SUMMARY
αβ T cell receptors (TCRs) bind specifically to foreign antigens presented by major histocompatibility complex proteins (MHC) or MHC-like molecules. Accumulating evidence indicates that the germline-encoded TCR segments have features that promote binding to MHC and MHC-like molecules, suggesting co-evolution between TCR and MHC molecules. Here, we assess directly the evolutionary conservation of αβ TCR specificity for MHC. Sequence comparisons showed that some Vβs from distantly related jawed vertebrates share amino acids in their complementarity determining region 2 (CDR2). Chimeric TCRs containing amphibian, bony fish or cartilaginous fish Vβs can recognize antigens presented by mouse MHC class II and CD1d (an MHC-like protein), and this recognition is dependent upon the shared CDR2 amino acids. These results indicate that features of the TCR that control specificity for MHC and MHC-like molecules were selected early in evolution and maintained between species that last shared a common ancestor over 400 million years ago.
INTRODUCTION
In jawed vertebrates, foreign antigens are recognized by B cells and T cells. Both B cells and T cells generate diverse antigen receptors using random rearrangements of germline-encoded gene segments. While B cell antigen receptors bind directly to many types of antigens, T cells bearing αβ T cell receptors (TCRs) recognize antigen bound to major histocompatibility complex (MHC) or MHC-like molecules on cell surfaces (Hansen et al., 2007; Hunig and Bevan, 1981; Kappler et al., 1981; Marrack et al., 2008; Rudolph et al., 2006; Zinkernagel and Doherty, 1974). The TCR binding site is composed of the combination of peptide fragments of antigens and MHCI or MHCII molecules (Babbitt et al., 1985; Bjorkman et al., 1987; Townsend et al., 1985) or of lipids or glycolipids and MHC-like molecules such as CD1d (Bendelac et al., 1995; Bendelac et al., 2007; Cui et al., 1997; Silk et al., 2008).
The fact that αβ T cells are constrained by their requirement for recognition of MHC as well as antigen has long mystified scientists. Certainly, such a requirement ensures that T cells will always react with other cells, an important attribute. However, the phenomenon remains poorly understood. This property of the αβ TCR is partly determined during T cell development in the thymus where thymocytes are allowed to mature only if they express a TCR that can bind MHC or MHC-like molecules (Bevan, 1977b; Fink and Bevan, 1978; Kisielow et al., 1988; Sha et al., 1988; Zinkernagel et al., 1978). Thymic selection therefore shapes the random TCR repertoire in favor of receptors that can recognize self-MHC (Bevan, 1977b; Goldrath and Bevan, 1999).
However, several observations have suggested that the specificity of the random, unselected repertoire of TCRs is also biased in favor of binding MHC (Blackman et al., 1986; Jerne, 1971; Marrack et al., 2008; Merkenschlager et al., 1997; Zerrahn et al., 1997). For example, at least 1 in 5 randomly created TCRs can recognize MHC (Merkenschlager et al., 1997; Zerrahn et al., 1997). This idea is supported by structural analyses of TCR-MHC interactions (Garcia et al., 2009; Marrack et al., 2008). Although there are exceptions, different mouse and human TCRs frequently interact with MHC in a similar, diagonal, orientation and, in the complexes, the angled position places the germline-encoded CDR1 and CDR2 loops of the TCR Vα and Vβ segments over the MHC α-helices such that certain CDR1 and CDR2 amino acids often contribute to, and are important in, engagement of MHC (Marrack et al., 2008; Rudolph et al., 2006).
Some αβ T cells recognize antigens presented by MHC-like molecules such as those of the CD1 family and MR1 (Bendelac et al., 1995; Hansen et al., 2007; Silk et al., 2008). Among the MHC-like molecules, detailed analysis of the interaction between the TCR expressed by natural killer T (NKT) cells and glycolipid antigens presented by CD1d revealed that, by comparison with TCR interactions with MHC, the NKT TCR is oriented differently when bound to CD1d. Nevertheless, the TCR recognizes jointly the complex of antigen and CD1d and certain TCR CDR amino acids contribute consistently to engagement of CD1d (Borg et al., 2007; Mallevaey et al., 2009; Pellicci et al., 2009; Scott-Browne et al., 2007).
These ideas lead to the hypothesis that germline-encoded TCR gene segments might have been selected by evolution to promote recognition of cell-associated antigens bound to MHC and MHC-like molecules. TCR and MHC genes are present in all jawed vertebrates (Flajnik and Kasahara, 2010; Litman et al., 2010; Rast et al., 1997), therefore such a hypothesis is reasonable. However, even if the hypothesis is correct, the predisposition of TCRs and MHC to interact with each other may not be conferred by the same amino acids in every species. Indeed MHC genes are very polymorphic within species, and many different TCR variable gene sequences can be found in every species examined, suggesting that this might not be the case. However, careful comparison of jawed vertebrate TCR β-chain variable (Vβ) sequences revealed that, despite many amino acid differences, a motif shown to dictate the TCR bias for MHC in mice and humans is also present in some Vβs of these species. Here we show that these Vβs can contribute to MHC + peptide and CD1d + glycolipid ligand recognition by TCRs that are otherwise mouse in origin. These properties depend on the amino acids of the conserved motif. Thus, features of the TCR that promote binding to MHC were present early during vertebrate evolution and have been maintained in several species that last shared a common ancestor more than 400 million years ago.
RESULTS
A common complementarity determining region-2 (CDR2) motif in jawed vertebrate Vβs
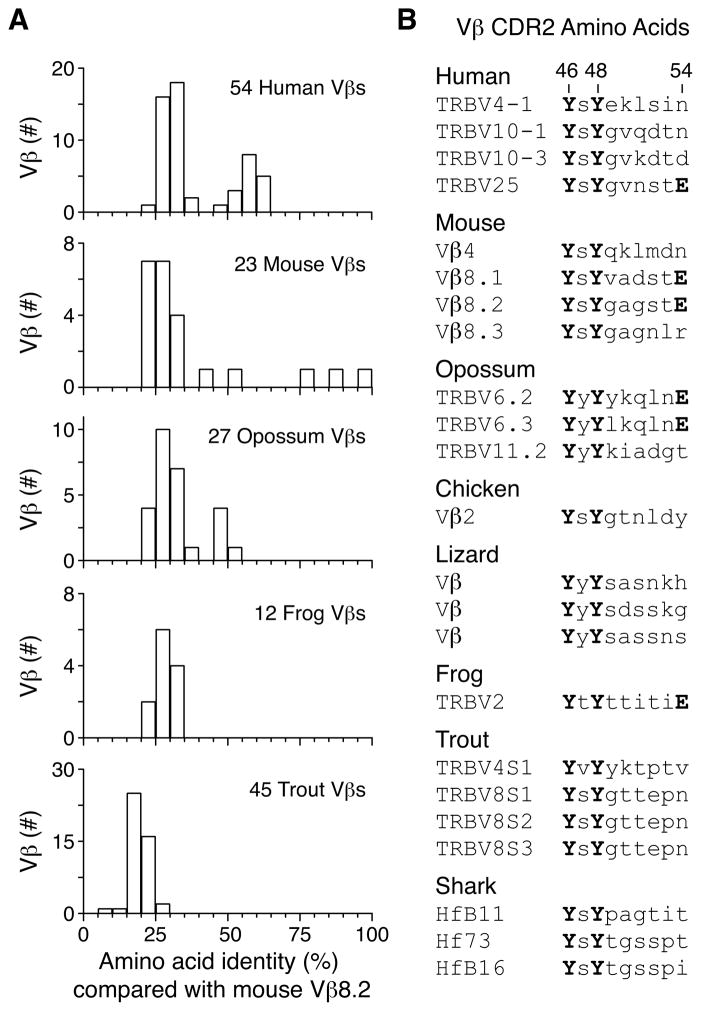
To understand how evolutionary pressures may have selected TCR variable (V) regions toward specificity for MHC, we compared the amino acid sequence of Mus musculus (house mouse) Vβ8.2 with those of Vβ domains from several species of major jawed vertebrate lineages. Amino acid sequences for Vβs in mouse, Homo sapiens (human), Xenopus laevis (African clawed frog) and Oncorhynchus mykiss (rainbow trout) were isolated from the ImMunoGeneTics information system database (Lefranc et al., 2009), while the predicted amino acid sequences of Monodelphis domestica (opossum) Vβs were generated via translation of genomic DNA sequences (Parra et al., 2008). When the full-length Vβ sequences from these species were aligned, most Vβ domains had less than 30% amino acid identity with mouse Vβ8.2 (Figure 1A). A few human and mouse Vβs were highly similar to Vβ8.2, but none of the frog or trout Vβs were substantially more similar to mouse Vβ8.2 than the average (Figure 1A).
Figure 1. Conserved CDR2 motif present in jawed vertebrate Vβ despite low primary Vβ sequence conservation.
A. Amino acid sequence identity between mouse Vβ8.2 and human, mouse, opossum, frog, and trout Vβ regions from pairwise alignments in ClustalW2 using the PAM scoring matrix. B. Predicted CDR2 sequences of jawed vertebrate Vβ regions containing amino acids Y46 and Y48. Those that also contain E54 are indicated.
Because TCR V regions bind MHC via amino acids in their CDR loops, we hypothesized that, in order to preserve this function, motifs in these regions might have been specifically conserved. This idea is supported by the crystallographic and mutational analyses of TCRs, which have shown that some germline-encoded TCR amino acids contribute consistently to binding MHC (Garcia et al., 2009; Marrack et al., 2008). For example, in 8 complexes of TCRs containing mouse Vβ8 family members bound to MHCII, CDR2 amino acids Y46, Y48, and E54 bind MHCII at a common site (Figure S1A)(Dai et al., 2008; Feng et al., 2007; Maynard et al., 2005). For TCR-MHCI complexes, Vβ8 CDR2 amino acids Y46 and Y48 are also positioned over a similar site on the MHCI α1 α-helix with at least one of them making direct contact with the MHCI molecules in all the structures examined (Figure S1B)(Buslepp et al., 2003; Colf et al., 2007; Yin et al., 2011). Furthermore, in one TCR-MHCI complex, these CDR2 amino acids from a mouse TCR bind human MHCI HLA-A2 suggesting that this interaction is shared between the species (Figure S1B)(Buslepp et al., 2003). Thus, these Vβ CDR2 amino acids are required for recognition of antigens presented by MHCI, MHCII, and CD1d (Borg et al., 2007; Dai et al., 2008; Feng et al., 2007; Manning et al., 1998; Rubtsova et al., 2009; Scott-Browne et al., 2007; Scott-Browne et al., 2009) and also control MHC-dependent thymic selection of a broad repertoire of T cells (Scott-Browne et al., 2009).
We searched for Vβ CDR2 amino acids Y46 and Y48 in the appropriate positions in Vβs from human, mouse, opossum, frog and trout, as well as from Gallus gallus domesticus (chicken), Anolis carolinensis (Carolina anole lizard), and Heterodontus fransisci (horn shark)(Hawke et al., 1996). Y46 and Y48 were jointly present in 4 human Vβs, 4 mouse Vβs, 3 opossum Vβs, 1 chicken Vβ, 3 predicted lizard Vβs, 1 frog Vβ, 4 trout Vβs, and 3 shark Vβs (Figure 1B). In addition, E54, an amino acid that is found in the mouse Vβ8 family, and that consistently engages MHC class II (Figure S1A), was found in combination with Y46 and Y48 in the human TRBV25, opossum TRBV6, and frog TRVB2 (Figure 1B). These observations identified a common motif shared between Vβs from different jawed vertebrate species that last shared a common ancestor more than 400 million years ago.
Conserved CDR2 amino acids control frog, fish, and shark Vβ binding to mouse MHCII
To assess the function of these Vβs, we replaced the mouse Vβ8.2 region of the DO-11.10 TCRβ (White et al., 1983) with frog TRBV2, trout TRBV8S2, or shark Vβ3-Hf73 (Table 1 and Figure S2). Only 30% of the amino acids in the sequence of these Vβs are shared with mouse Vβ8.2 and most identical amino acids are in regions that stabilize the Vβ domain or form the interface with Vα (Table 1 and Figure S2A). The amino acids in the CDR1 loops of frog TRBV2, trout TRBV8S2, and shark Vβ3-Hf73 are similar to each other, but these amino acids are dissimilar with those in mouse Vβ8.2 (Table 1 and Figure S2A). Among the amino acids in the CDR2 loop, only Y46 and Y48 are present in all four Vβs while E54 is present in both mouse Vβ8.2 and frog TRBV2 (Table 1). For the most part, the sequences vary at the other CDR2 positions, except for the presence of a Ser or Thr at position 49, which has been observed to be important for a properly folded protein (Manning et al., 1998; Scott-Browne et al., 2007). To refer to the DO-11.10 TCRβ containing the different Vβs we will use the common name of the species of origin (mouse DOβ, frog DOβ, trout DOβ and shark DOβ).
Table 1.
Comparison of mouse, frog, trout, and shark Vβ region sequences.
| Species | Common Name | Class | Vβ | CDR1 | CDR2 | Identity (with Vβ8.2) |
|---|---|---|---|---|---|---|
| M. musculus | House mouse | Mammalia | Vβ8.2 | NNHNN | YSYGAGSTE | - |
| X. laevis | African clawed frog | Amphibia | TRBV2 | DSSKVS | YTYTTITIE | 27% |
| O. mykiss | Rainbow trout | Osteichthyes | TRBV8S2 | DSNLLV | YSYGTTEPN | 20% |
| H. francisci | Horn shark | Chondrichthyes | Vβ3-Hf73 | DSSKSV | YSYTGSSPT | 33% |
We independently expressed mouse DOβ, frog DOβ, trout DOβ or shark DOβ with the full-length mouse TCRβ constant region (Cβ) in hybridomas with the mouse DO-11.10 TCRα (Figure 2A). The cells stained comparably with an antibody specific for the TCRβ constant region, regardless of the source of their Vβ (Figure S3A), suggesting that these various Vβ chains were able to fold and pair properly with the mouse TCRα chain.
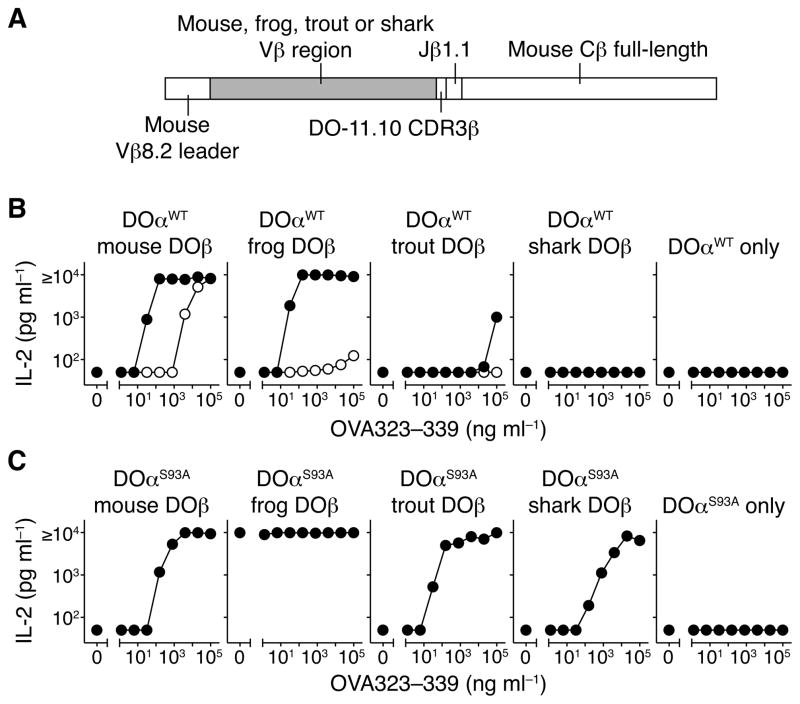
Figure 2. Hybridomas expressing mouse DO-11.10 TCRα plus DO-11.10 TCRβ with mouse, frog, trout, and shark Vβ respond to cells expressing mouse MHCII IAd plus OVA peptide.
A. Schematic of chimeric TCRβ construction replacing mouse Vβ8.2 with frog, trout, and shark Vβ in the DO-11.10 TCRβ with full-length mouse TCRβ constant region for expression in hybridomas. (B and C) IL-2 production of 5KC-73.8.20 hybridomas expressing wild-type DOα (B) or S93A DOα (C) plus mouse DOβ, frog DOβ, trout DOβ, shark DOβ, or no TCRβ after stimulation with IAd-expressing A20.2J cells plus the indicated concentration of OVA323–339 peptide. Filled circles indicate wild-type TCRβ and open circles indicate Y48A TCRβ. Data in (B) and (C) are mean of 2 independent experiments, with a lower limit of detection of IL-2 at 50 pg ml−1.
We used these cells to assess antigen recognition by TCRs containing the different TCRβ chains. The TCR composed of the natural DOα plus mouse DOβ recognizes mouse MHCII IAd plus a chicken ovalbumin peptide (IAd-OVA) (White et al., 1983). Hybridomas expressing DOα with the frog DOβ or the trout DOβ also responded to IAd-OVA (Figure 2B), although high concentrations of OVA peptide were necessary to stimulate cells bearing the trout DOβ. Cells expressing the DOα plus shark DOβ did not respond to any concentration of peptide (Figure 2B).
To determine whether the conserved amino acids in Vβ CDR2 were involved in this recognition, we generated hybridomas expressing mutants of mouse DOβ, frog DOβ and trout DOβ in which Y48 was substituted with Ala (Y48A). In all cases, the Y48A substitution reduced responses to IAd-OVA (Figure 2B), while all hybridomas responded comparably to anti-CD3ε stimulation (Figure S3B). Thus the evolutionarily conserved Vβ Y48 amino acid is involved in recognition of IAd-OVA in all 3 cases.
We have previously identified a natural variant of DOα, with a Ser to Ala (S93A) change in the CDR3 (Scott-Browne et al., 2009). Hybridomas expressing mouse DOβ plus DOαS93A stain with IAd-OVA tetramer better than cells expressing DOαWT, indicating that this CDR3 change increases TCR affinity for IAd-OVA (Figure S3C). We tested whether the S93A substitution in DOα would also improve IAd-OVA recognition by TCRs that contained the various species’ Vβs, and whether it would reveal a reactivity of shark DOβ-containing TCRs for IAd-OVA. This turned out to be true, as hybridomas expressing DOαS93A plus mouse DOβ or frog DOβ or trout DOβ, or even shark DOβ, all responded to IAd-OVA (Figure 2C). In fact, the hybridoma bearing DOαS93A plus frog DOβ responded to the IAd-expressing cells even in the absence of OVA peptide (Figure 2C). An IAd-specific antibody blocked this auto-reactivity and the cells did not respond when cultured alone or with H-2k antigen presenting cells (Figure S3D). Altogether, these results suggest that the autoreactive response of the hybridoma is mediated by a self-peptide presented by a H-2d MHCII molecule.
We next measured directly the binding of the various TCRs to IAd-OVA. Because the DO-11.10 TCR is of somewhat low affinity for IAd-OVA, fluorescently labeled IAd-OVA tetramers stain the T cell hybridomas poorly (Figure S3C). Because MHC tetramer staining intensity is proportional to the amount of TCR expressed on the cell surface, we developed a system to express TCRs at higher concentrations on the surface on insect cells. In this setting, the TCRα and TCRβ transmembrane domains are replaced by acidic and basic leucine zippers, respectively (Figure 3A) (Kappler et al., 1994) and the transmembrane domain of the baculoviral gp64 protein is fused to the COOH-terminus of the TCRβ chain to retain the protein on the insect cell surface (Figure 3A) (Huseby et al., 2006; Stadinski et al., 2010).
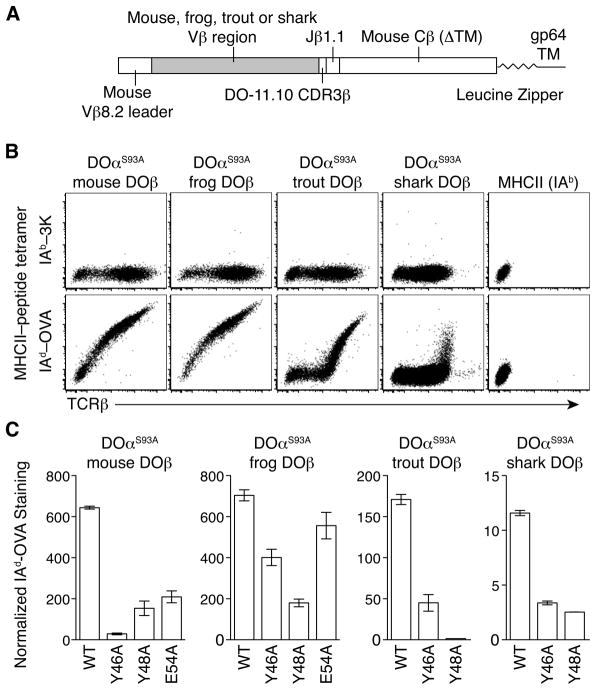
Figure 3. TCRs containing mouse, frog, trout, and shark V β bind mouse MHCII using conserved Vβ CDR2 amino acids.
A. Schematic of chimeric TCRβ construction replacing mouse Vβ8.2 with frog, trout, and shark Vβ in the DO-11.10 TCRβ for expression in insect cells. The mouse TCRβ constant region was truncated just before the transmembrane domain (ΔTM) and linked to a leucine zipper and baculovirus gp64 protein transmembrane (TM) domain. B. IAb-3K or IAd-OVA tetramer staining and TCRβ expression on SF9 insect cells expressing DOαS93A plus mouse DOβ, frog DOβ, trout DOβ, or shark DOβ compared with SF9 cells expressing surface-bound MHCII IAb plus linked 3K peptide. Plots are gated on live cells and are representative of at least 2 independent experiments. C. Geometric mean fluorescence intensity (gMFI) of normalized IAd-OVA tetramer staining on SF9 insect cells expressing DOαS93A plus wild-type (WT) mouse DOβ, frog DOβ, trout DOβ, or shark DOβ and indicated CDR2β amino acid Ala-substitutions. Tetramer staining was normalized by dividing gMFI of IAd-OVA staining by gMFI of background IAb-3K staining from a narrow gate of TCR expression. Data are mean ± range from 2 independent experiments.
Using this system, we expressed DOαS93A plus mouse DOβ, frog DOβ, trout DOβ or shark DOβ on the surface of SF9 cells using baculoviruses. All TCR pairs were expressed on the cell surface and stained specifically with IAd-OVA tetramers. No staining was detected with control IAb-3K or IAd-CLIP tetramers (Figures 3B and S4A). To assess the contribution of the conserved Vβ CDR2 amino acids to this reactivity, we repeated the experiment with Ala-substitutions of Y46 or Y48 in all of the Vβs and Ala-substitution of E54 in mouse DOβ and frog DOβ. For all TCRs, the Y46A and Y48A changes reduced IAd-OVA tetramer staining compared to the wild-type TCRβ of the same species (Figures 3C and S4A). The E54A mutation in mouse DOβ impaired the interaction with IAd-OVA, but this change in frog DOβ only slightly reduced staining compared to wild-type frog DOβ (Figures 3C and S4A).
To assess whether these CDR2β mutations might have altered the expression or folding of the TCR, we expressed the TCRβ chains using retroviruses containing an internal ribosome entry site linked to green fluorescent protein (IRES-GFP) as a reporter in T cell hybridomas together with the Vα2-containing B3K0508 TCRα chain (Huseby et al., 2005). Cells expressing each of the TCRβ chains stained similarly with antibodies specific for the TCRβ constant region compared to the expression of GFP (Figure S4B). Similarly, the staining with a Vα2-specific antibody for the partner TCRα chain was similar for all TCRβ chains, indicating that the mutations did not affect the overall structure of the TCR (Figure S4B).
Altogether, these observations indicate that TCRs containing frog, trout and shark Vβ regions can all bind mouse MHCII. This cross-species interaction occurs despite differences at 70% of the amino acids, including those in the CDR1 loop in the Vβs compared to mouse Vβ8.2 (Table 1). Instead, in all cases, two conserved amino acids (Y46 and Y48) in CDR2 contribute substantially to the TCR-MHC interaction.
Frog Vβ CDR2 amino acid Y48 controls thymic selection in mice
We have shown previously that efficient thymic selection of a diverse repertoire of TCRs in mice depends on the Vβ CDR2 amino acids that comprise the MHC-binding motif, particularly the tyrosine at position 48 (Scott-Browne et al., 2009). To determine whether frog TRBV2 could promote T cell development in mice and whether thymic selection in mice was dependent on the evolutionarily conserved Vβ CDR2 amino acid Y48, we generated mice expressing wild-type frog DOβ or Y48A frog DOβ. Bone marrow chimeras were generated using donor cells transduced with retroviruses expressing the TCRβ plus IRES-GFP as a reporter (Figure 4A).
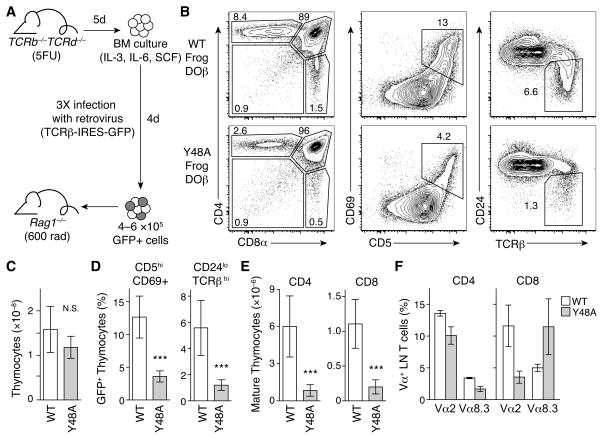
Figure 4. Thymic selection of mouse cells expressing a frog Vβ containing TCRβ is controlled by CDR2 amino acid Y48.
A. Schematic of retroviral mediated expression of frog DOβ in mice using bone marrow chimeras. B. Flow cytometry analysis of CD4 and CD8 expression, CD5 and CD69 expression, or CD24 and TCRβ expression on thymocytes from mice expressing WT or Y48A frog DOβ. Plots are gated on live, GFP positive cells and are representative of two independent experiments. C. Total number of thymocytes in mice expressing WT or Y48A frog DOβ. D. Frequency of activated (CD5hiCD69+) or mature (CD24loTCRβhi) cells among GFP-expressing thymocytes in chimera expressing WT or Y48A frog DOβ. E. Total number of mature CD24loTCRβhi CD4SP (left) or CD8SP (right) thymocytes from mice expressing WT or Y48A frog DOβ. F. Frequency of Vα2 and Vα8.3 expressing CD4+ (left) or CD8+ (right) LN T cells from mice expressing WT frog DOβ (white bars) or Y48A frog DOβ (grey bars). Data in (C–F) are mean ± s.d. and include all mice from two independent experiments (n=8 for WT frog DOβ, n=9 for Y48A frog DOβ); ***P<0.001; N.S., not significant.
We observed the characteristic developmental subsets (Figure 4B) in the thymuses of mice expressing each of the TCRβ chains indicating that a TCR containing a frog Vβ can initiate β- and positive selection in mice. Additionally, mice expressing wild-type or Y48A frog DOβ had comparable thymic cellularity (Figure 4C) with similar GFP expression and TCRβ expression profiles (Figure S5). In mice expressing Y48A frog DOβ, the frequency of immature CD4+CD8+ double-positive thymocytes was increased, while the frequencies of CD4 single positive (CD4SP) and CD8 single positive (CD8SP) thymocytes were reduced, compared to mice expressing the wild-type frog DOβ (Figure 4C). This reduction in positive selection effected by the Y48A mutation in the frog Vβ was confirmed by the lower frequencies of positively selected, activated (CD5+CD69+) and mature (CD24loTCRβhi) thymocytes in chimeras expressing Y48A frog DOβ compared to those expressing wild-type frog DOβ (Figure 4D). As an estimation of thymic output, we calculated the total numbers of mature CD4SP and CD8SP thymocytes. These were reduced in mice expressing Y48A versus wild-type frog DOβ (Figure 4E), indicating that recognition of both MHC class II and MHC class I during thymic selection is impaired by mutation of this CDR2 amino acid.
Although the Y48A substitution in mouse DOβ greatly impairs thymic selection, some T cells bearing the mutant TCRβ can nevertheless be positively selected (Scott-Browne et al., 2009). These T cells use a different TCRα repertoire than cells bearing the wild-type mouse DOβ, perhaps because cells expressing Y48A mouse DOβ require a greater contribution of TCRα to MHC binding (Scott-Browne et al., 2009). We tested whether the TCRα repertoire was also altered in T cells bearing Y48A frog DOβ versus wild-type frog DOβ. The frequency of CD4+ T cells expressing Vα2 or Vα8.3 was reduced, by comparison with wild-type frog DOβ, in Y48A frog DOβ chimeras (Figure 4F). Vα2-expressing CD8+ T cells were also reduced, while the frequency of Vα8.3-expressing CD8+ cells was increased in Y48A frog DOβ chimeras (Figure 4F). Interestingly, these changes in Vα use are similar to results obtained in comparable experiments using variants of mouse DOβ (Scott-Browne et al., 2009).
Conserved CDR2 amino acids are required for frog, fish, and shark Vβ to bind mouse CD1d
Some αβ T cells recognize MHC-like molecules, such as the mammalian CD1 family, which present lipid antigens (Bendelac et al., 1995; Silk et al., 2008). Genes encoding CD1-like proteins are present in birds, but not in amphibians or bony or cartilaginous fish (Dascher, 2007). CD1d is found only in mammals (Dascher, 2007) and a special αβ T cell population, the natural killer T (NKT) cells, recognizes glycolipid antigens presented by CD1d (Bendelac et al., 1995; Bendelac et al., 2007; Cui et al., 1997). In mice, most NKT cells express a restricted TCR repertoire with an invariant TCRα (iNKTα) (Lantz and Bendelac, 1994) and a biased but diverse repertoire of TCRβs that often involve Vβ8.2. In fact, pairing the DO-11.10 TCRβ with iNKTα allows reaction with CD1d plus multiple glycolipids (Scott-Browne et al., 2007). Although the orientation of the NKT TCR on CD1d differs markedly from that of conventional αβ TCRs bound to MHCI and MHCII, the Vβ8.2 CDR2 amino acids Y46, Y48, and E54 also bind CD1d (Borg et al., 2007; Pellicci et al., 2009) and are required for antigen recognition (Mallevaey et al., 2009; Scott-Browne et al., 2007).
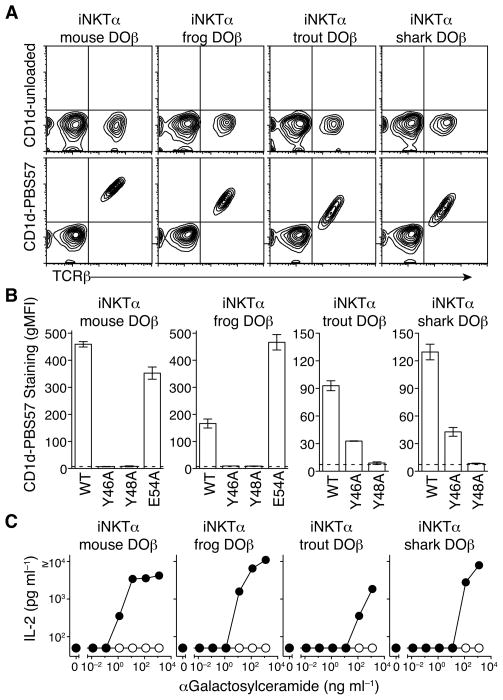
To find out if non-mammalian Vβs could participate in CD1d binding we expressed mouse DOβ, frog DOβ, trout DOβ or shark DOβ (Figure 2A) in hybridomas with the mouse iNKTα. TCRs containing any of these Vβs stained specifically with mouse CD1d tetramers loaded with the synthetic glycolipid antigen PBS57 (Liu et al., 2006) and did not stain with the same tetramer without addition of antigen (Figure 5A). To determine if the Vβ CDR2 amino acids Y46, Y48, or E54 were involved in antigen recognition we used hybridomas expressing Ala-substitutions of Y46 and Y48 in each of the Vβs and Ala substitutions for E54 in mouse DOβ or frog DOβ. Y46 and Y48 contributed to CD1d binding for all four TCRs, as the Y46A and Y48A substitutions in mouse DOβ and frog DOβ and Y48A substitution in trout DOβ and shark DOβ completely eliminated interaction with CD1d-PBS57 tetramers (Figures 5B and S6). Trout DOβ and shark DOβ had a slightly different pattern, since mutation of Y46 reduced, but did not completely eliminate, staining with CD1d-PBS57 tetramers (Figures 5B and S6). Finally, hybridomas expressing iNKTα plus wild-type mouse, frog, trout, and shark TCRβ chains secreted IL-2 after stimulation with α-Galactosylceramide presented by CD1d-expressing APCs, while cells expressing TCRβ chains with the Y48A substitution were unresponsive at all concentrations of antigen (Figure 5C). Thus, a common motif in jawed vertebrate TCR Vβ contributes to recognition of antigens presented by both MHC and CD1d.
Figure 5. NKT TCRs containing mouse, frog, trout, and shark Vβ bind mouse CD1d using a conserved motif.
A. Flow cytometry analysis of TCRβ expression and CD1d tetramer staining on 5KC-73.8.20 hybridomas expressing wild-type Vα14-Jα18 NKT TCRα (iNKTα) plus mouse DOβ, frog DOβ, trout DOβ, or shark DOβ. B. gMFI of CD1d-PBS57 tetramer staining on 5KC-73.8.20 hybridomas expressing wild-type iNKTα plus WT mouse DOβ, frog DOβ, trout DOβ, or shark DOβ and indicated CDR2β amino acid Ala-substitutions. Data are gMFI ± range calculated from a narrow gate of TCRβ expression from 2 independent experiments. The dotted line indicates average MFI of CD1d-PBS57 tetramer staining on 5KC-73.8.20 hybridomas expressing only the iNKTα and no TCRβ. C. IL-2 secretion from 5KC-73.8.20 hybridomas expressing iNKTα plus wild-type (filled circles) or Y48A (open circles) mouse DOβ, frog DOβ, trout DOβ, or shark DOβ after stimulation with A20 cells expressing mouse CD1d plus the indicated concentration of α-GalCer. Data are mean of 2 independent experiments, with a lower limit of detection of IL-2 at 50 pg ml−1.
DISCUSSION
The genes encoding TCRα, TCRβ, MHCI, MHCII, and MHC-like molecules have been identified in all lineages of jawed vertebrates, including the cartilaginous fishes, which last shared a common ancestor with mammals over 400 million years ago (Cooper and Alder, 2006; Flajnik and Kasahara, 2010; Hawke et al., 1996; Litman et al., 1999; Litman et al., 2010; Rast et al., 1997; Rast et al., 1995). While the αβ T cell recognition of MHC has primarily been characterized in mammals, evidence for MHC-dependent T cell immunity has been observed in other vertebrate lineages as well (Litman et al., 1999).
These observations led us to investigate the features that control the specificity of αβ TCRs for MHC. Based on the many functional, mutational, and crystallographic analyses, we and others have observed that germline-encoded features of some mammalian αβ TCR V regions promote recognition of MHC and MHC-like molecules (Borg et al., 2007; Dai et al., 2008; Feng et al., 2007; Jerne, 1971; Lee et al., 2000; Mallevaey et al., 2009; Manning et al., 1998; Marrack et al., 2008; Maynard et al., 2005; Newell et al., 2011; Rudolph et al., 2006; Scott-Browne et al., 2007; Scott-Browne et al., 2009; Sim et al., 1996). This leads directly to the hypothesis that evolutionary pressures have selected these regions for interaction with MHC (Jerne, 1971; Marrack et al., 2008), thereby promoting T cell recognition of cell-associated antigens.
We and others have previously identified a motif in the CDR2 loop of some mouse and human Vβs. Structural and biochemical studies have shown that the amino acids that define this motif are consistently involved in engagement of MHCI, MHCII, and the MHC-like protein, CD1d (Borg et al., 2007; Dai et al., 2008; Feng et al., 2007; Jerne, 1971; Lee et al., 2000; Manning et al., 1998; Marrack et al., 2008; Maynard et al., 2005; Rudolph et al., 2006; Scott-Browne et al., 2007; Scott-Browne et al., 2009). We found that this Vβ CDR2 motif was also present in some Vβs from jawed vertebrate species distantly related to mammals. Even though the overall sequence of these Vβs was only 30% similar to those of mouse and human, these distant species’ Vβs could nevertheless substitute for the mouse Vβ in recognition of mouse MHCII + peptide. More amazingly, the Vβs from the distant species could substitute for mouse Vβ in recognition of mouse CD1d + glycolipid, even though CD1d-like proteins are not thought to exist in amphibians, cartilaginous fish, or bony fish and even though TCRs engage CD1d in a configuration that is completely different from that of their engagement of conventional MHC. In every case recognition of MHC or CD1d was determined by at least one amino acid in the motif.
Thus, a motif in CDR2 exists, and functions similarly, in some Vβ gene segments from mammals, amphibians, bony and cartilaginous fishes; species that last shared a common ancestor more than 400 million years ago (Flajnik and Kasahara, 2010). These experiments suggest that the predilection of at least some TCR Vβs to interact with MHC was determined at least 400 million years ago, very early in vertebrate evolution. Moreover, it is likely that when CD1d and NKT cells evolved (Dascher, 2007), the immune system took advantage of Vβs with a preexisting motif to produce TCRs that could react in a non-conventional way with CD1d plus glycolipid ligands.
There are some exceptions to the idea that amino acid sequences in the germline-encoded CDR loops of TCR variable regions confer the bias for MHC reactivity on T cells. For example, for some TCRs, the non-germline-encoded CDR3 loops dominate the interaction with their MHC-peptide ligand (Borg et al., 2005; Tynan et al., 2005). Additionally, some TCRs bind MHC in atypical orientations (Sethi et al., 2011), while other TCRs that bind ligands other than MHC have been found (Hanada et al., 2011). These examples suggest that the idea that germlime-encoded TCR regions have amino acids that promote interaction with MHC might be incorrect. However, the αβ TCR repertoire is extraordinarily diverse and the highly selective pressures of clonal expansion, thymic selection or autoimmune responses may, in some cases, pick out cells expressing TCRs that may not use conserved mechanisms for antigen specificity. In spite of these exceptions, a large amount of crystallographic and mutational data indicates that there is a CDR2 motif in a group of jawed vertebrate Vβs that controls TCR interaction with MHCI, MHCII, and CD1d (Borg et al., 2007; Dai et al., 2008; Feng et al., 2007; Jerne, 1971; Lee et al., 2000; Manning et al., 1998; Marrack et al., 2008; Maynard et al., 2005; Rudolph et al., 2006; Scott-Browne et al., 2007; Scott-Browne et al., 2009).
While the group of jawed vertebrate TCR Vβ regions with the tandem CDR2 tyrosines is somewhat small, there are several Vβ regions that contain similar amino acids at these positions. Both tyrosine and phenylalanine are commonly found at these positions in TCR Vβs. For example, Y46 (but not Y48) is found in a family that is related to Vβ8.2 including mouse Vβ6 and mouse Vβ14 (Lefranc et al., 2009; Marrack et al., 2008). We have observed previously that Vβ6 CDR2 Y46 contributes substantially to both specific MHCII-peptide recognition and enhances the efficiency of development of thymocytes expressing TCRs with this Vβ (Scott-Browne et al., 2009). Similarly, in crystal structures of mouse Vβ7 (which also has the CDR2 Y48 amino acid) and Vβ8.2 containing Vα14i NKT TCRs bound to αGalCer-CD1d, the Vβ CDR2 Y48 side chain binds the CD1d α1 α-helix in a similar orientation and is required for antigen recognition (Mallevaey et al., 2009; Pellicci et al., 2009).
Thus, although some of these CDR2 features are shared with a larger group of Vβs, other Vβs in these species do not contain these amino acids (Lefranc et al., 2009). It is possible that other germline-encoded amino acids in these Vβs may have been selected to promote binding to MHC or MHC-like proteins. Indeed, TCRs that contain mouse Vβ3, which does not share this CDR2 motif, have been observed to bind to MHCII-peptide complexes using a different motif and this interaction is shared between different TCRs (Newell et al., 2011).
The evolution of TCR and their MHC ligands has long been a mystery. It seems that neither could exist usefully without the other and the two seem to have evolved almost simultaneously. Moreover, if the idea that TCR variable regions have been selected to engage MHC is right, how can this be achieved when the TCR V region families are so large? How can natural selection operate on individual members of the families? Of course infectious disease is a powerful selective force, and individual variable regions might each have a role to play in effective responses to the many invading organisms that have plagued us and our ancestors.
The data presented here support the co-evolution point of view, since, for at least some Vβs, the motif that predisposes recognition of MHC appears to have existed in a common ancestor of sharks and mammals, a common ancestor that existed at least 400 million years ago. Evolution has found other ways to solve the problem of clone specific recognition of antigen, such as the variable lymphocyte receptors expressed in lampreys (Alder et al., 2005; Pancer and Cooper, 2006). In the jawed vertebrates, T cell receptors and MHC, together with the extremely variable antibody molecules, are firmly entrenched. The problem of how TCRs and MHC appeared within a relatively short space of time remains unsolved.
The idea that TCR and MHC proteins have been evolutionarily selected to bind one another (Jerne, 1971; Marrack et al., 2008) may explain the high frequency of T cells that react with allogeneic MHC. Likewise, the observations presented here may also account for the relatively high frequency of xenoreactive TCRs. However, neither the data presented here nor elsewhere suggest that TCR recognition of MHC is peptide independent. Rather, the combined results indicate that TCR V region motifs bias TCRs for generic recognition of MHC (Bevan, 1977a; Colf et al., 2007), and that, usually, specific recognition of peptide, mostly by TCR CDR3 regions, is needed to lift the affinity of the TCR interaction with MHC-peptide interaction to trigger T cell activation (Colf et al., 2007; Macdonald et al., 2009; Matzinger and Bevan, 1977; Morris et al., 2011; Rubtsova et al., 2009).
EXPERIMENTAL PROCEDURES
TCR Vβ sequence analysis
Sequences of the *01 allele of human, mouse, frog, and trout TCR Vβ gene segments (lacking the signal peptide) were downloaded from the IMGT database (Lefranc et al., 2009). DNA sequences of opossum TCR Vβ gene segments were isolated from the genome sequence (MonDom5) using published coordinates (Parra et al., 2008) and electronically translated in reading frame 1. The region of the Vβ surrounding CDR2 from in human, mouse, opossum, frog, and trout gene segments were isolated and searched for a YxYxxxxxx motif using the 3of5 web application (Seiler et al., 2006).
Horned shark Vβ sequences were searched manually for the CDR2 motif from the list of published TCRβ sequences (Hawke et al., 1996). We also searched for Vβ sequences with similarity to mouse Vβ8.2 in the genome of chicken (WASHUC2) and anole lizard (AnoCar2.0) with BLAST. Hits that were in the same locus as TCRβ constant gene segments were searched manually for the CDR2 motif when aligned to mouse Vβ8.2.
αβ TCR constructs
Plasmids encoding wild-type and mutant TCRα and TCRβ chains for the DO-11.10 TCR were described previously (Scott-Browne et al., 2009). DNA fragments of frog TRBV2 (Lefranc et al., 2009) and shark Vβ3-Hf73 (Hawke et al., 1996) gene segments linked to mouse Vβ8.2 signal peptide (plus Ala-substitutions of Y46, Y48, and E54) were synthesized by GenScript (Piscataway, NJ, USA) based on published amino acid sequences. The trout TRBV8S2 gene segment was amplified by PCR from genomic DNA isolated from rainbow trout fillet (Shamrock Foods, Commerce City, CO, USA) and extended with overlapping primers to encode the mouse Vβ8.2 signal peptide. The sequence of the Vβ gene segment matched the published nucleotide sequence (AJ517978.1) (Lefranc et al., 2009).
For retroviral expression, TCR constructs were cloned in MSCV-based plasmids with natural mouse TCR α or β chain constant region plus an IRES and GFP as a reporter, as described previously (Scott-Browne et al., 2009). For expression in insect cells, TCR α and β chains were cloned in a baculovirus transfer plasmid containing mouse TCRα and TCRβ chain constant regions, both truncated just before the transmembrane domain and linked to complementary acidic or basic leucine zippers, as described previously (Crawford et al., 2004; Kappler et al., 1994). The TCRα chain constant region contained a single amino acid substitution N81D, to eliminate a potential N-linked glycoslation site. The TCRβ chain constant region contained three amino acid substitutions (N5D, C70S, N115D), to eliminate two potential N-linked glycosylation sites and to remove an unpaired cysteine. The transmembrane region of the baculovirus gp64 protein was added to the COOH-terminus of the TCRβ chain to retain the protein on the surface (Crawford et al., 2004).
Retroviral packaging
Retroviral plasmids were co-transfected into Phoenix cells with pCLEco accessory plasmid using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), as described previously (Scott-Browne et al., 2009).
Hybridomas
TCR constructs were expressed by retroviral transduction into a hybridoma that lacks TCRα and TCRβ (5KC-73.8.20) (White et al., 1993), as described previously (Scott-Browne et al., 2009). After spin-infection, expanded hybridomas were sorted on a MoFlo XDP cell sorter (Beckman-Coulter) on the basis of retroviral reporter and TCRβ expression.
For all stimulations, 5×104 hybridoma cells were cultured overnight with indicated stimuli. For OVA323–339 peptide stimulation, hybridomas were cultured with 5×104 IAd-expressing A20.2J cells plus the indicated concentration of peptide. For αGalCer stimulation, hybridomas were cultured with 5×104 A20 cells expressing mouse CD1d (Brossay et al., 1998) plus the indicated concentration of glycolipid. Hybridoma responses were measured by an ELISA for IL-2, using standard protocols.
Insect cells
SF9 insect cells were maintained as described previously (Kappler et al., 1994). Genes encoding αβ TCRs or MHCII molecules were introduced into baculoviruses by cotransfection of the transfer plasmid plus linear Sapphire baculovirus DNA (Orbigen, San Diego, CA, USA) using calcium phosphate in SF9 cells (Kappler et al., 1994). After 7–10 days, culture supernatant was collected and expanded to high-titer stock by reinfection of SF9 cells, as described previously (Kappler et al., 1994).
Tetramers
To generate IAd-OVA protein, the IAd α chain and the IAd β chain were cloned in a baculovirus transfer vector, as described previously (Crawford et al., 1998; Stadinski et al., 2010). The OVA peptide (GHAAHAEINEA) was linked to the NH2-terminus of the IAd β chain with by a flexible linker (GCGGTSGGGSGGS), as described previously (Stadinski et al., 2010). The IAd α chain has an N72C substitution and forms a disulfide bond with the cysteine in the linker between the peptide and the IAd β chain (Stadinski et al., 2010).
IAd-OVA, IAd-CLIP and IAb-3K tetramers were generated with phycoerythrin-labeled streptavidin plus biotinylated MHCII protein, as described previously (Crawford et al., 1998). Phycoerythrin-labeled mouse CD1d tetramers loaded with PBS57 or without added antigen were provided by the NIH tetramer facility.
Tetramer staining
T cell hybridomas or SF9 insect cells (infected 3 days earlier with baculovirus) were stained with 20 μg ml−1 phycoerythrin-labeled MHCII tetramer at 37°C for 90 minutes in anti-CD16/CD32 antibody producing hybridoma supernatant (clone 2.4G2), at which point allophycocyanin-labeled anti-TCRβ (clone H57–597) was added and stained for an additional 20 minutes at 25°C. For CD1d tetramer staining, T cell hybridomas were co-stained with phycoerythrin-labeled CD1d tetramer and allophycocyanin-labeled anti-TCRβ in 2.4G2 supernatant for 30 minutes at 25°C.
Bone marrow chimeras
Bone marrow chimeras were generated as described previously (Scott-Browne et al., 2009). Briefly, bone marrow cells were harvested from 5-fluorouracil treated TCRb−/−TCRd−/− double deficient mice and cultured in medium containing IL-3, IL-6, and stem cell factor. Cells were retrovirally infected on days 1, 2, and 3 of in vitro culture and on day 4, ~5.0×105 GFP expressing cells were injected into irradiated Rag1−/−mice (600 RAD). Mice were analyzed at days 35–39 post reconstitution. Mice that reconstituted poorly (those with less than 3.0×107 total thymocytes) were excluded from the analysis. All experiments with mice were performed under protocols approved by the Institutional Animal Care and Use Committee at National Jewish Health.
Antibodies
Prior to staining, T cell hybridomas and ex vivo cells were incubated in supernatant from an anti-CD16/CD32 producing hybridoma (clone 2.4G2). Cells were stained under saturating conditions with antibodies to mouse TCRβ (clone H57–597), CD4 (clone GK1.5), CD8 (clone 53–6.7), CD5 (clone 53–7.3), CD69 (clone H1.2F3), CD24 (clone M1/69), B220 (clone RA3-6B2), Vα2 (clone B20.1), and Vα8.3 (clone B21.14) purchased from eBiosciences (San Diego, CA, USA), BD Pharmingen (San Diego, CA, USA), or generated in house.
Flow cytometry
Cells were analyzed by flow cytometry on an LSR II (BD Biosciences, San Diego, CA, USA) and data were analyzed in FlowJo (Treestar, Ashland, OR, USA).
Statistical Analysis
An unpaired, two-tailed Student’s t-test was used to compare log10-transformed data (GraphPad Prism, La Jolla, CA, USA).
Supplementary Material
Acknowledgments
We thank members of the Kappler/Marrack and Gapin labs for thoughtful discussion and Drs. Peter Henson (NJH) and Mitchell Kronenberg (LIAI) for comments on the manuscript. We thank the flow cytometry facility at NJH for technical assistance with cell sorting and Ella Kushnir and Dr. Megan MacLeod (NJH) for assistance generating bone marrow chimeras. We thank Dr. Yosef Refaeli (NJH) for providing stem cell cytokine containing conditioned medium. Rainbow trout fillet was generously provided by Juan Campos (NJH). This work was supported by grants from the National Institutes of Health (AI18785 and AI22295 to P.M. and J.W.K. and AI078246, AI076463 and AI092108-01A1 to L.G.). J.P.S.-B. and M.H.Y. were supported by an NIH training grant (T32 AI07405).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alder MN, Rogozin IB, Iyer LM, Glazko GV, Cooper MD, Pancer Z. Diversity and function of adaptive immune receptors in a jawless vertebrate. Science. 2005;310:1970–1973. doi: 10.1126/science.1119420. [DOI] [PubMed] [Google Scholar]
- Babbitt BP, Allen PM, Matsueda G, Haber E, Unanue ER. Binding of immunogenic peptides to Ia histocompatibility molecules. Nature. 1985;317:359–361. doi: 10.1038/317359a0. [DOI] [PubMed] [Google Scholar]
- Bendelac A, Lantz O, Quimby ME, Yewdell JW, Bennink JR, Brutkiewicz RR. CD1 recognition by mouse NK1+ T lymphocytes. Science. 1995;268:863–865. doi: 10.1126/science.7538697. [DOI] [PubMed] [Google Scholar]
- Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- Bevan MJ. High determinant density may explain the phenomenon of alloreactivity. Immunology Today. 1977a;5:128–130. doi: 10.1016/0167-5699(84)90233-0. [DOI] [PubMed] [Google Scholar]
- Bevan MJ. In a radiation chimaera, host H-2 antigens determine immune responsiveness of donor cytotoxic cells. Nature. 1977b;269:417–418. doi: 10.1038/269417a0. [DOI] [PubMed] [Google Scholar]
- Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature. 1987;329:512–518. doi: 10.1038/329512a0. [DOI] [PubMed] [Google Scholar]
- Blackman M, Yague J, Kubo R, Gay D, Coleclough C, Palmer E, Kappler J, Marrack P. The T cell repertoire may be biased in favor of MHC recognition. Cell. 1986;47:349–357. doi: 10.1016/0092-8674(86)90591-x. [DOI] [PubMed] [Google Scholar]
- Borg NA, Ely LK, Beddoe T, Macdonald WA, Reid HH, Clements CS, Purcell AW, Kjer-Nielsen L, Miles JJ, Burrows SR, et al. The CDR3 regions of an immunodominant T cell receptor dictate the ‘energetic landscape’ of peptide-MHC recognition. Nat Immunol. 2005;6:171–180. doi: 10.1038/ni1155. [DOI] [PubMed] [Google Scholar]
- Borg NA, Wun KS, Kjer-Nielsen L, Wilce MC, Pellicci DG, Koh R, Besra GS, Bharadwaj M, Godfrey DI, McCluskey J, et al. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448:44–49. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- Brossay L, Tangri S, Bix M, Cardell S, Locksley R, Kronenberg M. Mouse CD1-autoreactive T cells have diverse patterns of reactivity to CD1+ targets. J Immunol. 1998;160:3681–3688. [PubMed] [Google Scholar]
- Buslepp J, Wang H, Biddison WE, Appella E, Collins EJ. A correlation between TCR Valpha docking on MHC and CD8 dependence: implications for T cell selection. Immunity. 2003;19:595–606. doi: 10.1016/s1074-7613(03)00269-3. [DOI] [PubMed] [Google Scholar]
- Colf LA, Bankovich AJ, Hanick NA, Bowerman NA, Jones LL, Kranz DM, Garcia KC. How a single T cell receptor recognizes both self and foreign MHC. Cell. 2007;129:135–146. doi: 10.1016/j.cell.2007.01.048. [DOI] [PubMed] [Google Scholar]
- Cooper MD, Alder MN. The evolution of adaptive immune systems. Cell. 2006;124:815–822. doi: 10.1016/j.cell.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Crawford F, Huseby E, White J, Marrack P, Kappler JW. Mimotopes for alloreactive and conventional T cells in a peptide-MHC display library. PLoS Biol. 2004;2:E90. doi: 10.1371/journal.pbio.0020090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford F, Kozono H, White J, Marrack P, Kappler J. Detection of antigen-specific T cells with multivalent soluble class II MHC covalent peptide complexes. Immunity. 1998;8:675–682. doi: 10.1016/s1074-7613(00)80572-5. [DOI] [PubMed] [Google Scholar]
- Cui J, Shin T, Kawano T, Sato H, Kondo E, Toura I, Kaneko Y, Koseki H, Kanno M, Taniguchi M. Requirement for Vα14 NKT cells in IL-12-mediated rejection of tumors. Science. 1997;278:1623–1626. doi: 10.1126/science.278.5343.1623. [DOI] [PubMed] [Google Scholar]
- Dai S, Huseby ES, Rubtsova K, Scott-Browne J, Crawford F, Macdonald WA, Marrack P, Kappler JW. Crossreactive T Cells spotlight the germline rules for alphabeta T cell-receptor interactions with MHC molecules. Immunity. 2008;28:324–334. doi: 10.1016/j.immuni.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dascher CC. Evolutionary biology of CD1. Curr Top Microbiol Immunol. 2007;314:3–26. doi: 10.1007/978-3-540-69511-0_1. [DOI] [PubMed] [Google Scholar]
- Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon’. Nat Immunol. 2007;8:975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- Fink PJ, Bevan MJ. H-2 antigens of the thymus determine lymphocyte specificity. J Exp Med. 1978;148:766–775. doi: 10.1084/jem.148.3.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet. 2010;11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia KC, Adams JJ, Feng D, Ely LK. The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat Immunol. 2009;10:143–147. doi: 10.1038/ni.f.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldrath AW, Bevan MJ. Selecting and maintaining a diverse T-cell repertoire. Nature. 1999;402:255–262. doi: 10.1038/46218. [DOI] [PubMed] [Google Scholar]
- Hanada KI, Wang QJ, Inozume T, Yang JC. Molecular identification of an MHC-independent ligand recognized by a human αβ T-cell receptor. Blood. 2011 doi: 10.1182/blood-2010-11-317743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TH, Huang S, Arnold PL, Fremont D. Patterns of nonclassical MHC antigen presentation. Nat Immunol. 2007;8:563–568. doi: 10.1038/ni1475. [DOI] [PubMed] [Google Scholar]
- Hawke NA, Rast JP, Litman GW. Extensive diversity of transcribed TCR-beta in phylogenetically primitive vertebrate. J Immunol. 1996;156:2458–2464. [PubMed] [Google Scholar]
- Hunig T, Bevan MJ. Specificity of T-cell cones illustrates altered self hypothesis. Nature. 1981;294:460–462. doi: 10.1038/294460a0. [DOI] [PubMed] [Google Scholar]
- Huseby ES, Crawford F, White J, Marrack P, Kappler JW. Interface-disrupting amino acids establish specificity between T cell receptors and complexes of major histocompatibility complex and peptide. Nat Immunol. 2006;7:1191–1199. doi: 10.1038/ni1401. [DOI] [PubMed] [Google Scholar]
- Huseby ES, White J, Crawford F, Vass T, Becker D, Pinilla C, Marrack P, Kappler JW. How the T cell repertoire becomes peptide and MHC specific. Cell. 2005;122:247–260. doi: 10.1016/j.cell.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Jerne NK. The somatic generation of immune recognition. Eur J Immunol. 1971;1:1–9. doi: 10.1002/eji.1830010102. [DOI] [PubMed] [Google Scholar]
- Kappler J, White J, Kozono H, Clements J, Marrack P. Binding of a soluble αβ T-cell receptor to superantigen/major histocompatibility complex ligands. Proc Natl Acad Sci U S A. 1994;91:8462–8466. doi: 10.1073/pnas.91.18.8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappler JW, Skidmore B, White J, Marrack P. Antigen-inducible, H-2-restricted, interleukin-2-producing T cell hybridomas. Lack of independent antigen and H-2 recognition. J Exp Med. 1981;153:1198–1214. doi: 10.1084/jem.153.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisielow P, Teh HS, Bluthmann H, von Boehmer H. Positive selection of antigen-specific T cells in thymus by restricting MHC molecules. Nature. 1988;335:730–733. doi: 10.1038/335730a0. [DOI] [PubMed] [Google Scholar]
- Lantz O, Bendelac A. An invariant T cell receptor α chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4−8− T cells in mice and humans. J Exp Med. 1994;180:1097–1106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PU, Churchill HR, Daniels M, Jameson SC, Kranz DM. Role of 2C T cell receptor residues in the binding of self- and allo-major histocompatibility complexes. J Exp Med. 2000;191:1355–1364. doi: 10.1084/jem.191.8.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc MP, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F, Wu Y, Gemrot E, Brochet X, Lane J, et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37:D1006–1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litman GW, Anderson MK, Rast JP. Evolution of antigen binding receptors. Annu Rev Immunol. 1999;17:109–147. doi: 10.1146/annurev.immunol.17.1.109. [DOI] [PubMed] [Google Scholar]
- Litman GW, Rast JP, Fugmann SD. The origins of vertebrate adaptive immunity. Nat Rev Immunol. 2010;10:543–553. doi: 10.1038/nri2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Goff RD, Zhou D, Mattner J, Sullivan BA, Khurana A, Cantu C, 3rd, Ravkov EV, Ibegbu CC, Altman JD, et al. A modified α-galactosyl ceramide for staining and stimulating natural killer T cells. J Immunol Methods. 2006;312:34–39. doi: 10.1016/j.jim.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Macdonald WA, Chen Z, Gras S, Archbold JK, Tynan FE, Clements CS, Bharadwaj M, Kjer-Nielsen L, Saunders PM, Wilce MC, et al. T cell allorecognition via molecular mimicry. Immunity. 2009;31:897–908. doi: 10.1016/j.immuni.2009.09.025. [DOI] [PubMed] [Google Scholar]
- Mallevaey T, Scott-Browne JP, Matsuda JL, Young MH, Pellicci DG, Patel O, Thakur M, Kjer-Nielsen L, Richardson SK, Cerundolo V, et al. T cell receptor CDR2β and CDR3β loops collaborate functionally to shape the iNKT cell repertoire. Immunity. 2009;31:60–71. doi: 10.1016/j.immuni.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning TC, Schlueter CJ, Brodnicki TC, Parke EA, Speir JA, Garcia KC, Teyton L, Wilson IA, Kranz DM. Alanine scanning mutagenesis of an αβ T cell receptor: mapping the energy of antigen recognition. Immunity. 1998;8:413–425. doi: 10.1016/s1074-7613(00)80547-6. [DOI] [PubMed] [Google Scholar]
- Marrack P, Scott-Browne JP, Dai S, Gapin L, Kappler JW. Evolutionarily Conserved Amino Acids That Control TCR-MHC Interaction. Annu Rev Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzinger P, Bevan MJ. Hypothesis: why do so many lymphocytes respond to major histocompatibility antigens? Cell Immunol. 1977;29:1–5. doi: 10.1016/0008-8749(77)90269-6. [DOI] [PubMed] [Google Scholar]
- Maynard J, Petersson K, Wilson DH, Adams EJ, Blondelle SE, Boulanger MJ, Wilson DB, Garcia KC. Structure of an autoimmune T cell receptor complexed with class II peptide-MHC: insights into MHC bias and antigen specificity. Immunity. 2005;22:81–92. doi: 10.1016/j.immuni.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Merkenschlager M, Graf D, Lovatt M, Bommhardt U, Zamoyska R, Fisher AG. How many thymocytes audition for selection? J Exp Med. 1997;186:1149–1158. doi: 10.1084/jem.186.7.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GP, Ni PP, Allen PM. Alloreactivity is limited by the endogenous peptide repertoire. Proc Natl Acad Sci U S A. 2011;108:3695–3700. doi: 10.1073/pnas.1017015108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell EW, Ely LK, Kruse AC, Reay PA, Rodriguez SN, Lin AE, Kuhns MS, Garcia KC, Davis MM. Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k) Journal of immunology. 2011;186:5823–5832. doi: 10.4049/jimmunol.1100197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancer Z, Cooper MD. The evolution of adaptive immunity. Annu Rev Immunol. 2006;24:497–518. doi: 10.1146/annurev.immunol.24.021605.090542. [DOI] [PubMed] [Google Scholar]
- Parra ZE, Baker ML, Hathaway J, Lopez AM, Trujillo J, Sharp A, Miller RD. Comparative genomic analysis and evolution of the T cell receptor loci in the opossum Monodelphis domestica. BMC Genomics. 2008;9:111. doi: 10.1186/1471-2164-9-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicci DG, Patel O, Kjer-Nielsen L, Pang SS, Sullivan LC, Kyparissoudis K, Brooks AG, Reid HH, Gras S, Lucet IS, et al. Differential recognition of CD1d-α-galactosyl ceramide by the Vβ8.2 and Vβ7 semi-invariant NKT T cell receptors. Immunity. 2009;31:47–59. doi: 10.1016/j.immuni.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rast JP, Anderson MK, Strong SJ, Luer C, Litman RT, Litman GW. α, β, γ, and δ T cell antigen receptor genes arose early in vertebrate phylogeny. Immunity. 1997;6:1–11. doi: 10.1016/s1074-7613(00)80237-x. [DOI] [PubMed] [Google Scholar]
- Rast JP, Haire RN, Litman RT, Pross S, Litman GW. Identification and characterization of T-cell antigen receptor-related genes in phylogenetically diverse vertebrate species. Immunogenetics. 1995;42:204–212. doi: 10.1007/BF00191226. [DOI] [PubMed] [Google Scholar]
- Rubtsova K, Scott-Browne JP, Crawford F, Dai S, Marrack P, Kappler JW. Many different Vβ CDR3s can reveal the inherent MHC reactivity of germline-encoded TCR V regions. Proc Natl Acad Sci U S A. 2009;106:7951–7956. doi: 10.1073/pnas.0902728106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- Scott-Browne JP, Matsuda JL, Mallevaey T, White J, Borg NA, McCluskey J, Rossjohn J, Kappler J, Marrack P, Gapin L. Germline-encoded recognition of diverse glycolipids by natural killer T cells. Nat Immunol. 2007;8:1105–1113. doi: 10.1038/ni1510. [DOI] [PubMed] [Google Scholar]
- Scott-Browne JP, White J, Kappler JW, Gapin L, Marrack P. Germline-encoded amino acids in the αβ T-cell receptor control thymic selection. Nature. 2009;458:1043–1046. doi: 10.1038/nature07812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler M, Mehrle A, Poustka A, Wiemann S. The 3of5 web application for complex and comprehensive pattern matching in protein sequences. BMC Bioinformatics. 2006;7:144. doi: 10.1186/1471-2105-7-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi DK, Schubert DA, Anders AK, Heroux A, Bonsor DA, Thomas CP, Sundberg EJ, Pyrdol J, Wucherpfennig KW. A highly tilted binding mode by a self-reactive T cell receptor results in altered engagement of peptide and MHC. J Exp Med. 2011;208:91–102. doi: 10.1084/jem.20100725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha WC, Nelson CA, Newberry RD, Kranz DM, Russell JH, Loh DY. Positive and negative selection of an antigen receptor on T cells in transgenic mice. Nature. 1988;336:73–76. doi: 10.1038/336073a0. [DOI] [PubMed] [Google Scholar]
- Silk JD, Salio M, Brown J, Jones EY, Cerundolo V. Structural and functional aspects of lipid binding by CD1 molecules. Annu Rev Cell Dev Biol. 2008;24:369–395. doi: 10.1146/annurev.cellbio.24.110707.175359. [DOI] [PubMed] [Google Scholar]
- Sim BC, Zerva L, Greene MI, Gascoigne NR. Control of MHC restriction by TCR Vα CDR1 and CDR2. Science. 1996;273:963–966. doi: 10.1126/science.273.5277.963. [DOI] [PubMed] [Google Scholar]
- Stadinski BD, Zhang L, Crawford F, Marrack P, Eisenbarth GS, Kappler JW. Diabetogenic T cells recognize insulin bound to IAg7 in an unexpected, weakly binding register. Proc Natl Acad Sci U S A. 2010;107:10978–10983. doi: 10.1073/pnas.1006545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend AR, Gotch FM, Davey J. Cytotoxic T cells recognize fragments of the influenza nucleoprotein. Cell. 1985;42:457–467. doi: 10.1016/0092-8674(85)90103-5. [DOI] [PubMed] [Google Scholar]
- Tynan FE, Burrows SR, Buckle AM, Clements CS, Borg NA, Miles JJ, Beddoe T, Whisstock JC, Wilce MC, Silins SL, et al. T cell receptor recognition of a ‘super-bulged’ major histocompatibility complex class I-bound peptide. Nat Immunol. 2005;6:1114–1122. doi: 10.1038/ni1257. [DOI] [PubMed] [Google Scholar]
- White J, Haskins KM, Marrack P, Kappler J. Use of I region-restricted, antigen-specific T cell hybridomas to produce idiotypically specific anti-receptor antibodies. J Immunol. 1983;130:1033–1037. [PubMed] [Google Scholar]
- White J, Pullen A, Choi K, Marrack P, Kappler JW. Antigen recognition properties of mutant V beta 3+ T cell receptors are consistent with an immunoglobulin-like structure for the receptor. J Exp Med. 1993;177:119–125. doi: 10.1084/jem.177.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Huseby E, Scott-Browne J, Rubtsova K, Pinilla C, Crawford F, Marrack P, Dai S, Kappler JW. A Single T Cell Receptor Bound to Major Histocompatibility Complex Class I and Class II Glycoproteins Reveals Switchable TCR Conformers. Immunity. 2011;35:23–33. doi: 10.1016/j.immuni.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerrahn J, Held W, Raulet DH. The MHC reactivity of the T cell repertoire prior to positive and negative selection. Cell. 1997;88:627–636. doi: 10.1016/s0092-8674(00)81905-4. [DOI] [PubMed] [Google Scholar]
- Zinkernagel RM, Callahan GN, Althage A, Cooper S, Klein PA, Klein J. On the thymus in the differentiation of “H-2 self-recognition” by T cells: evidence for dual recognition? J Exp Med. 1978;147:882–896. doi: 10.1084/jem.147.3.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkernagel RM, Doherty PC. Immunological surveillance against altered self components by sensitised T lymphocytes in lymphocytic choriomeningitis. Nature. 1974;251:547–548. doi: 10.1038/251547a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.