Abstract
eIF5A is highly conserved from archaea to mammals, essential for cell viability and the only protein known to contain the essential amino acid residue hypusine, generated by a unique posttranslational modification. eIF5A was originally identified as a translation initiation factor due to its ability to stimulate the formation of the first peptide bond. However, recent studies have shown that depletion of eIF5A causes a significant decrease in polysome run-off and an increase in the ribosome transit time, suggesting that eIF5A is actually involved in the elongation step of protein synthesis. We have previously shown that the depletion mutant tif51A-3 (eIF5AC39Y/G118D) shows a sicker phenotype when combined with the dominant negative mutant eft2H699K of the elongation factor eEF2. In this study, we used the eIF5AK56A mutant to further investigate the relationship between eIF5A and eEF2. The eIF5AK56A mutant is temperature sensitive and has a defect in protein synthesis, but instead of causing depletion of the eIF5A protein, this mutant has a defect in hypusine modification. Like the mutant tif51A-3, the eIF5AK56A mutant is synthetic sick with the mutant eft2H699K of eEF2. High-copy eEF2 not only improves cell growth of the eIF5AK56A mutant, but also corrects its increased cell size defect. Moreover, eEF2 suppression of the eIF5AK56A mutant is correlated with the improvement of total protein synthesis and with the increased resistance to the protein synthesis inhibitor hygromycin B. Finally, the polysome profile defect of the eIF5AK56A mutant is largely corrected by high-copy eEF2. Therefore, these results demonstrate that eIF5A is closely related to eEF2 function during translation elongation.
Keywords: eIF5A, Hypusine, eEF2, Translation elongation
Introduction
The putative eukaryotic translation initiation factor 5A (eIF5A) is a highly conserved and essential protein present in all organisms from archaea to mammals, but not in eubacteria (Schnier et al. 1991; Chen and Liu 1997). eIF5A is the only protein known to contain the unusual amino acid residue hypusine (Chen and Liu 1997; Wolff et al. 2007). Hypusine is formed by the addition of a 4-aminobutyl moiety from spermidine to a specific lysine residue of eIF5A, followed by its hydroxylation. Hypusine is essential for eIF5A function and several studies have aimed to characterize the mechanisms of the hypusine modification enzymes and the development of specific inhibitors (Wolff et al. 2007; Kerscher et al. 2010; Cano et al. 2010). This specific lysine converted to hypusine is located at the tip of a ~ 10 amino acid loop in the N-terminus domain of the protein and is the most conserved region of eIF5A through evolution (Park et al. 2010). eIF5A is organized in two independently folded domains (Stiuso et al. 1999): an N-terminal SH3-like barrel, found in other proteins related to translation; and a C-terminal OB-fold, present in nucleic acid-binding proteins (http://www.rcsb.org/pdb/).
Despite being highly conserved and essential, the critical role of eIF5A in the cell remains unclear. Originally purified from ribosomes isolated from reticulocyte lysates, eIF5A was shown to stimulate methionyl-puromycin synthesis, indicating a role in the formation of the first peptide bond (Benne and Hershey 1978). However, eIF5A depletion in yeast caused a small decrease in cell protein synthesis, arguing against a role as a general translation initiation factor. Curiously, eIF5A depletion induced an increased number of G1-arrested yeast cells (Kang and Hershey 1994). This result, together with the fact that inhibitors of hypusine formation caused cell-cycle arrest in mammalian cells (Hanauske-Abel et al. 1994), led to the hypothesis that eIF5A may be important for translation of mRNAs encoding specific proteins required for cell-cycle progression (Park et al. 1997). In support of this idea, eIF5A function is necessary for polarized cell growth, a process essential for the G1/S transition in yeast (Zanelli and Valentini 2005). Furthermore, synthetic lethality was revealed between mutants of eIF5A and of Ypt1, which is a protein essential for vesicular trafficking and also for proper polarized growth in yeast (Frigieri et al. 2008). However, no specific subset of mRNAs whose translation is controlled by eIF5A has been identified so far.
eIF5A has also been implicated in nuclear export of HIV-1 Rev and mRNA decay, but these findings are controversial and may reflect secondary effects of eIF5A function (Huang et al. 2007). More specifically, the effect of eIF5A on mRNA decay seems to be secondary, as the arrest of cell growth in eIF5A temperature-sensitive mutants does not directly correlate with mRNA accumulation (Valentini et al. 2002).
More recently, it has been demonstrated that eIF5A interacts physically with the 80S ribosome as well as with the translation elongation factors eEF1A and eEF2 (Jao and Chen 2006; Zanelli et al. 2006). eIF5A was shown to co-fractionate with monosomes in a translation-dependent manner. Moreover, eIF5A mutant strains show accumulation of polysomes instead of polysome run-off and an increase in the average time necessary for ribosomes to transit along mRNAs (Gregio et al. 2009; Saini et al. 2009). Therefore, these results not only support a role for eIF5A in translation, but also suggest that this factor plays a role in translation elongation rather than initiation. Furthermore, a function for eIF5A in translation elongation would explain the secondary effect of mRNA stabilization and a block in the formation of P bodies, observed in eIF5A mutants as a result of mRNA trapping in polysomes (Valentini et al. 2002; Gregio et al. 2009).
Herein, we use the eIF5AK56A mutant to further investigate the relationship between eIF5A and eEF2.
Materials and methods
Yeast strains, plasmids and standard procedures
Procedures for cell growth and genetic manipulations were carried out according to standard protocols (Guthrie and Fink 1991). Saccharomyces cerevisiae strains used in this study are: wild type eIF5A (VZL838—MATα leu2 ura3 trp1 his3 tif51A::HIS3 tif51B::KanMX4 [TIF51A/TRP1/CEN]), eIF5AK56A mutant (VZL987—MATα leu2 ura3 trp1 his3 tif51A::HIS3 tif51B::KanMX4 [tif51A-K56A/TRP1/CEN]) and eIF5AP83S (tif51A-1) mutant (VZL904—MATα leu2 ura3 trp1 his3 tif51A::HIS3 tif51B::KanMX4 [tif51A-1/TRP1/CEN]) (Valentini et al. 2002). The plasmids used are: high-copy vectors pRS426 and pRS425, high-copy eEF2 (pSV262—EFT2/URA3/2μ), high-copy Pkc1 (pSV181—PKC1/URA3/2μ) and dominant negative eft2H699K allele of eEF2 (pVZ1104—eft2H699K/LEU2/2μ) (Ortiz and Kinzy 2005).
Flow cytometry analysis
Strains were grown to mid-log phase (OD600nm = 0.5) at 25°C in 5 mL of Synthetic Complete medium lacking uracil (SC-ura) and then were shifted to the restrictive temperature (38°C) for 4 h. The graph under each micrograph was obtained by flow cytometry analysis of granulosity (side light scatter or SSC—y axis) in function of cell volume (forward light scatter or FSC—x axis) using a BD FACSC-anto apparatus and the BD FACSDiva software. The percentages under the dot plot graphs represent the relative number of cells (dots) inside the area delimited in each graph. The delimited area was determined as the area representing the cell dispersion of the wild type strain (eIF5A).
Protein synthesis measurement
Cells were grown to mid-log phase (OD600nm = 0.5) at 25°C in 5 mL of synthetic complete medium lacking uracil (SC-ura). [3H]leucine (PerkinElmer, MA, USA) was added to the medium to a final concentration of 2 μCi/mL and the cultures were shifted to the restrictive temperature (38°C) for 4 h, harvested at 4°C and frozen at −80°C. All frozen cell pellets were resuspended in 15% cold trichloroacetic acid solution and incubated on ice for 15 min. The samples were heated at 72°C for 30 min and then incubated on ice for 15 min. Trichloroacetic acid precipitates were collected by centrifugation (15,000 g, 4°C, 10 min) and washed four times with 10% trichloroacetic acid to remove free [3H]leucine. The final washed pellets were resuspended in 100 μL of 0.2 M NaOH, and 50 μL aliquots were used to determine the radioactivity incorporated into protein synthesis in a Beckman Scintillation Counter (Beckman Coulter, Brea, CA, USA). Total protein concentration was determined by the BCA protein assay (Thermo Scientific, IL, USA), using 5 μL aliquots of each sample. The amount of total protein synthesis was calculated for each sample as c.p.m. μg−1 total protein and then expressed as percentage of total protein synthesis with the wild type (eIF5A) strain arbitrarily set to 100% (Table 1).
Table 1.
Measurement of total protein synthesis
| Samples | Relative values (%) ± SD |
|---|---|
| eIF5A | 100.0 ± 1.0 |
| eIF5A (K56A) + vector | 59.6 ± 2.6 |
| eIF5A (K56A) + eEF2 | 85.5 ± 2.5 |
SD standard deviation
Polysome profiling
Cells from 100 mL cultures (OD600nm = 0.3–0.6) were shifted to 38°C for 4 h and used for each sucrose gradient. At the time of harvesting, the translational machinery was blocked by the addition of 0.1 mg/mL cycloheximide for 5 min at 38°C. Briefly, 10 A260nm units of cell lysates were layered onto 10–50% sucrose gradients and centrifuged for 3 h at 39,000g at 4°C in a Beckman SW41-Ti rotor. The gradients were then fractionated by upward displacement with 60% (w/v) sucrose using a gradient fractionator connected to a Control Unit UV-1 monitor (Amersham Pharmacia Biotech) for continuous measurement of the absorbance at 254 nm. The polysomal profile fractions were quantified with the aid of NIH Image J software.
Results and discussion
High-copy eEF2 suppresses growth and cell size defects of yeast eIF5AK56A mutant
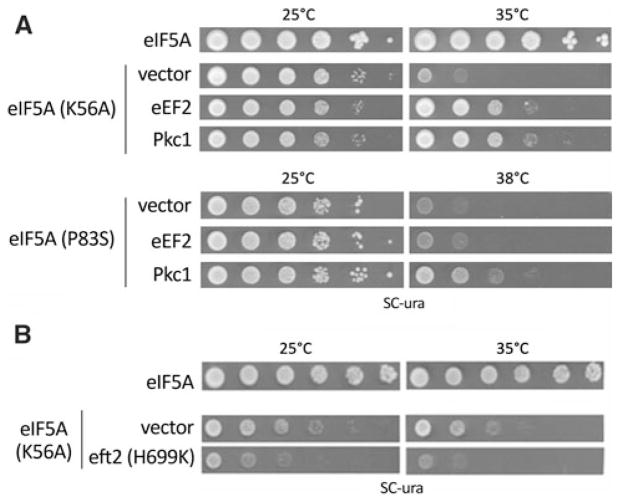
In order to better understand the function of eIF5A during protein synthesis, more specifically during translation elongation, we searched for yeast eIF5A temperature-sensitive mutants which would show genetic interactions with the elongation factor 2 (eEF2). Interestingly, we found that the temperature sensitivity of the eIF5AK56A mutant was suppressed by high-copy EFT2, the gene encoding the major form of eEF2 in yeast. As shown in Fig. 1a, both eIF5AP83S (tif51A-1) and eIF5AK56A mutants are suppressed by Pkc1. Overexpression of PKC1 suppresses eIF5A mutant temperature sensitivity through overactivation of the cell integrity pathway and resultant bypass of the secondary effect of eIF5A in cell-cycle progression (Zanelli and Valentini 2005). Therefore, Pkc1 is a more general suppressor of eIF5A, since it overcomes the secondary effects caused by eIF5A loss of function in the cell (Zanelli and Valentini 2007). On the other hand, only the eIF5AK56A mutant is suppressed by eEF2, demonstrating an allele-specific genetic interaction. This suggests that a specific defect in eIF5A function impaired in the eIF5AK56A mutant is compensated by high-copy eEF2, as discussed in the next paragraph.
Fig. 1.
Genetic interactions between eIF5A and eEF2. a Tenfold serial dilutions of wild type (eIF5A) cells harboring the vector alone, the mutant strains eIF5AK56A or eIF5AP83S harboring the vector alone, or high-copy eEF2 or high-copy Pkc1 plasmid were plated onto SC-ura to determine the growth at permissive (25°C) or non-permissive temperatures (35°C to eIF5AK56A and 38°C to eIF5AP83S). b The wild type strain harboring the vector alone or the eIF5AK56A mutant strain harboring the vector alone or together with eEF2H699K dominant negative mutant plasmid were assayed as described in (a). The plates were photographed after 3–4 days of growth
The eIF5AK56A mutant was previously shown to be defective in protein synthesis, similarly to other yeast eIF5A mutants (Dias et al. 2008; Gregio et al. 2009; Saini et al. 2009). Lysine 56 (K56) is the last residue of the amino acids stretch forming a hypusine-containing loop in the eIF5A N-terminal domain (Dias et al. 2008). It was shown that the eIF5AK56A mutant protein is less effectively modified to the hypusine form than the wild type protein (roughly 50% of the wild type), while the eIF5AK56D mutant protein is not modified at all (Dias et al. 2008). Besides, the eIF5AK56A mutant does not show reduced levels of eIF5A protein due to degradation at the restrictive temperature, as displayed by most of the temperature sensitive eIF5A mutants described so far, including the eIF5AP83S (tif51A-1) mutant (Dias et al. 2008; Saini et al. 2009). Since eIF5AK56A is the only eIF5A mutant defective in hypusine modification and the only one suppressed by high-copy eEF2, we believe that high-copy eEF2 is able to compensate for the decreased availability of functional hypusine-containing eIF5A.
We also tested whether impairment in the function of eEF2 would interfere with eIF5AK56A mutant. For that purpose, we used the eEF2 dominant negative mutant eft2H699K (Ortiz and Kinzy 2005). As shown in Fig. 1b, growth of eIF5AK56A mutant strain is significantly reduced upon coexpression of the eEF2 dominant negative mutant protein eft2H699K. This synthetic sick phenotype is similar to that previously observed for the tif51A-3 mutant of eIF5A (Gregio et al. 2009). Besides that, the genetic interaction between eIF5A and eEF2 mutants is correlated with the fact that another eIF5A mutant was shown to be sensitive to the eEF2 inhibitor drug sordarin (Saini et al. 2009). Therefore, the data presented here further support a close functional link between eIF5A and eEF2.
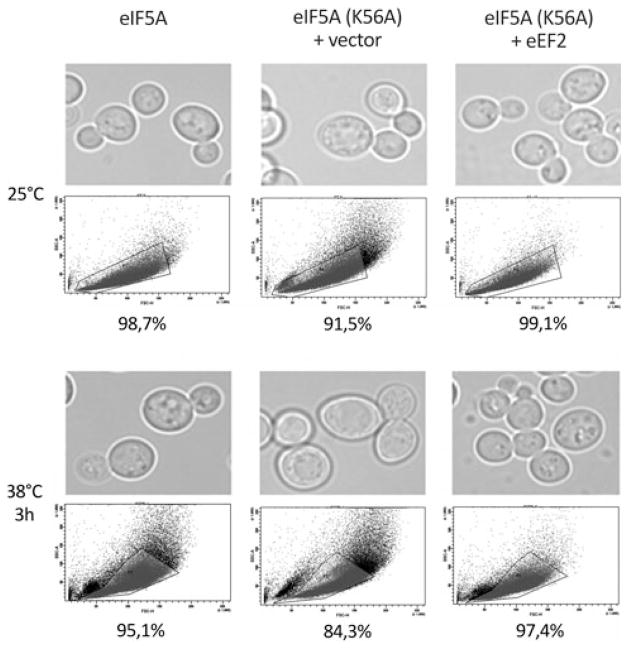
It has already been demonstrated that various yeast eIF5A mutants display a morphological phenotype of increased cell size (Kang and Hershey 1994; Zanelli and Valentini 2005), which is correlated with a defect in cell cycle events such as actin cytoskeleton polarization (Zanelli and Valentini 2005). As expected, the eIF5AK56A mutant also shows increased cell size at both temperatures (Fig. 2, microscopy images). To better quantify this effect, we used flow cytometry analysis to determine the alterations in cell volume (Fig. 2, dot plot graphs). However, when high-copy eEF2 is present, the morphology is remarkably similar to the wild type (eIF5A) strain at both permissive and restrictive temperatures. These results further implicate a role for eIF5A in cell cycle progression (a secondary effect) and demonstrate that this function of eIF5A is also linked to eEF2 function.
Fig. 2.
Suppression of cell size defect of the eIF5AK56A mutant by high-copy eEF2. Wild type (eIF5A) harboring the vector alone and the eIF5AK56A mutant strain harboring the vector alone or a high-copy eEF2 plasmid were grown at 25°C in SC-ura to mid-log phase and half of the cultures was then shifted to 38°C for 3 h. The microscopic images were then captured using a Nikon TE300 inverted microscope and a CCD camera. The graph under each micrograph was obtained by flow cytometry analysis of granulosity in function of cell volume using a BD FACSCanto and the BD FACSDiva software. The percentages under the dot plot graphs represent the relative number of cells (dots) inside the area delimited in each graph. The delimited area was determined as the area representing the cell dispersion of the wild type strain (eIF5A)
Translation elongation defect in the eIF5AK56A mutant is also suppressed by high-copy eEF2
Since eEF2 has a well established function in translation elongation and eIF5A has recently been shown to have a role in protein synthesis as well, we tested whether high-copy eEF2 can also improve the levels of protein synthesis in the eIF5AK56A mutant strain. We have previously shown that the eIF5AK56A mutant has a defect in protein synthesis, which is correlated with its temperature sensitivity for growth (Dias et al. 2008). As shown in Fig. 3a and Table 1, the defect in protein synthesis displayed by the eIF5AK56A mutant is considerably ameliorated by high-copy eEF2. This result strongly suggests that the phenotypic suppression promoted by high-copy eEF2 in the eIF5AK56A mutant is the result of a partial restoration of protein synthesis.
Fig. 3.
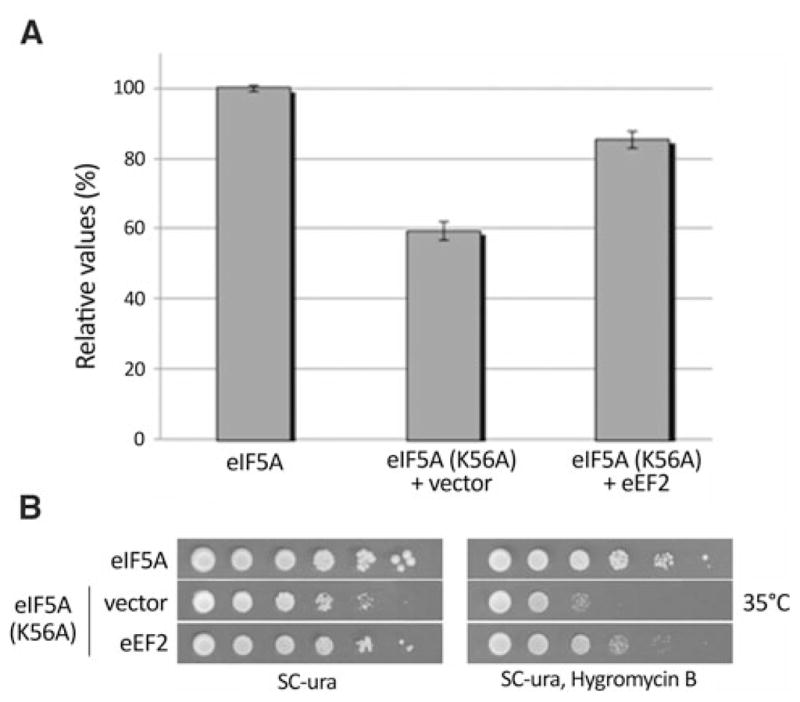
Improvement of eIF5AK56A mutant protein synthesis by high-copy eEF2. a Total protein synthesis was measured as described and the amount of [3H]leucine incorporation into proteins is shown relatively to the wild type (100%). b Tenfold serial dilutions of wild type and eIF5AK56A harboring the vector alone or high-copy eEF2 were plated onto SC-ura or SC-ura containing 200 μg/mL of hygromycin B and incubated at 35°C for 3–4 days
Mutants of factors involved in protein synthesis are expected to be more sensitive to translation inhibitors. In fact, several mutants of eIF5A are more sensitive to different antibiotics targeting translation (Zanelli et al. 2006) (and data not shown). We found that the eIF5AK56A mutant is hypersensitive to the aminoglycoside hygromycin B, which inhibits eEF2-dependent translocation of the ribosome (Gonzalez et al. 1978). Therefore, we tested the ability of high-copy eEF2 to attenuate hygromycin B sensitivity of the eIF5AK56A mutant and the result was positive as the growth was improved in the presence of high-copy eEF2 (Fig. 3b). These findings are in agreement with the idea that high-copy eEF2 acts through improvement in the process of protein synthesis to suppress the eIF5AK56A mutant.
Since eIF5A is known to be involved in translation elongation (Gregio et al. 2009; Saini et al. 2009), we decided to analyze the polysome profile of the eIF5AK56A mutant strain. As shown in Fig. 4, the polysome profile of the eIF5AK56A mutant shows increased polysome peaks and a decreased 80S peak at the restrictive temperature when compared with the wild type strain. Besides the evident difference between the shapes of wild type and eIF5AK56A mutant polysome profile graphs, quantification of the area of the peaks clearly demonstrates the accumulation of polysomes in the eIF5AK56A mutant as the ratio polysome/monosome area (P/M) was increased. An increase in the ratio of polysome to monosome in the mutant strain is due to continuous translation initiation and reduced rates of polysome run-off (Peltz et al. 1992). This result is in accordance with the previously published data (Gregio et al. 2009; Saini et al. 2009). On the other hand, the polysome profile of the eIF5AK56A mutant harboring high-copy eEF2 is strikingly similar to the wild type strain. These results once more support a function for eIF5A in translation elongation and suggest that this function is closely related to eEF2.
Fig. 4.
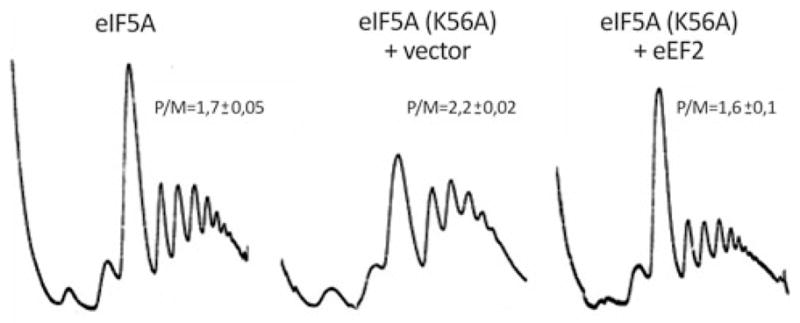
Recovery of polysome profile defects of the eIF5AK56A mutant by high-copy eEF2. Extracts prepared from the wild type harboring the vector alone and eIF5AK56A mutant strain harboring the vector alone or the high-copy eEF2 plasmid were fractionated by ultracentrifugation in a sucrose density gradient. Optical scans (OD254nm) of the gradients are shown. The ratios of P/M were calculated by comparing the areas of the 80S and polysome peaks
Although the precise mechanism of action of eIF5A has not been elucidated so far, its involvement in the elongation step of translation has been well documented recently (Gregio et al. 2009; Saini et al. 2009; Patel et al. 2009; Li et al. 2010; Landau et al. 2010). The observation that increase in eEF2 function cooperates with the role of eIF5A during the elongation step of protein synthesis, reported herein for the first time, strongly supports the hypothesis that eIF5A interacts functionally with this elongation factor. Whether these proteins interact physically and where on the ribosome eIF5A binds are still open questions and the answers will conclusively help to elucidate the mechanistic aspects underlying the essential role of eIF5A in the cell.
Acknowledgments
This work was supported by grants to S.R.V. from Fundação de Amparo à Pesquisa do Estado de São Paulo (FA-PESP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and PADC from Faculdade de Ciências Farmacêuticas, UNESP. We also thank FAPESP, CNPq and CAPES for fellowships awarded to most of the authors (C. A. O. D.; A. P. B. G.; D. R.; F. C. G. and T. F. W.).
Abbreviations
- eIF5A
Eukaryotic translation initiation factor 5A
- eEF2
Eukaryotic translation elongation factor 2
- TIF51A
Gene encoding eIF5A in yeast
- EFT2
Gene encoding eEF2 in yeast
- PKC1
Gene encoding protein kinase C in yeast
- HIV
Human immunodeficiency virus
- [3H]leucine
Tritium-radioactive leucine
- c.p.m
Counts per minute
Contributor Information
Camila A. O. Dias, Department of Biological Sciences, School of Pharmaceutical Sciences, UNESP - Univ Estadual Paulista, Araraquara, SP, Brazil
Ana Paula Borges Gregio, Department of Biological Sciences, School of Pharmaceutical Sciences, UNESP - Univ Estadual Paulista, Araraquara, SP, Brazil.
Danuza Rossi, Department of Biological Sciences, School of Pharmaceutical Sciences, UNESP - Univ Estadual Paulista, Araraquara, SP, Brazil.
Fábio Carrilho Galvão, Department of Biological Sciences, School of Pharmaceutical Sciences, UNESP - Univ Estadual Paulista, Araraquara, SP, Brazil.
Tatiana F. Watanabe, Department of Biological Sciences, School of Pharmaceutical Sciences, UNESP - Univ Estadual Paulista, Araraquara, SP, Brazil
Myung Hee Park, Oral and Pharyngeal Cancer Branch, National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD, USA.
Sandro R. Valentini, Department of Biological Sciences, School of Pharmaceutical Sciences, UNESP - Univ Estadual Paulista, Araraquara, SP, Brazil
Cleslei F. Zanelli, Email: zanellicf@fcfar.unesp.br, Department of Biological Sciences, School of Pharmaceutical Sciences, UNESP - Univ Estadual Paulista, Araraquara, SP, Brazil
References
- Benne R, Hershey JW. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J Biol Chem. 1978;253(9):3078–3087. [PubMed] [Google Scholar]
- Cano VS, Medrano FJ, Park MH, Valentini SR. Evidence for conformational changes in the yeast deoxyhypusine hydroxylase Lia1 upon iron displacement from its active site. Amino Acids. 2010;38(2):479–490. doi: 10.1007/s00726-009-0407-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KY, Liu AY. Biochemistry and function of hypusine formation on eukaryotic initiation factor 5A. Biol Signals. 1997;6(3):105–109. doi: 10.1159/000109115. [DOI] [PubMed] [Google Scholar]
- Dias CA, Cano VS, Rangel SM, Apponi LH, Frigieri MC, Muniz JR, Garcia W, Park MH, Garratt RC, Zanelli CF, Valentini SR. Structural modeling and mutational analysis of yeast eukaryotic translation initiation factor 5A reveal new critical residues and reinforce its involvement in protein synthesis. Febs J. 2008;275(8):1874–1888. doi: 10.1111/j.1742-4658.2008.06345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigieri MC, Joao Luiz MV, Apponi LH, Zanelli CF, Valentini SR. Synthetic lethality between eIF5A and Ypt1 reveals a connection between translation and the secretory pathway in yeast. Mol Genet Genomics. 2008;280(3):211–221. doi: 10.1007/s00438-008-0357-y. [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Jimenez A, Vazquez D, Davies JE, Schindler D. Studies on the mode of action of hygromycin B, an inhibitor of translocation in eukaryotes. Biochim Biophys Acta. 1978;521(2):459–469. doi: 10.1016/0005-2787(78)90287-3. [DOI] [PubMed] [Google Scholar]
- Gregio AP, Cano VP, Avaca JS, Valentini SR, Zanelli CF. eIF5A has a function in the elongation step of translation in yeast. Biochem Biophys Res Commun. 2009;380(4):785–790. doi: 10.1016/j.bbrc.2009.01.148. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GRE. Guide to yeast genetics. Academic Press; New York: 1991. [Google Scholar]
- Hanauske-Abel HM, Park MH, Hanauske AR, Popowicz AM, Lalande M, Folk JE. Inhibition of the G1-S transition of the cell cycle by inhibitors of deoxyhypusine hydroxylation. Biochim Biophys Acta. 1994;1221(2):115–124. doi: 10.1016/0167-4889(94)90003-5. [DOI] [PubMed] [Google Scholar]
- Huang Y, Higginson DS, Hester L, Park MH, Snyder SH. Neuronal growth and survival mediated by eIF5A, a polyamine-modified translation initiation factor. Proc Natl Acad Sci USA. 2007;104(10):4194–4199. doi: 10.1073/pnas.0611609104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao DL, Chen KY. Tandem affinity purification revealed the hypusine-dependent binding of eukaryotic initiation factor 5A to the translating 80S ribosomal complex. J Cell Biochem. 2006;97(3):583–598. doi: 10.1002/jcb.20658. [DOI] [PubMed] [Google Scholar]
- Kang HA, Hershey JW. Effect of initiation factor eIF-5A depletion on protein synthesis and proliferation of Saccharomyces cerevisiae. J Biol Chem. 1994;269(6):3934–3940. [PubMed] [Google Scholar]
- Kerscher B, Nzukou E, Kaiser A. Assessment of deoxyhypusine hydroxylase as a putative, novel drug target. Amino Acids. 2010;38(2):471–477. doi: 10.1007/s00726-009-0406-9. [DOI] [PubMed] [Google Scholar]
- Landau G, Bercovich Z, Park MH, Kahana C. The role of polyamines in supporting growth of mammalian cells is mediated through their requirement for translation initiation and elongation. J Biol Chem. 2010;285(17):12474–12481. doi: 10.1074/jbc.M110.106419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CH, Ohn T, Ivanov P, Tisdale S, Anderson P. eIF5A promotes translation elongation, polysome disassembly and stress granule assembly. PLoS One. 2010;5(4):e9942. doi: 10.1371/journal.pone.0009942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz PA, Kinzy TG. Dominant-negative mutant phenotypes and the regulation of translation elongation factor 2 levels in yeast. Nucleic Acids Res. 2005;33(18):5740–5748. doi: 10.1093/nar/gki882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, Lee YB, Joe YA. Hypusine is essential for eukaryotic cell proliferation. Biol Signals. 1997;6(3):115–123. doi: 10.1159/000109117. [DOI] [PubMed] [Google Scholar]
- Park MH, Nishimura K, Zanelli CF, Valentini SR. Functional significance of eIF5A and its hypusine modification in eukaryotes. Amino Acids. 2010;38(2):491–500. doi: 10.1007/s00726-009-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel PH, Costa-Mattioli M, Schulze KL, Bellen HJ. The Drosophila deoxyhypusine hydroxylase homologue nero and its target eIF5A are required for cell growth and the regulation of autophagy. J Cell Biol. 2009;185(7):1181–1194. doi: 10.1083/jcb.200904161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltz SW, Donahue JL, Jacobson A. A mutation in the tRNA nucleotidyltransferase gene promotes stabilization of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12(12):5778–5784. doi: 10.1128/mcb.12.12.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini P, Eyler DE, Green R, Dever TE. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009;459(7243):118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnier J, Schwelberger HG, Smit-McBride Z, Kang HA, Hershey JW. Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1991;11(6):3105–3114. doi: 10.1128/mcb.11.6.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiuso P, Colonna G, Ragone R, Caraglia M, Hershey JW, Beninati S, Abbruzzese A. Structural organization of the human eukaryotic initiation factor 5A precursor and its site-directed variant Lys50 → Arg. Amino Acids. 1999;16(1):91–106. doi: 10.1007/BF01318888. [DOI] [PubMed] [Google Scholar]
- Valentini SR, Casolari JM, Oliveira CC, Silver PA, McBride AE. Genetic interactions of yeast eukaryotic translation initiation factor 5A (eIF5A) reveal connections to poly(A)-binding protein and protein kinase C signaling. Genetics. 2002;160(2):393–405. doi: 10.1093/genetics/160.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff EC, Kang KR, Kim YS, Park MH. Posttranslational synthesis of hypusine: evolutionary progression and specificity of the hypusine modification. Amino Acids. 2007;33(2):341–350. doi: 10.1007/s00726-007-0525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanelli CF, Valentini SR. Pkc1 acts through Zds1 and Gic1 to suppress growth and cell polarity defects of a yeast eIF5A mutant. Genetics. 2005;171(4):1571–1581. doi: 10.1534/genetics.105.048082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanelli CF, Valentini SR. Is there a role for eIF5A in translation? Amino Acids. 2007;33(2):351–358. doi: 10.1007/s00726-007-0533-0. [DOI] [PubMed] [Google Scholar]
- Zanelli CF, Maragno AL, Gregio AP, Komili S, Pandolfi JR, Mestriner CA, Lustri WR, Valentini SR. eIF5A binds to translational machinery components and affects translation in yeast. Biochem Biophys Res Commun. 2006;348(4):1358–1366. doi: 10.1016/j.bbrc.2006.07.195. [DOI] [PubMed] [Google Scholar]