Summary
It is believed that the introns are removed from pre-mRNAs during transcription, while the pre-mRNA is still tethered to the gene locus via RNA polymerase. However, during alternative splicing it is important that splicing be deferred until all of the exons and introns involved in the choice have been synthesized. We have developed an in situ RNA imaging method with single-molecule sensitivity to define the intracellular sites of splicing. Using this approach, we found that the normally tight coupling between transcription and splicing is broken in situations where the intron’s polypyrimidine tract is sequestered within strong secondary structures. We also found that in two cases of alternative splicing, in which certain exons are skipped due to the activity of the RNA binding proteins Sxl and PTB, splicing is uncoupled from transcription. This uncoupling occurs only on the perturbed introns, while the preceding and succeeding introns are removed co-transcriptionally.
Introduction
Where in the nucleus, and at what point during mRNA biogenesis, does splicing occur, are questions that have held the attention of cell biologists for many years (Han et al., 2011; Moore and Proudfoot, 2009; Neugebauer, 2002; Oesterreich et al., 2011; Perales and Bentley, 2009). When splicing factors were first found concentrated in subnuclear structures called “speckles” (Lamond and Spector, 2003), it was suggested that these might correspond to splicing centers (Fu and Maniatis, 1990). However, subsequent studies indicated that splicing generally does not occur in speckles (Huang et al., 1994; Zhang et al., 1994). Instead it proceeds while nascent mRNAs are still tethered to the DNA via RNA polymerase II (Bauren and Wieslander, 1994; Beyer and Osheim, 1988; Listerman et al., 2006; Pandya-Jones and Black, 2009; Singh and Padgett, 2009; Zhang et al., 1994).
While it is nearly universally accepted that transcription and splicing are coupled, two views concerning the mechanism of coupling prevail: structural coupling and kinetic coupling. According to the structural coupling model, splicing factors are pre-positioned on the RNA polymerase II C-terminal domain and hop on to the introns as they emerge from polymerase (Das et al., 2007; Fong and Bentley, 2001; Yuryev et al., 1996). The kinetic coupling model, on the other hand, suggests that, owing to their high concentration and mobility (Phair and Misteli, 2000), splicing factors directly assemble on the nascent introns into productive spliceosomes, as fast as the RNA polymerase can synthesize them (Neugebauer, 2002; Oesterreich et al., 2011). The rate-limiting step is not splicing, but rather it is the completion of mRNA synthesis, 3’-end processing and release. Support for the kinetic coupling model comes from the finding that exon inclusion is promoted by an intrinsically slow RNA polymerase, or by the nucleosomes impending the progress of the polymerase (Batsche et al., 2006; de la Mata et al., 2003). Furthermore, there is evidence that the rates of the two processes are sometimes coordinated to ensure that only fully spliced mRNAs are released (Alexander et al., 2010; Carrillo Oesterreich et al., 2010; Custodio et al., 1999).
The processive removal of introns immediately after their synthesis provides an attractive mechanism for ensuring fidelity in joining constitutively spliced exons in the proper sequential order. However, during alternative splicing (Black, 2003; Chen and Manley, 2009; Han et al., 2011), the splicing must be slowed down until all of the splice sites involved in the choice have been synthesized. Is processing just delayed briefly until alternative splice sites are generated, or does alternative splicing result instead in the uncoupling of splicing from transcription, so that it is concluded post-transcriptionally? The former has been found to be the case for several alternatively spliced transcripts (Dutertre et al., 2010; Pandya-Jones and Black, 2009; Waks et al., 2011). However, how RNA binding splicing regulators impact the splicing-transcription coupling when they impose strict tissue and developmental stage specific alternative splicing patterns remains to be explored.
We have developed and used a single-molecule imaging approach to explore how splicing of constitutively or alternatively spliced introns-exons is coupled to transcription. Our results show that the processing of constitutively spliced introns-exons is generally tightly coupled to transcription. However, it is possible to uncouple the transcription and processing of a constitutively spliced intron by introducing mutations that interfere with the recognition of key splice signals by the splicing machinery. Using the same approach, we also examined two well studied examples of regulated splicing: the sex-specific splicing of Sxl pre-mRNAs in Drosophila and polypyrimidine tract binding (PTB) protein mediated exclusion of exon 10 during alternative splicing of neuronal PTB pre-mRNA. Significantly, transcriptional coupling depends upon whether the pre-mRNA is spliced in the default or regulated pattern. When the alternatively spliced intron-exon cassette is spliced in the default pattern by the basal splicing machinery, splicing is co-transcriptional. By contrast, when the appropriate regulatory factors are present and impose the regulated pattern of splicing, processing is delayed until after transcription has been completed. The uncoupling of processing and transcription appears to be an intrinsic property of the regulated cassette, as the splicing of other, constitutively spliced introns in the same pre-mRNA remains co-transcriptional.
Results
Imaging individual molecules of pre-mRNA, mRNA and introns
Our approach involves the attachment of multiple fluorophores to target mRNAs via hybridization probes (depositing 32–384 fluorescent moieties depending upon the construct and the probe), in order to render them so intensely fluorescent that each molecule can be seen as a diffraction-limited spot with a wide-field microscope (Raj et al., 2006; Raj et al., 2008; Vargas et al., 2005). When a pair of distinctly labeled probe sets, one against the coding sequence and the other against an intron, is used in combination, spliced mRNA molecules become visible in one channel and the free introns in the other, whereas pre-mRNA molecules are visible in both channels, allowing us to image the precursor and products of splicing with single-molecule sensitivity. Evidence for single-molecule sensitivity and high specificity of this method is reviewed in the supplementary material.
We first examined the coupling of splicing to transcription utilizing a pair of GFP mRNA reporters that have a tandemly repeated sequence, array 3, inserted in their 3’-UTR and one of two tandem arrays (1 or 2) inserted into an artificial intron (with canonical splice sites) placed in the middle of the GFP coding sequence (Fig.1A). The tandem arrays of arbitrarily selected sequences (Supplementary Online Material) were used to achieve single-molecule sensitivity with just one oligonucleotide probe and to permit imaging in live cells. Doxycycline-controlled versions of the two genes were stably integrated into the genome of Chinese hamster ovary cell lines. We isolated clones that integrated 8 and 19 copies of these reporter genes at one locus in each cell (Fig. S1A). Upon induction by removing doxycycline from the culture medium, both cell lines produced appropriately spliced mRNAs and exhibited GFP fluorescence.
Fig. 1.
Imaging the intracellular distribution of single-molecules of pre-mRNAs and their spliced products expressed from a pair of reporter genes integrated into the genome of CHO cell lines. A. Schematic depiction of two reporter genes. Array 1 contains 32-repeats of a 49 nucleotide sequence, array 2 contains 96 repeats of a 50 nucleotide sequence, and array 3 contains 32 repeats of a 80 nucleotide sequence. B. Images of cells from clones expressing the reporter gene in two fluorescence channels. The targets of the probes are indicated on the top of the panels, and the array within the intron is shown on the left. All images in this report are maximum intensity merges of 3-dimensional optical slices. In the composite images, red represents either the 3’-UTR or the coding sequence, and green represents the introns. C. Identification of RNA species using our image-processing algorithm. Circles of different colors are drawn around each detected spot. D. Percentage of three different species in the nuclei of individual cells from two cell lines. Examples of regions from which the counts were obtained are indicated by blue circles in C. Error bars represent 95% confidence intervals. Images from 15–50 cells were analyzed to obtain the data in this report. The probability of obtaining the indicated (or higher) percentage of unspliced pre-mRNA of array 2 by chance is < 10−6. Scale bars are 5 µm in all figures in this report.
The pre-mRNAs and their spliced products were imaged by fixing the cells after hours of induction, followed by in situ hybridization with fluorescently labeled probes against array 3 and either array 1, or array 2 repeats. We observed three classes of diffraction-limited spots. Two of these corresponded to single-molecules containing either the intron array or the 3’-UTR array (Fig. 1B). The third class consisted of unspliced molecules containing both the intron and the 3’-UTR arrays (Fig. 1B). When the center of a spot seen in one channel was located within 0.25 µm of the center of a spot seen in the other channel, we considered the spots to be colocalized and to be representing an unspliced pre-mRNA (Fig. 1C and D). This criterion for identifying single-molecules was obtained empirically by measuring the distances between spots produced by two sets of probes bound to two halves of the same mRNA (Fig. S1B).
Divergent modes of splicing depending on whether array 1 and array 2 is present within the intron
Transcripts expressed from the two reporter genes exhibited striking differences in how they coordinate transcription and splicing. In the case of array 1, high levels of pre-mRNA accumulated at the gene locus while pre-mRNA was rarely seen elsewhere in the nucleoplasm. While splicing and transcription were tightly coupled for array 1 transcripts, this was not true of array 2 transcripts. Most array 2 pre-mRNA molecules were scattered throughout the nucleoplasm with little retention at the transcription site. In addition, the spliced introns from array 1 and 2 transcripts degraded differently. Only a few spliced array 1 intron molecules diffuse away from the gene locus, whereas, a large number of array 2 intron molecules were found scattered in the nucleoplasm. For both constructs, the spliced mRNAs were exported efficiently into the cytoplasm, while the pre-mRNAs and the intron were retained within the nucleus (Fig. 1B–D).
To show that the array 2 pre-mRNAs that are dispersed into the nucleoplasm are substrates for splicing, we induced the reporter for a short period (2 hours) and then after turning off the reporter we monitored the fate of the previously synthesized pre-mRNAs. In cells fixed immediately after 2 hours of induction, many pre-mRNA molecules were seen scattered within the nucleoplasm with little accumulation of spliced mRNA molecules in the cytoplasm (Fig. 2A). As predicted if the dispersed array 2 pre-mRNAs are splicing competent, there was a decrease in the proportion of pre-mRNA molecules in cells fixed after the chase period, with a concomitant increase in spliced mRNA molecules in the cytoplasm (Fig. 2B). While there were almost no spliced mRNA molecules in the cytoplasm in the beginning, they came to dominate the RNA population after chasing for 2 hour.
Fig. 2.
Demonstration that pre-mRNA molecules dispersed in the nucleus are capable of being spliced. A. Upper panels show composite images of cells in which the gene containing array 2 intron within an intron was induced for a brief period (2 hour). Most molecules are present within the nuclei. Lower panels show images from the same batch of cells as above, but in which induction was followed by a period of suppression (2 hour). Raw images are shown on the left and overlays with colored balls identifying the RNA species are presented on the right. B. Percentage of three different RNA species in individual cells as a function of time after the addition of doxycycline. Doxycycline turns off new RNA synthesis within several minutes. Even though the proportion of spliced mRNAs continues to increase after 3 hour, the overall number of RNAs declines due to degradation.
The splicing behavior of array 1 and array 2 remained the same irrespective of the chromatin context in which the gene was integrated or the location of the intron within the GFP coding sequence (Fig. S2).
Visualization of splicing in live cells
To explore where in the nucleus the pre-mRNAs of the array 2 construct are spliced, we imaged it in live cells using a pair of molecular beacons against array 2 and array 3 in live cells. Molecular beacons are probes that become fluorescent upon hybridization (Bratu et al., 2003) and allow detection and tracking of individual RNA molecules containing tandemly repeated sequences (Vargas et al., 2005). We found that the pre-mRNA molecules exhibited three distinct behaviors: freely diffusing as individual molecules; diffusing as clusters of a few molecules; and congregating around, but never entering, relatively stationary circular bodies in the nucleus (Movie S1). Suspecting that these structures correspond to speckles, the nuclear bodies with a high concentration of splicing factors, we stained fixed cells with antibodies raised against SC35, a key marker for speckles (Lamond and Spector, 2003), while simultaneously probing them with probes specific for array 2. The resulting images, presented as z-sections through a nucleus, confirm that RNAs containing the intron tend to encircle the speckles without entering them (Movie S2). As the splicing factors rapidly shuttle in and out of speckles (Phair and Misteli, 2000), it is possible that this clustering reflects splicing factors in the process of entering the speckles while being bound to the pre-mRNAs. A similar tendency was observed in the case of a β-globin pre-mRNA expressed from a plasmid that was microinjected into a nucleus (Dias et al., 2010).
Two introns are removed in their characteristic manner irrespective of their order in the gene
The striking differences between array 1 and array 2 introns in how their splicing and transcription are coordinated caused us to wonder what would happened if both introns were included in the same pre-mRNA. Would array 1 impose co-transcriptional splicing on the chimeric pre-mRNA, or would array 2 delay the splicing of the array 1 intron until transcription is finished? To explore this issue we constructed a pair of reporter genes, “array1–array2” and “array2-array1”, in which the two arrays are present in the same pre-mRNA but in different order (surrounded by the same splice sites as before) (Fig. 3A). The first intron was placed towards the 5’ end and the second towards the middle of the GFP coding sequence. Since it is difficult to construct genes with three arrays, we did not insert an array in the 3’-UTR and the detection of spliced mRNA molecules was accomplished using 48-labelled oligonucleotides complementary to the GFP coding sequence. The two methods of visualizing the RNA yield equivalent results (Fig. S1B–E). Cells expressing these reporters were imaged for the two intronic arrays and the GFP coding sequence in three different fluorescence channels and molecules corresponding to each of the seven possible permutations were computationally identified (Table S1).
Fig. 3.
Introns containing array 1 and 2 are spliced co-transcriptionally and post-transcriptionally irrespective of the order of their appearance in the gene. A. Schematic representation of two genes that contain the two introns in a different order. Both array 1 and array 2 have 32 repeats each, and they are embedded within the same splice sites. B. Raw, composite, and interpreted images of a cell expressing the construct array2-array1 in the three different channels that correspond to the coding sequence, array 1, and array 2. The composite shows the GFP coding sequence in red, array 1 in blue, and array 2 in green. The color key on the right lists each of the seven combinations of spliced and unspliced RNA species that can occur. Images for the construct array1–array2 are not shown as it yielded similar results (Table S1).
For both constructs, the partially spliced array2-GFP pre-mRNA was one of the most abundant species, and was found scattered throughout the nucleus (Fig. 3 and Table S1). By contrast, the other partially spliced product, array1-GFP, and the unspliced array1-array2-GFP or array2-array1-GFP pre-mRNAs were rarely detected except at the gene locus. These observations indicate that, irrespective of their order in the transcript, array 1 is spliced co-transcriptionally and array 2 post-transcriptionally. Significantly, in the case of the array2-array1 transcript, array 2 was not spliced at the gene locus, even though the splicing apparatus assembled on the downstream array 1 intron and spliced it co-transcriptionally.
Sequestration of intronic polypyrimidine tract within a secondary structure promotes post-transcriptional splicing
The post-transcriptional splicing of array 2 is either an intrinsic property of the array sequence or it arises from interactions between array 2 and the splice sites we used in our GFP reporter construct. To explore the former possibility, we tested whether array 2 can confer post-transcriptional splicing on an otherwise co-transcriptionally spliced natural intron. Before selecting a host gene, we studied the splicing pattern of two different endogenous genes, c-Fos and FKBP5, using sets of 48-probes each for one of their introns and the coding sequences. Consistent with prevailing understanding, we found that intron 1 of the c-Fos pre-mRNA and intron 4 of the FKBP5 pre-mRNA were spliced co-transcriptionally at their respective gene loci (Fig. S3A).
Focusing on c-Fos, we placed the human c-Fos gene under a doxycycline controlled promoter, inserted either array 1 or array 2 into the first intron and then integrated the recombinant genes into the HeLa cell genome (Fig. S3B). Since the endogenous gene is expressed only transiently after the addition of serum, we could keep it silent by growing the cells continuously in the presence of serum, and turn on the reporter genes by removing doxycycline from the culture medium. When we examined the transcripts produced by the two recombinant transcripts, we found that neither array 2, nor array 1, had any influence on the splicing behavior of the c-Fos intron. For both pre-mRNAs, the intron was spliced co-transcriptionally, just like the pre-mRNA of the endogenous gene (Fig. S3C).
As this result suggests that post-transcriptional splicing is not an intrinsic property of array 2, we reasoned that array 2 must specifically interfere with one of the splice signals in our GFP reporter. Modeling of the folding patterns of the full length GFP-array2 pre-mRNA predicted that the polypyrimidine tract (a key intron recognition element that is situated towards the 3’ end of introns) is sequestered in a double stranded region (Fig. S4A). This hypothesis was validated by ribonuclease T1 and ribonuclease V1 mapping of an RNA segment containing a 3’ proximal repeat from array 2 and the 3’ intron recognition elements (Fig. S4B).
Mutations that impair the function of splice site recognition elements uncouple splicing from transcription
If sequestration of the polypyrimidine tract causes post-transcriptional splicing in transcripts containing array 2, then would sequestration of the polypyrimidine tract in the array 1 reporter cause its transcripts to behave in a similar manner? To test this possibility we modified the array 1 intron sequence upstream of the polypyrimidine tract so that it would be occupied in a strong stem (Fig. 4A and S4B fourth panel, S5A). The cell line expressing this construct exhibited an increased number of unspliced pre-mRNAs in the nucleoplasm compared to the parent construct (Fig. 4B and Fig. S5B).
Fig. 4.
Engagement of an intron’s polypyrimidine tract in a strong double-stranded structure increases its tendency to splice post-transcriptionally. A. Sequences upstream of the branch point sites (red) in the array 1 construct and c-Fos intron 1 were altered so that they would form a 13-nucleotide stem with their respective polypyrimidine tracts. B. Percentage of pre-mRNAs in the nucleus for three array 1 constructs: unmodified array 1; array 1 in which an upstream sequence forms a hairpin stem with the polypyrimidine tract; and array 1 in which the polypyrimidine tract is modified by replacing two thymidine residues with adenosines. C. Distribution of c-Fos pre-mRNA and its spliced products resulting from different sequence modifications. The upper panels show merged raw images with green representing the signal from intron probes, and red representing the signal from exon probes. The lower panels identify the molecules on DIC images. D. Number of pre-mRNA (excluding the clusters at the gene loci) as a percentage of total cellular RNA in cell lines of different constructs. Representative images corresponding to the insertions of the arrays into intron 1 are shown in Fig. S3.
The uncoupling of transcription and splicing likely arises because the splicing factor U2AF, has reduced or slower access to the polypyrimidine tract. This hypothesis suggests that other means of reducing the U2AF polypyrimidine interaction may result in the same effect. We tested that proposition by weakening the polypyrimidine tract by converting two pyrimidines residues into purines (Fig. S5). This perturbation resulted in the release of unspliced pre-mRNA molecules in the nucleoplasm (Fig. 4B and S5B).
We also explored if sequestering the polypyrimidne tract within a strong hairpin stem results in the post-transcriptional splicing of intron 1 in c-Fos. We modified a portion of the sequence upstream of the polypyrimidine tract in intron 1 so that it would form a 13-basepair stem that includes a portion of it’s polypyrimidine tract (Fig. 4A and S4B). As predicted, masking the c-Fos polypyrimidine tract within a hairpin stem resulted in a striking increase in the number of pre-mRNAs that were dispersed away from the gene locus (Fig. 4C and D). When splicing was allowed to proceed without the synthesis of new RNA by the addition of doxycycline, no pre-mRNA remained in the nucleus, indicating that the dispersed pre-mRNA molecules are precursors to mRNAs (data not shown). These results indicate that when the polypyrimidine tract of a natural intron is masked, it tends to be spliced post-transcriptionally.
Regulated splicing in Sxl and nPTB pre-mRNAs occurs post-transcriptionally
In alternative splicing, spliceosomal complexes that are assembled on newly synthesized splice sites must remain inactive until the transcription of all of the introns and exons involved in the choice has been completed. Moreover, the recognition signals associated with alternatively spliced introns-exons are often suboptimal. These features of alternative splicing led us to wonder whether the processing of regulated alternatively spliced introns-exons is also uncoupled from transcription? To address this question we turned to two canonical examples of regulated splicing: sex-specific splicing of the pre-mRNA of Drosophila gene Sex-lethal (Sxl) (Bell et al., 1988) and polypyrimidine tract binding protein (PTB) modulated splicing of the neuronal PTB pre-mRNA (Boutz et al., 2007; Spellman et al., 2007).
Default and regulated Sxl pre-mRNA splicing
Drosophila gene Sxl controls sex determination by regulating the splicing of several pre-mRNAs including its own. In males, where Sxl is off and there is no Sxl protein, the pre-mRNAs are spliced in the default pattern to include a translation-terminating male-specific exon, exon 3 (Fig. 5). In females, Sxl protein binds to multiple polyuridine tracts in introns 2 and 3, forces the splicing machinery to skip exon 3, thereby linking exon 2 directly to exon 4 (Black, 2003; Sakamoto et al., 1992; Samuels et al., 1991)(Fig.5). Translation of the resulting mRNAs into Sxl protein establishes a positive feedback loop that serves to maintain female identity.
Fig. 5.
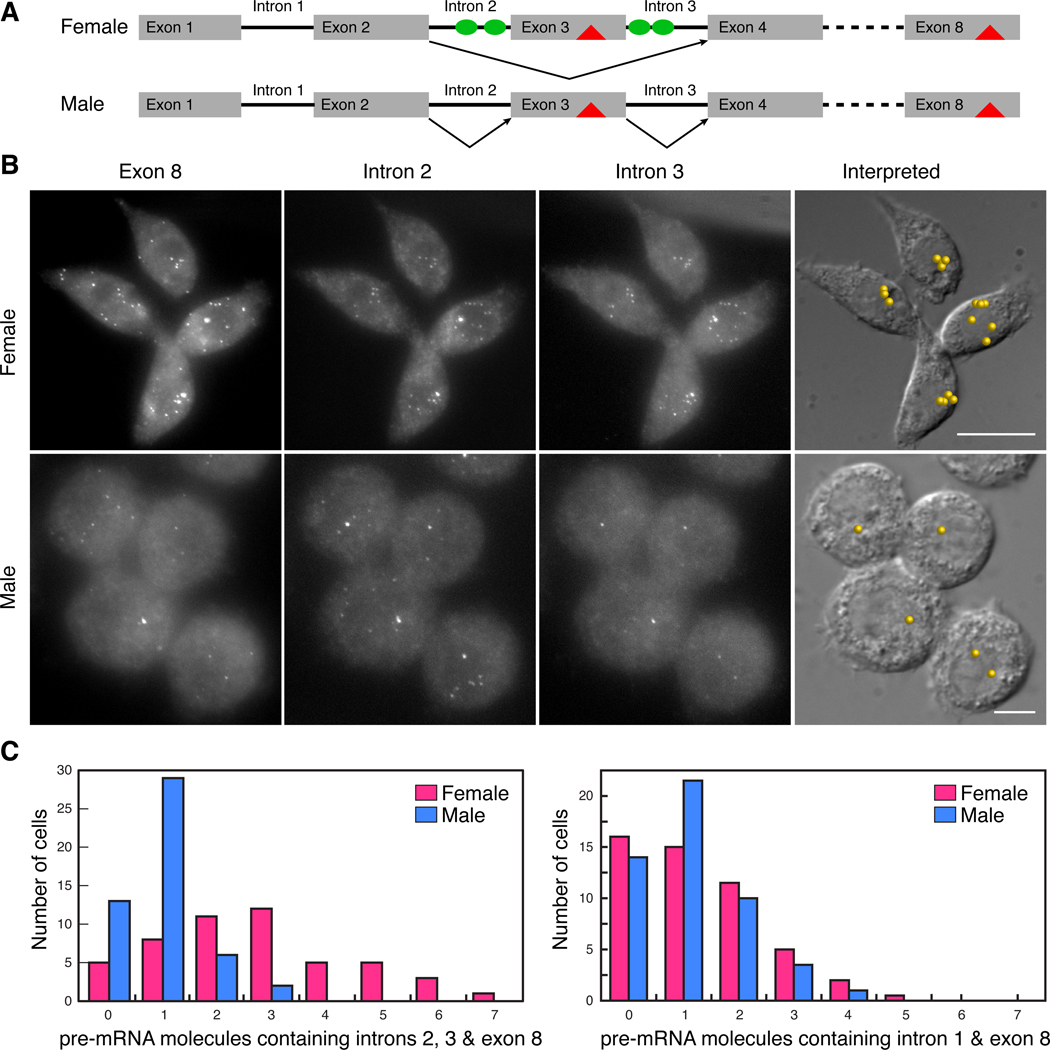
Sex-specific alternative splicing of Sxl pre-mRNA occurs post-transcriptionally in female Drosophila cells. A. Sxl protein (green oval), present only in females, binds to polypyrimidine tracts in introns 2 and 3 and results in a splicing pattern in which intervening exon 3, which contains a stop codon (red triangle) is not included in the final spliced product. Males have no Sxl protein and constitutive splicing yields a transcript possessing a premature termination codon. B. Images of spots produced by probes against the indicated components of Sxl transcripts in female (Kc167) and male (S2) cells. When the centers of spots in all three channels were within the molecular co-localization distance (Fig. S2), they were interpreted to be originating from a single pre-mRNA molecule containing intron 2, intron 3 and exon 8. The locations of the identified molecules are shown on DIC images. C. Frequency with which the given numbers of pre-mRNA molecules were found in a group of 50 cells. After a 30 minute exposure to actinomycin D no pre-mRNAs could be detected in the cells.
To examine the coordination between transcription and splicing, we took advantage of Drosophila male and female cell lines. The sex-specific patterns of Sxl pre-mRNA splicing are faithfully recapitulated in these lines, and they have been used to study the mechanisms of Sxl-dependent splicing regulation (Penalva et al., 2001; Sakamoto et al., 1992). In the first experiment we examined the coupling of transcription and splicing of the first Sxl intron, using sets of distinctly labeled fluorescent probes for this intron and for the downstream exon 8. This intron is about the same size as the regulated intron2-exon3-intron3 cassette, but is spliced in the same pattern in both sexes. There was, on average, about one molecule of pre-mRNA containing intron 1 and exon 8 in both male (1.12 ±0.09) and female (1.27±0.09) cells. This frequency suggests that this constitutively spliced intron is generally processed co-transcriptionally in both sexes. Moreover, though both cell lines are reported to have extra X and other chromosomes, the number of Sxl genes being transcribed at any one time is about the same whether the cell is male or female.
We next examined the transcriptional coupling of the regulated intron2-exon3-intron3 cassette in male cells using distinctly labeled probes specific to intron2, intron3 and exon8. As was seen for the constitutively spliced intron 1, only about one pre-mRNA molecule per cell containing the intron 2, intron 3 and exon 8 sequences was detected in nuclei from the male cell line (Fig. 5B and C). Thus, in spite of the fact that the splice sites of the male exon are sub-optimal (Horabin and Schedl, 1993; Penalva et al., 2001), the default splicing machinery joins the regulated cassette exons 2, 3, and 4 together co-transcriptionally.
Strikingly, a quite different result is obtained in female cells. We found that pre-mRNAs containing intron 2, intron 3, and exon 8 sequences are dispersed throughout the nuclei of female cells (Fig. 5B). On average, there were about 3 pre-mRNA molecules containing both of the introns and exon 8 in female nuclei, with many nuclei having 5–7 molecules of these incompletely spliced pre-mRNAs (Fig. 5C). Thus, unlike the processing of the constitutively spliced intron 1 which is co-transcriptional, the splicing of the regulated Sxl intron2-exon3-intron3 cassette is uncoupled from transcription in female cells. Moreover, since the presence of suboptimal splice signals is not sufficient to impose post-transcriptional splicing, it would appear that uncoupling requires the active participation of Sxl, and may be important for this regulatory mechanism. The presence of incompletely spliced pre-mRNAs in female cells also explains previous biochemical studies, which showed that polyadenylated transcripts containing intron 2 and 3 sequences, but not the other intron sequences, can be readily detected in RNA isolated from a mixed sex population (Samuels et al., 1991). Presumably, these partially spliced pre-mRNAs are derived from female and not male flies.
PTB controlled alternative splicing in nPTB pre-mRNA also occurs post-transcriptionally
Similar to Sxl, PTB also promotes exon skipping events by binding to pyrimidine-rich sequences associated with the alternatively spliced exons in many pre-mRNAs (Boutz et al., 2007; Spellman et al., 2007; Xue et al., 2009). This results in a particularly interesting splicing regulation in its neuronal paralog, nPTB (Boutz et al., 2007; Spellman et al., 2007). In non-neuronal cells, where PTB is abundantly expressed and is localized in the nucleus, PTB binds to exon 10 of nPTB and to surrounding introns and promotes its exclusion from the final mRNA. The resulting mRNA harbors a premature termination codon and is degraded by the non sense mediated decay (NMD) machinery. In neuronal cells, however, where PTB expression is lower, constitutive splicing produces functional nPTB mRNAs containing exon 10 with the normal stop codon (Fig. 6A).
Fig. 6.
nPTB pre-mRNA molecules containing intron 9 are released into the nucleoplasm more frequently from its gene locus than other introns in the same gene. A. Non-neuronal cells express high levels of PTB (blue ovals), which bind to pyrimidine-rich sequences in and around exon 10 and cause the skipping of exon 10, resulting in an mRNA species that contains a premature termination codon and is a substrate of non sense mediated decay. In neuronal cells, there is little PTB, and only constitutive splicing can occur, which produces larger amounts of functional mRNA (Fig. S6.). B. Representative images from HeLa cells showing the locations of pre-mRNAs containing the indicated intron and 3’ terminal exon 13. The panels on the right show the location of each type of molecular species. The average number of molecules per cell are provided in Table 1. C. Kinetics of splicing and degradation of pre-mRNAs containing intron 9 and exon 13 after the addition of actinomycin D to the culture medium. The yellow line is an exponential decay curve fitted to the observed decrease in the pre-mRNA population. The red line is a fitted curve based on modeling of reaction: pre-mRNA → mRNA → Φ.
We examined the splicing of nPTB introns 1, 8, 9, and 11 in pair wise combinations with the 3’ terminal exon 13 in HeLa cells. The results indicate that, while there was only a low frequency of pre-mRNAs containing introns 1 or 11 paired with exon13, there were a large number of pre-mRNAs containing intron 9 and exon 13 dispersed throughout the nucleoplasm (Fig. 6B, Table 1 and Movie S3). The average numbers of intron 8 and exon 13-containing pre-mRNAs were at an intermediate level, perhaps reflecting the facts that this intron is associated with another alternative splicing event in which the 3’ splice site is selected from two different alternatives (Rahman et al., 2002) and that the PTB binds to a region near its 3’ splice site (Xue et al., 2009).
Table 1.
Numbers of nPTB pre-mRNA molecules containing the indicated intron and exon 13 and their splicing products in HeLa cells. The numbers are mean values ± 95% confidence interval obtained from at least 20 cells.
| Intron | pre-mRNA | Spliced mRNA | Spliced Intron |
|---|---|---|---|
| 1 | 0.32±0.20 | 18.18±2.51 | 0.39±0.20 |
| 8 | 4.75±0.75 | 20.13±1.99 | 0.65±0.34 |
| 9 | 9.28±0.88 | 25.78±4.84 | 1.78±0.59 |
| 11 | 2.33±0.48 | 22.98±1.96 | 0.37±0.19 |
To confirm that the dispersed intron 9 pre-mRNAs are processed, we treated the cells with actinomycin D. In short incubations with this drug, we found that there was a decrease in the number of pre-mRNA molecules per cell and an accompanying increase in spliced mRNAs. After longer periods of incubation with actinomycin D, the overall population of all three RNA species declined due to decay (Fig. 6C). We fitted the data to a simple model consisting of a pair of first order sequential reactions, in which pre-mRNA first converts into mRNA, which then decays over time. The results show that these pre-mRNA molecules are converted into mRNA with a half-life of about 24 minutes, whereas, the half-life of the processed mRNA is 230 minutes.
To show that dispersed pre-mRNAs containing intron 9 arise from the regulatory activity of PTB, we decreased the expression of PTB by RNAi. After checking that PTB was effectively knocked down (Fig. S6A), we counted the number of molecules of each RNA species. We found that the number of pre-mRNAs decreased from 9.3 to 3.8 molecules per cell while the number of spliced mRNAs increased from 26 to 111 molecules per cell (Fig. S6B). This confirms that PTB represses the expression of nPTB. In order to show that the repression of nPTB mRNA stems from NMD, we treated the cells with the protein synthesis inhibitor cyclohexamide, which prevents the NMD. This treatment also resulted in an increase in spliced mRNA (to 100 molecules per cell); however, it did not result in a decrease in pre-mRNAs (8.0 molecules per cell). These observations indicate that like Sxl, PTB action is necessary for the release of unspliced pre-mRNA from gene locus.
Discussion
The experiments reported here support the idea that co-transcriptional splicing is the default mechanism. In all of the constitutively spliced introns we examined using single-molecule imaging, splicing was completed prior to transcription termination and transcript release. One likely mechanism for coordinating transcription and splicing is suggested by recent studies in yeast. These studies showed that there are regulated pauses in polymerase elongation at each 3’ splice site and in the 3’ most exon, which function to ensure that splicing is completed before the transcript is released (Alexander et al., 2010; Carrillo Oesterreich et al., 2010). Equivalent checkpoints that prevent the release of unspliced transcripts are also likely to exist in higher eukaryotes (Custodio et al., 1999; Rigo and Martinson, 2009), and would account for the co-transcriptional splicing of constitutively spliced introns that we observed.
However, there are circumstances in which it is possible to overcome or escape whatever checkpoints exist to ensure that transcripts are fully processed before RNA polymerase terminates transcription and releases the transcript. One of these is in artificial and naturally occurring introns that have functionally impaired splice signals. For example, interference with polypyrimidine tract recognition in our reporter transcripts delayed splicing until after transcription is finished, and high levels of unprocessed pre-mRNAs accumulated in the nucleoplasm. In these transcripts, the binding of U1 snRNP to the 5’ splice site will be normal, but the binding of U2AF, U2 snRNP, and other factors to the 3’ splice site will be rate limiting and may not occur co-transcriptionally. In this situation, the signals that normally trigger pausing might not be properly activated, and instead of pausing, the polymerase would transcribe through the termination signals and release the incompletely processed transcript. Once a functional complex is assembled on the defective 3’ splice site, the remaining processing steps should proceed unimpeded. In these instances, functionally compromised splicing signals are, by themselves, sufficient to uncouple splicing from transcription.
The other circumstances in which splicing is uncoupled from transcription occur during the alternative splicing of Sxl and nPTB pre-mRNAs. However, the uncoupling seen in these regulated events is different from that observed when the 3’ splice site is functionally compromised. When the alternatively spliced Sxl cassette is spliced in the default pattern, as it occurs in male flies, splicing is co–transcriptional, just like the constitutively spliced introns in the same transcript. Thus, even though the splicing signals in the regulated Sxl cassette are suboptimal, this is not sufficient to uncouple default splicing and transcription in males. A plausible explanation is that these suboptimal sites differ from the functionally compromised signals in that they are capable of directing the association of the needed splicing factors on at least a subset of the regulated splice sites while the mRNA is being actively transcribed. This allows them to signal to the polymerase to pause until the default splicing of the regulated cassette is complete. On the other hand, in female flies, Sxl bound at the regulated cassette is somehow able to disrupt this signaling and cause the release of the partially processed transcript.
How does splicing become uncoupled from transcription during regulated splicing? In the case of Sxl, the female splicing pattern is absolutely dependent upon Sxl protein, thus Sxl must be responsible, either directly or indirectly. Since Sxl binds to its own pre-mRNAs co-transcriptionally (Samuels et al., 1994), it could potentially block the association of U2 and U1 snRNPs with the male exon 3’ and 5’ splice sites. However, this model for uncoupling seems unlikely, since the key Sxl target sites are located more than 200 nucleotides from the male exon splice sites. Therefore, Sxl probably can’t prevent U2 and U1 snRNPs from interacting with these splice sites (Horabin and Schedl, 1993; Sakamoto et al., 1992). Moreover, even if Sxl could block these sites, functional U1 and U2 complexes should still be able to assemble at the flanking female-specific splice sites (the exon 2 5’ site and the exon 4 3’ site) (see figure 5A) by the time the polymerase has reached the end of the Sxl gene. Thus, Sxl would have to act on transcripts that have, at the minimum, U1/U2 spliceosomal complexes associated with the incompletely spliced cassette. While it remains to be determined how Sxl promotes the uncoupling of splicing and transcription, the fact that an analogous switch also occurs in the case of PTB, suggests that the mechanism may be prevalent.
A second question is whether uncoupling is important for changing the splicing pattern. In the case of splicing regulators, such as Sxl and PTB, which use a blockage mechanism, the rapid release of transcripts from the gene locus where spliceosomes are being assembled de novo would help them prevent functional spliceosomal complexes from forming on splice sites that should not have such complexes. In fact, the longer the transcript release is delayed, the greater the chance that components of the default splicing machinery could circumvent the regulatory effects of these factors. Further support for the idea that uncoupling plays an important role in alternative splicing comes from recent genetic studies, which indicate that a translation factor, eIF4E (the cap binding protein), functions as a co-factor in Sxl regulation of Sxl and male specific lethal-2 pre-mRNA splicing in females (Graham et al., 2011). eIF4E is normally thought to associate with mRNAs only when they are ready for export; however, these studies showed that in nuclear extracts eIF4E is bound to Sxl pre-mRNAs that haven’t yet spliced the regulated cassette and is present in complexes with Sxl, a Sxl co-factor, Fl(2)d, U2AF and components of the U1 and U2 snRNPs.
One of the key principles that emerge from this study is that when transcription and splicing are uncoupled, uncoupling is restricted to the affected intron and the preceding and succeeding introns continue to be removed co-transcriptionally. We found that no matter how introns containing array 1 and array 2 sequences are arranged within our GFP splicing reporter, splicing of the array 1 intron is co-transcriptional while splicing of the array 2 intron remains post-transcriptional. Likewise, the processing of intron 1 and the regulated cassette in Sxl pre-mRNA and the processing of the three constitutive introns and the regulated cassette in nPTB pre-mRNA were independent of each other. This indicates that spliceosomes assemble at each intron independently of the surrounding introns, and they catalyze the splicing reaction of each intron with its own unique kinetics.
Does post-transcriptional splicing have other biologically useful roles? A number of alternative splicing events occur in response to external stimuli, such as electric stimuli in neurons and signal transduction cascades in other cells (Nilsen and Graveley, 2010). A pool of unspliced mRNAs in the nucleus could provide a mechanism for readily producing the needed isoforms upon receipt of the stimuli. Furthermore, cells may be able to exert additional controls on splicing by coupling it with downstream processes such as RNA export (Han et al., 2011).
While this study demonstrates that splicing and transcription are uncoupled in two canonical cases of regulated alternative splicing, many questions remain. Do Sxl and PTB act directly on the transcriptional machinery to cause the uncoupling or is the uncoupling simply a consequence of interfering with the functioning of splice sites in the regulated cassettes? Is the uncoupling of splicing and transcription a general feature of splicing regulators that function by a blockage mechanism, or is it true for only a special subset? Do splicing factors that activate rather than block splice sites have the opposite effect on coupling, promoting a delay in transcript release until splicing is complete? Our results suggest that constitutively spliced introns are processed co-transcriptionally; however, we only examined a limited sample and it is possible that there is a group of constitutively spliced introns that are processed only after transcription termination. If so, what distinguishes these introns from constitutive introns that are processed co-transcriptionally?
Although this study clearly demonstrates several cases of post-transcriptional splicing, given the diversity of mechanisms regulating alternative splicing (Nilsen and Graveley, 2010), there are likely to be instances in which splicing is not only uncoupled from transcription, but also where this uncoupling is important for facilitating the regulatory activities of tissue, developmental, or sex-specific factors that orchestrate alternative splicing. Genome-wide studies are needed to uncover such examples, and indeed, to understand the full extent of the post-transcriptional splicing.
Experimental Procedures
Probes and in situ hybridization
We achieve single-molecule sensitivity by either having tandemly repeated sequences in the target to which a single probe binds multiple times (as for constructs with arrays), or by targeting unique sequences in the RNA with multiple probes (for all natural genes). Array repeats were detected using probes that either had a single fluorophore (molecular beacons) or had five fluorophores that were attached to their thymidine residues. The natural unrepeated targets were detected with sets of about 50 different oligonucleotides, each about 20 nucleotides long, and each labeled with a single fluorescent moiety. Thus, depending upon the target and the probe, 32 to 384 fluorophore moieties were bound to each target molecule, which creates sufficient fluorescence for each molecule to become visible as a diffraction-limited spot in a fluorescence microscope. The length and number of probes required necessitates that the target be at least 800 to 1000 nunleotides long. The sequences of the probes that we used are compiled in a supplementary spread sheet (Table S2).
For in situ hybridization, cells were attached to thin coverslips which were fixed with 4% formaldehyde, permeabilized with 70% alcohol and hybridized overnight with the probe sets in 2X SSC supplemented with 10% formamide. The coverslips were washed and mounted in a special deoxygenated medium that limits photobleaching, and then imaged in a wide-field microscope.
Image analysis
For each image, 10 to 30 optical slices, with 0.2 µm separation between them were acquired in each fluorescence channel with 1 second exposure. These stacks were analyzed using custom computer programs written in the Matlab programming environment. These programs enhance the stack of images using a Laplacian filter optimized for the size of spots that we expect, permit users to select a threshold based on a 3-dimentional display of intensity in spots, segment the image based on the provided threshold, and produce a list of coordinates of the centers of all spots in 3-dimentions in each channel. The programs can also determine the distances between spots in two or three fluorescence channels, identify co-localized spots based on provided distance limits, draw circles to produce overlays on the raw images, and count the number of spots in a user-defined region. The scripts of the programs are available upon request.
The spot-detection algorithm works well for spots that do not overlap with each other. However, in some regions of a cell, such as at the gene locus, where RNA molecules are crowded together, the algorithm performs poorly. For this reason, when we observed an RNA cluster at the gene locus, we restricted the analysis to regions that exclude the cluster. The accuracy of detection of co-localized spots is demonstrated in Movie S3.
Supplementary Material
Acknowledgements
We thank Ben Gold and Fred Russell Kramer for their contributions. This work was supported by National Institutes of Health grants MH 079197 (ST) and GM 043432 (PS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
The details of experimental procedures, including a review of the evidence for single-molecule sensitivity, the sequences of repeats and their probes, structures of constructs, cloning procedures, cell culture conditions, and a description of the imaging set up are provided in the supplementary information.
References
- Alexander RD, Innocente SA, Barrass JD, Beggs JD. Splicing-dependent RNA polymerase pausing in yeast. Mol Cell. 2010;40:582–593. doi: 10.1016/j.molcel.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batsche E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol. 2006;13:22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- Bauren G, Wieslander L. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994;76:183–192. doi: 10.1016/0092-8674(94)90182-1. [DOI] [PubMed] [Google Scholar]
- Bell LR, Maine EM, Schedl P, Cline TW. Sex-lethal, a Drosophila sex determination switch gene, exhibits sex-specific RNA splicing and sequence similarity to RNA binding proteins. Cell. 1988;55:1037–1046. doi: 10.1016/0092-8674(88)90248-6. [DOI] [PubMed] [Google Scholar]
- Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M, Jr, Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratu DP, Cha BJ, Mhlanga MM, Kramer FR, Tyagi S. Visualizing the distribution and transport of mRNAs in living cells. Proc Natl Acad Sci U S A. 2003;100:13308–13313. doi: 10.1073/pnas.2233244100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo Oesterreich F, Preibisch S, Neugebauer KM. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol Cell. 2010;40:571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custodio N, Carmo-Fonseca M, Geraghty F, Pereira HS, Grosveld F, Antoniou M. Inefficient processing impairs release of RNA from the site of transcription. Embo J. 1999;18:2855–2866. doi: 10.1093/emboj/18.10.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Dias AP, Dufu K, Lei H, Reed R. A role for TREX components in the release of spliced mRNA from nuclear speckle domains. Nat Commun. 2010;1:97. doi: 10.1038/ncomms1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre M, Sanchez G, De Cian MC, Barbier J, Dardenne E, Gratadou L, Dujardin G, Le Jossic-Corcos C, Corcos L, Auboeuf D. Cotranscriptional exon skipping in the genotoxic stress response. Nat Struct Mol Biol. 2010;17:1358–1366. doi: 10.1038/nsmb.1912. [DOI] [PubMed] [Google Scholar]
- Fong N, Bentley DL. Capping, splicing, and 3' processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev. 2001;15:1783–1795. doi: 10.1101/gad.889101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Graham PL, Yanowitz JL, Penn JKM, Deshpande G, Schedl P. The Translation Initiation Factor eIF4E Regulates the Sex-Specific Expression of the Master Switch Gene Sxl in Drosophila melanogaster. PLoS Genetics. 2011;7:e1002185. doi: 10.1371/journal.pgen.1002185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Xiong J, Wang D, Fu XD. Pre-mRNA splicing: where and when in the nucleus. Trends Cell Biol. 2011;21:336–343. doi: 10.1016/j.tcb.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horabin JI, Schedl P. Sex-lethal autoregulation requires multiple cis-acting elements upstream and downstream of the male exon and appears to depend largely on controlling the use of the male exon 5' splice site. Mol Cell Biol. 1993;13:7734–7746. doi: 10.1128/mcb.13.12.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Deerinck TJ, Ellisman MH, Spector DL. In vivo analysis of the stability and transport of nuclear poly(A)+ RNA. J Cell Biol. 1994;126:877–899. doi: 10.1083/jcb.126.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Listerman I, Sapra AK, Neugebauer KM. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol. 2006;13:815–822. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Neugebauer KM. On the importance of being co-transcriptional. J Cell Sci. 2002;115:3865–3871. doi: 10.1242/jcs.00073. [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterreich FC, Bieberstein N, Neugebauer KM. Pause locally, splice globally. Trends Cell Biol. 2011;21:328–335. doi: 10.1016/j.tcb.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Pandya-Jones A, Black DL. Co-transcriptional splicing of constitutive and alternative exons. Rna. 2009;15:1896–1908. doi: 10.1261/rna.1714509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penalva LO, Lallena MJ, Valcarcel J. Switch in 3' splice site recognition between exon definition and splicing catalysis is important for sex-lethal autoregulation. Mol Cell Biol. 2001;21:1986–1996. doi: 10.1128/MCB.21.6.1986-1996.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales R, Bentley D. "Cotranscriptionality": the transcription elongation complex as a nexus for nuclear transactions. Mol Cell. 2009;36:178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- Rahman L, Bliskovski V, Reinhold W, Zajac-Kaye M. Alternative splicing of brain-specific PTB defines a tissue-specific isoform pattern that predicts distinct functional roles. Genomics. 2002;80:245–249. doi: 10.1006/geno.2002.6826. [DOI] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigo F, Martinson HG. Polyadenylation releases mRNA from RNA polymerase II in a process that is licensed by splicing. RNA. 2009;15:823–836. doi: 10.1261/rna.1409209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto H, Inoue K, Higuchi I, Ono Y, Shimura Y. Control of Drosophila Sex-lethal pre-mRNA splicing by its own female-specific product. Nucleic Acids Res. 1992;20:5533–5540. doi: 10.1093/nar/20.21.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels ME, Bopp D, Colvin RA, Roscigno RF, Garcia-Blanco MA, Schedl P. RNA binding by Sxl proteins in vitro and in vivo. Mol Cell Biol. 1994;14:4975–4990. doi: 10.1128/mcb.14.7.4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels ME, Schedl P, Cline TW. The complex set of late transcripts from the Drosophila sex determination gene sex-lethal encodes multiple related polypeptides. Mol Cell Biol. 1991;11:3584–3602. doi: 10.1128/mcb.11.7.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Padgett RA. Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol. 2009;16:1128–1133. doi: 10.1038/nsmb.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman R, Llorian M, Smith CW. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol Cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas DY, Raj A, Marras SA, Kramer FR, Tyagi S. Mechanism of mRNA transport in the nucleus. Proc Natl Acad Sci U S A. 2005;102:17008–17013. doi: 10.1073/pnas.0505580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waks Z, Klein AM, Silver PA. Cell-to-cell variability of alternative RNA splicing. Mol Syst Biol. 2011;7:506. doi: 10.1038/msb.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuryev A, Patturajan M, Litingtung Y, Joshi RV, Gentile C, Gebara M, Corden JL. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci U S A. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Taneja KL, Singer RH, Green MR. Localization of pre-mRNA splicing in mammalian nuclei. Nature. 1994;372:809–812. doi: 10.1038/372809a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.