Abstract
The endocannabinoid anandamide is removed from the synaptic space by a selective transport system, expressed in neurons and astrocytes, which remains molecularly uncharacterized. Here we describe a partly cytosolic variant of the intracellular anandamide-degrading enzyme, fatty acid amide hydrolase-1 (FAAH-1), termed FAAH-like anandamide transporter (FLAT), which lacks amidase activity but binds anandamide with low micromolar affinity and facilitates its translocation into cells. Known anandamide transport inhibitors, such as AM404 and OMDM-1, block these effects. Additionally, we identify a competitive antagonist of the interaction of anandamide with FLAT, the phthalazine derivative ARN272, which prevents anandamide internalization in vitro, interrupts anandamide deactivation in vivo, and exerts profound analgesic effects in rodent models of nociceptive and inflammatory pain, which are mediated by CB1 cannabinoid receptors. The results identify FLAT as a critical molecular component of anandamide transport in neural cells and a potential target for therapeutic drugs.
INTRODUCTION
Brain cells release a variety of lipid mediators, which act in close proximity of their site of production to modulate synaptic plasticity and neural development1. The reliability of this highly localized form of neural communication depends on the existence of deactivation mechanisms that ensure the rapid termination of lipid-mediated signaling, but few such mechanisms have been discovered so far. Anandamide is an arachidonic acid derivative that regulates ion-channel activity and neurotransmitter release by engaging CB1 cannabinoid receptors on axon terminals2. There is evidence that the intensity and duration of anandamide signaling are controlled by a two-step elimination process in which the substance is first internalized by neurons and astrocytes3–5 and then hydrolyzed by the intracellular membrane-bound amidases, FAAH-1 and FAAH-26–8. Removal of anandamide from the extracellular space exhibits several identifying features of a carrier-mediated facilitated diffusion process4,9,10: (i) it is saturable and displays low micromolar affinity for anandamide (apparent Michaelis constant, KM, 1.2 μM in rat cortical neurons)3; (ii) it preferentially recognizes anandamide over similar molecules, including the non-cannabinoid FAAH substrates oleoylethanolamide (OEA) and palmitoylethanolamide (PEA)3,11; (iii) it is inhibited in a competitive and stereoselective manner by substrate mimics10; and (iv) it does not require cellular energy3,4. Inhibitors of anandamide transport - which include the compounds AM404 and OMDM-110 - increase the levels of this endocannabinoid substance in vivo and produce a spectrum of CB1-mediated responses that only partially overlap with those elicited by FAAH blockade, presumably owing to the different kinetic properties and substrate preferences of the two deactivation mechanisms10. These data indicate that carrier-mediated transport may play an important role in terminating the actions of anandamide and might represent a potential drug target10. Nevertheless, the molecular entity (or entities) involved in anandamide translocation is still unknown and the mechanistic bases of this process remain controversial5,12. Here, we identify a partly cytosolic variant of FAAH-1, termed FLAT, which lacks amidase activity but binds anandamide with low micromolar affinity and confers anandamide transport to cells that are engineered to express it. AM404 and other anandamide transport inhibitors suppress these effects. Moreover, we disclose a small-molecule competitive inhibitor of the interaction of anandamide with FLAT, the compound ARN272, and show that this agent suppresses anandamide translocation in vitro and interrupts anandamide deactivation in vivo.
RESULTS
FLAT is an intracellular anandamide-binding protein
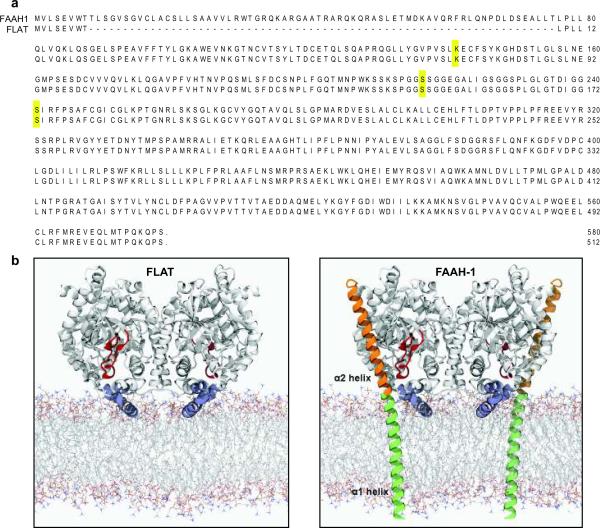
We isolated total RNA from brain and other rat tissues, and amplified products of the faah-1 gene using reverse-transcriptase polymerase chain reaction (RT-PCR). One of the complementary DNA products obtained was identical to faah-1 except that it lacked a 2 0 4 base-pair segment encoding for amino-acid residues 9–76 (Fig. 1a, Supplementary Fig. 1a–c). Ribonuclease protection assays and Southern blot analyses of RT-generated cDNA confirmed the normal occurrence of FLAT mRNA in rat brain and liver tissue (Supplementary Fig. 1d,e). Quantitative RT-PCR measurements showed that FLAT is unevenly transcribed in the rat brain, with highest levels in neocortex and hippocampus and lowest levels in brainstem and hypothalamus (Supplementary Fig. 1f). Detectable levels of FLAT mRNA were also found in rat primary astrocytes in cultures, rat neuroblastoma cells, and human astrocytoma cells (Supplementary Fig. 1c), which were previously shown to express anandamide transport3,11,13. An antibody raised against the C-terminus of FAAH-1 identified in brain cytosolic and membrane fractions obtained from wild-type mice, but not in those obtained from FAAH-1-deficient mice, a band with an apparent molecular weight of 56 kDa, which is consistent with the calculated molecular weight of FLAT (56,008 Da) (Supplementary Fig. 1g). This suggests that FLAT might be a product of the faah-1 gene generated by alternative splicing at non-canonical sites14. The predicted structure of FLAT lacks most of FAAH-1's α1 helix, which spans the lipid bilayer of intracellular membranes, and the entire α2 helix, which flanks the globular body of the protein exposed to the cytosol (Fig. 1b)15.
Fig. 1.
Structural properties of FLAT. (a) Predicted amino acid sequences of FLAT and FAAH-1; residues comprising the catalytic triad of FAAH-1 (Lys142, Ser217 and nucleophile Ser241) are highlighted. (b) Model of rat FLAT (left) based on the structure of FAAH-ΔTM (right), a FAAH-1 mutant lacking the α1 helix15 which is redrawn in green for illustration purposes. Most of the α1 helix and the entire α2 helix (orange) of FAAH-1 are absent in FLAT. Both FAAH-1 and FLAT contain a membrane-binding domain (blue, FLAT residues 343–367) and an 'α2-interacting loop' (red, FLAT residues 187–210), which may interact with the α2 helix and help shield the enzyme's catalytic pocket from water. The membrane model was generated using Molecular Dynamics simulations of a 1,2-dioleoyl-sn-glycerol-3-phosphorylcholine bilayer39.
When expressed in Hek293 cells, FLAT displayed no detectable amidase activity toward anandamide or OEA (Supplementary Fig. 2a,b), suggesting that the protein is catalytically defective. Computational studies identified two factors that might contribute to this loss of activity: (i) increased flexibility of regions proximal to the missing α1 and α2 helices - such as the 'α2-interacting loop' (Fig. 1b) - could facilitate access of water to the catalytic site buried inside the enzyme's hydrophobic core (Supplementary Fig. 3)7; and (ii) deletion of the α2 helix, which carries a positively charged surface, could lower the electrostatic potential in the region surrounding the catalytic triad component Lys74 (corresponding to Lys142 in FAAH-1)7 (Supplementary Fig. 4). Both factors are expected to impair amidase activity by interrupting the proton transfer from Ser173 (corresponding to catalytic Ser241 in FAAH-1) to neutral Lys74. Though critical for the amidase activity of FAAH-1, Lys142 does not influence the ability of this enzyme to cleave ester substrates16. Consistent with those data and the model proposed here, we found that recombinant FLAT effectively hydrolyzes the fatty acyl ester, 2-oleoyl-sn-glycerol (Supplementary Fig. 2c).
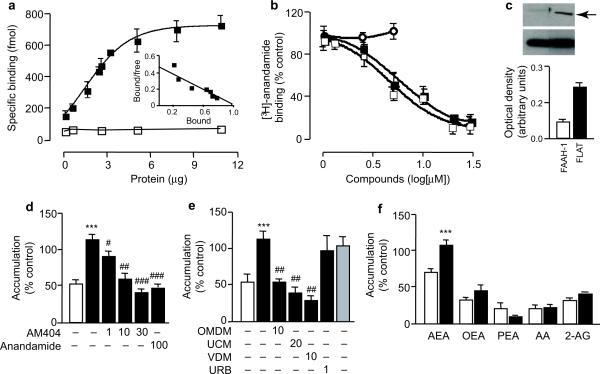
Like FAAH-1, recombinant purified FLAT forms homodimers in aqueous solution (Supplementary Fig. 5). To test whether FLAT also retains the ability to ligate anandamide, we expressed the protein fused with glutathione-S-transferase (GST) in E. coli and purified it by affinity chromatography. Saturation binding studies showed that [3H]-anandamide associates with FLAT-GST (dissociation constant, Kd=2 μM), but not with GST alone (Fig. 2a). The binding of [3H]-anandamide to FLAT is displaced by the anandamide transport inhibitors AM404 and OMDM-1 (Fig. 2b), with median inhibitory concentrations similar to those required for the inhibition of neuronal [3H]-anandamide internalization (IC50: AM404, 5.3 μM; OMDM-1, 4.8 μM)10,17. By contrast, the covalent FAAH inhibitor URB597 had no such effect (Fig. 2b), likely because the productive interaction of this compound with the Ser241 nucleophile of FAAH-1 requires a fully functional catalytic triad18. Collectively, the experiments described above indicate that FLAT lacks amidase activity, but binds anandamide with low micromolar affinity.
Fig. 2.
FLAT binds to anandamide and facilitates its transport into cells. (a) Specific binding of [3H]-anandamide to rat FLAT-glutathione-S-transferase (GST) (closed squares) or GST alone (open squares). The inset shows a Scatchard transformation (bound - [bound/free], in nmol) of binding data. (b) AM404 (closed squares) and OMDM-1 (closed squares) antagonize [3H]-anandamide binding to FLAT-GST, whereas URB597 (open circles) has no effect. (c) Cytosolic fractions of FLAT-expressing Hek293 cells contain detectable amounts of FLAT (arrow); a corresponding fraction from FAAH-1-expressing cells is shown for comparison. The lower band is β-actin. (d) [3H]- Anandamide accumulation in control cells (vector-transfected, open bar) or FLAT-expressing Hek293 cells (closed bars) incubated with vehicle (0.01%dimethylsulfoxide), AM404 or non-radioactive anandamide (concentrations in μM). (e) The anandamide transport inhibitors OMDM-1, UCM-707 and VDM-11 suppress [3H]-anandamide accumulation in FLAT-expressing Hek293 cells. This process is not affected by URB597 or mutation of catalytic Ser242 (shaded bar). (f) Accumulation of labeled lipids in control (open bars) or FLAT-expressing Hek293 cells (closed bars). Abbreviations: [3H]-anandamide, AEA; [3H]-oleoylethanolamide, OEA; [3H]-palmitoylethanolamide, PEA; [3H]-arachidonic acid, AA; [3H]-2-arachidonoyl-sn-glycerol, 2-AG. Results are expressed as mean±SEM of 3-7 experiments. ***P<0.01, versus vector-transfected cells, Student's t test; #P<0.05; ##, P<0.01; ###, P<0.001 versus vehicle, one-way ANOVA followed by Dunnett's test.
A role for FLAT in anandamide transport
We detected significant amounts of FLAT in cytosol fractions prepared from mouse brain (Supplementary Fig. 1g) or transfected Hek293 cells (Fig. 2c), and found that the protein can be readily detached from Hek293 cell membranes by incubation with sodium carbonate (0.1 M) (Supplementary Fig. 6)19. These properties, along with the observation that AM404 and OMDM-1 antagonize the binding of [3H]-anandamide to FLAT (Fig. 2b), are suggestive of a role in anandamide translocation. Such a role was further implied by the reduced [3H]-anandamide accumulation observed in cultures of brain neurons obtained from faah-1−/− mice (Supplementary Fig. 9)20,21, which lack both FAAH-1 and FLAT (Supplementary Fig. 1g). We examined therefore whether heterologous expression of FLAT might increase anandamide transport in Hek293 cells. We incubated control and FLAT-expressing cells for 5 min at 37°C in a buffer containing [3H]-anandamide, and measured cell-associated radioactivity after removal of excess tracer3,11. Compared to controls, FLAT-expressing cells displayed a significantly higher level of [H3]-anandamide accumulation, which (i) was prevented by AM404 (IC50 = 4 μM) and other transport inhibitors (OMDM-1, VDM11, UCM707), as well as by non-radioactive anandamide (100 μM) (Fig. 2D,E); and (ii) was selective for anandamide compared to four structurally related lipids: the FAAH substrates [3H]-OEA and [3H]-PEA, the eicosanoid precursor [3H]-arachidonic acid, and the endocannabinoid fatty acyl ester [3H]-2-arachidonoyl-sn-glycerol (2-AG) (Fig. 2f). [3H]-Anandamide accumulation in FLAT-expressing Hek293 cells may not be attributed to passive diffusion driven by FAAH-mediated hydrolysis22, because the very low amidase activity present in native Hek293 cells was not increased by FLAT expression (Supplementary Fig. 2a,b). Moreover, treatment with a maximally active concentration of the FAAH inhibitor URB597 (1 μM) or mutation of Ser173 (corresponding to the nucleophile Ser241 in FAAH-1) did not affect [3H]-anandamide uptake by FLAT-expressing Hek293 cells (Fig. 2e). URB597 did not affect [3H]-anandamide uptake at any of the concentration tested (up to 10 μM: 103±5% of vehicle control).
In addition to internalizing anandamide, the anandamide transport system may also facilitate the release of this lipid mediator from cells4,23. To test whether FLAT contributes to this process, we over-expressed the protein in mouse Neuro-2a cells and measured, by liquid chromatography/mass spectrometry, the levels of endogenously produced anandamide in the incubation medium. Consistent with a role in anandamide release, FLAT over-expression was accompanied by a significant elevation in the extracellular levels of anandamide, but not 2-AG or OEA (Supplementary Fig. 7). The results suggest that FLAT facilitates anandamide translocation through a mechanism that is selective for anandamide, independent of amidase activity, and prevented by known inhibitors of anandamide transport.
Discovery of a competitive FLAT inhibitor
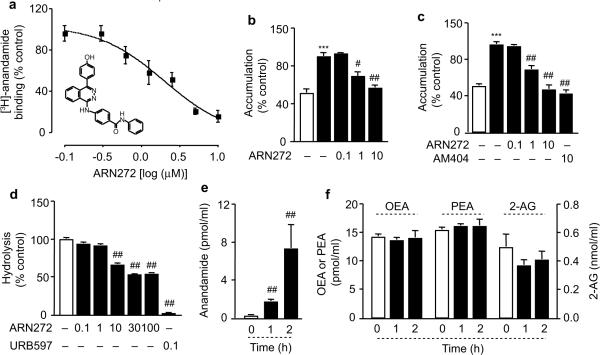
To further investigate the functions of FLAT, and differentiate them from those of FAAH-1, we searched for small drug-like molecules that might selectively interfere with FLAT's ability to sequester anandamide. We subjected a virtual 4.3 million compound library to a ligand-screening campaign structured in multiple steps of progressively increasing stringency (Supplementary Fig. 8). The campaign returned a set of 46 structurally diverse compounds, which were tested for their interaction with FLAT in vitro. One of them, the substituted phthalazine ARN272 (Fig. 3a), competitively antagonized [3H]-anandamide binding to purified FLAT (IC50=1.8 μM; Fig. 3a) and inhibited [3H]-anandamide accumulation in both FLAT-expressing Hek293 cells (IC50≈3 μM; Fig. 3b) and primary cultures of cortical neurons prepared from rats (Fig. 3c) or wild-type mice (Supplementary Fig. 9). By contrast, ARN272 exerted no significant effect on the residual [3H]-anandamide accumulation observed in cortical neurons of faah-1−/− mice (Supplementary Fig. 9).
Fig. 3.
ARN272 is a competitive FLAT inhibitor. (a) Effects of ARN272 on [3H]-anandamide binding to FLAT-GST. The inset shows the chemical structure of ARN272. (b) Effects of ARN272 (concentrations in μM) on [3H]-anandamide accumulation in FLAT-expressing Hek-293 cells (closed bars); the open bar represents vector-transfected cells. (c) Effects of vehicle (open bar), ARN272 and AM404 (closed bars) on [3H]-anandamide accumulation in rat cortical neurons in cultures. (d) Effects of ARN272 and URB597 on FAAH activity in rat brain membranes. (e, f) Effects of ARN272 (1 mg-kg−1, i.p.) on plasma levels of (e) anandamide or (f) OEA, PEA and 2-AG in mice. Results are expressed as mean±SEM of 3–7 experiments. ***, P<0.001 versus vector-transfected cells, Student's t test; #, P<0.05 and ##, P<0.01 versus vehicle; one-way ANOVA followed by Dunnett's test.
FLAT inhibition by ARN272 appeared to be selective, because the compound had little or no inhibitory activity on several endocannabinoid-metabolizing enzymes, including N-acylphosphatidyl-ethanolamine-selective phospholipase D, diacylglycerol lipase-α, monoacylglycerol lipase, and N-acylethanolamide-hydrolyzing acid amidase (Supplementary Fig. 10a–d). Moreover, ARN272 produced only a weak and incomplete inhibition of rat brain FAAH activity (Fig. 3d), and was not significantly hydrolyzed after incubation with recombinant human FAAH-1 (≈5% hydrolysis after 24h at 37°C). Consistent with these observations, administration of ARN272 (1 mg-kg−1, intraperitoneal, i.p.) in mice increased plasma levels of anandamide (Fig. 3e) without changing the levels of 2-AG, OEA or PEA (Fig. 3f). The inhibitory effects of ARN272 on anandamide internalization in vitro and anandamide deactivation in vivo, along with the diminished anandamide accumulation observed in faah-1−/− mice, suggest that FLAT plays an important role in the membrane translocation of this endocannabinoid transmitter.
ARN272 attenuates nociceptive and inflammatory pain
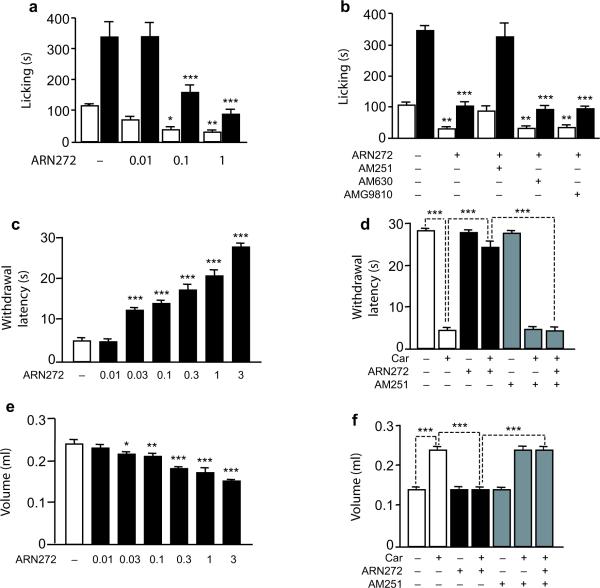
Anandamide transport inhibitors produce a variety of CB1-mediated responses, which include analgesia in animal models of nociceptive and inflammatory pain24,25. We tested therefore whether ARN272 might alleviate pain-related behaviors elicited in mice by intraplantar injection of the chemical irritant, formalin. Systemic administration of ARN272 (0.01–1 mg-kg−1, i.p.) caused a dose-dependent reduction of formalin-induced pain behavior (Fig. 4a). Substantial antinociceptive activity was observed on both the first phase of formalin pain, which involves acute activation of sensory C fibers, and the second phase of formalin pain, in which sensory fiber activity is accompanied by inflammation and central sensitization (Fig. 4a). Similar effects were observed when ARN272 (0.01−3 μg per animal) was injected into the cerebral ventricles (Supplementary Fig. 11). The CB1 antagonist AM251 suppressed the antinociceptive actions of systemic ARN272, whereas the CB2 antagonist AM630 and the transient receptor potential vanilloid-1 (TRPV-1) antagonist AMG9810 were ineffective (all drugs administered at 1 mg-kg−1, i.p) (Fig. 4b). The CB1-mediated antinociceptive effects demonstrated by ARN272 in the formalin test are similar to those previously reported for anandamide transport inhibitors such as AM404, OMDM-1 and UCM70724.
Fig. 4.
ARN272 produces CB1-dependent antinociception in mice. Intraplantar injection of formalin (5%, 20 μL) elicited two temporally distinct phases of nocifensive behavior in mice: phase I (0–5 min, open bars) and phase II (5–45 min, closed bars). (a) ARN272 (doses in mg-kg−1, i.p.) decreased nocifensive behavior in both phases. (b) The CB1 antagonist AM251 (1 mg-kg−1, i.p.) abolished the antinociceptive effects of ARN272, whereas the CB2 antagonist AM630 and the TRPV1 antagonist AMG9810 did not. (c–f) Intraplantar injection of carrageenan (car) elicited a local inflammatory response in mice. ARN272 (mg-kg−1, i.p.) decreased (c) thermal hyperalgesia (withdrawal latency, in seconds), and (e) edema (volume, in ml). The CB1 antagonist AM251 (1 mg-kg−1, i.p.) suppressed the effects of ARN272 on (d) thermal hyperalgesia and (f) edema. Results are expressed as the mean±SEM of 6 mice per group. *, P<0.05; **, P<0.01; ***, P<0.001 versus vehicle-injected controls; two-way ANOVA followed by Bonferroni's test.
In another set of experiments, we evaluated the ability of ARN272 to alleviate thermal hyperalgesia and paw edema elicited in mice by the pro-inflammatory polysaccharide, carrageenan. Similarly to FAAH inhibitors26,27, ARN272 (0.01–3 mg-kg−1, i.p.) exerted anti-hyperalgesic (Fig. 4c and Supplementary Fig. 12) and anti-inflammatory effects (Fig. 4e, Supplementary Fig. 13). These were suppressed by blockade of CB1, but not CB2 or TRPV-1 receptors (Fig. 4d,f and Supplementary Fig. 14). ARN272 did not evoke any detectable nocifensive response when administered alone into the mouse paw (Supplementary Table 1), confirming its inability to interact productively with TRPV-1. Finally, ARN272 did not significantly alter the binding of the cannabinoid ligands [3H]-CP55940 or [3H]-anandamide to rat brain membranes (Supplementary Fig. 10e). Together, the findings indicate that ARN272 elicits profound analgesic effects in mice, which result from inhibition of anandamide transport.
DISCUSSION
The functional properties of FLAT suggest that this protein is a key molecular component of the anandamide transport system in neural cells, and a potential target for therapeutic drugs. The findings presented here indicate that FLAT selectively binds to and internalizes anandamide, and that several known inhibitors of anandamide translocation - AM404, OMDM-1, U C M-707 and VDM-1110 - interfere with these properties. Moreover, our results show that ARN272, a small-molecule inhibitor of the interaction of anandamide with FLAT, suppresses anandamide accumulation by rat brain neurons in vitro and reproduces two key effects of transport blockade in vivo: elevation of plasma anandamide levels, and analgesia in models of nociceptive and inflammatory pain3. Consistent with these data and previous reports3,20,21,23, deletion of the faah-1 gene substantially reduced anandamide transport in mouse cortical neurons, whereas acute pharmacological blockade of FAAH activity failed to do so. While implying an important role for FLAT in anandamide transport, our findings do not rule out the possibility that additional components of the endocannabinoid transport system remain to be discovered. In this context, it is important to point out that FLAT expression did not confer [3H]-2-AG or [3H]-OEA transport to Hek293 cells, and administration of the FLAT inhibitor ARN272 did not increase plasma levels of 2-AG or OEA in mice, which indicates that the translocation of these lipid mediators4,28,29 may be independent of FLAT. Because of its ability to inhibit anandamide deactivation selectively, ARN272 may be useful to differentiate the functional roles of anandamide from those of other lipid amides that are substrates for FAAH (e.g., OEA and PEA).
Multicellular organisms utilize protein carriers to coordinate the traffic of functionally important lipids, and target these biomolecules toward specific cells and subcellular compartments. Two main types of lipid-carrier proteins are employed for this task: integral membrane transporters, such as CD3630 and PGT31, and lipid chaperones, such as aP2 and mal1 (fatty acid-binding protein-4 and 5, respectively)32. For example, membrane-bound CD36 in small-intestinal enterocytes captures dietary oleic acid and directs it toward the intracellular biosynthesis of OEA, an important gut hormone33. On the other hand, cytosolic aP2 in adipocytes encapsulates fatty acids derived from the circulation and partitions them toward appropriate cellular sites for storage or oxidative metabolism32. Our experiments suggest that, similarly to a lipid chaperone, FLAT might function by desorbing anandamide from the cell membrane and delivering it to intracellular organelles where FAAH-1 is located34 (Supplementary Fig. 15). It is also possible, though remains to be fully tested, that FLAT might contribute to anandamide release by facilitating the intracellular transfer of this lipophilic molecule from its as-yet-unknown site of biosynthesis to the cell membrane.
Despite its similarities with other lipid chaperones, FLAT appears to be functionally unique in at least two ways. First, its substrate preference and sensitivity to pharmacological agents distinguish it from other carriers for lipophilic ligands, such as serum albumin and fatty acid-binding proteins, which are known to sequester anandamide35–38. Second, our studies suggest that the capacity of FLAT to ligate anandamide may be based on structural modifications that silence the amidase activity of FAAH-1 without compromising its anandamide-binding function. This mechanism provides an elegant example of phylogenetic parsimony and raises the possibility that other lipid transporters might have evolved following similar principles.
Supplementary Material
Acknowledgements
We thank Giampiero Colombano, Janet Kim, Sine Mandrup-Bertozzi, Dean Thongkham and Drs. Guillermo Moreno-Sanz and Rita Scarpelli for help with experiments and discussions about the project. This research was supported by grants from the National Institute on Drug Abuse and the National Institute on Alcohol Abuse and Alcoholism (DA-012413 to D.P.) and the National Institute on General Medicine (to W.R.). The support provided by the Agilent/UCI Analytical Discovery Facility is gratefully acknowledged.
Footnotes
Methods and associated references are available in the online version of the paper at http://www.nature.com/natureneuroscience.
Supplementary information is linked to the online version of the paper at http://www.nature.com/neuro/index.html
Author contributions J.F. conducted the molecular biological, biochemical and pharmacological experiments in vitro and analyzed data. G.B. conducted the virtual ligand screening campaign. G.B., W.R., M.M., A.L. and A.C. performed the computational studies. T.B. provided chemical expertise. O.S., R.B. and A.R. conducted the pharmacological experiments in vivo and analyzed data. A.G. measured lipid levels in vivo and analyzed data. A.A. and G.G. conducted analytical studies on purified recombinant FLAT. D.P. conceived and designed the experiments, oversaw the project and wrote the manuscript with assistance from J.F., G.B., W.R., M.M., A.L., and A.C.
Author information The authors declare the following competing financial interest: a patent was filed by the University of California, Irvine, and the Italian Institute of Technology on behalf of D.P., J.F., G.B., and A.C.
References
- 1.Piomelli D, Astarita G, Rapaka R. A neuroscientist's guide to lipidomics. Nat Rev Neurosci. 2007;8:743–54. doi: 10.1038/nrn2233. [DOI] [PubMed] [Google Scholar]
- 2.Devane WA, et al. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science. 1992;258:1946–9. doi: 10.1126/science.1470919. [DOI] [PubMed] [Google Scholar]
- 3.Beltramo M, et al. Functional role of high-affinity anandamide transport, as revealed by selective inhibition. Science. 1997;277:1094–7. doi: 10.1126/science.277.5329.1094. [DOI] [PubMed] [Google Scholar]
- 4.Hillard CJ, Edgemond WS, Jarrahian A, Campbell WB. Accumulation of N-arachidonoylethanolamine (anandamide) into cerebellar granule cells occurs via facilitated diffusion. J Neurochem. 1997;69:631–8. doi: 10.1046/j.1471-4159.1997.69020631.x. [DOI] [PubMed] [Google Scholar]
- 5.Hillard CJ, Shi L, Tuniki VR, Falck JR, Campbell WB. Studies of anandamide accumulation inhibitors in cerebellar granule neurons: comparison to inhibition of fatty acid amide hydrolase. J Mol Neurosci. 2007;33:18–24. doi: 10.1007/s12031-007-0045-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cravatt BF, et al. Supersensitivity to anandamide and enhanced endogenous cannabinoid signaling in mice lacking fatty acid amide hydrolase. Proc Natl Acad Sci USA. 2001;98:9371–6. doi: 10.1073/pnas.161191698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McKinney MK, Cravatt BF. Structure and function of fatty acid amide hydrolase. Annu Rev Biochem. 2005;74:411–32. doi: 10.1146/annurev.biochem.74.082803.133450. [DOI] [PubMed] [Google Scholar]
- 8.Wei BQ, Mikkelsen TS, McKinney MK, Lander ES, Cravatt BF. A second fatty acid amide hydrolase with variable distribution among placental mammals. J Biol Chem. 2006;281:36569–78. doi: 10.1074/jbc.M606646200. [DOI] [PubMed] [Google Scholar]
- 9.Hillard CJ, Jarrahian A. Cellular accumulation of anandamide: consensus and controversy. Br J Pharmacol. 2003;140:802–808. doi: 10.1038/sj.bjp.0705468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Marzo V. Targeting the endocannabinoid system: to enhance or reduce? Nat Rev Drug Discov. 2008;7:438–55. doi: 10.1038/nrd2553. [DOI] [PubMed] [Google Scholar]
- 11.Piomelli D, et al. Structural determinants for recognition and translocation by the anandamide transporter. Proc Natl Acad Sci U S A. 1999;96:5802–7. doi: 10.1073/pnas.96.10.5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glaser ST, et al. Evidence against the presence of an anandamide transporter. Proc Natl Acad Sci U S A. 2003;100:4269–74. doi: 10.1073/pnas.0730816100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobsson SO, Fowler CJ. Characterization of palmitoylethanolamide transport in mouse Neuro-2a neuroblastoma and rat RBL-2H3 basophilic leukaemia cells: comparison with anandamide. Br J Pharmacol. 2001;132:1743–54. doi: 10.1038/sj.bjp.0704029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–98. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 15.Bracey MH, Hanson MA, Masuda KR, Stevens RC, Cravatt BF. Structural Adaptations in a Membrane Enzyme That Terminates Endocannabinoid Signaling. Science. 2002;298:1793–1796. doi: 10.1126/science.1076535. [DOI] [PubMed] [Google Scholar]
- 16.Patricelli MP, Cravatt BF. Fatty acid amide hydrolase competitively degrades bioactive amides and esters through a nonconventional catalytic mechanism. Biochemistry. 1999;38:14125–30. doi: 10.1021/bi991876p. [DOI] [PubMed] [Google Scholar]
- 17.Ortar G, Ligresti A, De Petrocellis L, Morera E, Di Marzo V. Novel selective and metabolically stable inhibitors of anandamide cellular uptake. Biochem Pharmacol. 2003;65:1473–81. doi: 10.1016/s0006-2952(03)00109-6. [DOI] [PubMed] [Google Scholar]
- 18.Alexander JP, Cravatt BF. Mechanism of carbamate inactivation of FAAH: implications for the design of covalent inhibitors and in vivo functional probes for enzymes. Chem Biol. 2005;12:1179–87. doi: 10.1016/j.chembiol.2005.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Speers AE, Wu CC. Proteomics of integral membrane proteins--theory and application. Chem Rev. 2007;107:3687–714. doi: 10.1021/cr068286z. [DOI] [PubMed] [Google Scholar]
- 20.Ortega-Gutierrez S, Hawkins EG, Viso A, Lopez-Rodriguez ML, Cravatt BF. Comparison of anandamide transport in FAAH wild-type and knockout neurons: evidence for contributions by both FAAH and the CB1 receptor to anandamide uptake. Biochemistry. 2004;43:8184–90. doi: 10.1021/bi049395f. [DOI] [PubMed] [Google Scholar]
- 21.Kathuria S, et al. Modulation of anxiety through blockade of anandamide hydrolysis. Nat Med. 2003;9:76–81. doi: 10.1038/nm803. [DOI] [PubMed] [Google Scholar]
- 22.Yates ML, Barker EL. Inactivation and biotransformation of the endogenous cannabinoids anandamide and 2-arachidonoylglycerol. Mol Pharmacol. 2009;76:11–7. doi: 10.1124/mol.109.055251. [DOI] [PubMed] [Google Scholar]
- 23.Ligresti A, et al. Further evidence for the existence of a specific process for the membrane transport of anandamide. Biochem J. 2004;380:265–72. doi: 10.1042/BJ20031812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.La Rana G, et al. Modulation of neuropathic and inflammatory pain by the endocannabinoid transport inhibitor AM404 [N-(4-hydroxyphenyl)-eicosa-5,8,11,14-tetraenamide] J Pharmacol Exp Ther. 2006;317:1365–71. doi: 10.1124/jpet.105.100792. [DOI] [PubMed] [Google Scholar]
- 25.Maione S, et al. Antinociceptive effects of tetrazole inhibitors of endocannabinoid inactivation: cannabinoid and non-cannabinoid receptor-mediated mechanisms. Br J Pharmacol. 2008;155:775–82. doi: 10.1038/bjp.2008.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lichtman AH, et al. Reversible inhibitors of fatty acid amide hydrolase that promote analgesia: evidence for an unprecedented combination of potency and selectivity. J Pharmacol Exp Ther. 2004 doi: 10.1124/jpet.104.069401. [DOI] [PubMed] [Google Scholar]
- 27.Russo R, et al. The fatty acid amide hydrolase inhibitor URB597 (cyclohexylcarbamic acid 3'-carbamoylbiphenyl-3-yl ester) reduces neuropathic pain after oral administration in mice. J Pharmacol Exp Ther. 2007;322:236–42. doi: 10.1124/jpet.107.119941. [DOI] [PubMed] [Google Scholar]
- 28.Beltramo M, Piomelli D. Carrier-mediated transport and enzymatic hydrolysis of the endogenous cannabinoid 2-arachidonylglycerol. Neuroreport. 2000;11:1231–5. doi: 10.1097/00001756-200004270-00018. [DOI] [PubMed] [Google Scholar]
- 29.Bisogno T, et al. The uptake by cells of 2-arachidonoylglycerol, an endogenous agonist of cannabinoid receptors. Eur J Biochem. 2001;268:1982–1989. doi: 10.1046/j.1432-1327.2001.02072.x. [DOI] [PubMed] [Google Scholar]
- 30.Niot I, Poirier H, Tran TT, Besnard P. Intestinal absorption of long-chain fatty acids: evidence and uncertainties. Prog Lipid Res. 2009;48:101–15. doi: 10.1016/j.plipres.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 31.Kanai N, et al. Identification and characterization of a prostaglandin transporter. Science. 1995;268:866–9. doi: 10.1126/science.7754369. [DOI] [PubMed] [Google Scholar]
- 32.Reese-Wagoner A, Thompson J, Banaszak L. Structural properties of the adipocyte lipid binding protein. Biochim Biophys Acta. 1999;1441:106–16. doi: 10.1016/s1388-1981(99)00154-7. [DOI] [PubMed] [Google Scholar]
- 33.Schwartz GJ, et al. The lipid messenger OEA links dietary fat intake to satiety. Cell Metab. 2008;8:281–8. doi: 10.1016/j.cmet.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gulyas AI, et al. Segregation of two endocannabinoid-hydrolyzing enzymes into pre- and postsynaptic compartments in the rat hippocampus, cerebellum and amygdala. Eur J Neurosci. 2004;20:441–58. doi: 10.1111/j.1460-9568.2004.03428.x. [DOI] [PubMed] [Google Scholar]
- 35.Bojesen IN, Hansen HS. Binding of anandamide to bovine serum albumin. J Lipid Res. 2003;44:1790–4. doi: 10.1194/jlr.M300170-JLR200. [DOI] [PubMed] [Google Scholar]
- 36.Kaczowka SJ, et al. Probing the stability of native and activated forms of alpha2-macroglobulin. Int J Biol Macromol. 2008;42:62–7. doi: 10.1016/j.ijbiomac.2007.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maccarrone M, Dainese E, Oddi S. Intracellular trafficking of anandamide: new concepts for signaling. Trends Biochem Sci. 2010;35:601–8. doi: 10.1016/j.tibs.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 38.Oddi S, et al. Molecular identification of albumin and Hsp70 as cytosolic anandamide-binding proteins. Chem Biol. 2009;16:624–32. doi: 10.1016/j.chembiol.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 39.Rosso L, Gould IR. Structure and dynamics of phospholipid bilayers using recently developed general all-atom force fields. J Comput Chem. 2008;29:24–37. doi: 10.1002/jcc.20675. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.