Abstract
Crystallographic, computational and functional analyses of LeuT have revealed details of the molecular architecture of Na+-coupled transporters and the mechanistic nature of ion/substrate coupling, but the conformational changes that support a functional transport cycle have yet to be described fully. We have used site-directed spin labeling and EPR analysis to capture the dynamics of LeuT in the region of the extracellular vestibule associated with the binding of Na+ and Leu. The results outline the Na+-dependent formation of a dynamic outward-facing intermediate that exposes the primary substrate binding site, and the conformational changes that occlude this binding site upon subsequent binding of the Leu substrate. Furthermore, the binding of the transport inhibitors Trp, clomipramine and octyl-glucoside is shown to induce structural changes that distinguish the resulting inhibited conformation from the Na+/Leu bound state.
In the central nervous system, chemical neurotransmission is dependent on the termination of signaling mediated by the clearance of neurotransmitters from the synaptic cleft by plasma membrane transport proteins1,2. The neurotransmitter:sodium symporter (NSS) family (SLC6A) includes biogenic amine transporters for serotonin, norepinephrine, and dopamine, the targets of widely prescribed therapeutic drugs (e.g., selective serotonin re-uptake inhibitors (SSRIs) and tricyclic antidepressants (TCAs)) and drugs of abuse (e.g., cocaine, amphetamine)3. The NSS proteins catalyze the thermodynamically uphill transport of their respective substrates powered by the electrochemical Na+ gradient across the plasma membrane4,5. Despite the elucidation of high-resolution crystal structures of LeuT6,7, a bacterial homolog of mammalian NSS, and intensive efforts to understand the dynamics of ion-coupled substrate translocation8–16, many important aspects of the conformational changes related to individual steps in the transport cycle are still lacking.
The structure of LeuT revealed a novel fold6, which is now known to be shared by several Na+-dependent and Na+-independent transport proteins without obvious sequence similarity to NSS17–22. In this structure (PDB 2A65)6, Leu and two Na+ (Na1, Na2) are positioned in close proximity within a dehydrated protein core occluded from both the extracellular and intracellular sides. Partially unwound regions of transmembrane helix (TM) 1 and TM6 contribute main-chain atoms and helix dipoles to the formation of the Leu and Na+ binding sites, and the carboxylate of Leu participates in the direct coordination of Na1. Highly conserved charged and aromatic residues in these two TMs on both the extracellular and intracellular sides likely play key roles in forming the “gates” that allow access to this substrate binding site6,23,24.
In the context of an alternating access mechanism25, this static snapshot of LeuT was interpreted as a conformational intermediate in which both the extracellular and intracellular gates are closed, i.e. in an occluded conformation, which prevents access to the bound substrates6. Subsequent crystal structures of LeuT revealed that several other amino acids can also occupy the Leu binding site7 and produce an essentially identical occluded conformation when compared to the Leu-bound LeuT crystal structure6. In contrast, Trp acts as a competitive inhibitor and, at high concentrations, wedges the transporter into a putative “open-to-out” conformation7.
Additional structures were solved with the TCA inhibitors, clomipramine, imipramine, and desipramine26,27, or the SSRIs, sertraline and fluoxetine28, bound in an extracellular vestibule about 11 Å above the Leu-occupied binding site (subsequently referred to as the primary (S1) binding site). These inhibitor-bound structures were nearly identical to that of the Na+/Leu-bound structure6. Subsequent crystallographic evidence showed that a molecule of n-octyl-β-D-glucopyranoside (OG), the detergent used for all LeuT crystallization procedures, occupies this extracellular vestibule in the absence of TCAs or SSRIs29.
Based on this structural information, a number of models have been proposed for the mode of substrate transport. One model posits formation of outward- and inward-facing states facilitated by a “rocking bundle” of transmembrane helices from the topologically inverted repeats13,30. Another model envisions two gates controlling access to the central binding site8,31, implying that the doubly occluded crystal structure is an obligatory intermediate between outward- and inward facing conformations. A more detailed functional model emerged from the application of steered molecular dynamics (SMD) simulations in conjunction with radiotracer binding. These experiments identified a second high affinity substrate binding site (S2) in the extracellular vestibule of LeuT and established that Leu can bind simultaneously to the S1 and S2 sites. Additionally, these studies showed that binding of Leu to the S2 site allosterically triggers release of Na+ and Leu from the S1 site, enabling Na+-coupled substrate symport. Disturbing the integrity of the S2 site by mutations or occupation of the S2 site by a TCA, SSRI or OG impairs this transport mechanism23,26–29. These findings rationalize the lack of significant differences between the OG (not identified in the structure with PDB 2A65)-bound, TCA-bound and SSRI-bound structures, as they all are likely to represent inhibited conformational intermediates despite having Na+/Leu (or other amino acid substrate) bound in the S1 site.
To provide the first direct measurements of functional dynamics for a NSS family member that complement the static crystallographic snapshots of LeuT, here we used systematic spin labeling and electron paramagnetic resonance (EPR) analysis which has been shown to be sensitive to conformational changes in membrane transport proteins32–37. The focus of these studies was on the structural dynamics enabling ligand permeation from the extracellular milieu to the binding sites within the protein core during distinct conformational states in the transport cycle. Spin label mobility, accessibility to paramagnetic reagents (NiEDDA and O2) and inter-spin label distances were interpreted as constraints on local helix packing and global structural organization. Specifically, we have identified novel Na+-dependent conformational changes in and near the extracellular vestibule and have contrasted the effects of subsequent substrate (Leu) binding to those induced by inhibitors.
Results
In order to introduce nitroxide spin labels for the EPR studies, residues in LeuT wild-type (WT) were substituted for Cys at selected positions to generate 41 single and 6 double Cys mutants; it should be noted that WT LeuT has no endogenous Cys residues. The selection of single Cys positions was guided, in part, by predictions from previous SMD simulations of LeuT in which the substrate was pulled to the extracellular side to identify the extracellular transport pathway23. The Cys residues were subsequently reacted with the thiol-specific nitroxide spin label MTSSL. Analysis of these mutants by scintillation proximity assay (SPA) confirmed that each of the LeuT variants exhibited Na+-dependent Leu binding activity38. Spin labeling had no substantial effect on binding in nearly all of the Cys mutants (Supplementary Table 1, Supplementary Figure 1).
Na+ increases accessibility of the extracellular vestibule
In the absence of both Na+ and Leu (the Apo form), spin labels attached to specific Cys residues introduced in or near the extracellular permeation pathway of LeuT exhibited low accessibility to the water-soluble reagent NiEDDA (Table 1) and to membrane soluble molecular oxygen (O2, data not shown). For example, a spin label attached to the S2 site mutant I111C had less than 2% of the accessibility of a spin label attached to the surface-exposed A309C in extracellular loop (EL) 4a. The differences in the absolute accessibilities reflect the larger local concentrations and diffusion rates of NiEDDA at surface-exposed residues39. On the other hand, the spin label at the S2 site mutant L400C23 exhibited higher absolute accessibility to NiEDDA, relative to I111C, in the absence of Na+. These results indicate that in the Apo form of LeuT reconstituted into proteoliposomes a tightly packed structural organization prevents solvent permeation into some portions of the extracellular vestibule, but not others.
Table 1.
NiEDDA accessibility of MTSSL-labeled LeuT single Cys mutants
| Position | Π (Apo) | Π (NaCl) | Π (Na/Leu) | |
|---|---|---|---|---|
| TM1 | 25 | 0.11 ± 0.01 | 0.14 ± 0.02 | 0.14 ± 0.01 |
| 29 | 0.09 ± 0.02 | 0.09 ± 0.01 | 0.06 ± 0.02 | |
| 33* | 0.13 ± 0.03 | 0.42 ± 0.02 | 0.17 ± 0.01 | |
| 34 | 1.08 ± 0.07 | 1.15 ± 0.06 | 1.13 ± 0.07 | |
|
| ||||
| TM3 | 111* | 0.12 ± 0.01 | 0.37 ± 0.03 | 0.23 ± 0.01 |
| 114 | 0.12 ± 0.02 | 0.16 ± 0.02 | 0.11 ± 0.03 | |
|
| ||||
| EL2 | 136 | 7.39 ± 0.30 | 8.03 ± 0.35 | 8.38 ± 0.38 |
| 137 | 2.80 ± 0.09 | 2.40 ± 0.07 | 2.51 ± 0.11 | |
| 138 | 4.71 ± 0.18 | 3.73 ± 0.26 | 4.85 ± 0.14 | |
| 139 | 6.38 ± 0.33 | 6.06 ± 0.34 | 5.84 ± 0.40 | |
| 140 | 1.05 ± 0.05 | 0.83 ± 0.05 | 0.50 ± 0.03 | |
| 141 | 1.68 ± 0.07 | 1.78 ± 0.05 | 1.66 ± 0.07 | |
| 142 | 7.02 ± 0.26 | 6.77 ± 0.26 | 6.58 ± 0.31 | |
| 143 | 3.00 ± 0.08 | 2.54 ± 0.09 | 2.81 ± 0.13 | |
| 144 | 0.28 ± 0.02 | 0.22 ± 0.01 | 0.31 ± 0.02 | |
| 145 | 1.18 ± 0.06 | 1.09 ± 0.06 | 1.10 ± 0.07 | |
| 146 | 4.15 ± 0.22 | 4.03 ± 0.18 | 3.79 ± 0.18 | |
| 147 | 0.49 ± 0.12 | 0.22 ± 0.03 | 1.73 ± 0.11 | |
| 148 | 0.56 ± 0.07 | 0.35 ± 0.06 | 0.49 ± 0.10 | |
| 149 | 0.86 ± 0.02 | 0.96 ± 0.04 | 0.94 ± 0.04 | |
| 150 | 5.40 ± 0.23 | 5.33 ± 0.22 | 5.24 ± 0.31 | |
|
| ||||
| TM6 | 243 | 0.46 ± 0.05 | 0.44 ± 0.06 | 0.29 ± 0.07 |
| 246 | 0.43 ± 0.02 | 0.38 ± 0.02 | 0.63 ± 0.03 | |
| 249 | 0.13 ± 0.01 | 0.18 ± 0.01 | 0.23 ± 0.04 | |
| 253* | 0.14 ± 0.01 | 0.11 ± 0.01 | 0.12 ± 0.02 | |
|
| ||||
| EL4 | 305 | 2.05 ± 0.09 | 1.85 ± 0.06 | 2.12 ± 0.09 |
| 307 | 0.04 ± 0.01 | 0.04 ± 0.01 | 0.06 ± 0.01 | |
| 309 | 7.11 ± 0.34 | 7.27 ± 0.38 | 6.98 ± 0.28 | |
| 311 | 0.21 ± 0.01 | 0.35 ± 0.03 | 0.24 ± 0.02 | |
| 314 | 2.38 ± 0.08 | 2.73 ± 0.11 | 2.55 ± 0.07 | |
| 315* | 0.14 ± 0.02 | 0.36 ± 0.04 | 0.09 ± 0.01 | |
| 317 | 3.15 ± 0.16 | 3.19 ± 0.19 | 3.86 ± 0.31 | |
| 320* | 0.21 ± 0.02 | 0.29 ± 0.01 | 0.21 ± 0.02 | |
| 324 | 0.11 ± 0.02 | 0.10 ± 0.03 | 0.17 ± 0.02 | |
| 325 | 0.61 ± 0.06 | 0.83 ± 0.10 | 0.58 ± 0.08 | |
| 333 | 2.79 ± 0.12 | 3.54 ± 0.16 | 3.57 ± 0.16 | |
|
| ||||
| TM10 | 397 | 1.35 ± 0.15 | 1.57 ± 0.12 | 1.27 ± 0.08 |
| 400* | 0.85 ± 0.09 | 0.74 ± 0.06 | 0.19 ± 0.04 | |
| 404 | 0.20 ± 0.03 | 0.17 ± 0.02 | 0.12 ± 0.02 | |
| 405 | 1.05 ± 0.04 | 1.32 ± 0.05 | 0.62 ± 0.04 | |
|
| ||||
| EL6 | 480 | 4.16 ± 0.15 | 4.16 ± 0.16 | 4.06 ± 0.22 |
Values are reported with confidence intervals for a single experiment.
indicates the mean ± S.D. obtained from three independently determined Π values.
Remarkably, Na+ binding increased accessibility at positions in TM1b, TM3 and EL4 that project into the extracellular vestibule above the S1 site. Spin labels at these positions (highlighted for V33C, I111C, A315C in Figure 1a, b) reported an increase in mobility and steady state collision frequency with NiEDDA in the presence of Na+, indicating looser packing and increased hydration. Mapping the Na+-induced changes in NiEDDA accessibility onto the structure of LeuT illustrates that positions of increased accessibility cluster within the putative permeation pathway in close proximity to EL4 (Figure 1c). V33C in TM1b, which is located directly under EL4, reported more than a factor of three increase in the NiEDDA accessibility parameter Π in the presence of Na+. A similar increase in accessibility was observed at A315C, a site in EL4 that projects into the nearby environment of V33C. Spin labels attached to other buried positions of EL4 (e.g. A311C) also reported significant increases in accessibility relative to the Apo intermediate, whereas at surface residues (e.g. A317C) accessibility changes were typically less than 20%, reflecting insignificant changes in the local NiEDDA concentration. Accessibilities at other positions found in TM10 and TM6a that are in proximity to EL4 remained relatively unchanged upon Na+ binding. This distinct pattern of accessibility changes suggests that the presence of Na+ induces a movement of EL4 that increases NiEDDA penetration into regions that are buried in the Apo state, and thus allows water and substrate to access sites within the protein core. The movement of EL4 also alters the accessibility of residues in EL2 lining the packing interface. Upon Na+ binding, we observed concomitant decreases in the accessibilities at positions F147C and L148C (red side chains in EL2 in Figure 1c) and in the relative proximity between EL2 and EL4 (Supplementary Figure 2).
Figure 1.
Na+ binding establishes an outward-facing conformation and Leu binding closes access to the extracellular vestibule. Representative EPR spectra (a) and the corresponding NiEDDA accessibility profiles (b) in proteoliposomes demonstrate that increased water penetration correlates with increased spin label dynamics (arrows, panel a). Leu binding leads to a decrease in spin label dynamics which corresponds with a decrease in solvent accessibility. The low field changes in EPR lineshape for V33C are highlighted in the boxed inset in a. Data in (b) are shown as mean ± S.D from three independent measurements. (c,d) Residue-specific mapping of the change in NiEDDA accessibility upon Na+ binding (relative to the Apo state) and Leu binding (relative to the Na+ state) onto the LeuT crystal structure (PDB 2A65)6. Na+ ions are depicted as green spheres and Leu is colored yellow in a space-filling model. Positions exhibiting an increase in accessibility are shaded in blue according to the color scale. Structures were generated using UCSF Chimera43.
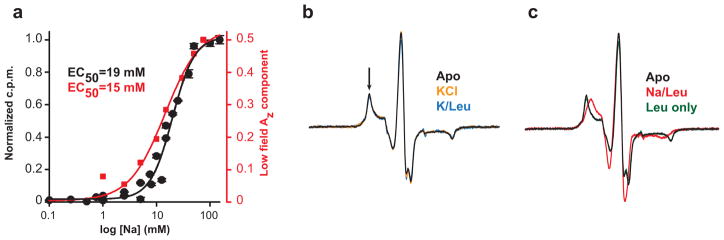
The Na+-induced loosening of packing within the extracellular permeation pathway that leads to water penetration in this region was also evident as an overall increase in spin label mobility, which refers to the rotational isomerization of the nitroxide around the tethering bonds to the protein40 (Figure 1a). Increased mobility at site I111C (Figure 1a) was Na+-concentration dependent with an , similar to that obtained for Na+ binding by SPA (Figure 2a). K+ failed to elicit a change in spin label dynamics consistent with the functional dependence of LeuT on Na+ (Figure 2b). Overall, the increase in mobility and accessibility of multiple positions in the extracellular vestibule supports a model of Na+-dependent enhancement of water entry and the development of an “outward-facing” transporter conformation poised to bind substrate.
Figure 2.
Changes in the EPR spectrum are dependent upon Na+ binding. (a) The [Na+]-dependent increase in probe mobility at 111C (Figure 1a) as monitored by the intensity of the low field Az component (arrow in b) revealed an similar to that obtained by Na+-stimulated binding of 100 nM 3H-Leu23. (b–c) Leu-dependent changes in the EPR spectrum at I111C required the presence of Na+. In the absence of Na+ or the presence of K+, Leu did not induce changes in the EPR spectrum relative to the Apo state.
Leu binding reduces accessibility of the extracellular vestibule
Addition of Leu to the Na+-bound state reduced spin label accessibilities to NiEDDA and dampened the overall dynamics of LeuT - effects opposite to those produced by Na+ binding (Figure 1). Mapping the accessibility changes relative to the Na+-bound state onto the crystal structure (PDB 2A65)6 revealed a steric restriction of water access to the extracellular vestibule as shown in Figure 1d. Concomitant changes in spin label mobility were Na+-dependent, as neither the absence of Na+ nor the presence of K+ supported the Leu-induced EPR lineshape changes (Figure 2b–c).
Spin labels monitoring the entrance pathway to the S1 site from the extracellular milieu, such as L400C in TM10, exhibited the type of restricted motion characteristic of positions in hydrophobic cores of proteins40. A similar pattern of lineshape changes - from mobile to highly restricted - was observed for labels at other positions along TM10 ranging from the extracellular milieu toward the S1 site (Supplementary Figure 3). While these positions did not demonstrate Na+-dependent changes in NiEDDA accessibility, they reported a significant decrease in accessibility upon Leu binding (Figure 1b,d, Table 1). Together, these Leu-dependent site-specific changes suggest a substrate-dependent conformational change of TM10, which may be similar to the substrate-dependent inward movement of TM10 in the NCS1 benzyl-hydantoin transporter, Mhp117. Similar changes in the EPR lineshape were observed at L400C in the presence of other LeuT substrates such as Ala, Val, and Ile7, but not Glu, corresponding with the ability of these amino acids to compete with 3H-Leu binding (Supplementary Figure 4). Overall, the EPR parameters are consistent with reduced access of the extracellular permeation pathway induced by Leu binding.
The addition of Leu affected in a similar way the NiEDDA accessibility of probes attached to target positions in the S2 site (I111C and L400C), and also to other positions that are in close proximity but that do not impair binding of Leu to this site (V33C and A315C). Thus, even in the absence of a directional ion gradient, Na+-dependent substrate binding to the S1 site alone appears to alter the aqueous accessibility of the extracellular permeation pathway. This observation also emphasizes that, unlike uphill transport, Leu binding does not require the presence of a Na+ gradient. Indeed, when LeuT-I111C and -L400C were reconstituted into proteoliposomes in the presence of Na+ and Leu (identical concentrations on the outside and inside of the liposome), the resulting EPR lineshapes and accessibilities were nearly identical to those obtained from external addition of Na+ and Leu to Apo LeuT (data not shown).
Binding of substrate dehydrates the S1 site
The changes in accessibility observed in the permeation pathway with Na+ and Leu binding were attenuated near the S1 site reflecting a steric exclusion of NiEDDA, not necessarily of water, from the vicinity of spin labels. Indeed spin labeled F253C (Figure 3a) reported low accessibility of the relatively bulky NiEDDA (~13 times larger than water) in all states (Figure 3b), precluding a structural interpretation of changes in water activity at the S1 site. The spin label was also highly immobilized, which is not unexpected given the ordered environment of the S1 site in the LeuT crystal structure (PDB 2A65)6 (Figure 3a,c). Although the F253C mutation prevents Na+-coupled substrate transport (Figure 3d), Leu binds with high affinity and a stoichiometry of 1:1 (Supplementary Table 1, Figure 3e) consistent with disruption of the S1 binding site and the proposed symport mechanism23.
Figure 3.
Substrate binding induces dehydration of the S1 site. (a) Representation of the LeuT crystal structure (PDB 2A65)6 highlighting the bound Leu in the S1 site (Leu is shown in a yellow space-filling model), the position of a key residue, F253 (red stick representation), and the distribution of water molecules (shown as blue spheres). Note that there are no water molecules in the S1 site. (b) Na+-dependent Ala uptake (Methods) was greatly impaired in the F253C mutant. (c) Na+-dependent binding of 100 nM 3H-Leu was reduced ~50% for the F253C mutant relative to WT. Data shown are mean ± S.E.M. (d) Accessibility to both 50mM NiEDDA and 20% O2 of F253C in proteoliposomes was low in all states, consistent with a buried location. Equimolar Leu binding decreased the EPR spectral breadth at both room temperature (e) and –53°C (f) indicating that water becomes excluded from the primary binding site (S1). The structure in (a) was generated using PyMOL.
To monitor water activity at the S1 site, we used a sensitive indicator of environmental polarity in the immediate vicinity of the spin label by analyzing the splitting of hyperfine extrema of the EPR spectrum, which decreases in low dielectric conditions41. At room temperature, the EPR spectrum of the spin label at site F253C decreased in breadth by 8 gauss upon the addition of Leu (Figure 3c) or Ala (data not shown). That these spectral changes unequivocally reflect a reduction in water activity was confirmed by a similar Leu-induced change in spectral breadth at low temperature (−53°C) that freezes out the confounding contribution of probe dynamics to the spectral width (Figure 3f). Therefore, the reduction in the local dielectric underlying this observation is consistent with dehydration of the S1 site upon Leu binding.
Na+-binding induces an outward-facing conformation
The accessibility changes described above implicate Na+/substrate-dependent movements of EL4 and possibly EL2, which occlude access to the S1 and S2 sites in the crystal structure. To define specifically the conformational changes that alter accessibility of the permeation pathway, we used Double Electron-Electron Resonance (DEER) spectroscopy to measure distances between pairs of spin labels in the loops surrounding the extracellular vestibule42. Introduced at surface exposed positions, these spin labels did not alter significantly the affinity or the stoichiometry of Leu binding (Supplementary Figure 5). Although LeuT crystallized as a dimer with an interface between TM9 and TM126, neither EPR lineshapes (Supplementary Figure 6) nor DEER analysis (data not shown) of spin labeled TM12 mutants showed evidence of spin-spin dipolar coupling. This suggests that LeuT is predominantly a monomer under our conditions (DDM and proteoliposomes) and that the distances measured between labels arise from intramolecular dipolar interactions.
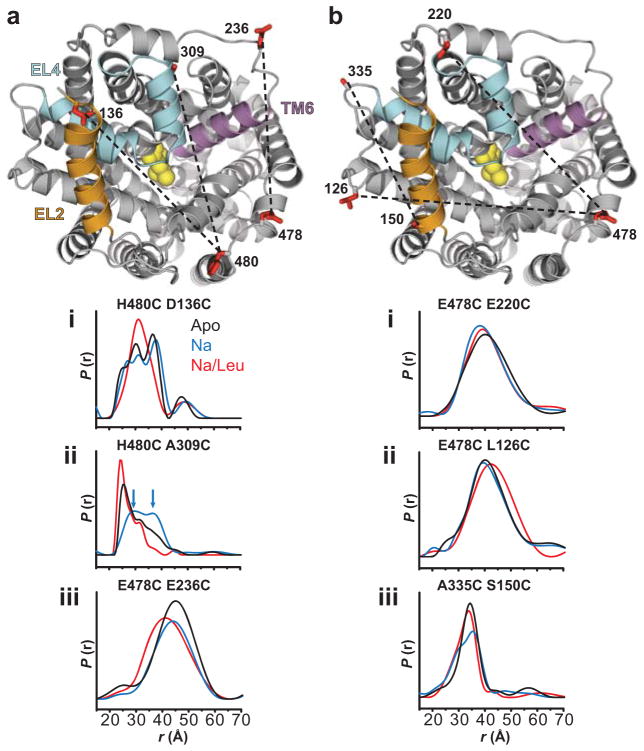
The spin label pairs were positioned to monitor distances from reference positions in EL6 (H480C or E478C) to positions in each of EL4a (A309C), EL2 (D136C and L126C), and EL3 (E220C and E236C, at the two ends of the loop; Glu220 is positioned behind EL4 as shown in Figure 4). Additionally, the distance between the N-terminus of TM8 and the C-terminus of EL2 was probed with the A335C/S150C mutant. In general, the distance distributions (P(r)), which describe the probability of a given distance between the two labels, were either broad or consisted of multiple populations reflecting the intrinsic dynamics of the protein backbone. In these cases, only a minor fraction of spin label pairs are separated by the weighted average distance. Therefore, our analysis focused on changes in the width of the distributions and/or shifts in relative distance components (Figure 4, Supplementary Table 2 and Supplementary Figure 7).
Figure 4.
Distance measurements reveal Na+- and Leu-dependent spatial rearrangements. (a) Na+ binding enhanced an outward-facing conformation characterized by movement of EL4 as suggested by the H480C/A309C pair distance measures. Blue arrows in the distance distribution of H480C/A309C indicate two unresolved distance populations in the Na+ intermediate. Leu binding to the Na+ intermediate decreased the distance between the probes for both H480C/A309C and E478C/E236C pairs. Furthermore, Leu binding increased structural homogeneity as indicated by a narrowing of the distance distribution for both H480C/D136C and H480C/A309C pairs. (b) Limited distance changes were observed for pairs sampling protein structure outside of the vestibule. The LeuT structure was obtained from PDB 2A656. The structures were generated using PyMOL.
We found that Na+ binding led to a right-shift in the distance distribution of H480C/A309C, suggesting an increase in distance between EL6 and EL4a, relative to the Apo and the Na+/Leu-bound states (Figure 4aii). The broadened distribution in the Na+-bound intermediate showed evidence of a new component at longer distances (arrows, Figure 4aii). Decomposition of P(r) into multiple Gaussians reveals two distance populations of near equal intensity centered about 28.5(±3.1)Å and 36.7(±3.8)Å (Supplementary Table 2, Supplementary Figure 7). The lack of significant changes in spin label mobility upon Na+ binding (Supplementary Figure 7) suggests that the origin of multiple distance populations is an equilibrium between distinct LeuT conformers, which are likely the result of dynamic fluctuations of EL4a. Na+ binding also produced an additional component in the distribution of the A335C/S150C mutant, consistent with increased dynamic fluctuations at the C-terminus of EL2 (Figure 4biii, Supplementary Table 2). In contrast, only negligible changes in average distance or relative populations were observed between EL6/EL3 (E478C/E220C, E478C/E236C) and EL6/EL2 (H480C/D136C) in the presence of Na+ (Figure 4a,b). These results support a model wherein an outward Na+-dependent movement of EL4 exposes the extracellular permeation pathway, including the S2 site, thus rationalizing the accessibility profiles discussed above for positions located within, and in close proximity to, EL4 (Figure 1).
The shape of the distance distribution for H480C/A309C in the Apo state also suggests an equilibrium between a prominent population of short distance (~26Å), and other population(s) sampling longer distances (30–40Å). This pattern of distances may reflect closed and open conformers of LeuT, in which the closed conformer (short distance) dominates in the absence of Na+ –consistent with the notion that Na+ binding enhances an outward-facing conformation resulting in part from movement of EL4. The establishment of such an open conformation relative to the Na+/Leu bound state is further supported by results from corresponding MD simulations of LeuT in the presence of both Na1 and Na2, but without Leu, which reveal an outward movement of TM6a. This conformational rearrangement is coordinated by Na+ binding and by changes in the configuration of an aromatic cluster in its vicinity, as described below.
Leu shifts the conformational preference to an occluded form
As described above, Leu binding to Na+-bound LeuT restored the tight packing in the extracellular vestibule. The left-shift in the distance distribution of H480C/A309C relative to the Na+-bound outward-facing conformation, and its narrowing by elimination of the longest distance component (Supplementary Table 2), imply an effective decrease in distance between EL6 and EL4a (Figure 4aii). A similar observation was made for H480C/D136C and A335C/S150C, in which broad, multi-component distributions were reduced to a predominately one component distribution upon Leu binding (Figure 4ai, 4biii, Supplementary Table 2). Indeed, the three modeled Gaussian populations of H480C/D136C in the Apo and Na+ intermediates coalesced into a single dominant (86%) component in the presence of Leu, suggesting a decrease in conformational heterogeneity (Supplementary Table 2). A left-shift in the distance distribution of E478C/E236C also indicated a decrease in distance between EL6 and EL3 upon Leu binding (Figure 4aiii). These distance changes were observed in both DDM micelles and in proteoliposomes (Supplementary Figure 8). In combination with the mobility and accessibility analysis, these results lead us to the conclusion that LeuT in the Na+/Leu-bound state adopts a structurally more homogeneous conformation characterized by a closed extracellular region. This is in contrast to the more open, heterogeneous ensemble of the Na+-bound intermediates, which promotes increased access to the S1 and/or S2 site.
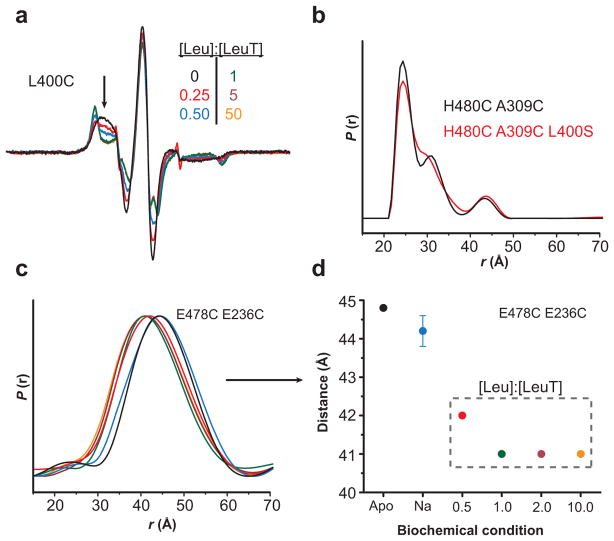
Consistent with the accessibility analysis, in the absence of a Na+ gradient, Leu binding in the S1 site appears sufficient to reduce access to the extracellular permeation pathway, regardless of the occupancy of the S2 site. The Leu titration of spin labeled L400C, which supports binding in the S1 site only, completely restricted probe motion at a 1:1 molar ratio of Leu to LeuT (Figure 5a). Also, the incorporation of a similar S2-disrupting mutation, L400S, into the H480C/A309C mutant did not alter the Na+/Leu-bound distance distribution (Figure 5b, Supplementary Figure 5). Furthermore, the distance changes between EL6 and each of EL4 (data not shown) and EL3 (Figure 5c–d) were saturated with equimolar Leu binding, consistent with an S1-dependent conformational change. Therefore, substrate binding to the S2 site is not required to induce the conformational changes that occlude the extracellular permeation pathway.
Figure 5.
Leu binding to the S1 site is the primary determinant of Leu-dependent conformational changes. (a) The decrease in spin label mobility at L400C was dependent on the [Leu]. The Leu titration of LeuT-L400C indicated a maximum change in EPR lineshape (arrow) at a 1:1 molar ratio of Leu-to-LeuT. Because the L400C mutation disrupts binding to the S2 site23, these spectral changes result from binding in the S1 site. (b) The distance distributions of LeuT-H480C/A309C (2:1 Leu-to-LeuT stoichiometry) and –H480C/A309C/L400S (1:1 stoichiometry) are superimposable, suggesting that the Leu-dependent change in distance is due to Leu occupancy in the S1 site. (c–d) The change in distance between LeuT-E478C/E236C (2:1 stoichiometry) is saturated at a 1:1 ratio, consistent with S1-driven conformational changes. The distances in (d) were obtained from the peak probability of the distance distributions in (c).
Inhibitors induce distinct LeuT conformations
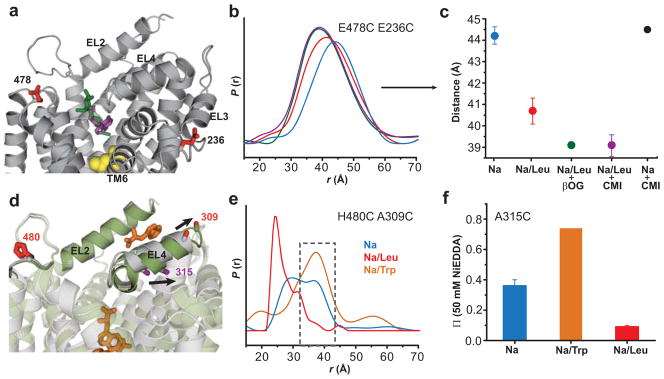
Functional and crystallographic evidence suggests that the LeuT structure determined in OG detergent represents an inhibitor-bound structure29. Furthermore, comparative molecular dynamics simulations of OG or TCA inhibitor binding in the S2 site indicate structural changes distinct from those produced by the substrate Leu29. Experimentally, we found that addition of OG or CMI to LeuT induced a small but reproducible left-shift in the distance distribution of E478C/E236C relative to the Na+/Leu bound state, consistent with a decrease in the average distance between EL6 and EL3 (Figure 6a–c). The addition of CMI to the Na+ intermediate did not alter the distance distribution, suggesting that the decrease in distance between EL6 and EL3 was dependent on the presence of Leu, presumably occupying the S1 site (Figure 6c). Because this distance change occurred in the Na+/Leu bound state, the distributions are likely to reflect rearrangements associated with the binding of these inhibitors (CMI and OG) to the extracellular vestibule. CMI binding to the Na+/Leu intermediate suggests that occlusion of the permeation pathway to S1 is dynamic so that the conformational fluctuations allowing access to the extracellular vestibule for substrate and inhibitors are substantially reduced, but not eliminated. Thus, inhibitor binding to the extracellular vestibule appears to lead to a state with structural features distinct from the Leu-bound conformation. Furthermore, molecular dynamics simulations with an occupied S2 site had demonstrated distinct substrate and inhibitor-dependent conformational rearrangements of TM6a23,29.
Figure 6.
Binding of inhibitors lead to structural changes distinct from the Na+/Leu bound conformation. (a) Both OG (green stick) and CMI (purple stick) can occupy the extracellular vestibule with Leu (yellow sphere) in the S1 site. For clarity, OG (from PDB 3GJC29) and CMI (from PDB 2Q6H27) have been mapped onto the Na+/Leu bound LeuT crystal structure (PDB 2A65)6. (b–c) OG and CMI binding produce similar decreases in distance between the probes. Addition of 1mM CMI to LeuT in the presence of Na+ alone did not shift the distance distribution. The mean ± S.D. of three repeated distances measurements in (c) were obtained from the peak probability of the distance distributions in (b). (d) Trp (orange stick) competitively inhibits transport by binding to the S1 site and displacing Leu7. (e) 20mM Trp increases the average distance between H480C/A309C, selectively enhancing the outer-most component of the Na+ intermediate (gray dashed box). For comparison, the Trp-bound LeuT crystal structure (green, PDB 3F3A) overlays the Na+/Leu crystal structure (gray, PDB 2A65) illustrating the altered conformation EL4 adopts in the presence of Trp. Arrows in (d) indicate the position change of A309 and A315 in the LeuT-Trp structure. (f) Trp binding doubles the NiEDDA accessibility of the A315C mutant in proteoliposomes (relative to the Na+-bound intermediate), consistent with altering the conformation of EL4. Data are shown as mean ± S.D. The structures in (a) and (d) were generated using PyMOL
The binding of Trp, which was also shown to inhibit transport7, was found in our experiments to increase the distances between spin labels in the H480C/A309C (Figure 6d,e) and E478C/E236C pairs (data not shown) relative to the Na+/Leu bound state. Also, the distance distribution in the presence of Na+/Trp was narrower than in the Na+-bound intermediate, and overlapped its most extended component (gray box, Figure 6e), consistent with the stabilization of an extreme Na+-induced conformer. The larger distance relative to the Na+/Leu-bound conformation is in agreement with the expansion of the extracellular vestibule, in part through movement of EL4a, that was seen in the crystal structure of LeuT in the presence of 50 mM Trp (PDB 3F3A)7, and has been interpreted as an “open-to-out” conformation. Indeed, the Trp-induced expansion of the structure also increased NiEDDA accessibility at positions in the vestibule, as illustrated by residue A315C (Figure 6d,f). Furthermore, the spectral breadth of F253C, which reported Leu-induced S1 reorganization (described above), also indicated that Trp impaired the dehydration of the S1 site (data not shown).
These experimental findings concur with MD simulation of the effects of Na+ binding (Figure 7). In these simulations, TM6a adopts a strongly outward tilt (Figure 7c) similar to that observed in the LeuT structure with Trp in the S1 site (Figure 7b, PDB 3F3A)7. Additionally, we observed rotamer changes of Phe252 and Phe253 (which interacts with the substrate in the S1 site) that are associated with those of Tyr107 and Tyr108 in TM3. The orientations of these four interrelated aromatic residues are affected by the occupancy of the S1 site. When S1 is occupied, Phe252 is the only one showing a rotamer difference (gauche vs trans) in comparing Leu-bound to Trp-bound structures. Thus, the trans rotamer of Phe252 appears to be involved in stabilizing the outward tilting of TM6a (Figure 7d–e). When S1 is not occupied, but in the presence of Na+, the dynamics showed the Phe253 side chain rotating away from its initial orientation so as to reside in the same space that is occupied by the indole ring of the Trp bound near the extracellular vestibule in the crystal structure (Trp602, Figure 7d). Thus, the effect of the inhibitor Trp may be viewed as trapping LeuT in an outward-facing conformation by preventing the configurational changes that need to occur in the aromatic cluster upon ligand binding to S1.
Figure 7.
Configuration changes of the TM3–TM6 aromatic cluster associated with the conformational transition simulated in the 660 ns MD trajectory. The starting conformation was the LeuT crystal structure6, in which the Leu molecule found in the S1 site (PDB 2A65) was removed (a) and (b) are the crystal structures with substrate (Leu)6 or inhibitor (Trp)7 bound in the S1 site, respectively. Y107 and Y108 from TM3, and F252 and F253 from TM6 are rendered in sticks. The ligands are in space-filling representation. (c) The outward movement of TM6 (with Na+ only bound) indicated by a white arrow, overlaps the one evident in the LeuT-Trp structure (PDB 3F3A)7, which has been proposed to represent the ‘open-to-out’ conformation. (d) Superimposed view of the aromatic cluster in the context of the Na1 site. The side chains are colored corresponding to the backbone colors in a–c. The overlap between F253 (in the Na-only simulation) and the Trp in the extracellular vestibule of the Trp-bound crystal structure (Trp602 of PDB 3F3A), is indicated by a dotted circle. Only the ligand and Na+ ions from the Trp-bound structure are shown. (e) Changes in Χ1 rotamer values for the aromatic cluster residues along the MD trajectory.
Discussion
Mechanistic implications for Na+-coupled substrate symport
Our results, compiled from the combined experimental and computational approaches described here, provide a unique perspective on the conformational dynamics in the functional cycle of NSS. Fundamental to this cycle is a dynamic outward-facing state induced by the binding of Na+ and achieved, substantially, by movement of EL4. In the cellular environment, Na+-induced outward-facing conformations are likely to predominate in the absence of substrate due to the high extracellular Na+ concentration. This outward-facing state exposes the substrate binding sites and indeed we find that Leu binding is facilitated by Na+ (Figure 2b–c). Leu binding to the S1 site efficiently engages local and global conformational changes that reverse accessibility to the extracellular permeation pathway, providing unequivocal evidence of distinct Na+- and substrate-dependent structural rearrangements. In contrast, we found that the binding of inhibitors in the extracellular vestibule altered the conformation of LeuT in a manner different from the changes induced by Leu, which supports the notion that the current set of LeuT crystal structures represent inhibited conformations29.
We find that the dynamics of the outward facing conformation involves fluctuations of the extracellular loops and extends into the permeation pathway (vestibule) where the looser packing relative to the Apo state facilitates water and substrate access to the S1 site. Subsequent Leu binding reduces the dynamics of the extracellular loops and constricts the permeation pathway thereby occluding access to the binding sites. Hints of movements in other protein segments besides EL4 such as TM6a and TM10 were also evident. A role for TM6 (and TM1) was originally proposed based on the discontinuity (unwound segments of TM1 and TM6 that contribute to substrate binding) of the helix near the S1 site, suggesting that flexibility of this helix may confer dynamic properties during transport6. In our findings, movement of TM6 is suggested by a Leu-dependent decrease in distance between E478C/E236C, in which Glu236 is just four residues away from the N-terminus of TM6. The pattern of mobility and accessibility of TM10 residues in the presence of Leu suggests movement similar to that captured by the crystal structure of Mph1 where TM10 bends toward an outward-facing cavity to occlude the substrate binding site17.
Substrate exit on the intracellular side must entail conformational changes related to those we have observed in the extracellular permeation pathway. Several models have been proposed for substrate permeation8,13,23 but the nature and amplitude of the underlying movement on the intracellular side have not been measured. Further spectroscopic experiments that investigate the dynamics of the intracellular region of LeuT and the nature of the coupling between the extracellular and intracellular portions are required to elucidate the corresponding effects on the dynamics of the inward permeation pathway.
Methods
Mutagenesis, expression, purification, labeling and reconstitution of LeuT
Cysteine residues were introduced, individually or in combination, in recombinant LeuT possessing an N-terminal 10-His tag23 using standard molecular biological methods. DNA sequencing confirmed the presence of the desired mutation and absence of unwanted changes. LeuT mutants were expressed in E. coli C41(DE3) as previously described38. Harvested E. coli membranes were washed three times in 200 mM Tris/Mes pH 7.5, 20% glycerol buffer to remove endogenous Leu bound to LeuT during expression. 40 mM (2% w/v) DDM (Anatrace) was used to extract LeuT mutants from native membranes. Solubilized mutants were purified by Ni2+ affinity chromatography, immediately labeled for 6 h at 23°C with MTSSL (Toronto Research Chemicals, Inc) and placed on ice overnight. To generate the Apo intermediate and remove unbound spin label, LeuT mutants were subjected to size exclusion chromatography on a Shodex KW-803 column and subsequent desalting on a HiTrap desalting column (GE Healthcare) into 200 mM Tris/Mes pH 7.2 buffer containing 0.025% (w/v) DDM and 20% (v/v) glycerol. Protein concentration was determined using an extinction coefficient of 1.91 cm2/mg at 280 nm. Apo-LeuT mutants were reconstituted into 200 nm unilamellar liposomes made from a 3:1 (w/w) ratio of E. coli polar lipids and phosphatidylcholine (Avanti Polar Lipids, Inc) loaded with 0.1 M KPi pH 6.5 buffer using a 5000:1 (mol/mol) lipid-to-DDM ratio. Detergent was removed during reconstitution using SM-2 bio-beads (Bio-Rad). Harvested proteoliposomes were resuspended in 200 mM Tris/Mes pH 7.2 buffer. For EPR lineshape and solvent accessibility analysis, single cysteine mutants were reconstituted with a 1000:1 (mol/mol) lipid-to-protein ratio. For DEER analysis, the lipid-to-protein ratio was increased to 2000:1 to reduce non-specific intermolecular dipolar coupling.
Functional analysis
Binding studies were performed by scintillation proximity assay (SPA) with 100 nM 3H-Leu (112 Ci mmol−1) and 50 mM NaCl 23,38, with and without spin label. The Leu equilibrium binding constant ( ) and molar stoichiometry were determined by varying the [3H]-Leu concentration from 0.5 – 1000 nM. For transport activity (Figure 3), the time course of 1 μM 3H-Ala uptake was performed with proteoliposomes containing LeuT-WT or –F253C and with control liposomes in the presence of an inwardly-directed Na+ electrochemical gradient (25 mM external NaCl) or by dissipation of the gradient with gramicidin, yielding the total accumulation and specific binding, respectively. Specific substrate transport was determined by the difference between total accumulation and specific binding.
EPR spectroscopy
EPR spectra were collected at room temperature on a Bruker EMX spectrometer (X-band) at an incident power of 10 mW and 1.6 Gauss modulation amplitude. Substrates were allowed to incubate with LeuT mutants on ice for at least 20 minutes before collection of EPR spectra. The addition of 200 mM NaCl to the Apo intermediate was used to create the Na+ state. The addition of gramicidin was without effect, consistent with the equilibration of Na+ on both sides of the proteoliposomes during the extended incubation. The Na+/Leu state was produced by adding Leu to the Na+ state. For solvent accessibility, samples were loaded into a gas permeable TPX capillary tube and subjected to either 20% O2 (g) or 50 mM Ni(II)ethylenediaminediacetic acid (NiEDDA) equilibrated with N2 (g) on a Bruker Elexsys E500 spectrometer to generate power saturation curves39. The accessibility parameter Π was determined from a nonlinear least-squares fit of the power saturation curves in the program Origin (OriginLab Inc.). The experimental error was less than 20% based on the mean ± S.D. of three independently determined Π values (denoted as (*) in Table 1).
DEER Spectroscopy
Distance measurements were carried out on a Bruker 580 pulsed EPR spectrometer operating at X-band frequency (9.5 GHz) using double electron electron resonance (DEER) with a standard four-pulse protocol44. Glycerol was added to 30% (w/w) prior to cooling for both detergent and liposome samples. Deuterated buffer and glycerol was used to enhance the phase memory time for liposome samples. All DEER experiments were performed at 83K. The DEER signals were analyzed by Tikhonov regularization45,46 to yield the distance distributions.
Molecular dynamics (MD) simulations
Based on our established simulation protocols and the molecular system detailed in previous studies23,29, the “Na-only” simulation was carried out with NAMD47. Briefly, the all-atom simulations used the CHARMM27-CMAP force field with improved backbone treatment48,49. Constant temperature (310 K) was maintained with Langevin dynamics, and 1 atm constant pressure was achieved with the hybrid Nosé-Hoover Langevin piston method50 applied to a flexible periodic cell, with orthogonal pressure components computed independently to account for system anisotropy. Long-range electrostatic effects were evaluated with Particle Mesh Ewald. A time step of 1 fs was used for the first 30 ns, and was then increased to 2 fs for the rest of the simulation.
Supplementary Material
Acknowledgments
This work was financially supported by National Institutes of Health grants DA022413 and DA17293 to JAJ, DA023694 to LS, and DA012408 to HW; unrestricted funds from Vanderbilt University to HSM; and a predoctoral National Research Service Award (F31NS063699) from the National Institute of Neurological Disorders and Stroke to DPC. Computations were performed on the Ranger at the Texas Advanced Computing Center (TG-MCB090022) and the Cofrin Center for Bioinformation of the Institute for Computational Biomedicine at Weill Cornell Medical College.
Abbreviations
- NSS, neurotransmitter sodium
symporters
- SSRI
selective serotonin reuptake inhibitor
- TCA
tricyclic antidepressant
- CMI
clomipramine
- OG
n-octyl-β-D-glucopyranoside
- DDM
n-dodecyl-β-D-maltopyranoside
- EPR
electron paramagnetic resonance
- NiEDDA
Ni(II)ethylenediaminediacetic acid
- MTSSL
1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl methanethiosulfonate
Footnotes
Author Contributions
DPC designed and performed the EPR experiments and analyzed the results; MQ designed, carried out, and analyzed the binding and transport experiments with the help of FC; LS designed, carried out, and analyzed the computational studies; FC helped construct, express, and purify membranes harboring LeuT mutations; HW, JAJ and HSM helped to design experiments and analyze data related to the computational, biochemical, and EPR studies, respectively; DPC, MQ, LS, HW, JAJ and HSM contributed to writing and editing the manuscript.
Author Information
The authors have no competing financial interests.
References
- 1.Nelson N. The family of Na+/Cl− neurotransmitter transporters. J Neurochem. 1998;71:1785–803. doi: 10.1046/j.1471-4159.1998.71051785.x. [DOI] [PubMed] [Google Scholar]
- 2.Wadiche JI, Kavanaugh MP. Macroscopic and microscopic properties of a cloned glutamate transporter/chloride channel. J Neurosci. 1998;18:7650–61. doi: 10.1523/JNEUROSCI.18-19-07650.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amara SG, Sonders MS. Neurotransmitter transporters as molecular targets for addictive drugs. Drug Alcohol Depend. 1998;51:87–96. doi: 10.1016/s0376-8716(98)00068-4. [DOI] [PubMed] [Google Scholar]
- 4.Rudnick G, editor. Mechanisms of biogenic amine neurotransmitter transporters. Humana Press Inc; Totowa, New Jersey: 2002. pp. 25–52. [Google Scholar]
- 5.Krause S, Schwarz W. Identification and selective inhibition of the channel mode of the neuronal GABA transporter 1. Mol Pharmacol. 2005;68:1728–35. doi: 10.1124/mol.105.013870. [DOI] [PubMed] [Google Scholar]
- 6.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature. 2005;437:215–23. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 7.Singh SK, Piscitelli CL, Yamashita A, Gouaux E. A competitive inhibitor traps LeuT in an open-to-out conformation. Science. 2008;322:1655–61. doi: 10.1126/science.1166777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krishnamurthy H, Piscitelli CL, Gouaux E. Unlocking the molecular secrets of sodium-coupled transporters. Nature. 2009;459:347–55. doi: 10.1038/nature08143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gouaux E, Mackinnon R. Principles of selective ion transport in channels and pumps. Science. 2005;310:1461–5. doi: 10.1126/science.1113666. [DOI] [PubMed] [Google Scholar]
- 10.Gouaux E. Review. The molecular logic of sodium-coupled neurotransmitter transporters. Philos Trans R Soc Lond B Biol Sci. 2009;364:149–54. doi: 10.1098/rstb.2008.0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noskov SY, Roux B. Control of ion selectivity in LeuT: two Na+ binding sites with two different mechanisms. J Mol Biol. 2008;377:804–18. doi: 10.1016/j.jmb.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noskov SY. Molecular mechanism of substrate specificity in the bacterial neutral amino acid transporter LeuT. Proteins. 2008;73:851–63. doi: 10.1002/prot.22108. [DOI] [PubMed] [Google Scholar]
- 13.Forrest LR, et al. Mechanism for alternating access in neurotransmitter transporters. Proc Natl Acad Sci U S A. 2008;105:10338–43. doi: 10.1073/pnas.0804659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Celik L, Schiott B, Tajkhorshid E. Substrate binding and formation of an occluded state in the leucine transporter. Biophys J. 2008;94:1600–12. doi: 10.1529/biophysj.107.117580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caplan DA, Subbotina JO, Noskov SY. Molecular mechanism of ion-ion and ion-substrate coupling in the Na+-dependent leucine transporter LeuT. Biophys J. 2008;95:4613–21. doi: 10.1529/biophysj.108.139741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quick M, et al. State-dependent conformations of the translocation pathway in the tyrosine transporter Tyt1, a novel neurotransmitter:sodium symporter from Fusobacterium nucleatum. J Biol Chem. 2006;281:26444–54. doi: 10.1074/jbc.M602438200. [DOI] [PubMed] [Google Scholar]
- 17.Weyand S, et al. Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science. 2008;322:709–13. doi: 10.1126/science.1164440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaffer PL, Goehring A, Shankaranarayanan A, Gouaux E. Structure and mechanism of a Na+-independent amino acid transporter. Science. 2009;325:1010–4. doi: 10.1126/science.1176088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ressl S, Terwisscha van Scheltinga AC, Vonrhein C, Ott V, Ziegler C. Molecular basis of transport and regulation in the Na+/betaine symporter BetP. Nature. 2009;458:47–52. doi: 10.1038/nature07819. [DOI] [PubMed] [Google Scholar]
- 20.Gao X, et al. Structure and mechanism of an amino acid antiporter. Science. 2009;324:1565–8. doi: 10.1126/science.1173654. [DOI] [PubMed] [Google Scholar]
- 21.Fang Y, et al. Structure of a prokaryotic virtual proton pump at 3.2 A resolution. Nature. 2009;460:1040–3. doi: 10.1038/nature08201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faham S, et al. The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science. 2008;321:810–4. doi: 10.1126/science.1160406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi L, Quick M, Zhao Y, Weinstein H, Javitch JA. The mechanism of a neurotransmitter:sodium symporter--inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol Cell. 2008;30:667–77. doi: 10.1016/j.molcel.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kniazeff J, et al. An intracellular interaction network regulates conformational transitions in the dopamine transporter. J Biol Chem. 2008;283:17691–701. doi: 10.1074/jbc.M800475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211:969–70. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- 26.Zhou Z, et al. LeuT-desipramine structure reveals how antidepressants block neurotransmitter reuptake. Science. 2007;317:1390–3. doi: 10.1126/science.1147614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh SK, Yamashita A, Gouaux E. Antidepressant binding site in a bacterial homologue of neurotransmitter transporters. Nature. 2007;448:952–6. doi: 10.1038/nature06038. [DOI] [PubMed] [Google Scholar]
- 28.Zhou Z, et al. Antidepressant specificity of serotonin transporter suggested by three LeuT-SSRI structures. Nat Struct Mol Biol. 2009;16:652–7. doi: 10.1038/nsmb.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quick M, et al. Binding of an octylglucoside detergent molecule in the second substrate (S2) site of LeuT establishes an inhibitor-bound conformation. Proc Natl Acad Sci U S A. 2009;106:5563–8. doi: 10.1073/pnas.0811322106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forrest LR, Rudnick G. The rocking bundle: a mechanism for ion-coupled solute flux by symmetrical transporters. Physiology (Bethesda) 2009;24:377–86. doi: 10.1152/physiol.00030.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lester HA, Cao Y, Mager S. Listening to neurotransmitter transporters. Neuron. 1996;17:807–10. doi: 10.1016/s0896-6273(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 32.Smirnova I, et al. Sugar binding induces an outward facing conformation of LacY. Proc Natl Acad Sci U S A. 2007;104:16504–9. doi: 10.1073/pnas.0708258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hubbell WL, Mchaourab HS, Altenbach C, Lietzow MA. Watching proteins move using site-directed spin labeling. Structure. 1996;4:779–83. doi: 10.1016/s0969-2126(96)00085-8. [DOI] [PubMed] [Google Scholar]
- 34.Dong J, Yang G, Mchaourab HS. Structural basis of energy transduction in the transport cycle of MsbA. Science. 2005;308:1023–8. doi: 10.1126/science.1106592. [DOI] [PubMed] [Google Scholar]
- 35.Borbat PP, et al. Conformational motion of the ABC transporter MsbA induced by ATP hydrolysis. PLoS Biol. 2007;5:e271. doi: 10.1371/journal.pbio.0050271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zou P, Mchaourab HS. Alternating access of the putative substrate-binding chamber in the ABC transporter MsbA. J Mol Biol. 2009;393:574–85. doi: 10.1016/j.jmb.2009.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zou P, Bortolus M, Mchaourab HS. Conformational cycle of the ABC transporter MsbA in liposomes: detailed analysis using double electron-electron resonance spectroscopy. J Mol Biol. 2009;393:586–97. doi: 10.1016/j.jmb.2009.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quick M, Javitch JA. Monitoring the function of membrane transport proteins in detergent-solubilized form. Proc Natl Acad Sci U S A. 2007;104:3603–8. doi: 10.1073/pnas.0609573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altenbach C, Froncisz W, Hemker R, Mchaourab H, Hubbell WL. Accessibility of nitroxide side chains: absolute Heisenberg exchange rates from power saturation EPR. Biophys J. 2005;89:2103–12. doi: 10.1529/biophysj.105.059063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mchaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35:7692–704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 41.Owenius R, Engstrom M, Lindgren M, Huber M. Influence of Solvent Polarity and Hydrogen Bonding on the EPR Parameters of a Nitroxide Spin Label Studied by 9-GHz and 95-GHz EPR Spectroscopy and DFT Calculations. The J Phys Chem A. 2001;105:10967–10977. [Google Scholar]
- 42.Borbat PP, Mchaourab HS, Freed JH. Protein structure determination using long-distance constraints from double-quantum coherence ESR: study of T4 lysozyme. J Am Chem Soc. 2002;124:5304–14. doi: 10.1021/ja020040y. [DOI] [PubMed] [Google Scholar]
- 43.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 44.Jeschke G. Distance measurements in the nanometer range by pulse EPR. Chemphyschem. 2002;3:927–32. doi: 10.1002/1439-7641(20021115)3:11<927::AID-CPHC927>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 45.Jeschke G, Chechik V, Ionita P, Godt A, Zimmermann H, Banham J, Timmel CR, Hilger D, Jung H. Deer Analysis 2006--a comprehensive software package for analyzing pulsed ELDOR data. Appl Magn Reson. 2006;30:473–498. [Google Scholar]
- 46.Chiang YW, Borbat PP, Freed JH. The determination of pair distance distributions by pulsed ESR using Tikhonov regularization. J Magn Reson. 2005;172:279–95. doi: 10.1016/j.jmr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 47.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.MacKerell AD, Jr, Feig M, Brooks CL., 3rd Improved treatment of the protein backbone in empirical force fields. J Am Chem Soc. 2004;126:698–9. doi: 10.1021/ja036959e. [DOI] [PubMed] [Google Scholar]
- 49.Mackerell AD., Jr Empirical force fields for biological macromolecules: overview and issues. J Comput Chem. 2004;25:1584–604. doi: 10.1002/jcc.20082. [DOI] [PubMed] [Google Scholar]
- 50.Feller SE, et al. Constant pressure molecular dynamics simulation: The Langevin piston method. The J Chem Phys. 1995;103:4613–4621. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.