Abstract
Tetraspanins have gained increased attention due to their functional versatility. But the universal cellular mechanism that governs such versatility remains unknown. Herein we present the evidence that tetraspanins CD81 and CD82 regulate the formation and/or development of cell membrane protrusions. We analyzed the ultrastructure of the cells in which a tetraspanin is either overexpressed or ablated using transmission electron microscopy. The numbers of microvilli on the cell surface were counted, and the radii of microvillar tips and the lengths of microvilli were measured. We found that tetraspanin CD81 promotes the microvillus formation and/or extension while tetraspanin CD82 inhibits these events. In addition, CD81 enhances the outward bending of the plasma membrane while CD82 inhibits it. We also found that CD81 and CD82 proteins are localized at microvilli using immunofluorescence. CD82 regulates microvillus morphogenesis likely by altering the plasma membrane curvature and/or the cortical actin cytoskeletal organization. We predict that membrane protrusions embody a common morphological phenotype and cellular mechanism for, at least some if not all, tetraspanins. The differential effects of tetraspanins on microvilli likely lead to the functional diversification of tetraspanins and appear to correlate with their functional propensity.
Keywords: microprotrusion, microvilllus, membrane curvature, membrane microdomain, adhesion zipper
INTRODUCTION
Tetraspanins are small, membrane-spanning proteins. Without having obvious signaling motif or module, tetraspanins engage surprisingly a wide variety of biological functions such as cell migration, cell-cell fusion, cell-cell adhesion, cell-matrix adhesion, cell spreading, cell proliferation, cell signaling, intracellular vesicle trafficking, peri-cellular proteolysis, viral entry and release, immune response, vascular morphogenesis and remodeling, tumor progression and metastasis, neurite navigation, and thrombosis (1–8). With the expanding repertoire of tetraspanin-involved, -regulated, or -required functions, the biochemical and/or biophysical nature that governs tetraspanins to engage these physiological and pathological events still remains basically unknown. Based on the same ancestry shared by tetraspanin genes and the sequence homology shared by tetraspanin proteins, we extrapolate the existence of a universal mechanism that governs most of tetraspanin-related functions.
We predict that the regulation of membrane protrusion morphogenesis is a cell biological mechanism by which tetraspanins engage various functions. This prediction is based on the repeated observations that the appearance or disappearance of membrane protrusion correlates with the gain, loss, or fluctuation of tetraspanin expressions. Tetraspanins such as CD82, CO-029, and Tspan-1 are found at the lumenal vesicles secreted from intestinal microvilli (9), strongly suggesting the presence of these tetraspanins in the microvilli. Tetraspanin CD9 is enriched at the microvilli on egg cells (10–12), and CD9 and tetraspanin CD63 are found on the microprotrusions of activated platelets (13, 14). Tetraspanin CD81 is localized at the microvilli at the inner surface of retinal pigment epithelial cells (15). Also, CD9 can be found in the microprotrusions that are localized at cell-cell contacts (16, 17). The association of tetraspanins with cell-cell contact is consistent with the co-emergence of tetraspanin and multi-cellular organism in evolution (18, 19). Tetraspanin CD151 and its associated integrin α3β1 regulate the foot process morphogenesis, evidenced by the foot process effacement in CD151-or integrin α3-null podocytes (20–22). Fungal tetraspanin PLS1 is required for the formation of penetration pegs (23), a cell wall-penetrative structure similar to invadopodia.
Thus, tetraspanins likely regulate the morphogenesis of membrane extrusions, and some tetraspanins promote while others inhibit it. To test these hypotheses, we examined the roles and explored the mechanism of CD81 and CD82 in microvillus morphogenesis.
MATERIALS AND METHODS
Cells
The WT and CD81−/− Pre-B cell lines were established as early described (24, 25). Peripheral blood mononuclear cells (PBMCs) were isolated from wild type and CD81-null mice by the Ficoll-Paque gradient centrifugation method. CD81-null mice (25) were backcrossed onto the background of 129X1/SvJ strain (Jackson Laboratory, Bar Harbor, Me), and this backcross progeny reached more than 99% homozygous for alleles derived from 129X1/SvJ. U937.7C2 cells, a subline of U937 histiocytic lymphoma cell line and lacking the expression of CD81, were used for establishing U937-GFP and U937-CD81-GFP stable transfectants (26).
PC3 and Du145 prostate cancer cells barely express endogenous CD82 proteins, and the Mock and CD82 stable transfectants were established as previously described (27, 28).
Transmission Electron Microscopy (TEM)
Cells in suspension were fixed, stained, thin-sectioned, and then examined with TEM as described in the early study (28). See “Supplement” for details.
Epifluorescent and Confocal Fluorescent Microscopy
Cells in attachment were fixed, incubated with antibodies or phalloidin, and then examined with light microscopy as described in early studies (16, 27, 28). See “Supplement” for details.
Data Analysis
The number and length of microprotrusions were quantified based on the TEM images by counting microprotrusions visually and measuring microprotrusion longitudinal section with PhotoShop software, respectively. The radius of microprotrusion tip was measured, based on the circle that fits the curve at the tip of a microprotrusion, with PhotoShop software.
All experiments were performed at least three times. Data are presented as mean±SD. Statistical analysis was performed by Student’s t-test, and P<0.05 is considered as statistically significant.
RESULTS
Tetraspanin CD81 promotes the morphogenesis of membrane protrusions
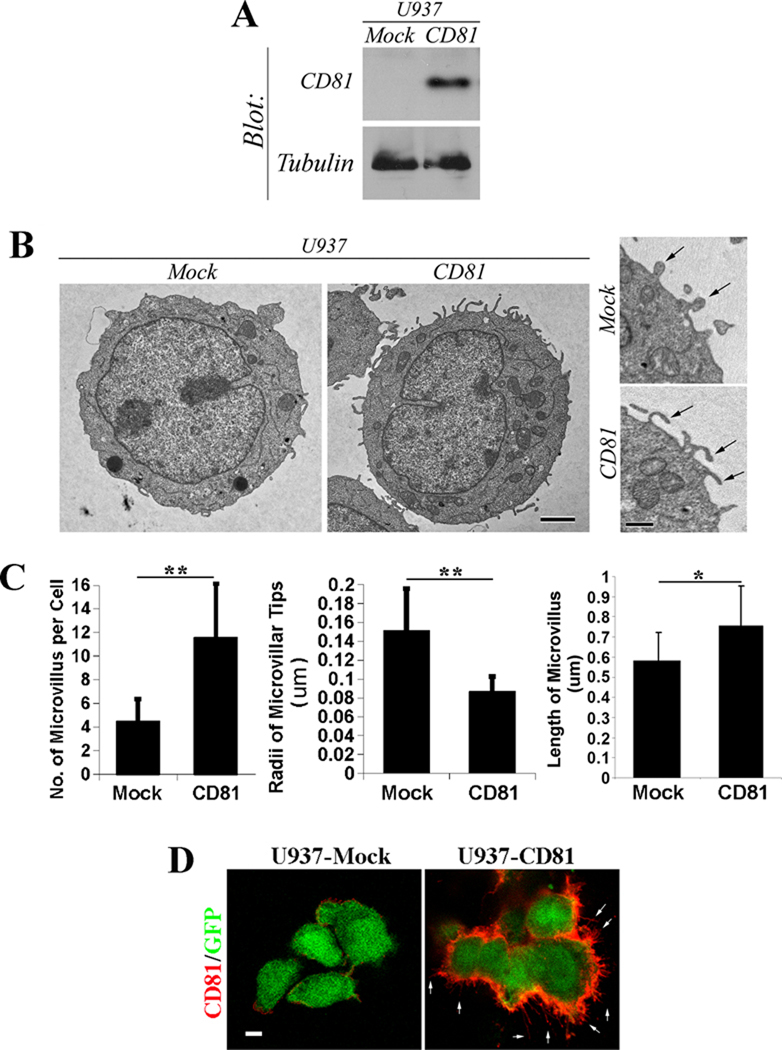
To determine the role of CD81 in regulating membrane protrusion, we examined the effect of CD81 overexpression on microvillus formation. U937-Mock cells express GFP, while U937-CD81 cells express CD81 and GFP (26). We confirmed that U937-Mock cells barely express any endogenous CD81 while U937-CD81 cells indeed express CD81 (Figure 1A). From TEM analysis, we found that CD81 promotes the formation of microvilli (Figure 1B). The number of microvilli was clearly increased when CD81 was expressed in U937 cells (Figure 1C). In addition, CD81 expression markedly increased the membrane curvature at the tips of microvilli, evidenced by thinner microvilli typically seen in CD81-expressing U937 cells (Figure 1C). Moreover, the average length of microvilli was significantly increased upon CD81 expression (Figure 1C).
Figure 1. The overexpression of tetraspanin CD81 promotes the formation and extension of microvilli and increases the curvature of microvilli.
A: The expressions of CD81 proteins in U937-Mock and -CD81 stable transfectant cells were examined by Western blot. The levels of tubulin proteins are used as loading control. B: The U937 transfectant cells were fixed, thin-sectioned, stained, and analyzed with TEM as described earlier (28). Images represent the results from one out of three individual experiments. Representative images of whole cells and microvilli from the U937 transfectants expressing either control construct or CD81 are presented. Scale bar, 2.0 µm. Arrow: microvillus. C. Quantitative analysis. The numbers of microvilli were counted from individual cells, the radii of microvillar tips were assessed using the transverse cross section of microvilli, and the lengths of microvilli were measured using the longitudinal cross section of microvilli. n=15~17 cells for Mock group and =17~20 cells for CD81 group. *, P<0.05; **, P< or =0.01. D: The localization of CD81 in U937 transfectant cells was examined using immunofluorescence under a confocal microscope. Arrows indicate the microvilli positive in CD81. Scale bar, 5.0 µm.
To assess whether CD81 proteins directly regulate microvillus morphogenesis, we analyzed the distribution of CD81 in U937-CD81 cells using immunofluorescence. CD81 proteins were indeed localized in microvilli (Figure 1D).
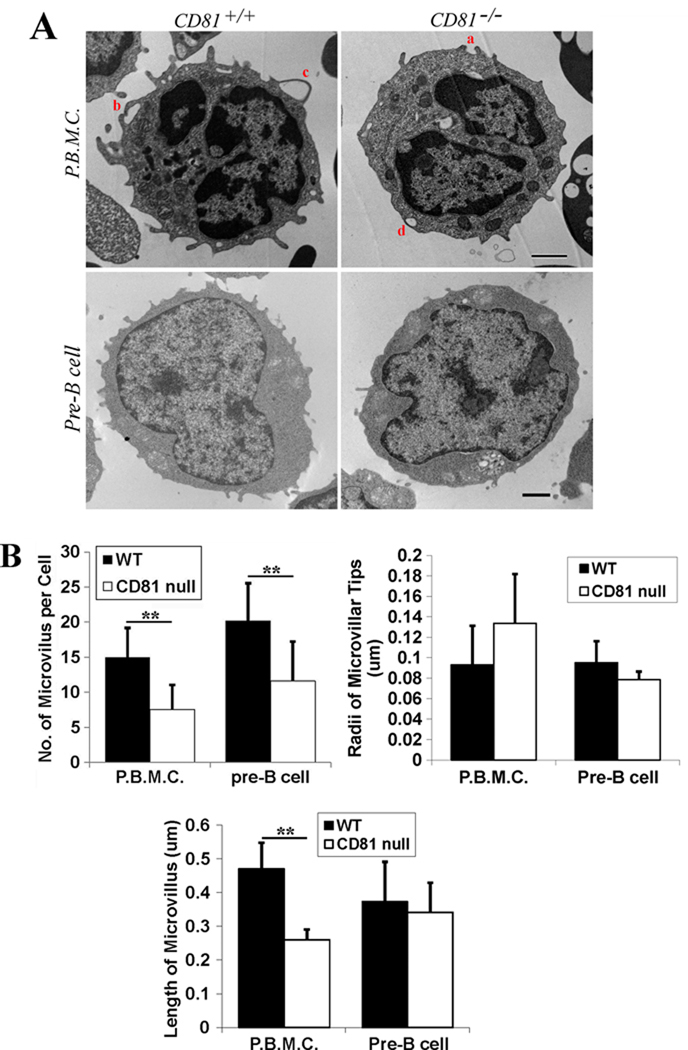
Conversely, microvillus formation became markedly reduced in the PBMCs isolated from CD81-null mice, compared with those derived from wild type mice (Figures 2A and 2B). However, we found no significant difference in the radii of microvillar tips between CD81-expressing and -null PBMCs (Figure 2B). But the lengths of microvilli in PBMCs were substantially reduced when CD81 is ablated (Figure 2B).
Figure 2. The ablation of tetraspanin CD81 reduces the formation and probably also extension of microvilli.
A: Cells were fixed, thin-sectioned, stained, and analyzed with TEM as described earlier (28). Images represent results from one out of three individual experiments. Representative images of the PBMCs isolated from and pre-B cells derived from the 129/SVJ strain of wild type or CD81-null mice are presented. Scale bar, 0.5 µm for PBMCs or 1.0 µm for pre-B cells. B: Quantitative analysis. The histograms display the mean± SE of 1) the numbers of microvillus per cell, 2) the radii of microvillar tips, and 3) the lengths of microvilli of the PBMCs isolated from wild type or CD81-null mice and the pre-B cell lines derived from wild type (2F3) or CD81-null (1C8) mice (43). For PBMCs, n=12~47 cells for wild type group and =12~39 cells for CD81-null group. For pre-B cells, n=13~45 cells for wild type group and =13~45 cells for CD81-null group. **, P<0.01 between wild type and CD81-null groups.
We also compared the microvillus formation in the pre-B lines established from wild type or CD81-null mice (Figure 2A). The number of microvilli in pre-B cells was obviously decreased without CD81 (Figure 2B), consistent with the observation made from PBMCs. However, we found no significant differences in the radii of microvillar tips between CD81-expressing and -null pre-B cells (Figure 2B). The lengths of microvilli in pre-B cells were also remained unchanged when CD81 is ablated (Figure 2B).
Hence, CD81 appears to function as a positive regulator for microvillus morphogenesis. Since microvilli are still found in CD81-null cells, CD81 is not required for the generation of microprotrusions.
Interestingly, the microvilli in U937 cells, PBMCs, and pre-B cells sometimes pair and become opposing with each other at the tips. One example is indicated by letter a in Figure 2A. These pairs of enfolding microprotrusions appear to fuse at the tips (see b in Figure 2A) and encompass the extracellular solutes (c in Figure 2A). This process specifies macropinocytosis, which likely results in the appearance of vesicles at the cell peripheral region (d in Figure 2A). The number of vesicles at the cell peripheral region, which is defined as the area within approximately 250 nm from the cell periphery, in CD81-null PBMCs was markedly reduced compared with the one in PBMCs from WT mice (Table 1), correlating with the difference in the numbers of microvilli at the cell surface between two groups (Figure 2B).
Table 1.
Microvesicles near the cell periphery*.
| No. of vesicles per cell: mean±SD | |
|---|---|
| Intracellular microvesicles near the cell periphery | |
| PBMCs from Wild type mice | 5.64±4.68(n=45)** |
| PBMCs from CD81-null mice | 2.03±2.33(n=34)** |
| Pericellular “microvesicles” near the cell periphery | |
| PC3-Mock cells | 5.50±1.29(n=14)** |
| PC3-CD82 cells | 1.75±0.58(n=14)** |
These microvesicles are likely derived from microvilli. Hence CD81 and/or CD82 are also likely present in these microvesicles.
P<0.01.
Tetraspanin CD82 inhibits microvillus morphogenesis
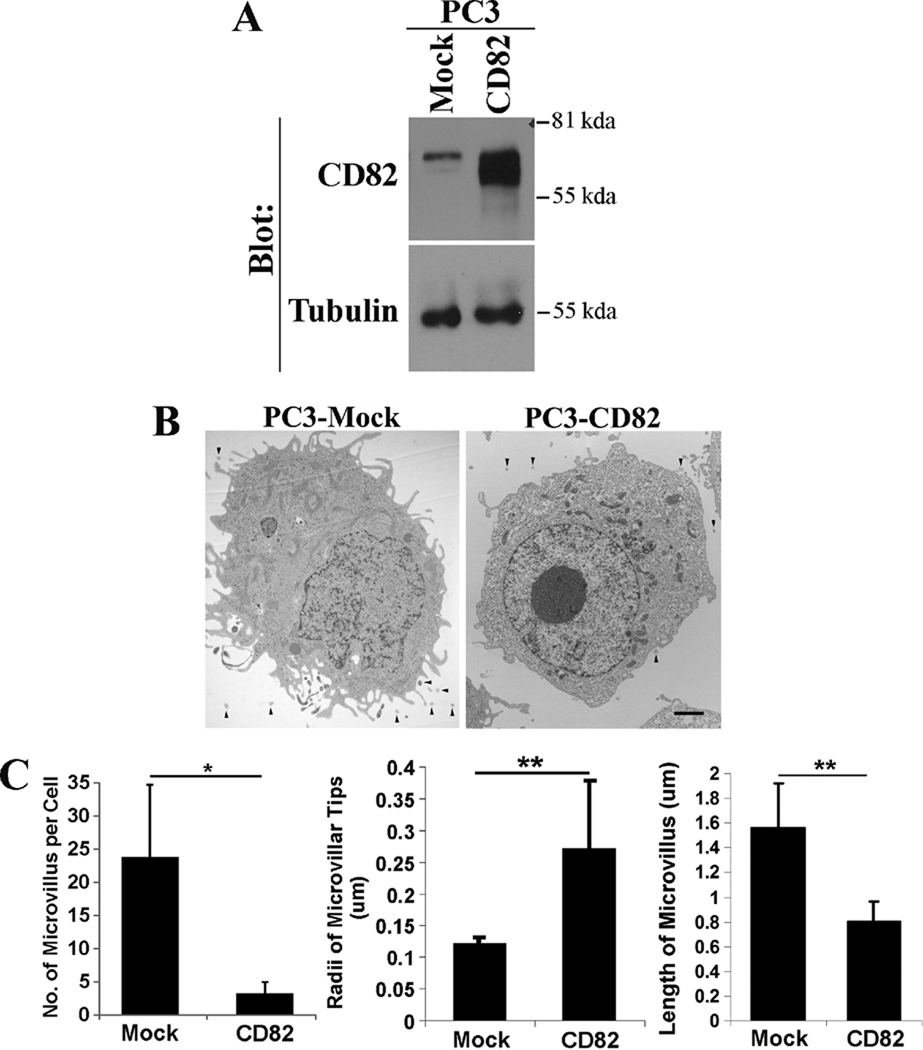
CD82 exerts just an opposite effect on membrane protrusive activities. In our earlier study, we found that, in Du145 cells, CD82 inhibits the formation and development of microvilli and reduces the number of pericellular “microvesicles” (28), which could be either the cross section of microvilli or microvillus-derived microvesicles. This observation can be generalized to other cells overexpressing CD82. For example, we generated the stable transfectant of CD82 in PC3 cells (Figure 3A). Consistently, CD82 overexpression also markedly reduces the numbers of microprotrusions (Figures 3B and 3C) and pericellular “microvesicles” (Table 1) in PC3 cells. PC3-CD82 cells also exhibited significantly decreased membrane curvature or increased radii of the microvillar tips, compared with the Mock cells (Figure 3C). Furthermore, the microvilli in PC3-CD82 cells are markedly shorter than those in PC3-Mock cells (Figure 3C). Thus, CD82 functions as a negative regulator for microvillus formation and/or development and for the outward curving of the plasma membrane.
Figure 3. The overexpression of tetraspanin CD82 inhibits the formation and extension of microvilli.
A. The expressions of CD82 proteins in PC3-Mock and -CD82 stable transfectant cells were analyzed with immunoprecipitation followed by immunoblot. Tubulin proteins were examined in Western blot and are used as loading control. B: PC3-Mock and -CD82 cells were fixed, thin-sectioned, stained, and analyzed with TEM as described earlier (28). Scale bar, 2.0 µm. Arrowheads indicate pericellular microvesicles. C. Quantitative analysis. The numbers of microvilli were counted from individual cells, the radii of microvillar tips were assessed using the longitudinal cross section of microvilli, and the lengths of microvilli were measured using the latitudinal cross section of microvilli. n=14 cells for each group. *, P<0.05; **, P<0.01.
Roles of membrane curvature and cortical actin meshwork in microvillus morphogenesis
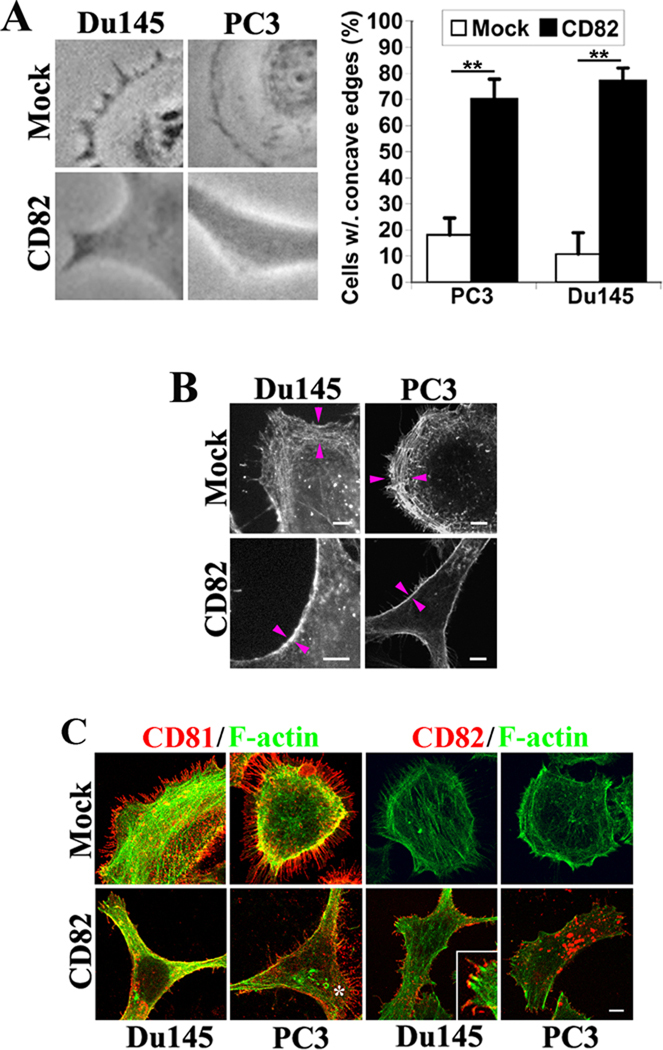
To explore the mechanism by which tetraspanins regulate microvillus morphogenesis, we further investigated the shape of cell peripheries. CD82 overexpression often induced cells to form concave peripheries, in contrast to the convex edges typically observed in the Mock cells (Figure 4A). This result, in addition to the enhancement in the radii of microvillus tips, further supports that CD82 diminishes the outward curvature of the plasma membrane.
Figure 4. Tetraspanin CD82 alters membrane curvature and cortical actin cytoskeleton.
A. The cell peripheries become concave upon CD82 expression. Du145-and PC3-Mock and CD82 transfectant cells were spread on fibronectin-coated substratum in serum-free media (for Du145) or cultured in complete media (for PC3). Phase contrast images were obtained under light microscope. CD82-expressing cells constantly formed concave edges, compared with the outward edges, i.e., lamellipodia, in Mock cells. The percentages of the cells with more than one concave edge were quantified. The histograms represent results from 3 experiments (mean ± SD). n=100 cells for each group. **, P<0.01. B. The Du145 and PC3 transfectant cells were fixed, permeabilized, and incubated with Alexa 488-conjugated α-phalloidin. The fluorescent images were acquired with a confocal microscope. Scale bars, 5.0 µm. Arrowheads indicated the cortical actin cytoskeleton. C: The localizations of CD81 and CD82 proteins to microvilli in Du145 and PC3 transfectant cells were examined using immunofluorescence. The cells were fixed, permeabilized, and incubated with Alexa 488-conjugated α-phalloidin and CD81 or CD82 mAb followed by the staining with Alexa 594-conjugated secondary Ab. The images were obtained with confocal microscopy. Scale bar, 5.0 µm. The asterisk indicates the local convex area of a PC3-CD82 transfectant cell.
We also analyze the role of actin cytoskeleton at the cell periphery in microvillus morphogenesis. The F-actin staining revealed that the convex edges of PC3- and Du145-Mock cells are typically associated with the well-developed cortical actin meshwork, the area between a pair of arrowheads (Figure 4B). While the concave edges in CD82-overexpressing PC3 and Du145 cells usually contain dense actin bundles (Figure 4B), reflecting the poorly developed cortical actin meshwork.
We then analyzed the distribution of microvilli at the peripheries of adherent cells. CD81-positive microvilli are largely distributed in the convex edges of PC3 and Du145 cells (Figure 4C). Because the Mock cells displayed more convex edges together with better developed cortical actin meshwork, more CD81-positive microvilli were found in Mock cells (Figure 4C). In PC3- and Du145-CD82 cells, cell peripheries were often concave and much fewer CD81-positive microvilli existed (Figure 4C). But even in these CD82-expressing cells, the convex edge usually contains relatively more CD81-positive microvilli, as indicated by the asterisk in the PC3-CD82 cell image of Figure 4C. These observations suggest that convex cellular edge and cortical actin meshwork correlate with the generation and development of CD81-positive microvilli.
We also examined the cellular distribution of CD82 relative to microvilli. We found that CD82 can be localized into microprotrusions (Figure 4C). First, the CD82-positive microprotrusions seem morphologically different from CD81-positive microvilli, namely shorter and thicker (Figure 4C). Second, unlike unanimous distribution of CD81 proteins throughout microvilli, CD82 proteins appear to concentrate at the tip or distal portion of microprotrusions, as shown in the inset in the Du145-CD82 cell image of Figure 4C. Third, CD82-positive microprotrusions can be found along the concave edges of cells (Figure 4C). These results suggest that CD82 inhibits microvillus morphogenesis by altering either membrane protrusive activity, cortical actin cytoskeleton organization, or both.
DISCUSSION
Tetraspanins differentially regulate membrane protrusive activity
To engage different cellular functions, the plasma membrane of cells forms protrusive structures (Supplemental Figure). The membrane protrusive structures may serve as sensor, effector, and communication device. Tetraspanins not only localize at various membrane protrusive structures such as microvilli and penetration peg but also regulate the protrusive activities of cell membrane. Using CD81 and CD82 as examples, our study underscored and strengthened this notion. Furthermore, we demonstrated that, for the morphogenesis of microvilli, CD81 facilitates it while CD82 attenuates it. Tetraspanins such as CD9 and CD151 may behave like CD81. For example, the development of microvilli at the surface of egg cells is impaired when CD9 is ablated (11, 12). CD151 facilitates the formation of adhesion zipper when cells are in contact (16, 29), and the engagement of CD151 with its Ab induces microprotrusions in isolated cells (29). Likewise, some tetraspanins may behave as CD82 because of functional similarity. Thus, different tetraspanins apparently play different roles in the morphogenesis of membrane protrusions. We predict that the regulation of membrane protrusive structures is a general property of tetraspanins, by which they affect various cellular behaviors.
Tetraspanins engage cellular functions probably by regulating membrane protrusive activity
Tetraspanins regulate cell adhesiveness, motility, and fusion (1–8). The membrane protrusive structures are likely involved in these cellular events, e.g., adhesion zipper during cell-cell adhesion and invadopodia during cell invasion. We propose that tetraspanins regulate cell adhesion and movement by modulating membrane protrusive activities. For example, fungal tetraspanin PSL1 directs the formation of cellular protrusive structure called penetration peg, which is important for fungus invasiveness (23). Drosophila tetraspanin Late Bloomer facilitates the transformation of invading axon growth cones into pre-synaptic arbors at neuromuscular junction (30) by functioning as a negative regulator of membrane protrusive activity. As aforementioned, CD82 inhibits the morphogenesis of microvilli and pericellular microvesicles, correlating with its motility-inhibitory activity (28). CD82 transmembrane polar residues are needed for the proper formations of these structures (28). Because CD81 is expressed in leukocytes, the proper formation and development of microvilli are likely required for cell-cell adhesion events such as the cell-cell engagement during immune response and leukocyte-endothelial interaction during inflammation.
Tetraspanins modulate endocytosis and exocytosis (6, 8). The protrusive activity of the plasma membrane might also reflect the endocytic and/or exocytic activities of cells. For example, microvilli could imply the early stage of macropinocytosis (Supplemental Figure), a form of endocytosis. CD81 appears to upregulate macropinocytosis because CD81 facilitates microvillus morphogenesis. Notably, the cell peripheral “microvesicles” are diminished upon CD81 ablation. These pericellular microvesicles also became substantially fewer when microvilli were reduced upon CD82 overexpression (28) or CD9 ablation (12), highlighting the roles of tetraspanins and micropvilli in excytosis or vesicle release (Supplemental Figure). Indeed, intestinal microvilli, in which tetraspanins CO-029, CD82, and Tspan1 are present, generate microvesicles (9). These observations are also consistent with the involvement of tetraspanin in viral entry and release (5, 31). Coincidently, one of the most frequent changes in viral infected cells is the formation of microvilli (32, 33), suggesting the contribution of microvilli to the viral budding or release process. For viral release, microprotrusions could be the location or device for membrane budding. Similarly, microprotrusion could facilitate the attachment of virus to the cell surface prior to viral entry. Hence, facilitating microvillus formation by CD81 is likely important for its function as a co-receptor for hepatitis C virus (34).
How tetraspanins regulate membrane protrusive activity
Mechanistically, tetraspanin may regulate the membrane protrusive activities by altering the membrane curvature (Supplemental Figure). For example, the tips of microvilli become more bending when CD81 is overexpressed, suggesting that CD81 promotes the outward curvature of the plasma membrane. In contrast, CD82 overexpression diminishes the outward curvature of the plasma membrane and often induces cell periphery to become concave. If other cellular factors such as cytoskeleton are not involved, more or thinner microvilli and more convex cell edge reflect higher outward intrinsic curving activity of the plasma membrane while fewer or thicker microvilli and more concave cell edge reflect relatively less outward or more inward activity.
Alternatively, tetraspanins may regulate membrane protrusive activities by altering membrane-dependent cytoskeletal reorganization. The outward actin polymerization activity can induce membrane protrusions (Supplemental Figure). Rho GTPases Cdc42, Rac, and Rho determine the formation of membrane protrusions such as lamellipodia and filopodia. Tetraspanins may modify actin reorganization through Rho GTPases since the signaling connection between tetraspanins and Rho GTPases are well recognized (5, 7, 8, 16). Especially, CD81 activates while CD82 inhibits Rac activation (34, our unpublished data), consistent with the roles of these tetraspanins in microvillus morphogenesis. In addition, ezrin/radixin/moesin (ERM) proteins are needed for microvillus formation (35, 36). Because tetraspanins such as CD81 were found to either directly or functionally associate with ERM proteins (15, 24, 37), ERM proteins could serve as the adaptors directly linking tetraspanins to actin cytoskeleton (Supplemental Figure) and thereby regulate membrane protrusion morphogenesis. Together, the roles of Rho GTPases and ERM proteins in CD81- and CD82-regulated morphogenesis of microvilli remains to be determined.
Because CD82 proteins are localized at the tips of microvilli and enlarge the radii of microvilli, CD82 proteins may directly affect membrane curvature. However, CD82 proteins are not enriched at the concave peripheries of the cells. The concave peripheries or the inward bending of the plasma membrane may largely result from the secondary effect of CD82 expression, e.g., the alteration in cortical actin cytoskeleton. Thus, tetraspanins regulate microvillus morphogenesis probably by affecting both cortical actin network and membrane curvature.
Highlights.
Tetraspanins regulate microvillus formation
Tetraspanin CD81 promotes microvillus formation
Tetraspanin CD82 inhibits microvillus formation
Based on this study, we extrapolated a general cellular mechanism for tetraspanins
Tetraspanins engage various functions by regulating membrane protrusion morphogenesis
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH Grant CA096991 (to X. A. Z.)
Abbreviations
- ERM
ezrin/radixin/moesin
- PBMC
peripheral blood mononuclear cell
- TEM
transmission electron microscopy
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Horejsi V, Vlcek C. Novel structurally distinct family of leucocyte surface glycoproteins including CD9, CD37, CD53 and CD63. FEBS Lett. 1991;288:1–4. doi: 10.1016/0014-5793(91)80988-f. [DOI] [PubMed] [Google Scholar]
- 2.Wright MD, Tomlinson MG. The ins and outs of the transmembrane 4 superfamily. Immunol. Today. 1994;15:588–594. doi: 10.1016/0167-5699(94)90222-4. [DOI] [PubMed] [Google Scholar]
- 3.Maecker HT, Todd SC, Levy S. The tetraspanin superfamily: molecular facilitators. FASEB J. 1997;11:428–442. [PubMed] [Google Scholar]
- 4.Boucheix C, Rubinstein E. Tetraspanins. Cell Mol. Life Sci. 2001;58:1189–1205. doi: 10.1007/PL00000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hemler ME. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 6.Berditchevski F, Odintsova E. Tetraspanins as regulators of protein trafficking. Traffic. 2007;8:89–96. doi: 10.1111/j.1600-0854.2006.00515.x. [DOI] [PubMed] [Google Scholar]
- 7.Richardson MM, Jennings LK, Zhang XA. Tetraspanins and Tumor Progression. Clinical Experimental Metastasis. 2011;28:261–270. doi: 10.1007/s10585-010-9365-5. [DOI] [PubMed] [Google Scholar]
- 8.Zhang F, Kotha JP, Jennings LK, Zhang XA. Tetraspanins and vascular function. Cardiovascular Res. 2009;83:7–15. doi: 10.1093/cvr/cvp080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McConnell RE, Higginbotham JN, Shifrin DA, Jr, Tabb DL, Coffey RJ, Tyska MJ. The enterocyte microvillus is a vesicle-generating organelle. J. Cell Biol. 2009;185:1285–1298. doi: 10.1083/jcb.200902147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaji K, Oda S, Shikano T, Ohnuki T, Uematsu Y, Sakagami J, Tada N, Miyazaki S, Kudo A. The gamete fusion process is defective in eggs of CD9-deficient mice. Nature Genetics. 2000;24:279–282. doi: 10.1038/73502. [DOI] [PubMed] [Google Scholar]
- 11.Runge KE, Evans JE, He Z-Y, Gupta S, McDonald KL, Stahlberg H, Primakoff P, Myles DG. Oocyte CD9 is enriched on the microvillar membrane and required for normal microvillar shape and distribution. Develop. Biol. 2007;304:317–325. doi: 10.1016/j.ydbio.2006.12.041. [DOI] [PubMed] [Google Scholar]
- 12.Miyado K, Yoshida K, Yamagata K, Sakakibara K, Okabe M, Wang X, Miyamoto K, Akutsu H, Kondo T, Takahashi Y, Ban T, Ito C, Toshimori K, Nakamura A, Ito M, Miyado M, Mekada E, Umezawa A. The fusing ability of sperm is bestowed by CD9-containing vesicles released from eggs in mice. Proc. Natl. Acad. Sci. USA. 2008;105:12921–12926. doi: 10.1073/pnas.0710608105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Israels SJ, McMillan-Ward EM. Platelet tetraspanin complexes and their association with lipid rafts. Thromb. Haemost. 2007;98:1081–1087. [PubMed] [Google Scholar]
- 14.Brisson C, Azorsa DO, Jennings LK, Moog S, Cazenave JP, Lanza F. Co-localization of CD9 and GPIIb-IIIa integrin on activated platelet pseudopods and alpha-granule membranes. Histochem. J. 1997;29:153–165. doi: 10.1023/a:1026437522882. [DOI] [PubMed] [Google Scholar]
- 15.Pan Y, Brown C, Wang X, Geisert EE. The developmental regulation of CD81 in rat retina. Mol. Vis. 2007;13:181–189. [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang F, Michaelson JE, Moshiach S, Sachs N, Zhao W, Sun Y, Sonnenberg A, Lahti JM, Huang H, Zhang XA. Tetraspanin CD151 maintains vascular stability by balancing the forces of cell adhesion and cytoskeletal tension. Blood. 2011;118:4274–4284. doi: 10.1182/blood-2011-03-339531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singethan K, Müller N, Schubert S, Lüttge D, Krementsov DN, Khurana SR, Krohne G, Schneider-Schaulies S, Thali M, Schneider-Schaulies J. CD9 clustering and formation of microvilli zippers between contacting cells regulates virus-induced cell fusion. Traffic. 2008;9:924–935. doi: 10.1111/j.1600-0854.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-España A, Chung PJ, Sarkar IN, Stiner E, Sun TT, Desalle R. Appearance of new tetraspanin genes during vertebrate evolution. Genomics. 2008;91:326–334. doi: 10.1016/j.ygeno.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 19.Huang S, Yuan S, Dong M, Su J, Yu C, Shen Y, Xie X, Yu Y, Yu X, Chen S, Zhang S, Pontarotti P, Xu A. The phylogenetic analysis of tetraspanins projects the evolution of cell-cell interactions from unicellular to multicellular organisms. Genomics. 2005;86:674–684. doi: 10.1016/j.ygeno.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 20.Kreidberg JA, Donovan MJ, Goldstein SL, Rennke H, Shepherd K, Jones RC, Jaenisch R. Alpha3beta1 integrin has a crucial role in kidney and lung organogenesis. Development. 1996;122:3537–3547. doi: 10.1242/dev.122.11.3537. [DOI] [PubMed] [Google Scholar]
- 21.Sachs N, Kreft M, van den Bergh Weerman MA, Beynon AJ, Peters TA, Weening J, Sonnenberg A. Kidney failure in mice lacking tetraspanin CD151. J. Cell Biol. 2006;175:33–39. doi: 10.1083/jcb.200603073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baleato RM, Guthrie PL, Gubler MC, Ashman LK, Roselli S. Deletion of CD151 results in a strain-dependent glomerular disease due to severe alterations of the glomerular basement membrane. Am. J. Pathol. 2008;173:927–937. doi: 10.2353/ajpath.2008.071149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clergeot PH, Gourgues M, Cots J, Laurans F, Latorse MP, Pepin R, Tharreau D, Notteghem JL, Lebrun MH. PLS1, a gene encoding a tetraspanin-like protein, is required for penetration of rice leaf by fungal pathogen Magnaporthe grisea. Proc. Natl. Acad. Sci. U S A. 2001;98:6963–6968. doi: 10.1073/pnas.111132998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coffey GP, Rajapaksa R, Liu R, Sharpe O, Kuo CC, Krauss SW, Sagi Y, Davis RE, Staudt LM, Sharman JP, Robinson WH, Levy S. Engagement of CD81 induces ezrin tyrosine phosphorylation and its cellular redistribution with filamentous actin. J. Cell Sci. 2009;122:3137–3144. doi: 10.1242/jcs.045658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shoham T, Rajapaksa R, Kuo CC, Haimovich J, Levy S. Building of tetraspanin web: distinct structural domains of CD81 function in different cellular compartments. Mol. Cell Biol. 2006;26:1373–1385. doi: 10.1128/MCB.26.4.1373-1385.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cocquerel L, Kuo CC, Dubuisson J, Levy S. CD81-dependent binding of hepatitis C virus E1E2 heterodimers. J. Virol. 2003;77:10677–10683. doi: 10.1128/JVI.77.19.10677-10683.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou B, Liu L, Reddivari M, Zhang XA. The palmitoylation of metastasis suppressoRKAI1/CD82 is important for its motility- and invasiveness-inhibitory activities. Cancer Res. 2004;64:7455–7463. doi: 10.1158/0008-5472.CAN-04-1574. [DOI] [PubMed] [Google Scholar]
- 28.Bari R, Zhang YH, Zhang F, Wang NX, Stipp CS, Zheng JJ, Zhang XA. The Transmembrane Domain Interactions Are Needed for KAI1/CD82-mediated Suppression of Cancer Invasion and Metastasis. Am. J. Path. 2009;174:647–660. doi: 10.2353/ajpath.2009.080685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shigeta M, Sanzen N, Ozawa M, Gu J, Hasegawa H, Sekiguchi K. CD151 regulates epithelial cell-cell adhesion through PKC-and Cdc42-dependent actin cytoskeletal reorganization. J. Cell Biol. 2003;163:165–176. doi: 10.1083/jcb.200301075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kopczynski CC, Davis GW, Goodman CS. A neural tetraspanin, encoded by late bloomer, that facilitates synapse formation. Science. 1996;271:1867–1870. doi: 10.1126/science.271.5257.1867. [DOI] [PubMed] [Google Scholar]
- 31.Nydegger S, Khurana S, Krementsov DN, Foti M, Thali M. Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. J. Cell Biol. 2006;173:795–807. doi: 10.1083/jcb.200508165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Volkman LE. Baculovirus infectivity and the actin cytoskeleton. Curr. Drug Targets. 2007;8:1075–1083. doi: 10.2174/138945007782151379. [DOI] [PubMed] [Google Scholar]
- 33.Gamliel H, Polliack A. Virus-cell interactions as seen by scanning electron microscopy. Isr J Med Sci. 1979;15:647–652. [PubMed] [Google Scholar]
- 34.Brazzoli M, Bianchi A, Filippini S, Weiner A, Zhu Q, Pizza M, Crotta S. CD81 is a central regulator of cellular events required for hepatitis C virus infection of human hepatocytes. J. Virol. 2008;82:8316–8329. doi: 10.1128/JVI.00665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niggli V, Rossy J. Ezrin/radixin/moesin: versatile controllers of signaling molecules and cortical cytoskeleton. Int. J. Biochem. Cell Biol. 2008;40:344–349. doi: 10.1016/j.biocel.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 36.Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Cell Biol. 2002;3:586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- 37.Sala-Valdes M, Ursa A, Charrin S, Rubinstein E, Hemler ME, Sanchez-Madrid F, Yanez-Mo M. EWI-2 and EWI-F link tetraspanin web to actin cytoskeleton through their direct association with ERM proteins. J. Biol. Chem. 2006;281:19665–19675. doi: 10.1074/jbc.M602116200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.