Abstract
An RNA silencing mechanism with broad regulatory capability has finally been demonstrated in one of the most widely used model organisms.
The budding yeast Saccharomyces cerevisiae has been instrumental in discovering molecular mechanisms of fundamental cellular processes in eukaryotes, including cell cycle control, transcription, chromatin structure, and signal transduction. Missing from this picture has been RNA interference (RNAi), a major regulatory pathway in eukaryotic cells that silences gene expression through small interfering RNAs (siRNAs) that bind target sequences (1–3). On page 544 of this issue, Drinnenberg et al. (4) show that RNAi is indeed present in some budding yeasts and can be restored in S. cerevisiae, opening exciting possibilities for studies focused on the biological functions and impact of RNAi in these organisms.
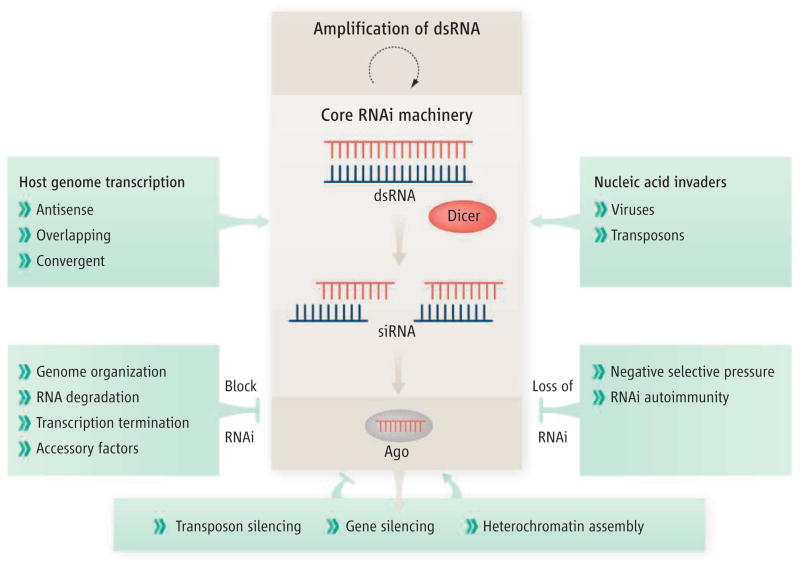
RNAi is an ancient mechanism present in plants, animals, and most fungi, but thought to have been lost during the evolution of budding yeasts. One of its major functions is defense against nucleic acids of invaders such as viruses, and against mobile segments of the host genome (transposons and retro-elements), and could be considered a type of genetic immune system (1, 2). The mechanism is triggered by the processing of double-stranded RNA (dsRNA) into siRNAs about 23 nucleotides in size by Dicer, an enzyme of the ribonuclease III (RNase III) family (1) (see the figure). siRNAs are assembled onto an effector protein called Argonaute (Ago), which uses base-pairing interactions to target and inactivate cognate RNAs or to repress transcription. In some systems, RNAi is amplified by RNA-dependent RNA polymerases that synthesize dsRNA. Another branch of the pathway uses small RNAs (microRNAs) and has been estimated to modulate the expression of up to a third of human genes at the post-transcriptional level (5). In plants, ciliates, some animal cells, and the fission yeast Schizosaccharomyces pombe, RNAi promotes the assembly of silent chromatin structures (3). How could a pathway with such broad functions be absent in budding yeast?
Figure. The core of silence.
Dicer and Ago form the core of RNAi mechanisms. RNA silencing may feed back on the pathways that generate siRNA to exert positive or negative effects. RNAi is also negatively regulated by factors that prevent dsRNA formation or modulate the activities of the core machinery. Inappropriate RNAi may target important cellular RNAs, resulting in RNAi autoimmunity. Negative selective pressure may have contributed to RNAi loss in some budding yeast species during evolution.
To address this question, Drinnenberg et al. focused on budding yeasts whose genomes encode an Ago homolog. Unlike their close relative S. cerevisiae, the budding yeasts Saccharomyces castelli, Kluyveromyces polysporus, and Candida albicans (an opportunistic human pathogen) have Ago homologs. High-throughput sequencing of small RNA libraries from these yeasts revealed abundant small RNAs about 23 nucleotides in size, similar to those associated with RNAi in other systems (4). In S. castelli, most of the sequenced RNAs mapped to both genomic DNA strands or to transcripts that form a hairpin structure, suggesting that they originated from dsRNA precursors. Furthermore, the presence of small RNAs correlated with the presence of an enzymatic activity in yeast extracts that cleaves dsRNA into discretely sized small RNA.
Although no Dicer homologs were identified in these yeast genomes, Drinnenberg et al. discovered a noncanonical Dicer with proper RNA cleaving activity. However, this variant (Dcr1) is closely related to RNase III enzymes involved in RNA processing pathways that are independent of RNAi (6). Most Dicers contain two RNase III domains and a PAZ domain, which acts as a molecular ruler by fixing one end of dsRNA relative to RNase III active sites (7). Dcr1 is unusual in that it contains only one RNase III domain and, like the S. pombe Dicer (8), lacks a PAZ domain and must use an alternative measuring mechanism. Dcr1 is distinguished from Dicer by the presence of a binding domain for dsRNA, and the authors propose that it acts as a homodimer so that each RNase III domain cleaves one strand of the dsRNA precursor. Thus, Dcr1 proteins define a previously unrecognized family of Dicer enzymes.
The authors identified about 1000 loci in the S. castelli genome that generate siRNAs, but the abundance of transcripts targeted by these molecules generally did not change upon deletion of the genes encoding Dcr1 and Ago. This suggests that for most S. castelli loci, the expression of siRNAs is below the threshold required to associate with Ago and target RNAs. Nevertheless, the authors show that the RNAi pathway is capable of robust gene silencing. For example, they observed strong silencing of a reporter gene when siRNA was produced from a hairpin RNA corresponding to the reporter.
Remarkably, introduction of S. castelli genes encoding Dcr1 and Ago into S. cerevisiae reconstituted RNAi and efficiently mediated silencing of reporter genes (4). Furthermore, unlike S. castelli, S. cerevisiae harbors active transposons in its genome. Reconstituting RNAi in S. cerevisiae silenced the transposition of a Ty1 transposon. This silencing appears to result from the processing of overlapping transcripts into Ty1 siRNAs, which in turn mediate the destruction of Ty1 mRNA. Inspection of all transcripts in S. cerevisiae revealed RNAs that were antisense to transposon elements. This suggests that a major role of RNAi in budding yeasts involves the silencing of transposons, but it is unclear whether other transcripts with the potential to form dsRNA exist and can be similarly silenced.
As proposed by Drinnenberg et al., loss of RNAi may result from a lack of positive selection, such as occurs during evolutionary periods in which transposons are dormant. Another possibility is that like any potent destructive force, RNAi is under negative selective pressure. This is probably evident in the radiation of mechanisms that modulate RNAi and restrict biogenesis of the dsRNA trigger (see the figure). For example, the evolution of gene arrangements that prevent formation of dsRNA, combined with RNA degradation mechanisms, curtail the ability to form dsRNA (9, 10). Restrictive mechanisms involving factors that limit siRNA production, such as siRNA ribonucleases, or proteins that control siRNA loading onto Ago, have also been observed (11, 12). In S. pombe, RNAi is limited to nascent hetero-chromatin-bound RNAs (3). And S. cerevisiae and some other budding yeast species appear to have gotten rid of RNAi altogether. Thus, RNAi is likely to have had a profound impact on the evolution of the eukaryotic genome and its potential to produce RNAs that can become double-stranded. It should now be possible to determine the price of reacquiring RNAi on S. cerevisiae fitness.
References
- 1.Malone CD, Hannon GJ. Cell. 2009;136:656. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghildiyal M, Zamore PD. Nat Rev Genet. 2009;10:94. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moazed D. Nature. 2009;457:413. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drinnenberg IA, et al. Science. 2009;326:544. doi: 10.1126/science.1176945. published online 10 September 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. Cell. 2004;116:281. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Macrae IJ, et al. Science. 2006;311:195. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 7.Colmenares SU, et al. Mol Cell. 2007;27:449. doi: 10.1016/j.molcel.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Houseley J, Tollervey D. Biochim Biophys Acta. 2008;1779:239. doi: 10.1016/j.bbagrm.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 9.Buhler M, et al. Nat Struct Mol Biol. 2008;15:1015. doi: 10.1038/nsmb.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kennedy S, Wang D, Ruvkun G. Nature. 2004;427:645. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- 11.Carthew RW, Sontheimer EJ. Cell. 2009;136:642. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]