Abstract
The ATPase Vps4 belongs to the type-I AAA family of proteins. Vps4 functions together with a group of proteins referred to as ESCRTs in membrane deformation and fission events. These cellular functions include vesicle formation at the endosome, cytokinesis and viral budding. The highly dynamic quaternary structure of Vps4 and its interactions with a network of regulators and co-factors have made the analysis of this ATPase challenging. Nevertheless, recent advances in the understanding of the cell biology of Vps4 together with structural information and in vitro studies are guiding mechanistic models of this ATPase.
Introduction
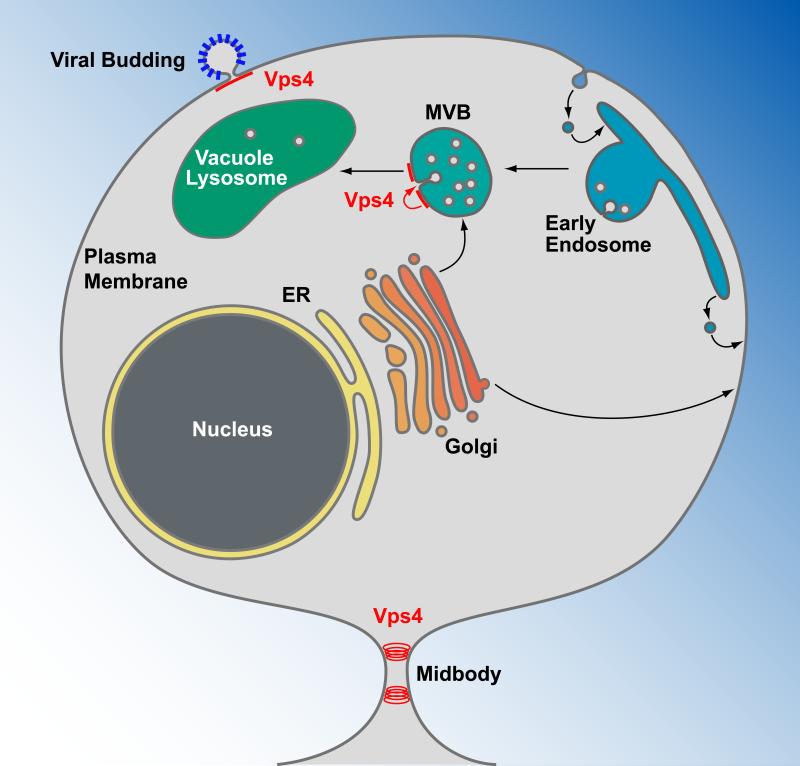
Vps4 is a type-1 AAA ATPase that functions together with the protein polymer ESCRT-III in the deformation and scission of membranes. This Vps4/ESCRT-III protein machinery represents an evolutionarily ancient system that is found in archaea as well as eukaryotes. Initially, Vps4 was characterized in baker's yeast, Saccharomyces cerevisiae. The VPS4 gene was identified through a genetic screen for factors that are involved in the transport of hydrolytic enzymes to the vacuole of yeast (Vacuole Protein Sorting gene 4) [1]. The yeast vacuole corresponds to the lysosomes in mammalian cells and functions in the degradation of macromolecules such as proteins and lipids. This degradative activity is executed by a number of hydrolases that are delivered to the vacuole via endoplasmic reticulum (ER), the Golgi network, and endosomes (Figure 1). Mutations in the yeast VPS4 gene result in impaired delivery of these hydrolases to the vacuole and the accumulation of endosomal organelles [2]. Additional analysis indicated that Vps4 is required for the formation of endosomal structures called Multi-Vesicular Bodies (MVBs) [3]. These endosomal structures invaginate parts of the limiting membrane, resulting in the formation of vesicles in the lumen of these endosomes, hence the name ‘multi-vesicular body’ (reviewed in [4, 5]. During MVB vesicle formation, transmembrane proteins destined for degradation are packaged into these vesicles. Upon fusion of the MVB with the vacuolar membrane, the vesicles are exposed to the hydrolytic lumen of the vacuole and both lipids and proteins are degraded. As such, the MVB pathway represents the major degradation pathway for membrane proteins in eukaryotic cells.
Figure 1.
Vps4 functions in the formation of MVB vesicles, viral budding and cytokinesis. Transmembrane proteins are synthesized at the endoplasmic reticulum (ER), transported through the Golgi network and delivered either to the plasma membrane or an endosomal compartment. Plasma membrane proteins destined for degradation are endocytosed and delivered to endosomes. At the multivesicular body (MVB), these proteins are sorted by Vps4 and the ESCRTs into MVB vesicles (red arrow). Upon fusion of the MVB with the vacuole/lysosome, the lumenal vesicles are exposed to a hydrolytic environment, causing the degradation of proteins and lipids of the vesicles. Vps4 together with a subset of the ESCRTs are recruited to the plasma membrane by newly forming enveloped viruses, such as HIV-1, where they function in the release of the virus particles. At the final stage of cytokinesis, Vps4 is recruited to the midbody where it functions together with ESCRT-III in the abscission of the membrane and the formation of two separate cells.
The MVB pathway plays an essential role in cellular physiology. For instance, inactivation of signaling pathways often depends on the rapid degradation of the signaling surface receptor via the MVB pathway (reviewed in [6-8]. Mutations that impair MVB formation have been shown to maintain prolonged activity of signaling cascades, which, in the case of higher eukaryotes, can result in uncontrolled cell proliferation and tumorigenesis. Furthermore, regulation of cell-surface activities such as nutrient uptake, cell-cell contacts and ion homeostasis are dependent on proper turnover of plasma membrane proteins, and thus dependent on a functional MVB pathway (reviewed in [9, 10]. As a consequence, impaired MVB activity has been implicated in numerous human diseases, including neurodegenerative disorders, cancer and cardiovascular disease (reviewed in [11, 12].
This review will summarize the current knowledge on the function of the AAA protein Vps4 and its structure. For simplicity, we will use yeast nomenclature when referring to proteins, unless stated otherwise.
Vps4 and the ESCRT Protein Complexes
In the formation of MVB vesicles Vps4 functions together with a group of four protein complexes called ESCRT-0 (Endosomal Sorting Complex Required for Transport-0), ESCRT-I, ESCRT-II and ESCRT-III (reviewed in [13]. These protein complexes are recruited from the cytoplasm to the endosomal membrane where they function in the sorting of membrane protein cargo and in the formation of MVB vesicles. In most cases, ubiquitination of membrane proteins serves as a signal for their sorting into the MVB pathway [14]. Recent studies are beginning to give insights into the structure of these ESCRT protein complexes and their interactions with each other, membranes and ubiquitinated protein cargo, although important details, including the overall assembly of the ESCRT machinery on endosomal membranes and the mechanism of MVB vesicle formation, remain elusive.
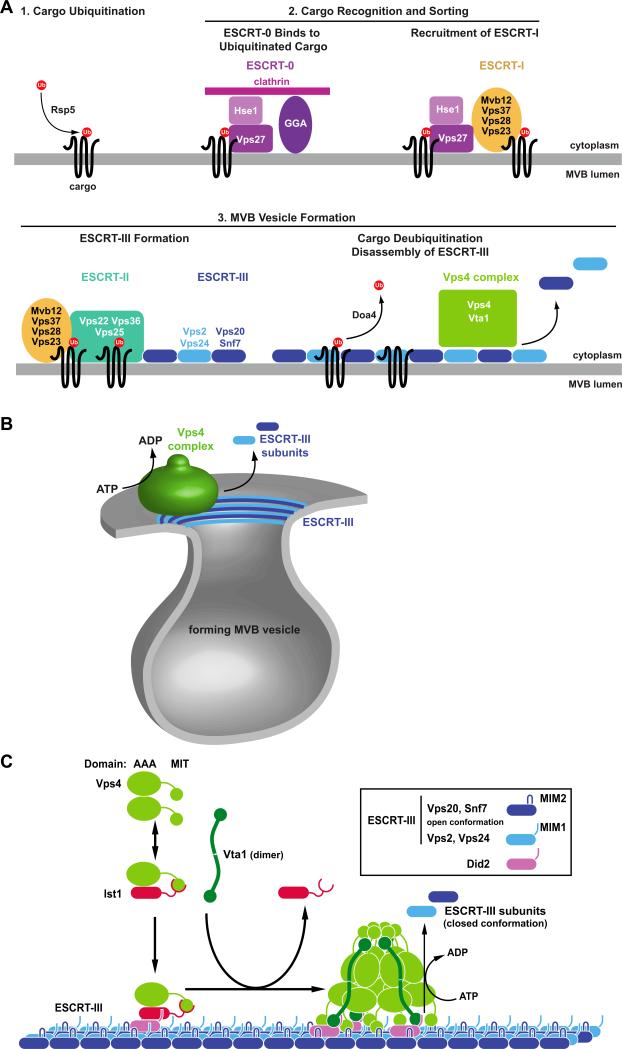
Based on epistasis experiments, a model for the sequential function of the ESCRTs has been proposed (Figure 2A). In this model, ESCRT-0 and ESCRT-I act as cargo-sorting systems that capture ubiquitinated proteins at the endosome and block their recycling back to the plasma membrane or Golgi. ESCRT-I interacts with ESCRT-II, which initiates the formation of ESCRT-III on the MVB. In yeast, ESCRT-III is composed of four functionally essential subunits and three subunits that seem to play a regulatory role in ESCRT-III activity. These soluble, cytoplasmic subunits are recruited to MVBs and oligomerize on the membrane into a polymer that seems to have the shape of concentric rings or spirals [15-18]. These ESCRT-III polymers are involved in the deformation of the membrane and/or the abscission of the vesicle neck. Finally, ESCRT-III recruits Vps4, which functions in the disassembly of the polymer, thereby recycling the ESCRT-III subunits for further rounds of MVB sorting [19, 20].
Figure 2.
Models for the function of Vps4 and the ESCRT protein complexes. (A) Epistasis model for the ubiquitin-dependent sorting of transmembrane proteins (cargo) into the MVB pathway. Flat clathrin together with GGA proteins and ESCRT-0 (Vps27-Hse1) form a protein network at the endosome that binds ubiquitinated cargo. Vps27 recruits ESCRT-I to the MVB, where it supports the cargo sorting function. ESCRT-I interacts with ESCRT-II, resulting in the formation of ESCRT-III. ESCRT-I, ESCRT-II and ESCRT-III are involved in the formation of MVB vesicles that contain the cargo proteins that have been deubiquitinated by Doa4. The disassembly of ESCRT-III by Vps4 drives abscission of the vesicle neck and recycles the ESCRT factors for further rounds of sorting. (B) Model for the function of the Vps4/ESCRT-III system in the abscission of the vesicle neck. ATP-driven disassembly of ESCRT-III by Vps4 constricts the ESCRT-III collar at the neck of the forming vesicle, which induces membrane fission and vesicle formation. (C) Model for the recruitment and assembly of Vps4 on ESCRT-III. Ist1 binds to Vps4, which, together with Did2, aids the recruitment of Vps4 to MVB-associated ESCRT-III. Vta1 dimers support the assembly of Vps4 into the active double-ring structure. The fully assembled Vps4 complex hydrolyses ATP in order to disassemble ESCRT-III.
One reason why the mechanism of vesicle formation by the ESCRTs is not understood lies in the unique topology of this membrane deformation event, in which the formation of MVB vesicles is directed away from the cytoplasm toward the lumen of the compartment. This topology requires a mechanism different from the well-characterized processes of clathrin- and COP-dependent vesicle formation, which use protein coats to induce the required membrane deformation. Therefore, concepts learned by these coat-dependent systems are not applicable to MVB vesicle formation.
Vps4 Functions Together with ESCRT-III in Membrane Fission
First insights into the mechanism of MVB vesicle formation came from studies that found a requirement for the ESCRT machinery for the release of newly formed retroviruses, such as HIV, from the plasma membrane (reviewed in [21, 22]. HIV budding shares the same topology as MVB vesicle formation in that the membrane deforms away from the cytoplasm (Figure 1). In cells mutated for ESCRT function, HIV particles assembled on the cell surface but failed to execute the final membrane scission step that releases the virus from the host cell. In light of the sequential action reported for different ESCRT complexes [20], these studies suggested that ESCRT-III and Vps4 are key components promoting the final membrane fission reaction [23-27].
The hypothesis that the ESCRT-III/Vps4 system acts as membrane-fission machinery was further supported by the observation that these ESCRT factors were essential for proper cytokinesis both in mammalian cells and certain archaea. During the final stage of cytokinesis, ESCRT-III and Vps4 localize to the midbody of the dividing mammalian cell [28-36]. Combination of fluorescence and electron microscopy of dividing cells indicated that ESCRT-III forms a spiral polymer at the site of membrane abscission [37]. Although the exact mechanism is not known, it seems likely that ESCRT-III and Vps4 drive the membrane fission that cause the formation of two daughter cells. Similarly, in archaea, homologues of ESCRT-III subunits and Vps4 assemble into a ring structure at the mid plane of the cell, which might contract during late phase of the cell cycle, thereby providing the mechanism for cell division [38, 39]. The general topology of the membrane fission reaction that occurs at the end of cytokinesis is similar to that necessary to release vesicles into the lumen of MVBs or bud newly formed virus particles at the plasma membrane (Figure 1). However, a major difference between these membrane-based reactions is the large diameter of the membrane neck that has to be constricted during cytokinesis in order to drive abscission, indicating that the ESCRT-III/Vps4 machinery is able to adapt to various membrane curvatures.
In summary, the present data indicate that ESCRT-III together with Vps4 forms a membrane deformation and abscission machinery that originally was developed for cell division. Later in evolution, ESCRT-III/Vps4 was integrated with other ESCRT factors to execute cargo sorting and vesicle formation at the MVB (Figure 2A). Viruses then usurped this function for their own propagation. Although electron microscopy has shown ESCRT-III to form spiral-shaped oligomers tightly associated with membranes, the mechanism of how this spiral constricts in order to sever the membrane neck is not clear. The ATPase Vps4 is an obvious candidate for the energy-providing system necessary to cause membrane constriction and although the only established function of Vps4 to date is the disassembly of the ESCRT-III complex, one attractive possibility is that Vps4 activity also drives membrane constriction (Figure 2B). A study using an in vitro vesicle budding assay found that the addition of ESCRT-III subunits initiated fission of the vesicle neck in absence of Vps4 [40]. This observation suggested that Vps4 functions exclusively in the recycling of ESCRT-III after vesicle formation has been completed. However, electron microscopy studies on yeast cells mutated for Vps4 co-factors showed the presence of enlarged MVB vesicles, which indicated that partial loss of Vps4 activity affected MVB vesicle morphology [41], perhaps because Vps4 plays a direct role in vesicle formation. In vivo localization studies demonstrated that Vps4 is recruited to the site of membrane abscission during cytokinesis and viral budding before membrane fission occurs, further supporting the notion that Vps4 functions as part of the membrane scission machinery [27, 36, 42]. Together the in vivo data suggest a model in which the disassembly of ESCRT-III by Vps4 causes constriction of the ESCRT-III polymer, which ultimately results in the fission of the associated membrane neck (Figure 2B).
Vps4 Structure
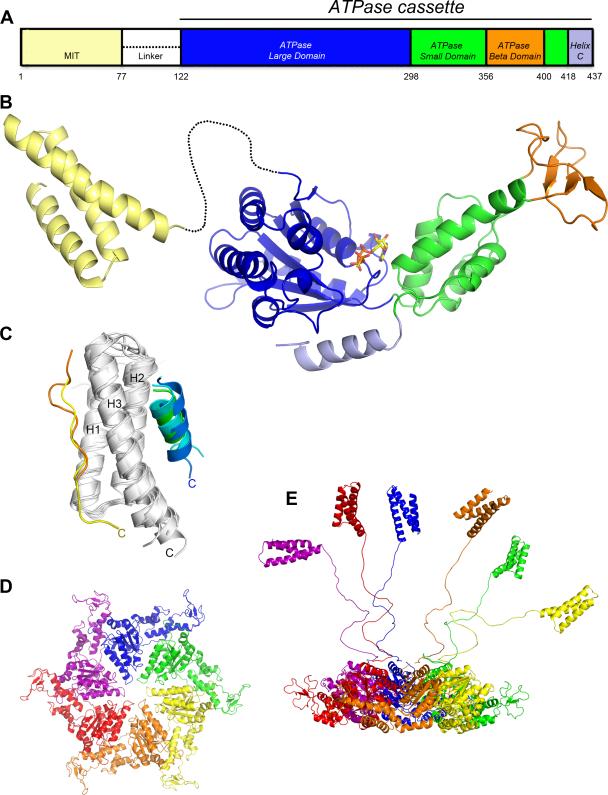
Vps4 exists as a single isoform in S. cerevisiae but can be present as two isoforms in higher eukaryotes such as human. Consistent with the high (60%) degree of sequence identity between the human and S. cerevisiae Vps4 proteins, their three-dimensional structures are also highly conserved. Vps4 proteins contain a single ATPase cassette that is preceded by an ~80 residue N-terminal MIT domain, which forms the initial interactions with the ESCRT-III substrate proteins, and a flexible linker segment of ~40 residues (Figure 3A).
Figure 3.
Structure of Vps4. (A) Schematic of the domain organization. (B) Composite ribbon diagram of the MIT domain and ATPase cassette. ATP-γ-S is bound at the ATP binding site between the large and small ATPase domains. (C) Overlap of independently determined structures of Vps4 MIT domains alone [45][47][48] and in complex with MIM1 peptides [48] blue, [47] green, [47] cyan or MIM2 peptides [24] yellow, [39] orange peptides. MIM1 and MIM2 peptides lie parallel to Helix 3 (C-termini are labeled), which places in them in an orientation to extend from the ESCRT-III complex and potentially bind the same MIT domain. (D) Model for the Vps4 hexamer based on the p97 D1 structure [49]. Top view showing only the ATPase cassettes. (E) Side view including MIT domains and linkers in a variety of potential relatively extended conformations. The extent to which linkers and MIT domains interact with the ATPase cassette hexamer is not currently defined.
First insights into the quaternary structure of Vps4 came from in vitro studies of yeast Vps4(E233Q), a mutant protein with very low ATPase activity [19]. In the presence of ADP, recombinant Vps4(E233Q) was found to form dimers. In contrast, the addition of ATP resulted in the assembly of Vps4(E233Q) into a large complex with at least ten subunits. Thus, Vps4 exhibits a dynamic structure that is regulated by the nucleotide-binding state. This study also found that the higher order Vps4 oligomer hydrolyses ATP at rate of 45 ATPs per minute per subunit, and that oligomerization of yeast Vps4 occurs at protein concentrations above ~0.5μM in vitro whereas the Vps4 concentration in vivo is ~0.2μM. These data suggest that in the cytoplasm Vps4 is dimeric and exhibits no ATPase activity, and that it is recruited to membrane-associated ESCRT-III where it oligomerizes into the active enzyme [19] (Figure 2C).
MIT Structure and its Interactions with ESCRT-III
MIT (contained within Microtubule-Interacting and Trafficking molecules) domains were named on the basis of their sequence conservation in a variety of proteins [43]. Like the N-terminal domains of many other AAA ATPase proteins, they function as the primary docking site for substrates, which in the case of Vps4 are ESCRT-III proteins [44]. As illustrated for the human VPS4A and VPS4B proteins, MIT domains adopt a three-helix bundle structure, in which helices 1 and 3 are parallel to each other and helix 2 is antiparallel [45][46](Figure 3B).
Vps4 MIT domains bind ESCRT-III subunits through two distinct MIT Interacting Motifs that are known as MIM1 and MIM2 (Figure 3C). Human VPS4A and VPS4B MIT domains were shown to bind ~30 residue MIM1 sequences located at the C-terminus of the CHMP1-3 ESCRT-III proteins, and structures of VPS4A MIT-CHMP1A and VPS4B MIT-CHMP2B showed that the MIM1 sequences bind in a helical conformation in a groove between helices 2 and 3 [47]. A parallel study showed that the yeast Vps2 and Did2 ESCRT-III proteins contain equivalent MIM1 sequences at their C-termini, and reported a crystal structure of the Vps2 MIM1 bound to the Vps4p MIT domain [48]. These human and yeast structures superimpose closely and show how the recognition elements include three MIM1 leucine side chains, and associated functional studies demonstrated the importance of these MIT-MIM1 interactions for endosomal sorting in yeast and HIV-1 budding in human cells. MIM2 sequences were subsequently identified in a different subset of human ESCRT-III subunits, including CHMP6, whose MIM2 structure was determined in complex with VPS4A and VPS4B MIT domains and shown to bind in an extended conformation along the groove between helices 1 and 3 [24]. A closely similar archaeal Vps4 MIT-ESCRT-III complex structure has also been reported [39].
MIM1 and MIM2 sequences both bind Vps4 MIT with the N-terminus toward the N-terminal end of helix 3, and the C-terminus toward the C-terminal end of the MIT domain. This orientation fits with the model that Vps4 can bind MIM1 and MIM2 sequences simultaneously and that the MIMs project out from an ESCRT-III assembly on the membrane. Indeed, both MIM1 and MIM2 interactions contribute to a range of Vps4 functions, including endosomal targeting, protein sorting, and HIV-1 budding, and some ESCRT-III proteins even display both MIM1 and MIM2 sequences [24].
Structure of the ATPase Cassette
Crystal structures revealed that mammalian [49][50] and yeast [51][52][53] Vps4 ATPase cassettes comprise four distinct structural elements: large and small AAA ATPase domains, a β domain that is inserted within the small ATPase domain, and a C-terminal helix (Figure 3B). Full-length mammalian proteins were crystallized, although the MIT and linker segment were not visible in the electron density maps, thereby supporting the model that MIT domains are highly mobile with respect to the ATPase cassette [49][50].
The large and small ATPase domains resemble those of other AAA ATPases, especially p97 [54] and spastin [55], whose ATPase domains share 41% and 54% sequence identity with Vps4. The large domain comprises a beta sheet, which is parallel except for the first strand, and is flanked on both sides by alpha helices, while the more C-terminal small ATPase domain comprises an antiparallel four-helix bundle. Like other AAA ATPases, Vps4 binds ATP at the interface between the large and small domains [51][50][53], where it makes contacts and adopts an antiparallel conformation that closely resembles other AAA ATPases such as the p97 D1 and NSF D2 domains [56][54][57][58]. The structures are consistent with the finding that mutation of the Walker A lysine residue to alanine impairs ATP binding, higher-order assembly, vacuolar protein sorting and HIV-1 budding [19][59][23][60]. Moreover, mutation of a highly conserved Walker B glutamate, which functions as the catalytic base that activates a nucleophilic water molecule, inhibits ATP hydrolysis without blocking ATP binding and thereby stabilizes the assembled state and inhibits ATP hydrolysis, vacuolar protein sorting [19][59][61][62] and HIV-1 budding [23][60].
The crystal structures are all very similar, but indicate an en bloc rotation of up to 20° between the large and small ATPase domain, and that nucleotide binding stabilizes a relatively closed conformation [51][53][50]. A second hinging motion is also apparent in the center of helix 8 in the small ATPase domain. This hinge angle can vary by up to 16°, and thereby allows the β domains (below) to project in different directions from the small ATPase domain.
The ~45-residue β domain comprises a three-stranded antiparallel beta sheet that is inserted between the third and fourth helices of the small ATPase domain. Its presence was not anticipated prior to structure determination, and its discovery allowed alignment of the small ATPase domain with other AAA ATPases. Indeed, structure-based sequence alignments indicate that all eukaryotic Vps4 proteins contain the β domain, but that it is absent in the other members of the meiotic clade of AAA ATPases and from the Vps4 protein of Crenarchaea. The β domain projects away from the rest of the Vps4 protein and makes little non-covalent interaction with even the small ATPase domain. As discussed below, the primary function of the β domain is to bind the Vps4 activator LIP5/Vta1.
The C-terminal helix appears to be unique to the meiotic AAA ATPases. It packs against the large ATPase domain, with which it forms an integral structural unit. The functional role of the C-terminal helix, beyond contributing to the structure of the large domain, has been unclear. One suggestion is that it mediates assembly [63] although another study failed to find mutations on the C-terminal helix that disrupted oligomerization without also disrupting the protein fold [49].
The Inactive Conformation
The configurations of the disassembled state in the cytosol and the assembled state bound to membrane-localized substrates have both been vigorously debated. Although the disassembled state is generally held to be dimeric [19][49][52][53][63], there have also been reports that it is monomeric [51][50]. One of the most complete analyses considered six interfaces seen in a variety of Vps4 crystal structures [53]. Using size exclusion chromatography and analytical equilibrium ultracentrifugation, it was shown that S. cerevisiae Vps4 is dimeric in the absence of ATP. The largest two-fold symmetric interface was discounted because mutation of interface residues did not disrupt the dimeric state, although see [52] for an alternative view. The only crystallographic interface whose mutation was found to result in monomeric Vps4 in the absence of ATP is an asymmetric association formed primarily by contacts between the large ATPase domain of one subunit and the small ATPase domain of the neighboring subunit. This interface propagates a six-fold helical stacking of Vps4 subunits that continues all the way through the crystals. The reason Vps4 forms a dimer in solution rather than an infinite helix is presumably because the crystallographic interface does not exactly represent the solution dimer, but that some adjustments occur in solution that make the ends of the dimer incompatible with continuation of the helix.
As discussed in the following section, a leading model is that the assembled state includes a hexamer whose primary interface resembles that of the crystallographic helix and hence also the proposed dimer. In support of these models, mutation of residues that are expected to be part of both the dimer and the hexamer interface resulted in monomeric protein in both the presence and absence of ATP, whereas mutation of a residue that is expected to be part of the dimer but not the hexamer interface was monomeric in the absence of ATP but assembled upon addition of ATP. This indicates that the dimer is not an obligate on-pathway intermediate for forming the Vps4 oligomer. The physiological importance of the dimer is unclear, and it is possible that different Vps4 homologs display different dimerization affinities.
Modeling the Assembled Hexamer
Most studies support the proposal that the assembled state includes a hexamer that resembles the p97 D1 domain structure [49]. This model explains how ATP binding promotes Vps4 assembly [19], as it does for p97 D1 [64], by defining the angle between large and small ATPase domains and allowing close approach of neighboring subunits. Notably, an “arginine finger” [65] residue that contacts the gamma phosphate of ATP bound to the neighboring subunit in the p97 hexamer is conserved in Vps4, and its mutation to alanine blocks assembly [50]. Moreover, many residues at the p97 hexamer interface are conserved in Vps4 and mutation of a conserved interface arginine residue to alanine allowed dimerization of S. cerevisiae Vps4 but blocked formation of the higher order assembly upon addition of ATP [53].
Like many AAA ATPases, an attractive model is that Vps4 translocates its ESCRT-III substrates through the central pore, with translocation driven by two loops near the center of the hexameric ring termed pore loop 1 and pore loop 2 [66][67][68][68]. A section of pore loop 2 is also known as the “arginine collar” owing to the presence of three conserved arginine residues [69][55][70]. Pore 1 motifs of AAA ATPases that act on peptide substrates display the consensus sequence of two large hydrophobic residues followed by a glycine [66][67][71]. The first of these residues is especially important for the protein unfolding/translocation activities of HslU [72], ClpB [71], ClpX [73], and FtsH [68]. Consistent with this model, mutation in the first Vps4 Pore 1 large hydrophobic residue, W206A, did not affect Vps4(E233Q) assembly in vitro but this mutation and other pore loop 1 mutations in human VPS4A or VPS4B inhibited HIV-1 release [49]. Pore loop 2 residues are also important in Vps4 because their mutation in human proteins also exerted a dominant negative effect that inhibited HIV-1 release [53] while corresponding mutations in yeast Vps4(E233Q) did not alter assembly.
The Fully Assembled Conformation
Although the p97-like hexamer model appears to be a component of the active state, it does not comprise the full active Vps4 oligomer, which is estimated to comprise 10-14 subunits [19][49][53][63].
Remarkably, electron microscopic analyses of S. cerevisiae Vps4 have generated three reconstructions that each comprise two rings, but are otherwise very different from each other. The Landsberg model [63] comprises two p97-like rings, the Yu model [57] comprises one p97-like ring and one other ring that is also six-fold symmetric but much more open, and the Hartmann model [52] comprises two seven-member rings that resemble an expanded p97 hexamer. The Hartmann model is a head-to-head configuration in which the two rings associate through symmetric interactions between the large ATPase domain, and the C-terminal helices are on the outer edges. In sharp contrast, the Landsberg model is a tail-to-tail configuration in which the rings associate through the C-terminal helices. All three studies note that their two rings are somewhat different from each other and in the Yu model the two ring configurations are radically different, with one p97-like hexamer, whose up/down orientation is not defined at the available resolution and another ring that has a much more open structure than p97 but again lacks the resolution to orient subunits. The asymmetry seen in all three reconstructions raises the possibility that the two rings are functionally distinct, as seen for the type II AAA ATPases that possess two different ATPase cassettes, such as NSF [74][75][76] and p97 [77].
It is important that the difference between these reconstructions is resolved. It is possible that some apparent discrepancies result from differences in sample preparation or in the constructs or nucleotides used. It is also possible that Vps4 undergoes substantial conformational changes during its reaction cycle and that the different reconstructions have captured different stages along the pathway. Unfortunately, none of the current reconstructions is at a resolution that allows definition of individual secondary structural elements. Urgent priorities include improving the resolution and defining the positions of MIT domains and their relationship to the ATPase cassettes. Studies on mammalian Vps4 indicated that in solution the MIT domain and linker region are auto-inhibitory and suppress the ATPase activity of Vps4, suggesting that these regions bind to the AAA domain thereby affecting the ATP hydrolysis activity [78]. Furthermore, interaction of the MIT domain with ESCRT-III subunits has been shown to enhance ATP hydrolysis by Vps4 [78, 79]. Together these data suggest a model in which interactions with ESCRT-III promote the MIT domain and linker region rearrangements during Vps4 assembly, thus releasing the inhibitory affect on the ATPase. Therefore, it might be necessary to structurally analyze a Vps4-ESCRT-III complex in order to determine the proper positioning of the MIT domain within the active Vps4 complex.
Structure of ESCRT-III
Yeast contains four different ESCRT-III subunits (Vps2, Vps20, Vps24, Snf7) and three ESCRT-III-like proteins (Vps60, Did2, Ist1) that function in the recruitment and regulation of Vps4. Crystal structure analyses of CHMP3/Vps24 and Ist1 showed that they share an N-terminal four-helical bundle that is thought to form the core of all ESCRT-III-type proteins [18, 25, 29]. A region that regulates the oligomeric state of the protein follows this core [18, 29, 80, 81]. In the ‘closed’ conformation this auto-inhibitory region interacts with the core, which maintains the protein in the monomeric state. Disruption of the interaction between the auto-inhibitory region and the core switches the ESCRT-III protein to the ‘open’ conformation, thereby promoting membrane association and oligomerization of the protein. The C-terminal region of the ESCRT-III proteins contains an MIM1- and/or MIM2-type Vps4-interaction motif, which is accessible in the open conformation of the protein [24, 47, 48].
ESCRT-III assembles into a large protein network on the MVB membrane by recruitment of monomeric subunits from the cytoplasm. Vps20 is a myristoylated protein that anchors ESCRT-III to the membrane [20, 82]. Furthermore, positive-charged surfaces of the ESCRT-III proteins are predicted to bind to negative-charged lipids [16, 83]. Together, these interactions ensure a stable association of ESCRT-III with the membrane. Studies of the composition of ESCRT-III suggested that Snf7 represents the main component of the complex [83], although the size of the complex is still under debate. Furthermore, the subunit arrangement and thus the shape of ESCRT-III are not known. Electron microscopic studies of cells overexpressing CHMP4/Snf7 indicated that this major ESCRT-III subunit is able to form spiral-shaped polymers on membranes [15]. Other studies demonstrated the ability of ESCRT-III subunits to assemble into tubes and sheets in vitro [16-18, 83]. However, the significance of these structures for the function of ESCRT-III in vivo is not clear. Furthermore, it remains an open question if the ESCRT-III complexes formed on MVBs and at the midbody are identical.
Regulators of Vps4 Assembly and Activity
Vps4 recruitment and assembly are regulated by a set of four proteins, called Ist1, Did2/CHMP1, Vta1/LIP5 and Vps60/CHMP5. Ist1 is related to the ESCRT-III subunits in that it contains the same alpha-helical core structure [18, 29]. Furthermore, Did2 and Vps60 share sequence homology with ESCRT-III subunits. The C-terminus of Ist1 contains both MIM1- and MIM2-type interaction motifs that bind simultaneously to the MIT domain of Vps4 [31]. In vitro, these interactions result in the formation of an Ist1-Vps4 heterodimer that impairs Vps4 assembly, suggesting that Ist1 is an inhibitor of Vps4 function [84]. The Vps4-Ist1 complex has not been observed in vivo, possibly because of the transient nature of this protein complex. However, overexpression of Ist1 in yeast has been shown to inhibit Vps4 function by blocking the recruitment of Vps4 to ESCRT-III [84]. This observation suggested that in the cytoplasm Ist1 is indeed able to bind Vps4, possibly acting as an inhibitor of spontaneous Vps4 oligomerization.
Ist1 is recruited to ESCRT-III via interaction with Did2/CHMP1 [29, 84, 85]. This observation led to the model that Ist1 together with Did2/CHMP1 might function to recruit Vps4 to ESCRT-III [86] (Figure 2C). This notion is supported by genetic studies in yeast that indicated a supportive role for Ist1 in ESCRT function at the MVB [84, 85]. Furthermore, during cytokinesis in mammalian cells, Ist1 and CHMP1/Did2 were shown to be necessary for the recruitment of Vps4 to the midbody, possibly mediated via the interaction of CHMP1 with midbody localized ESCRT-III [29-31]. Together, these data suggest that Ist1 binds to cytoplasmic Vps4 and, together with Did2/CHMP1, aids in the recruitment of Vps4 proteins to the different sites of ESCRT-III function (Figure 2C).
The Vta1/LIP5 protein contains two N-terminal MIT domains and a VSL domain at the C-terminus that is required for the dimerization of the protein [87]. The VSL domain also binds to the β domain of Vps4 [49][88]. The binding of Vta1 to Vps4 has been shown in vitro to promote both assembly and ATPase activity of Vps4 [89]. Biochemical analysis suggested that the active complex comprises Vps4:Vta1 subunits in a 2:1 stoichiometry [90][91][92][93][44]. These data support the model that Vta1 acts as an assembly factor that promotes formation of the active form of Vps4 on ESCRT-III (Figure 2C). Accordingly, the R352A mutation, which blocked formation of the higher order Vps4(E233Q) oligomer, also inhibited Vta1 binding [49]. Furthermore, in vitro studies have shown that Vta1 increases the maximal Vps4 ATPase activity by approximately 3-fold, suggesting that binding of Vta1 affects the active site of Vps4 [89, 91].
A crystal structure of Vta1 with a construct spanning the small ATPase and β domains has been reported [88]. This agrees closely with the VSL [87] and ATPase cassette structures determined in isolation. Vps4-Vta1 interface residues are conserved and their mutation eliminates binding in vitro and elicits an MVB sorting defect in yeast. Surprisingly, modeling of the Vta1 complex with a p97-like ring indicates that the two-fold axis of the Vta1 VSL domain is roughly parallel to the six-fold axis of the hexamer, which suggests that Vta1 might not crosslink the two hexameric rings of Vps4, but rather stabilizes an array of Vps4-Vta1 complexes for ESCRT-III disassembly. It is currently not clear how to reconcile this model with reports that Vta1 forms a stable complex that contains just one Vps4 dodecamer [90]. It has also been reported that the isolated VSL domain does not stimulate ATP hydrolysis as well as full-length Vta1 [89] indicating that other Vta1 regions likely also contact Vps4.
Vta1 not only binds to Vps4 but also interacts via one of its MIT domains with ESCRT-III associated Did2. This interaction might help coordinate the Vps4 recruiting function of Ist1-Did2 with the subsequent Vta1-supported assembly of Vps4 (Figure 2C). In addition, Vta1 recruits the fourth Vps4 regulator, Vps60/CHMP5, to the ESCRT machinery. Although the precise function of this ESCRT factor is not known, analysis of the VPS60 deletion strains suggested that this protein might aid the release of Vta1 from ESCRT-III containing membranes [79].
Together, Ist1, Did2, Vta1 and Vps60 function as a regulatory system that ensures proper recruitment and assembly of Vps4 on ESCRT-III. Furthermore, these four factors have been shown to directly or indirectly affect ATPase activity of Vps4, suggesting that they also regulate the Vps4 ESCRT-III disassembly activity. However, in yeast, mutations in Ist1, Did2, Vta1 or Vps60 result in only weak or intermediate MVB trafficking defects [84, 85, 89, 92, 94], indicating that these factors are not essential for Vps4 function, an observation that is consistent with the proposed regulatory role.
Vps4 as a disassembly factor of ESCRT-III
Formation of the active Vps4 complex on ESCRT-III results in the ATP-driven disassembly of the ESCRT-III polymer, an enzymatic reaction that is poorly understood. Based on insights to the mechanism of other AAA-type ATPases, it has been speculated that Vps4 engages the ESCRT-III substrate via the central pore. Mutations in conserved amino acids that are predicted to face the pore of the Vps4 oligomer impaired HIV virus release, indicating that the pore is functionally important [49]. However, a direct interaction between the Vps4 pore and ESCRT-III subunits has not been demonstrated. In contrast, the binding of the Vps4 MIT domain to ESCRT-III is well established and has been shown to be important not only for localization of Vps4 to ESCRT-III but also for ESCRT-III disassembly [19, 45-47, 95]. Studies in yeast have shown that although MIT-deleted Vps4 localizes to MVBs by binding to Vta1, this mutant ATPase is not able to disassemble ESCRT-III [86]. These observations suggest that the MIT domain might not only bind to ESCRT-III subunits in order to recruit Vps4 to the substrate but might also function together with the pore region in the ESCRT-III disassembly reaction. However, in part because of lack of knowledge of the ESCRT-III structure, the precise mechanism how Vps4 induces the disassembly is not understood. One possible scenario is that the binding of the MIT domain to MIM1 or MIM2 of a particular ESCRT-III subunit allows the interaction of Vps4 pore loops with that subunit. Based on studies of other AAA proteins, ATP hydrolysis is expected to move these loops further into the pore, thereby inducing a conformational change in the bound ESCRT-III subunit. This change in conformation would presumably break the interactions with other ESCRT-III subunits, thereby allowing the subunit to be released from the MVB. This dissociated subunit might be moved through the pore of the double-ring structure or stay associated with the membrane until the disassembly of ESCRT-III is complete. In any case, the disassembly reaction causes the ESCRT-III subunits to regain the monomeric conformational state, which is the high-energy state that is poised to reassemble again into the ESCRT-III oligomer for subsequent rounds of membrane scission.
Although the oligomeric structure of Vps4 is dependent on the nucleotide binding state, recent studies using an in vitro ESCRT-III disassembly system indicated that when assembled, Vps4 progresses through many ATP hydrolysis cycles without dissociation [95]. This suggests that the nucleotide exchange within the assembled Vps4 is faster than the ATP hydrolysis rate, and that a sufficient number of Vps4 subunits remain bound to ATP to stabilize the complex while other subunits bind ADP or are empty. For example, this could take the form of a highly coordinated ATPase reaction cycle within the Vps4 hexameric ring, such as the system found for the Escherichia coli AAA ATPase Rho [96]. In summary, the recent in vitro studies suggest that Vps4 assembles on ESCRT-III and performs multiple disassembly reactions until ESCRT-III dissociation has been completed.
Concluding Remarks
Although many structural and mechanistic questions about Vps4 are still open, the current data suggest that Vps4 and ESCRT-III represent an evolutionary ancient membrane remodeling system. Vps4 disassembles the ESCRT-III polymer, thereby changing the morphology of the underlying membrane. In this regard the Vps4/ESCRT-III machinery resembles the actin cytoskeleton, which uses polymerization and dissociation reactions at the cortex to induce plasma membrane deformation. Interestingly, Vps4 and ESCRT-III have recently been implicated in centrosome maintenance, a function that is not based on membrane deformation [97]. This suggests that future studies are likely to uncover additional cellular pathways that require the function of the Vps4/ESCRT-III system.
Highlights.
- Vps4 belongs to the type-I AAA family of ATPases and functions together with ESCRT-III in the deformation and fission of membranes.
- Vps4-mediated membrane fission plays an important role for MVB vesicle formation, cytokinesis and viral budding.
- Vps4 posses a dynamic quaternary structure that is regulated by the bound nucleotide and co-factors.
Acknowledgements
We thank Frank Whitby for assistance with Figures. The research conducted in the Hill and Babst laboratories on Vps4 is supported by NIH Grants P50 GM082545 and R01 GM074171, respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babst M, Sato TK, Banta LM, Emr SD. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 1997;16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Odorizzi G, Babst M, Emr SD. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- 4.Piper RC, Katzmann DJ. Biogenesis and Function of Multivesicular Bodies. Annu Rev Cell Dev Biol. 2007;23:519–547. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woodman PG, Futter CE. Multivesicular bodies: co-ordinated progression to maturity. Curr Opin Cell Biol. 2008;20:408–414. doi: 10.1016/j.ceb.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eden ER, White IJ, Futter CE. Down-regulation of epidermal growth factor receptor signalling within multivesicular bodies. Biochem Soc Trans. 2009;37:173–177. doi: 10.1042/BST0370173. [DOI] [PubMed] [Google Scholar]
- 7.Woodman P. ESCRT proteins, endosome organization and mitogenic receptor down-regulation. Biochem Soc Trans. 2009;37:146–150. doi: 10.1042/BST0370146. [DOI] [PubMed] [Google Scholar]
- 8.Wegner CS, Rodahl LM, Stenmark H. ESCRT Proteins and Cell Signalling. Traffic. 2011 doi: 10.1111/j.1600-0854.2011.01210.x. [DOI] [PubMed] [Google Scholar]
- 9.Luzio JP, Parkinson MD, Gray SR, Bright NA. The delivery of endocytosed cargo to lysosomes. Biochem Soc Trans. 2009;37:1019–1021. doi: 10.1042/BST0371019. [DOI] [PubMed] [Google Scholar]
- 10.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 11.Lee JA, Gao FB. Roles of ESCRT in autophagy-associated neurodegeneration. Autophagy. 2008;4:230–232. doi: 10.4161/auto.5384. [DOI] [PubMed] [Google Scholar]
- 12.Saksena S, Emr SD. ESCRTs and human disease. Biochem Soc Trans. 2009;37:167–172. doi: 10.1042/BST0370167. [DOI] [PubMed] [Google Scholar]
- 13.Hurley JH. The ESCRT complexes. Crit Rev Biochem Mol Biol. 2010;45:463–487. doi: 10.3109/10409238.2010.502516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piper RC, Luzio JP. Ubiquitin-dependent sorting of integral membrane proteins for degradation in lysosomes. Curr Opin Cell Biol. 2007;19:459–465. doi: 10.1016/j.ceb.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weissenhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghazi-Tabatabai S, Saksena S, Short JM, Pobbati AV, Veprintsev DB, Crowther RA, Emr SD, Egelman EH, Williams RL. Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24. Structure. 2008;16:1345–1356. doi: 10.1016/j.str.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 18.Bajorek M, Schubert HL, McCullough J, Langelier C, Eckert DM, Stubblefield WM, Uter NT, Myszka DG, Hill CP, Sundquist WI. Structural basis for ESCRT-III protein autoinhibition. Nat Struct Mol Biol. 2009;16:754–762. doi: 10.1038/nsmb.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. ESCRT-III: an endosome-associated heterooligomeric protein complex required for MVB sorting. Dev Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- 21.Bieniasz PD. The cell biology of HIV-1 virion genesis. Cell Host Microbe. 2009;5:550–558. doi: 10.1016/j.chom.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlton JG, Martin-Serrano J. The ESCRT machinery: new functions in viral and cellular biology. Biochem Soc Trans. 2009;37:195–199. doi: 10.1042/BST0370195. [DOI] [PubMed] [Google Scholar]
- 23.Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, Myszka DG, Sundquist WI. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 24.Kieffer C, Skalicky JJ, Morita E, De Domenico I, Ward DM, Kaplan J, Sundquist WI. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev Cell. 2008;15:62–73. doi: 10.1016/j.devcel.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 25.Muziol T, Pineda-Molina E, Ravelli RB, Zamborlini A, Usami Y, Gottlinger H, Weissenhorn W. Structural basis for budding by the ESCRT-III factor CHMP3. Dev Cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 26.Morita E, Sandrin V, McCullough J, Katsuyama A, Baci Hamilton I, Sundquist WI. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe. 2011;9:235–242. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat Cell Biol. 2011;13:394–401. doi: 10.1038/ncb2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Renvoise B, Parker RL, Yang D, Bakowska JC, Hurley JH, Blackstone C. SPG20 protein spartin is recruited to midbodies by ESCRT-III protein Ist1 and participates in cytokinesis. Mol Biol Cell. 2010;21:3293–3303. doi: 10.1091/mbc.E09-10-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao J, Chen XW, Davies BA, Saltiel AR, Katzmann DJ, Xu Z. Structural basis of Ist1 function and Ist1-Did2 interaction in the multivesicular body pathway and cytokinesis. Mol Biol Cell. 2009;20:3514–3524. doi: 10.1091/mbc.E09-05-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agromayor M, Carlton JG, Phelan JP, Matthews DR, Carlin LM, Ameer-Beg S, Bowers K, Martin-Serrano J. Essential role of hIST1 in cytokinesis. Mol Biol Cell. 2009;20:1374–1387. doi: 10.1091/mbc.E08-05-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bajorek M, Morita E, Skalicky JJ, Morham SG, Babst M, Sundquist WI. Biochemical analyses of human IST1 and its function in cytokinesis. Mol Biol Cell. 2009;20:1360–1373. doi: 10.1091/mbc.E08-05-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang D, Rismanchi N, Renvoise B, Lippincott-Schwartz J, Blackstone C, Hurley JH. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat Struct Mol Biol. 2008;15:1278–1286. doi: 10.1038/nsmb.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee HH, Elia N, Ghirlando R, Lippincott-Schwartz J, Hurley JH. Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science. 2008;322:576–580. doi: 10.1126/science.1162042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 35.Morita E, Sandrin V, Chung HY, Morham SG, Gygi SP, Rodesch CK, Sundquist WI. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elia N, Sougrat R, Spurlin TA, Hurley JH, Lippincott-Schwartz J. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc Natl Acad Sci U S A. 2011;108:4846–4851. doi: 10.1073/pnas.1102714108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guizetti J, Schermelleh L, Mantler J, Maar S, Poser I, Leonhardt H, Muller-Reichert T, Gerlich DW. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science. 2011;331:1616–1620. doi: 10.1126/science.1201847. [DOI] [PubMed] [Google Scholar]
- 38.Samson RY, Obita T, Hodgson B, Shaw MK, Chong PL, Williams RL, Bell SD. Molecular and structural basis of ESCRT-III recruitment to membranes during archaeal cell division. Mol Cell. 2011;41:186–196. doi: 10.1016/j.molcel.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in archaea. Science. 2008;322:1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nickerson DP, West M, Henry R, Odorizzi G. Regulators of Vps4 ATPase activity at endosomes differentially influence the size and rate of formation of intralumenal vesicles. Mol Biol Cell. 2010;21:1023–1032. doi: 10.1091/mbc.E09-09-0776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baumgartel V, Ivanchenko S, Dupont A, Sergeev M, Wiseman PW, Krausslich HG, Brauchle C, Muller B, Lamb DC. Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat Cell Biol. 2011;13:469–474. doi: 10.1038/ncb2215. [DOI] [PubMed] [Google Scholar]
- 43.Ciccarelli FD, Proukakis C, Patel H, Cross H, Azam S, Patton MA, Bork P, Crosby AH. The identification of a conserved domain in both spartin and spastin, mutated in hereditary spastic paraplegia. Genomics. 2003;81:437–441. doi: 10.1016/s0888-7543(03)00011-9. [DOI] [PubMed] [Google Scholar]
- 44.Yeo SC, Xu L, Ren J, Boulton VJ, Wagle MD, Liu C, Ren G, Wong P, Zahn R, Sasajala P, Yang H, Piper RC, Munn AL. Vps20p and Vta1p interact with Vps4p and function in multivesicular body sorting and endosomal transport in Saccharomyces cerevisiae. J Cell Sci. 2003;116:3957–3970. doi: 10.1242/jcs.00751. [DOI] [PubMed] [Google Scholar]
- 45.Scott A, Gaspar J, Stuchell-Brereton MD, Alam SL, Skalicky JJ, Sundquist WI. Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A. Proc Natl Acad Sci U S A. 2005;102:13813–13818. doi: 10.1073/pnas.0502165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takasu H, Jee JG, Ohno A, Goda N, Fujiwara K, Tochio H, Shirakawa M, Hiroaki H. Structural characterization of the MIT domain from human Vps4b. Biochem Biophys Res Commun. 2005;334:460–465. doi: 10.1016/j.bbrc.2005.06.110. [DOI] [PubMed] [Google Scholar]
- 47.Stuchell-Brereton MD, Skalicky JJ, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- 48.Obita T, Saksena S, Ghazi-Tabatabai S, Gill DJ, Perisic O, Emr SD, Williams RL. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449:735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- 49.Scott A, Chung HY, Gonciarz-Swiatek M, Hill GC, Whitby FG, Gaspar J, Holton JM, Viswanathan R, Ghaffarian S, Hill CP, Sundquist WI. Structural and mechanistic studies of VPS4 proteins. EMBO J. 2005;24:3658–3669. doi: 10.1038/sj.emboj.7600818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inoue M, Kamikubo H, Kataoka M, Kato R, Yoshimori T, Wakatsuki S, Kawasaki M. Nucleotide-dependent conformational changes and assembly of the AAA ATPase SKD1/VPS4B. Traffic. 2008;9:2180–2189. doi: 10.1111/j.1600-0854.2008.00831.x. [DOI] [PubMed] [Google Scholar]
- 51.Xiao J, Xia H, Yoshino-Koh K, Zhou J, Xu Z. Structural characterization of the ATPase reaction cycle of endosomal AAA protein Vps4. J Mol Biol. 2007;374:655–670. doi: 10.1016/j.jmb.2007.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hartmann C, Chami M, Zachariae U, de Groot BL, Engel A, Grutter MG. Vacuolar protein sorting: two different functional states of the AAA-ATPase Vps4p. J Mol Biol. 2008;377:352–363. doi: 10.1016/j.jmb.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 53.Gonciarz MD, Whitby FG, Eckert DM, Kieffer C, Heroux A, Sundquist WI, Hill CP. Biochemical and structural studies of yeast Vps4 oligomerization. J Mol Biol. 2008;384:878–895. doi: 10.1016/j.jmb.2008.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X, Shaw A, Bates PA, Newman RH, Gowen B, Orlova E, Gorman MA, Kondo H, Dokurno P, Lally J, Leonard G, Meyer H, van Heel M, Freemont PS. Structure of the AAA ATPase p97. Mol Cell. 2000;6:1473–1484. doi: 10.1016/s1097-2765(00)00143-x. [DOI] [PubMed] [Google Scholar]
- 55.Roll-Mecak A, Vale RD. Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin. Nature. 2008;451:363–367. doi: 10.1038/nature06482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brunger AT, DeLaBarre B. NSF and p97/VCP: similar at first, different at last. FEBS Lett. 2003;555:126–133. doi: 10.1016/s0014-5793(03)01107-4. [DOI] [PubMed] [Google Scholar]
- 57.Yu RC, Hanson PI, Jahn R, Brunger AT. Structure of the ATP-dependent oligomerization domain of N-ethylmaleimide sensitive factor complexed with ATP. Nat Struct Biol. 1998;5:803–811. doi: 10.1038/1843. [DOI] [PubMed] [Google Scholar]
- 58.Lenzen CU, Steinmann D, Whiteheart SW, Weis WI. Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein. Cell. 1998;94:525–536. doi: 10.1016/s0092-8674(00)81593-7. [DOI] [PubMed] [Google Scholar]
- 59.Bishop N, Woodman P. ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol Biol Cell. 2000;11:227–239. doi: 10.1091/mbc.11.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, Scott A, Krausslich HG, Kaplan J, Morham SG, Sundquist WI. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 61.Yoshimori T, Yamagata F, Yamamoto A, Mizushima N, Kabeya Y, Nara A, Miwako I, Ohashi M, Ohsumi M, Ohsumi Y. The mouse SKD1, a homologue of yeast Vps4p, is required for normal endosomal trafficking and morphology in mammalian cells. Mol Biol Cell. 2000;11:747–763. doi: 10.1091/mbc.11.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scheuring S, Rohricht RA, Schoning-Burkhardt B, Beyer A, Muller S, Abts HF, Kohrer K. Mammalian cells express two VPS4 proteins both of which are involved in intracellular protein trafficking. J Mol Biol. 2001;312:469–480. doi: 10.1006/jmbi.2001.4917. [DOI] [PubMed] [Google Scholar]
- 63.Landsberg MJ, Vajjhala PR, Rothnagel R, Munn AL, Hankamer B. Three-dimensional structure of AAA ATPase Vps4: advancing structural insights into the mechanisms of endosomal sorting and enveloped virus budding. Structure. 2009;17:427–437. doi: 10.1016/j.str.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 64.Wang Q, Song C, Yang X, Li CC. D1 ring is stable and nucleotide-independent, whereas D2 ring undergoes major conformational changes during the ATPase cycle of p97-VCP. J Biol Chem. 2003;278:32784–32793. doi: 10.1074/jbc.M303869200. [DOI] [PubMed] [Google Scholar]
- 65.Ogura T, Whiteheart SW, Wilkinson AJ. Conserved arginine residues implicated in ATP hydrolysis, nucleotide-sensing, and inter-subunit interactions in AAA and AAA+ ATPases. J Struct Biol. 2004;146:106–112. doi: 10.1016/j.jsb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 66.Wang J, Song JJ, Franklin MC, Kamtekar S, Im YJ, Rho SH, Seong IS, Lee CS, Chung CH, Eom SH. Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism. Structure. 2001;9:177–184. doi: 10.1016/s0969-2126(01)00570-6. [DOI] [PubMed] [Google Scholar]
- 67.Wang J, Song JJ, Seong IS, Franklin MC, Kamtekar S, Eom SH, Chung CH. Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU. Structure. 2001;9:1107–1116. doi: 10.1016/s0969-2126(01)00670-0. [DOI] [PubMed] [Google Scholar]
- 68.Yamada-Inagawa T, Okuno T, Karata K, Yamanaka K, Ogura T. Conserved pore residues in the AAA protease FtsH are important for proteolysis and its coupling to ATP hydrolysis. J Biol Chem. 2003;278:50182–50187. doi: 10.1074/jbc.M308327200. [DOI] [PubMed] [Google Scholar]
- 69.DeLaBarre B, Christianson JC, Kopito RR, Brunger AT. Central pore residues mediate the p97/VCP activity required for ERAD. Mol Cell. 2006;22:451–462. doi: 10.1016/j.molcel.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 70.White SR, Evans KJ, Lary J, Cole JL, Lauring B. Recognition of C-terminal amino acids in tubulin by pore loops in Spastin is important for microtubule severing. J Cell Biol. 2007;176:995–1005. doi: 10.1083/jcb.200610072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, Lee S, Zentgraf H, Weber-Ban EU, Dougan DA, Tsai FT, Mogk A, Bukau B. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 72.Song HK, Hartmann C, Ramachandran R, Bochtler M, Behrendt R, Moroder L, Huber R. Mutational studies on HslU and its docking mode with HslV. Proc Natl Acad Sci U S A. 2000;97:14103–14108. doi: 10.1073/pnas.250491797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Siddiqui SM, Sauer RT, Baker TA. Role of the processing pore of the ClpX AAA+ ATPase in the recognition and engagement of specific protein substrates. Genes Dev. 2004;18:369–374. doi: 10.1101/gad.1170304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Whiteheart SW, Rossnagel K, Buhrow SA, Brunner M, Jaenicke R, Rothman JE. N-ethylmaleimide-sensitive fusion protein: a trimeric ATPase whose hydrolysis of ATP is required for membrane fusion. J Cell Biol. 1994;126:945–954. doi: 10.1083/jcb.126.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nagiec EE, Bernstein A, Whiteheart SW. Each domain of the N-ethylmaleimide-sensitive fusion protein contributes to its transport activity. J Biol Chem. 1995;270:29182–29188. doi: 10.1074/jbc.270.49.29182. [DOI] [PubMed] [Google Scholar]
- 76.Whiteheart SW, Schraw T, Matveeva EA. N-ethylmaleimide sensitive factor (NSF) structure and function. Int Rev Cytol. 2001;207:71–112. doi: 10.1016/s0074-7696(01)07003-6. [DOI] [PubMed] [Google Scholar]
- 77.DeLaBarre B, Brunger AT. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat Struct Biol. 2003;10:856–863. doi: 10.1038/nsb972. [DOI] [PubMed] [Google Scholar]
- 78.Merrill SA, Hanson PI. Activation of human VPS4A by ESCRT-III proteins reveals ability of substrates to relieve enzyme autoinhibition. J Biol Chem. 2010;285:35428–35438. doi: 10.1074/jbc.M110.126318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Azmi IF, Davies BA, Xiao J, Babst M, Xu Z, Katzmann DJ. ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev Cell. 2008;14:50–61. doi: 10.1016/j.devcel.2007.10.021. [DOI] [PubMed] [Google Scholar]
- 80.Shim S, Kimpler LA, Hanson PI. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic. 2007;8:1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- 81.Lata S, Roessle M, Solomons J, Jamin M, Gottlinger HG, Svergun DI, Weissenhorn W. Structural basis for autoinhibition of ESCRT-III CHMP3. J Mol Biol. 2008;378:818–827. doi: 10.1016/j.jmb.2008.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yorikawa C, Shibata H, Waguri S, Hatta K, Horii M, Katoh K, Kobayashi T, Uchiyama Y, Maki M. Human CHMP6, a myristoylated ESCRT-III protein, interacts directly with an ESCRT-II component EAP20 and regulates endosomal cargo sorting. Biochem J. 2005;387:17–26. doi: 10.1042/BJ20041227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD. Functional reconstitution of ESCRT-III assembly and disassembly. Cell. 2009;136:97–109. doi: 10.1016/j.cell.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dimaano C, Jones CB, Hanono A, Curtiss M, Babst M. Ist1 regulates Vps4 localization and assembly. Mol Biol Cell. 2008;19:465–474. doi: 10.1091/mbc.E07-08-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rue SM, Mattei S, Saksena S, Emr SD. Novel Ist1-Did2 complex functions at a late step in multivesicular body sorting. Mol Biol Cell. 2008;19:475–484. doi: 10.1091/mbc.E07-07-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shestakova A, Hanono A, Drosner S, Curtiss M, Davies BA, Katzmann DJ, Babst M. Assembly of the AAA ATPase Vps4 on ESCRT-III. Mol Biol Cell. 2010;21:1059–1071. doi: 10.1091/mbc.E09-07-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xiao J, Xia H, Zhou J, Azmi IF, Davies BA, Katzmann DJ, Xu Z. Structural basis of Vta1 function in the multivesicular body sorting pathway. Dev Cell. 2008;14:37–49. doi: 10.1016/j.devcel.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang D, Hurley JH. Structural role of the Vps4-Vta1 interface in ESCRT-III recycling. Structure. 2010;18:976–984. doi: 10.1016/j.str.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Azmi I, Davies B, Dimaano C, Payne J, Eckert D, Babst M, Katzmann DJ. Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. J Cell Biol. 2006;172:705–717. doi: 10.1083/jcb.200508166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yu Z, Gonciarz MD, Sundquist WI, Hill CP, Jensen GJ. Cryo-EM structure of dodecameric Vps4p and its 2:1 complex with Vta1p. J Mol Biol. 2008;377:364–377. doi: 10.1016/j.jmb.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lottridge JM, Flannery AR, Vincelli JL, Stevens TH. Vta1p and Vps46p regulate the membrane association and ATPase activity of Vps4p at the yeast multivesicular body. Proc Natl Acad Sci U S A. 2006;103:6202–6207. doi: 10.1073/pnas.0601712103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shiflett SL, Ward DM, Huynh D, Vaughn MB, Simmons JC, Kaplan J. Characterization of Vta1p, a class E Vps protein in Saccharomyces cerevisiae. J Biol Chem. 2004;279:10982–10990. doi: 10.1074/jbc.M312669200. [DOI] [PubMed] [Google Scholar]
- 93.Ward DM, Vaughn MB, Shiflett SL, White PL, Pollock AL, Hill J, Schnegelberger R, Sundquist WI, Kaplan J. The role of LIP5 and CHMP5 in multivesicular body formation and HIV-1 budding in mammalian cells. J Biol Chem. 2005;280:10548–10555. doi: 10.1074/jbc.M413734200. [DOI] [PubMed] [Google Scholar]
- 94.Nickerson DP, West M, Odorizzi G. Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J Cell Biol. 2006;175:715–720. doi: 10.1083/jcb.200606113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Davies BA, Azmi IF, Payne J, Shestakova A, Horazdovsky BF, Babst M, Katzmann DJ. Coordination of substrate binding and ATP hydrolysis in Vps4-mediated ESCRT-III disassembly. Mol Biol Cell. 2010;21:3396–3408. doi: 10.1091/mbc.E10-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Thomsen ND, Berger JM. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell. 2009;139:523–534. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Morita E, Colf LA, Karren MA, Sandrin V, Rodesch CK, Sundquist WI. Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc Natl Acad Sci U S A. 2010;107:12889–12894. doi: 10.1073/pnas.1005938107. [DOI] [PMC free article] [PubMed] [Google Scholar]