL-10 polymorphisms on the breadth but not the magnitude of CD8+ T-cell immune response.
Our data suggest that in an African setting of chronic HIV-1 infection, IL-10 variants may influence the rate of disease progression. Our investigation into the role of IL-10 in HIV-1C pathogenesis showed that these IL-10-promoter polymorphisms that have been previously shown to be associated with differing levels of IL-10 expression [19, 22–24] significantly associate with differential IL-10 expression in an HIV setting. Low-IL-10-producing -592 variants showed a trend toward attenuated CD4+ T-cell loss. These data differ from earlier studies in European and African cohorts in which high-IL-10-producing genotypic variants were associated with attenuated CD4+ T-cell loss or progression to AIDS [18, 21]. A possible explanation for these differences might be that the effect of IL-10-promoter variants on HIV-1 pathogenesis is infection-phase dependent, as we have previously suggested [26]. This interpretation is also consistent with the observations of Shin and colleagues [21] who found that high-IL-10-producing genetic variants were protective against disease progression, particularly during late states of infection, and yet the genetic removal of or blockade of IL-10 in a mouse model of chronic viral infection has been shown to result in viral clearance [27, 28]. Our hypothesis is that high IL-10 production (and by extension high-IL-10-producing promoter genotypes) are detrimental during early HIV-1 infection because of downregulation of antiviral adaptive and innate effector mechanisms [30, 34, 35] but beneficial in late stages of infection because of anti-inflammatory effects of IL-10 and direct-inhibition viral replication in macrophages [36–38]. It is also possible that different strains of HIV-1 induce different levels of IL-10, as has been recently demonstrated for HIV-1 B versus C trans-activating proteins [39], although this explanation is unlikely to account for the different results observed in our study versus the other African study [18] since both were done in settings where HIV-1 subtype C predominates. Alternatively, IL-10 SNPs may have underlying undetermined epigenetic or environment modulatory factors, or these SNPs may be in linkage disequilibrium with other genes with a modulatory role on attenuated CD4+ T-cell loss. Further studies will be needed to discriminate between these possibilities.
Overall, IFN-γ, IL-2, IL-6, and TNF-α had a significant positive correlation with IL-10 expression. Showing that in an HIV-1C setting, both pro- and anti-inflammatory cytokines are upregulated. However, we found that the proportion of IL-10 expression seemed to dominate over the expression of the other cytokines. The dominance of IL-10 expression suggests that as the production of proinflammatory cytokines increases, the production of IL-10 also increases to reduce inflammation and activation.
The -592A variant, associated with low IL-10 production, significantly associated with a higher number of HIV-1 peptides targeted by CTLs. These data are consistent with results from mechanistic studies on the LCMV mouse model, which show that the removal or blockade of IL-10 enhance T-cell immune responses [27, 28]. Similarly, results from our study of IL-10 blockade in vitro resulted in restoration of proliferative and effector CD4 T-cell function [29]. Lower IL-10 levels may allow for the expression of HLA class I and II molecules, which in turn increases the presentation of pathogen-derived peptides on the cell surface of infected cells, for recognition by CD8+ T lymphocytes. However, as IL-10 levels did not correlate with plasma viral load, CD4+ T-cell count, or the breadth of immune responses, IL-10 polymorphisms may contribute to the quality of immune responses via a complex pathway that has yet to be elucidated. Alternatively, differential levels of IL-10 secretion that are critical for paracrine cell–cell interactions may not be reflected by differences in IL-10 plasma levels.
There are a number of limitations to this cohort study. First, the time since HIV-1 infection is unknown for study participants, which may have introduced a survivor bias in analysis of IL-10 polymorphisms since these have been shown to affect survival [18]. Second, since time of infection was unknown, we may be analyzing individuals at different phases of infection together, although we stratified our data according to viral loads and CD4 cell counts to partially mitigate for this limitation. This study also focused on 2 proximal IL-10-promoter polymorphisms, only a subset of IL-10 SNPs shown to affect IL-10 production. Finally, IL-10 and other cytokines can be induced by a plethora of pathogens, and we lacked data on coinfection status of participants in this study.
In conclusion, our study highlights the complex role that IL-10 and IL-10 genetic variants may play in HIV-1 pathogenesis. We show that IL-10-promoter polymorphisms may play a role in the rate of CD4 T-cell loss in chronic HIV infection, and affect IL-10 plasma levels and the breadth of anti-HIV CD8+ T-cell immune responses. However, to fully understand the effects and underlying mechanisms of IL-10 in HIV-1C pathogenesis, expanded analysis is required of the proximal and distal SNPs. Additional mechanistic studies will also be required in order to fully understand how best to target the IL-10 pathway for effective immunotherapy or a vaccine.
Notes
Acknowledgments.
We are grateful to the staff and management at McCord Hospital for their support and cooperation. We thank Drs Fundisiwe Chonco and Wendy Mphastse, Sisters Thandi Sikhakhane, Nonhlanhla Maphalala, Landiwe Nxele, and Nono Nkupiso, and many other clinic and laboratory staff members at HPP for their commitment to the Sinikithemba cohort study. Finally, we gratefully acknowledge the study participants without whom the Sinikithemba cohort study would not have been possible.
Financial support.
This work was supported by the National Institutes of Health (NIH, grants ROI-AI067073 and R01-AI46995, and contract #N01-AI-15422 and RO1 HL-092565), the International AIDS Vaccine Initiative (IAVI), and the South African AIDS Vaccine Initiative and the South African Department of Science and Technology through the National Research Foundation. Additional support was provided by the Mark and Lisa Schwartz Foundation.
Potential conflicts of interest.
All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.UNAIDS. AIDS epidemic update. UNAIDS; 2009. Available at: http://www.unaids.org/en/dataanalysis/epidemiology/2009aidsepidemicupdate/ [Google Scholar]
- 2.Buchbinder SP, Mehrotra DV, Duerr A, et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–93. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McElrath MJ, De Rosa SC, Moodie Z, et al. HIV-1 vaccine-induced immunity in the test-of-concept Step Study: a case-cohort analysis. Lancet. 2008;372:1894–905. doi: 10.1016/S0140-6736(08)61592-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trinvuthipong C. Thailand's Prime-boost HIV vaccine phase III. Science. 2004;303:954–5. doi: 10.1126/science.303.5660.954c. [DOI] [PubMed] [Google Scholar]
- 5.Carrington M, O'Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–51. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 6.Shrestha S, Strathdee SA, Galai N, et al. Behavioral risk exposure and host genetics of susceptibility to HIV-1 infection. J Infect Dis. 2006;193:16–26. doi: 10.1086/498532. [DOI] [PubMed] [Google Scholar]
- 7.Paxton WA, Martin SR, Tse D, et al. Relative resistance to HIV-1 infection of CD4 lymphocytes from persons who remain uninfected despite multiple high-risk sexual exposure. Nat Med. 1996;2:412–17. doi: 10.1038/nm0496-412. [DOI] [PubMed] [Google Scholar]
- 8.Kostrikis LG, Neumann AU, Thomson B, et al. A polymorphism in the regulatory region of the CC-chemokine receptor 5 gene influences perinatal transmission of human immunodeficiency virus type 1 to African-American infants. J Virol. 1999;73:10264–71. doi: 10.1128/jvi.73.12.10264-10271.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiepiela P, Leslie AJ, Honeyborne I, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–75. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 10.Fellay J, Shianna KV, Ge D, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–7. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Limou S, Le Clerc S, Coulonges C, et al. Genomewide association study of an AIDS-nonprogression cohort emphasizes the role played by HLA genes (ANRS Genomewide Association Study 02) J Infect Dis. 2009;199:419–26. doi: 10.1086/596067. [DOI] [PubMed] [Google Scholar]
- 12.Baştürk B, Tunali A, Karakus S. Interleukin-10 and interferon-gamma cytokine gene polymorphisms may be risk factors for chronic myelogenous leukemia. Turkish J Haemotol. 2005;22:191–6. [PubMed] [Google Scholar]
- 13.Lech-Maranda E, Baseggio L, Bienvenu J, et al. Interleukin-10 gene promoter polymorphisms influence the clinical outcome of diffuse large B-cell lymphoma. Blood. 2004;103:3529–34. doi: 10.1182/blood-2003-06-1850. [DOI] [PubMed] [Google Scholar]
- 14.Filippi CM, von Herrath MG. IL-10 and the resolution of infections. J Pathol. 2008;214:224–30. doi: 10.1002/path.2272. [DOI] [PubMed] [Google Scholar]
- 15.Middleton PG, Taylor PR, Jackson G, Proctor SJ, Dickinson AM. Cytokine gene polymorphisms associating with severe acute graft-versus-host disease in HLA-identical sibling transplants. Blood. 1998;92:3943–8. [PubMed] [Google Scholar]
- 16.Ness RB, Haggerty CL, Harger G, Ferrell R. Differential distribution of allelic variants in cytokine genes among African Americans and White Americans. Am J Epidemiol. 2004;160:1033–8. doi: 10.1093/aje/kwh325. [DOI] [PubMed] [Google Scholar]
- 17.Tedgui A, Mallat Z. Cytokines in atherosclerosis: pathogenic and regulatory pathways. Physiol Rev. 2006;86:515–81. doi: 10.1152/physrev.00024.2005. [DOI] [PubMed] [Google Scholar]
- 18.Erikstrup C, Kallestrup P, Zinyama-Gutsire RB, et al. Reduced mortality and CD4 cell loss among carriers of the interleukin-10 -1082G allele in a Zimbabwean cohort of HIV-1-infected adults. AIDS. 2007;21:2283–91. doi: 10.1097/QAD.0b013e3282f153ed. [DOI] [PubMed] [Google Scholar]
- 19.Gibson AW, Edberg JC, Wu J, Westendorp RG, Huizinga TW, Kimberly RP. Novel single nucleotide polymorphisms in the distal IL-10 promoter affect IL-10 production and enhance the risk of systemic lupus erythematosus. J Immunol. 2001;166:3915–22. doi: 10.4049/jimmunol.166.6.3915. [DOI] [PubMed] [Google Scholar]
- 20.Oleksyk TK, Shrestha S, Truelove AL, et al. Extended IL10 haplotypes and their association with HIV progression to AIDS. Genes Immun. 2009;10:309–22. doi: 10.1038/gene.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin HD, Winkler C, Stephens JC, et al. Genetic restriction of HIV-1 pathogenesis to AIDS by promoter alleles of IL10. Proc Natl Acad Sci USA. 2000;97:14467–72. doi: 10.1073/pnas.97.26.14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eskdale J, Gallagher G, Verweij CL, Keijsers V, Westendorp RG, Huizinga TW. Interleukin 10 secretion in relation to human IL-10 locus haplotypes. Proc Natl Acad Sci USA. 1998;95:9465–70. doi: 10.1073/pnas.95.16.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamali-Sarvestani E, Kiany S, Gharesi-Fard B, Robati M. Association study of IL-10 and IFN-gamma gene polymorphisms in Iranian women with preeclampsia. J Reprod Immunol. 2006;72:118–26. doi: 10.1016/j.jri.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Lan Q, Zheng T, Rothman N, et al. Cytokine polymorphisms in the Th1/Th2 pathway and susceptibility to non-Hodgkin lymphoma. Blood. 2006;107:4101–8. doi: 10.1182/blood-2005-10-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vasilescu A, Heath SC, Ivanova R, et al. Genomic analysis of Th1-Th2 cytokine genes in an AIDS cohort: identification of IL4 and IL10 haplotypes associated with the disease progression. Genes Immun. 2003;4:441–9. doi: 10.1038/sj.gene.6363983. [DOI] [PubMed] [Google Scholar]
- 26.Naicker DD, Werner L, Kormuth E, et al. Interleukin-10 promoter polymorphisms influence HIV-1 susceptibility and primary HIV-1 pathogenesis. J Infect Dis. 2009;200:448–52. doi: 10.1086/600072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brooks DG, Trifilo MJ, Edelmann KH, Teyton L, McGavern DB, Oldstone MB. Interleukin-10 determines viral clearance or persistence in vivo. Nat Med. 2006;12:1301–9. doi: 10.1038/nm1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ejrnaes M, Filippi CM, Martinic MM, et al. Resolution of a chronic viral infection after interleukin-10 receptor blockade. J Exp Med. 2006;203:2461–72. doi: 10.1084/jem.20061462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brockman MA, Kwon DS, Tighe DP, et al. IL-10 is up-regulated in multiple cell types during viremic HIV infection and reversibly inhibits virus-specific T cells. Blood. 2009;114:346–56. doi: 10.1182/blood-2008-12-191296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alter G, Kavanagh D, Rihn S, et al. IL-10 induces aberrant deletion of dendritic cells by natural killer cells in the context of HIV infection. J Clin Invest. 2010;120:1905–13. doi: 10.1172/JCI40913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiepiela P, Ngumbela K, Thobakgale C, et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 32.Goulder PJ, Addo MM, Altfeld MA, et al. Rapid definition of five novel HLA-A*3002-restricted human immunodeficiency virus-specific cytotoxic T-lymphocyte epitopes by ELISPOT and intracellular cytokine staining assays. J Virol. 2001;75:1339–47. doi: 10.1128/JVI.75.3.1339-1347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilkie AO. The molecular basis of genetic dominance. J Med Genet. 1994;31:89–98. doi: 10.1136/jmg.31.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herbein G, Varin A. The macrophage in HIV-1 infection: from activation to deactivation? Retrovirology. 2010;7:33. doi: 10.1186/1742-4690-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinic MM, von Herrath MG. Novel strategies to eliminate persistent viral infections. Trends Immunol. 2008;29:116–24. doi: 10.1016/j.it.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 36.Ancuta P, Bakri Y, Chomont N, Hocini H, Gabuzda D, Haeffner-Cavaillon N. Opposite effects of IL-10 on the ability of dendritic cells and macrophages to replicate primary CXCR4-dependent HIV-1 strains. J Immunol. 2001;166:4244–53. doi: 10.4049/jimmunol.166.6.4244. [DOI] [PubMed] [Google Scholar]
- 37.Bento CA, Hygino J, Andrade RM, et al. IL-10-secreting T cells from HIV-infected pregnant women downregulate HIV-1 replication: effect enhanced by antiretroviral treatment. AIDS. 2009;23:9–18. doi: 10.1097/QAD.0b013e328317461e. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Rice AP. Interleukin-10 inhibits HIV-1 LTR-directed gene expression in human macrophages through the induction of cyclin T1 proteolysis. Virology. 2006;352:485–92. doi: 10.1016/j.virol.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 39.Wong JK, Campbell GR, Spector SA. Differential induction of interleukin-10 in monocytes by HIV-1 clade B and clade C Tat proteins. J Biol Chem. 2010;285:18319–25. doi: 10.1074/jbc.M110.120840. [DOI] [PMC free article] [PubMed] [Google Scholar]