Abstract
The voltage-gated proton channel (Hv1) is homologous to the voltage-sensing domain (VSD) of voltage-gated potassium (Kv) channels but lacks a separate pore domain. The Hv1 monomer has dual functions: it gates the proton current and also serves as the proton conduction pathway. To gain insight into the structure and dynamics of the yet unresolved proton permeation pathway, we performed all-atom molecular dynamics simulations of two different Hv1 homology models in a lipid bilayer in excess water. The structure of the Kv1.2-Kv2.1 paddle-chimera VSD was used as template to generate both models, but they differ in the sequence alignment of the S4 segment. In both models, we observe a water wire that extends through the membrane, whereas the corresponding region is dry in simulations of the Kv1.2-Kv2.1 paddle-chimera. We find that the kinetic stability of the water wire is dependent upon the identity and location of the residues lining the permeation pathway, in particular, the S4 arginines. A measurement of water transport kinetics indicates that the water wire is a relatively static feature of the permeation pathway. Taken together, our results suggest that proton conduction in Hv1 may occur via Grotthuss hopping along a robust water wire, with exchange of water molecules between inner and outer ends of the permeation pathway minimized by specific water-protein interactions.
Keywords: Voltage-gated ion channels, Voltage-sensing domains, Membrane Proteins, Molecular Dynamics Simulations
1. Introduction
The Hv1 channel [1, 2] is found in a wide range of tissues and has been shown to serve a variety of important physiological functions. In neutrophils Hv1 activity is required for optimal reactive oxygen species (ROS) production by the NADPH oxidase during the oxidative burst [3–5]. In B lymphocytes, the activation of the Hv1 channel enhances the B-cell antigen receptor (BCR) mediated intracellular signaling cascade that leads to B-cell proliferation and differentiation [6]. In basophils, Hv1 has been proposed to play an important role in the stimulation of histamine release [7]. In mature human spermatozoa, Hv1 acts as a flagellar regulator of intracellular pH, promoting sperm capacitation [8]. In the respiratory system, Hv1 is involved in the pH homeostasis of the airway surface liquid, by contributing to acid extrusion from the airway epithelium [9].
In the plasma membrane, Hv1 is found as a homodimer that is stabilized by intracellular coiled-coil interactions in the C terminus [10]. Truncations in the C-terminus result in the formation of functional monomeric channels that have their own permeation pathway and voltage sensor [11, 12]. The Hv1 channel is made of four transmembrane helices (S1-S4), which are homologous to the voltage-sensing domain (VSD) of voltage-gated potassium (Kv) channels. Voltage sensitivity in the VSD is conferred by a series of highly conserved positions for charged residues. In particular, the S4 segment contains several triplet repeats composed of a basic side chain (mostly arginine) followed by two hydrophobic residues. In the case of Shaker, it has been shown that the first four arginines in S4 (termed R1 through R4) contribute most of the gating charge during activation [13–15]. Hv1 exhibits three of these repeats, and mutations of the S4 arginines have been shown to alter its activation kinetics [1, 2]. In addition, voltage-clamp fluorometry and state-dependent accessibility measurements in Hv1 channels report motion of S4 during activation [16, 17], as previously observed in Kv channels [18–21]. A helical hairpin made of portions of the S3 and S4 helices, called the paddle motif, can be transplanted from a variety of proteins, including Hv1, into a Kv channel without loss of function [22]. These findings establish a structural connection between Hv1 and nonconducting VSDs. However, the molecular mechanisms behind the specific functional features of Hv1, namely, proton permeation and selectivity, and regulation by pH, remain unknown. In addition, functional predictions at the molecular level are hindered by the lack of a high-resolution structure.
The ability to conduct ions through the VSD is not a unique feature of the Hv1 channel, since mutated VSDs of Kv channels [23–27] and voltage-gated sodium (Nav) channels [28–30] can also support ion permeation. The current view of cation permeation through these VSDs is that specific mutations of side chains lining the VSD interior remove an occlusion between the hydrated intracellular and extracellular crevices of the VSD structure [31], allowing ion flux. Most conducting Kv and Nav VSD variants involve the mutation of at least one of the basic side chains in S4 [23–26, 28], but proton conduction has also been reported in mutants of residues in S1 and S2 [27].
Only one Kv VSD mutant, Shaker R371H, has been reported to generate proton currents under a depolarizing potential, as in Hv1 [23]. R371 in Shaker corresponds to the fourth S4 arginine, which suggests a possible alignment of R1 through R3 in Hv1 to R1 through R3 in Shaker, leaving N214 in Hv1 at the R4 position in Kv channels. The N214R mutation has been reported to abolish the outward proton current in the human Hv1 [11, 32], while the similar mutation in the mouse channel (N210R) yields very low density currents and slow activation kinetics [33]. In addition, Hv1 proton currents have been shown to be reversibly blocked by intracellular guanidinium ions, while trimethylaminoethyl-methanethiosulfonate (MTSET) has been shown to block the current carried by the N214C mutant of the human Hv1 [11] or by the equivalent mutant of the Ciona intestinalis ortholog [16]. These results suggest that permeation through Hv1 may occur in a similar manner as in conducting VSD mutants of Kv and Nav channels.
An alternative view of the structural relationship between Kv VSDs and Hv1 could be derived from considerations of sequence conservation. Although most Kv channel families exhibit the same R1 through R4 charge distribution in the sequence of the S4 segment as in Shaker, conservation of the R1 position for a basic side chain is somewhat lower than for R2-R4 [34]. Thus, while functional data in Hv1 and Shaker suggest an alignment of the three S4 arginines in Hv1 to R1-R3 in Kv channels, multiple sequence alignment of Hv channels to the VSDs in the superfamily of voltage-gated ion channels could lead to an alignment of the three S4 arginines in Hv1 to positions R2-R4 in Kv channels [1].
Here, we use atomistic molecular dynamics (MD) simulations of two different homology models of Hv1 in a lipid bilayer in excess water to probe the structural and dynamical features of a putative proton permeation pathway. Both homology models were generated using the Kv1.2 paddle-chimera structure [35] as a template, but they were aligned differently in the region of the S4 segment, as described above. The simulations reveal a water wire connecting the intracellular and extracellular sides, suggesting proton conduction via a Grotthuss hopping mechanism. The kinetic stability of the water wire is dependent upon the interactions with a set of highly conserved side chains lining the permeation pathway. We find that persistent interactions with two of the S4 arginines, as part of a larger cluster of polar residues, as well as the formation of a well defined hydrophobic gap in the center of the protein are key to the formation of a robust water wire. In addition, a measurement of water transport kinetics indicates that the present model of Hv1 cannot be characterized as a water channel.
2. Methods
2.1 Molecular Dynamics Simulations
Using the crystal structure the Kv1.2-Kv2.1 paddle-chimera VSD [38] as a template, we constructed two different atomistic models for Hv1. In the first model, R1-Hv1, the three arginines in the S4 segment of Hv1 were aligned to positions R1 through R3 in the S4 segment of the paddle chimera (see Fig. 1). In the R2-Hv1 model, the Hv1 arginines were shifted three residues over to align with positions R2, R3 and R4 of the paddle chimera. These two sequence alignments (see Fig. 1A) were then used as input for MODELLER 9.8 [39] to build the initial three-dimensional configurations (shown in Figs. 1C and 1D).
Figure 1.
Hv1 homology modeling. A) Two sequence alignments of the human Hv1 transmembrane region to the rat Kv1.2-Kv2.1 paddle-chimera VSD (PadChVSD). In the Kv1.2-Kv2.1 paddle-chimera VSD structure (shown in B) the S3b-S4 paddle motif has been replaced by the one from the rat Kv2.1 channel [38]. The R1-Hv1 model (shown in C) was generated by matching all the highly-conserved positions in the four transmembrane segments that have been implicated in Kv VSD function (highlighted in grey and orange in A). The R2-Hv2 model (shown in D) differs from R1-Hv1 only in the alignment of the S4 segment. In R1-Hv1, the three Hv1 arginines were aligned to positions R1, R2, and R3 in the Kv1.2-Kv2.1 paddle-chimera, while in R2-Hv1 they were aligned to positions R2, R3, and R4. Notice that positions R1 through R4 (highlighted in orange in A) correspond to arginines in most Kv channel families, including Kv1. The transmembrane segments in the Kv1.2-Kv2.1 paddle-chimera VSD are indicated by the horizontal lines above the sequence. The S4 segment is shown in secondary structure representation in panels B, C and D (black, blue and red, respectively). Residues R1 through R4 in the paddle- chimera structure, as well as the three S4 arginines in Hv1, are shown in licorice representation in panels B, C and D.
Each of the resulting configurations were embedded in a POPC bilayer in excess water. Each system included one Hv1 monomer (residues 91 to 223), 282 lipids, 10,652 water molecules, and sufficient Cl− counterions to make the system charge neutral, for a total of 72,055 atoms. The initial bilayer configuration for each system was such that the water wires that are reported in this work were not present. We also performed a simulation of an N214R Hv1 mutant based on the R1-Hv1 configuration in an otherwise identical system. The initial configuration for the N214R simulation was taken from a snapshot of R1-Hv1 after 130 ns of simulation. Finally, we performed a simulation of the Kv1.2-Kv2.1 paddle-chimera VSD (residues 145 to 320) under similar conditions (282 lipids, 13,993 water molecules, 82,610 total atoms). The initial configuration of that simulation was taken from an equilibrated snapshot of the full Kv1.2-Kv2.1 paddle-chimera tetramer, such that the S4 arginines were already hydrated. To initialize each of the simulations, we first ran 2000 steps of conjugate-gradient energy minimization followed by 100 ps of simulation with the protein backbone atoms fixed at constant temperature (300 K) and volume. We then switched to constant pressure (1 atm) and gradually released the backbone in 100-ps steps, using 50, 20, 10, 5, 2, and 1 kcal mol−1 Å−2 restraints. The total unrestrained simulation lengths were roughly 200 ns for the R1- and R2-Hv1 simulations, 135 ns for the N214R R1-Hv1 simulation, and 275 ns for the Kv paddle chimera VSD simulation.
All simulations were performed with the NAMD 2.7b2 software package [40]. The CHARMM22 and CHARMM32 force fields [41, 42] were used for protein and lipids, respectively, and the TIP3P model was used for water [43]. The smooth particle mesh Ewald method [44, 45] was used to calculate electrostatic interactions. Short-range real-space interactions were cut off at 11 Å, employing a switching function. A reversible multiple time-step algorithm [46] was employed to integrate the equations of motion with a time step of 4 fs for electrostatic forces, 2 fs for short-range non-bonded forces, and 1 fs for bonded forces. All bond lengths involving hydrogen atoms were held fixed using the SHAKE [47] and SETTLE [48] algorithms. A Langevin dynamics scheme was used for temperature control and a Nosé-Hoover-Langevin piston was used for pressure control [49, 50]. Molecular graphics and trajectory analyses were performed using VMD 1.8.7 [51] over the last 120 ns of each trajectory.
2.2 Water dynamics analyses
We define a transmembrane water wire as any configuration of H-bonded waters in the channel connecting the extracellular side to the intracellular side. A H-bond was defined using conventional geometric criteria (donor-acceptor distance < 3.5 Å and donor-hydrogen-acceptor angle > 140 Å). Water wire formation along each simulation trajectory was modeled as a binary process, wire/no-wire, irrespective of the actual number of H-bonded configurations or the identity of the water molecules. The reported correlation times correspond to single-exponential fits to the binary process autocorrelation function and to the water wire survival function. The latter is defined as the time correlation function computed only over the configurations showing a wire
where H(t) = 1 if there is a water wire at time t = 0 and at time t, and H(t) = 0 otherwise.
The residence time for water molecules in the solvation shell of a given amino acid residue lining the pore of the R1-Hv1 channel were calculated as the time corresponding to the 1/e value of the water survival function in the solvation shell defined as
where h(t) = 1 if a given water molecule present in the residue coordination shell at t = 0 is also present at time t, and the average is over all waters.
The osmotic permeability coefficient for waters in the R1-Hv1 pathway was calculated using the method of Zhu et al. [52]. A collective coordinate for waters in the interior of the channel was calculated as the integral of
where the zi is the position of the ith water along the transmembrane direction over two consecutive configurations and L(t) is the instantaneous length of the stretch between D174 on the intracellular side and R208 on the extracellular side. The osmotic permeability coefficient (pf) is given by
where vw is the molecular volume of water and Dn is given by
3. Results
We performed two 200 ns all-atom MD simulations of two different structural models of the transmembrane region of Hv1 from Homo sapiens embedded in a POPC lipid bilayer in excess water. Both models where generated using the crystal structure of the Kv1.2-Kv2.1 paddle-chimera VSD as a template [38]. In the first model (hereafter termed R1-Hv1), the three arginines in the S4 segment of Hv1 were aligned to positions of the first three arginines (R1, R2, R3) in the S4 segment of the Kv1.2 VSD. In the second model (hereafter termed R2-Hv1), the Hv1 S4 arginines were shifted three residues over to align with R2, R3 and R4 of the Kv1.2 VSD. The remaining three transmembrane segments (S1 through S3) were aligned by matching all the highly-conserved positions that have been implicated in Kv VSD function to the Hv1 sequence (see Fig. 1).
3.1 Kinetic stability of water wires through Hv1
Both R1-Hv1 and R2-Hv1 develop an open pathway through the center of the channel that is readily occupied by water molecules (see Fig. 2). To characterize the kinetics of water wire formation along the permeation pathway, we consider the occurrence of a H-bonded water chain connecting the water-filled extracellular and intracellular crevices in the VSD structure, along the simulation trajectory, as a binary process (i.e., irrespective of identity of the water molecules involved). Block-averaged versions of the corresponding time series are shown in Fig. 2, and the corresponding correlation times for the binary process, as well as those of the water wire survival function (see Methods for details), are shown in Table 1. This time series analysis reveals that only R1-Hv1 is able to maintain a robust H-bonded water wire throughout the membrane. The differences in kinetic stability between R1-Hv1 and R2-Hv1 can be traced back to the configurations assumed by the water molecules occupying the permeation pathway (see Fig. 2).
Figure 2.
Transmembrane water wires in Hv1. A water wire is defined as any instantaneous configuration of H-bonded water molecules connecting the VSD intracellular and extracellular crevices. The frequency of water wire formation in 500-ps trajectory blocks as a function of time is shown in the left panels for R1-Hv1 (A), R2-Hv1 (B), and the N214R variant of R1-Hv1 (C). The red lines in the R1-Hv1 and R2-Hv1 frequency plots corresponds to the frequency of finding a water wire in uncorrelated blocks of 8 ns for R1-Hv1 and 4 ns for R2-Hv1, dashed lines are 95% confidence intervals. The central and right panels show configuration snapshots for the water molecules inside the VSD. The R1-Hv1 model (A) supports a robust water wire through a persistent cluster of water molecules located at the center of the permeation pathway. In the R2-Hv1 model (B), the frequency of water wire formation often goes to zero along the trajectory. The water wire typically breaks at the end of the intracellular crevice. The R1-Hv1 N214R variant (C) exhibits very little connectivity along the simulation trajectory, after a period of equilibration from the native configuration (first ~30 ns). The VSD internal waters assume a configuration similar to those observed in simulations of Kv VSDs [53–57]. Water molecules are colored by atom (red, oxygen; white, hydrogen). In the central panel, water molecules inside the VSD are shown in ball-and-stick representation, and those outside the VSD are shown as filled-spheres.
Table 1.
Water wire correlation times.
Correlation time from the autocovariance function binary time series (wire/no-wire).
Correlation time from the survival function of configurations showing a wire.
To verify that the water H bond connectivity through the VSD is a unique feature of the Hv1 channel sequence and not an artifact of the homology modeling, we performed a simulation of the R1-Hv1 N214R mutant from an end configuration of the R1-Hv1 trajectory. Consistent with the available experimental evidence [11, 32, 33], we find that formation of a water wire in N214R, after a period of initial equilibration, is sporadic in the time scale of the simulation trajectory (Fig. 2C). In addition, the configuration of the water molecules in the interior of the VSD resembles that of nonconducting VSDs [53–57] (Fig. 2C).
3.2 Water distribution and protein-water interactions along the permeation pathway
The water distribution along the transmembrane direction for the paddle-chimera VSD embedded in a lipid bilayer shows limited water penetration into the protein extracellular and intracellular crevices, and a dip to zero in the center of the membrane at the location of a network of internal salt-bridges between highly conserved acidic and basic side chains that is a hallmark of the VSD structure in nonconducting voltage-gated channels (Fig. 3) [15, 38, 58–60]. In contrast the presence of a permeation pathway in the Hv1 models is immediately evident in their water number density profiles (Fig. 3). Both models of Hv1 show a nonzero water density profile throughout the membrane, but they also reveal unique features around the protein core that, as suggested before, are presumably related to the differences in water wire kinetic stability between the models.
Figure 3.
Number density profiles for water in the Kv1.2-Kv2.1 paddle-chimera VSD (black), R1-Hv1 (blue) and R2-Hv1 (red). Both Hv1 models exhibit a fully hydrated permeation pathway. In contrast, in the Kv1.2-Kv2.1 paddle-chimera VSD, a central dry region separates the hydrated intracellular and extracellular crevices. R1-Hv1 exhibits a constriction region that starts roughly at the protein center and extends about 5 Å toward the extracelluar side (solid line). R2-Hv1 shows a non-uniform stretch of about 10 Å that extends from the protein intracellular crevice to the extracellular crevice (broken line).
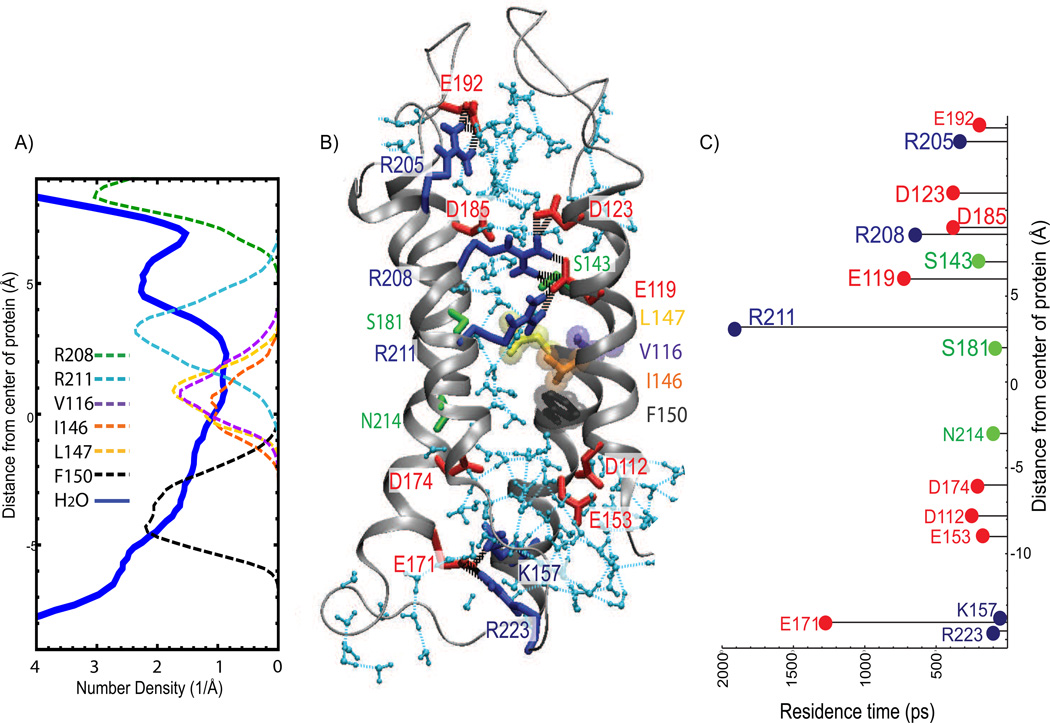
R1-Hv1 exhibits a constriction region that starts roughly at the protein center; it is bounded by F150 on the intracellular side, and extends about 5 Å along the transmembrane direction toward the extracellular side (Figs. 3 and 4). The constriction is formed by a cluster of hydrophobic side chains (V116, I146 and L147), at the center of protein close to the intracellular side, and by R211 (R3 in S4), which occupies the extracellular side of the constriction (Fig. 4). This particular distribution of side chains may account for the kinetic stability of the water wire in R1-Hv1. The cluster of hydrophobic side chains, together with F150 on the intracellular crevice, reduces the water H-bond connectivity to one or two single files, thereby limiting the water exchange between the intracellular and extracellular crevices. On the other side of the constriction, R211 forms part of a dynamic H-bonded network of polar side chains that occupies the extracellular crevice. This polar network features a salt-bridge chain between R211, E119, R208, and D123 that is coordinated by S181 and S143. Hydration of the network results in water molecules with residence times in the time scale of 100–1000 ps (see Fig. 4).
Figure 4.
R1-Hv1 permeation pathway structure and dynamics. (A) Number density profile of water oxygen (thick line) and heavy atoms of selected side chains (dashed lines). (B) Snapshot of R1-Hv1 model. Waters in the interior of the channel are colored light blue. Amino acid residues are shown in licorice representation. Basic residues are colored blue, acidic residues are in red, and polar residues are colored green. V116, I146, L147, and F150 are shown in purple, orange, yellow and dark grey, respectively. Water-water (light blue dashed lines) and protein-protein (black dashed lines) hydrogen bonds are shown; hydrogen bonds between waters and charged residues are omitted for clarity. (C) Residence times of water for selected residues along the water wire. The constriction region in the R1-Hv1 permeation pathway is formed by a cluster of hydrophobic residues (V116, I146 and L147) located, roughly, at the center of the protein, and by R211 in the extracellular cavity. F150 is the closest side chain to the constriction on the intracellular side. N214 is located approximately at the same position as F150 along the transmembrane direction but is away from the constriction. H-bonded networks of waters and polar side chains including S181, S143, and salt-bridge chains between R211, E119, R208 and D123 occupy the extracellular cavity. The elevated water residence times in the H-bonded network, especially around R208, E119, and R211, suggest that those residues have a role in stabilizing the water wire.
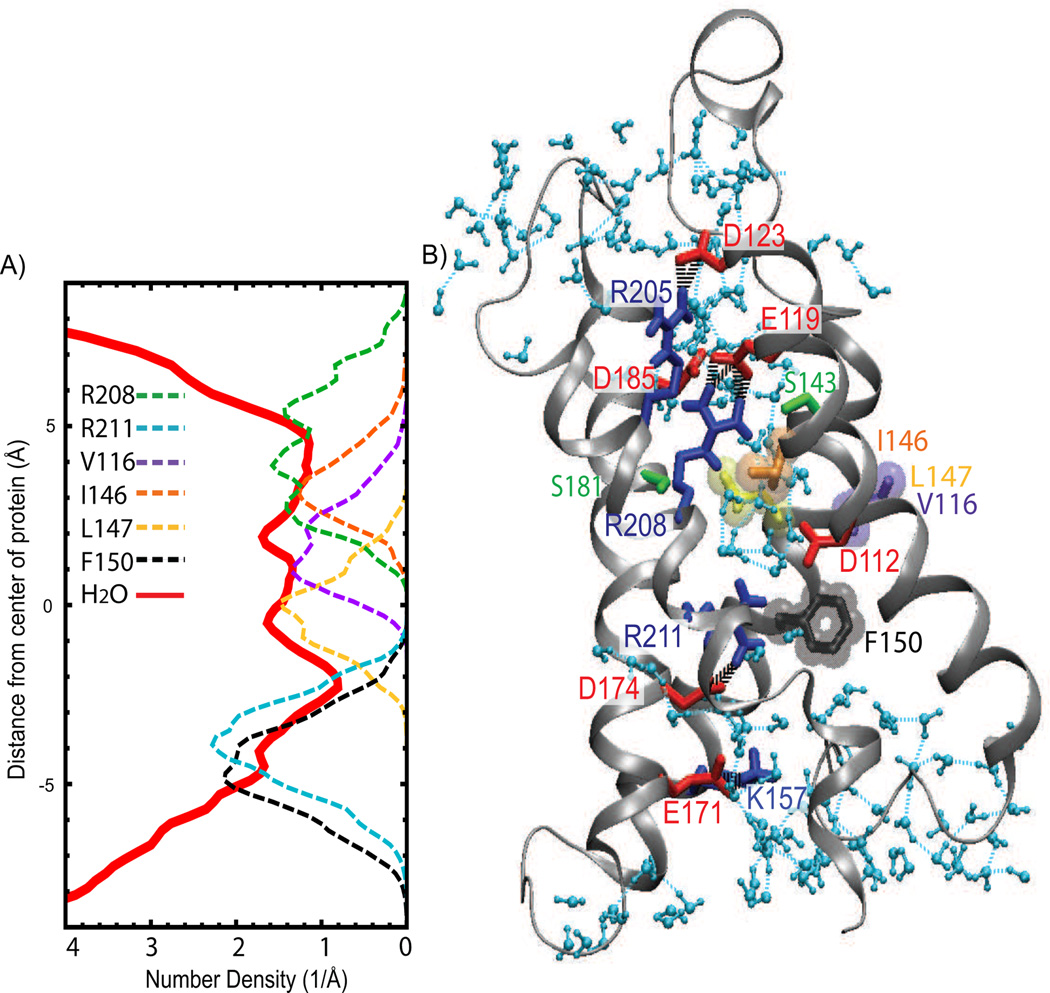
In R2-Hv1, the region of low water density along the transmembrane direction is not a single constriction as in R1-Hv1, but a non-uniform stretch of about 10 Å that extends from the protein intracellular crevice to the extracellular crevice (Figs. 3 and 5). The region of lowest density is located on the intracellular crevice and corresponds to a localized constriction formed by the R211-D112 salt-bridge and by F150. The three side chains are roughly at the same position along the transmembrane direction and reduce the H-bond water connectivity to a single file. This crowded arrangement frequently breaks the water wire. R211 is only partially solvated by waters since it also forms a salt-bridge with D174, which is away from the constriction. In contrast to R1-Hv1, there is no evidence of a central hydrophobic cluster or an extracellular polar cluster in R2-Hv1. The V116, I146 and L147 side chains are spread along the pathway constriction. Similar to R211 on the intracellular side, R208 occupies the water density minimum on the extracellular side where interacts with E119. However, a salt-bridge chain involving R211 and R208 with D112 and D174 is broken for most of the trajectory, with R208 separated from D112 by one or two waters. This arrangement may not be conducive to the longer water residence times in the extracellular crevice observed in R1-Hv1.
Figure 5.
R2-Hv1 permeation pathway structure. A) Number density profile of water oxygen (thick line) and heavy atoms of selected side chains (dashed lines). (B) Snapshot of R2-Hv1 model. Coloring scheme and H-bond representations are the same as Figure 4. R2-Hv1 exhibits an extended region of low water density between R211 on the intracellular side and R208 on the extracellular side, and there is not a central cluster of hydrophobic residues as in R1-Hv1. The typical breaking point of the water wire is located at the most constricted site along the permeation pathway formed by the R211-D112 salt-bridge and F150. R208 forms a persistent salt-bridge with E119 and, for most of the trajectory, is separated from the double salt-bridge formed between D174, R211 and D112 by one or two waters.
3.3 Water transport through R1-Hv1
We have shown that R1-Hv1 exhibits persistent H-bonded water connectivity between the intracellular and extracellular sides. The stability of these water wires can be accounted for by their interaction with the protein side chains lining the permeation pathway. However, it is plausible that long-lived water wires through the channel can occur concurrently with water permeation, so we calculated the osmotic permeability coefficient through the R1-Hv1 channel using the method of Zhu et al. [52]. We obtained a value of 3.4 × 10−28 cm3/s (effectively zero), while the values reported in the literature for simulations of aquaporin channels are between 10−13 and 10−15 cm3/s [61–64]. Thus, despite having a fully permeated interior, the R1-Hv1 model cannot be considered a water channel. A relatively static wire is in good agreement with Hv1’s high proton selectivity, because a fairly immobile single file wire would hinder water flux and disfavor permeation of heavy ions.
4. Discussion
We have modeled the open state of the Hv1 channel using the crystallographic structure of the Kv1.2-Kv2.1 paddle-chimera VSD as a template, relying on the assumption that the crystal structure of its “up-state” conformation is a good approximation for a VSD under depolarizing potentials. Despite the low sequence similarity between Hv1 and the VSD of the Kv1.2-Kv2.1 paddle-chimera, helices S1 through S3 could be aligned based on the positions of conserved residues known to play important roles in Kv VSD function (e.g., D183 in S1, F233 in S2 and D259 in S3). Even though S4 is the most conserved helix of the VSD, the alignment of different VSDs in the S4 region is not straightforward due to the periodicity of the arginine triplet repeats. Here we probed the two most likely alignments, one in which the first three arginine positions (R1-R3) of the Hv1 S4 correspond to the first three arginine positions of the Kv S4 (R1-Hv1 model), and one in which the R1-R3 positions of the Hv1 S4 correspond to the second, third and forth arginine positions (R2-R4) of the Kv S4 (R2-Hv1 model). Our results show R1-Hv1 maintains a robust water wire, suggesting it may be able to support the Grotthuss hopping mechanism. We note that the presence of a water wire is not sufficient in itself to conclude that proton conduction will occur along that pathway; the question of proton transfer, which must be addressed through the calculation of the charge transfer free energy profile (see, e.g., [35–37]) is beyond the scope of the present work.
State-dependent internal salt-bridge networks between the basic residues in S4 and highly conserved positions for acidic residues in S1-S3 are a hallmark of the VSD structure, and have shown to be key in folding, stability and function [15, 58–60]. Hv1 not only lacks one of the R1-R4 arginines in S4, but the number and distribution of conserved positions for acidic side chains in S1-S3 differ from the ones in nonconducting VSDs. The most constricted region in the up-state conformation of the Kv1.2-Kv2.1 paddle-chimera VSD is formed by R4 (R299), and three S2 side chains (E226, I230, and F233). These hydrophobic side chains, which are also highly conserved, have been shown to play a crucial role in the molecular mechanism of voltage-dependent activation in Kv VSDs [27, 65]. Our R1-Hv1 model preserves the hydrophobic component of this arrangement, while the two small polar side chains replacing the salt-bridge (N214 and S148, respectively) are away from the center of the channel, thereby eliminating the steric hindrance imposed by the salt-bridge pair. Despite the loss of the innermost pair, the extracellular salt-bridge network in Hv1, involving R2 and R3, remains in place due to an additional conserved acidic position in S1 (E119).
In contrast, in the R2-Hv1 model, the most constricted region of the channel is formed by F150 and the R3-D112 salt-bridge pair, which leads to a partially open pathway. In addition, the extracellular salt-bridge network is broken. Similarly, the introduction of an arginine side chain at the same location of F150 along the transmembrane direction, leads to a mostly closed pathway in the R1-Hv1 N214R variant.
Previously, Ramsey et al. reported homology models of the Hv1 channel using the Kv1.2-Kv2.1 paddle-chimera structure as a template and a similar alignment to our R2-Hv1 model [32]. Consistent with our results, their models show a fully hydrated pathway connecting the intracellular and extracellular sides in 20-ns long atomistic simulations. In the first 20 ns of our equilibrated trajectories, the VSDs of the R1-Hv1 and R2-Hv1 models displayed significant differences in core hydration compared to the VSD of the Kv1.2-Kv2.1 paddle-chimera. But, only with longer simulations it became apparent that the R1-Hv1 model could support a water wire more effectively than the R2-Hv1 model. This is consistent with the idea that in order to compare the overall stability of two water wires, the trajectories need to be long enough to capture a significant number of breaks in at least one wire.
The experimental evidence available to date on the biophysics of Hv channels [1, 2, 11, 16, 17, 32, 33] seems to align with the classical view that proton conduction through membranes occurs over H-bonded water chains [66, 67]. Furthermore, despite the abundance of titratable side chains lining the putative permeation pathway, no single residue has been identified that could account for proton selectivity [32, 33]. The existence of a fully-hydrated permeation pathway in Hv1 could be extrapolated from the available structural data on Kv channel VSDs, both crystallographic [38] and in membrane environments [53, 68]. Our molecular models for Hv1 in lipid membrane provide two additional insights. First, the formation of a robust water wire connecting both sides of the membrane is dependent on the same structural features that are associated with the VSD gating function. This is consistent with the notion that both gating and permeation occur through the same structural pathway. Second, the water wire through Hv1 is not accompanied by water transport. Since the energetic cost of dehydrating an ion is prohibitively large [69], the lack of water transport implies that there is also no heavy ion transport, suggesting that proton selectivity in Hv1 may also be a consequence of the VSD architecture.
Highlights.
Two homology models of the Hv1 proton channel are presented.
The models are based on two alignments with the voltage-sensing domain of a potassium channel of known structure.
Both models support water wires, but the wire is more stable in one of the models.
No water flux is observed in the model with the most robust water wire.
The model with the most robust wire is supported by experimental studies and modeling of a mutant.
Acknowledgements
This work was supported in part by the National Institutes of Health (grants GM74637 to SHW and GM72507 to SHW and DJT), the National Science Foundation (grant CHE-0750175 to DJT), and the American Heart Association (WSA grant 09BGIA2160044 to FT). Computer time was provided by National Science Foundation-supported TeraGrid resources at the Texas Advanced Computing Center, and the University of California Shared Research Computing Services (ShaRCS) pilot project. MLW is grateful for support from the UC Irvine Medical Scientist Training Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sasaki M, Takagi M, Okamura Y. A voltage-sensor domain protein is a voltage-gated proton channel. Science. 2006;312:589–592. doi: 10.1126/science.1122352. [DOI] [PubMed] [Google Scholar]
- 2.Ramsey IS, Moran MM, Chong JA, Clapham DE. A voltage-gated proton-selective channel lacking the pore domain. Nature. 2006;440:1213–1216. doi: 10.1038/nature04700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramsey SI, Ruchti E, Kaczmarek JS, Clapham DE. Hv1 proton channels are required for high-level NADPH oxidase-dependent superoxide production during the phagocyte respiratory burst. Proc. Natl. Acad. Sci. U.S.A. 2009;106:7642–7647. doi: 10.1073/pnas.0902761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okochi Y, Sasaki M, Iwasaki H, Okamura Y. Voltage-gated proton channel is expressed on phagosomes. Biochem. Biophys. Res. Commun. 2009;382:274–279. doi: 10.1016/j.bbrc.2009.03.036. [DOI] [PubMed] [Google Scholar]
- 5.El Chemaly A, Okochi Y, Sasaki M, Arnaudeau S, Okamura Y, Demaurex N. VSOP/Hv1 proton channels sustain calcium entry, neutrophil migration, and superoxide production by limiting cell depolarization and acidification. The Journal of Experimental Medicine. 2010;207:129–139. doi: 10.1084/jem.20091837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capasso M, Bhamrah MK, Henley T, Boyd RS, Langlais C, Cain K, Dinsdale D, Pulford K, Khan M, Musset B, Cherny VV, Morgan D, Gascoyne RD, Vigorito E, DeCoursey TE, MacLennan ICM, Dyer MJS. HVCN1 modulates BCR signal strength via regulation of BCR-dependent generation of reactive oxygen species. Nat. Immunol. 2010;11:265–U212. doi: 10.1038/ni.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Musset B, Morgan D, Cherny VV, MacGlashan DW, Thomas LL, Rios E, DeCoursey TE. A pH-stabilizing role of voltage-gated proton channels in IgE-mediated activation of human basophils. Proc. Natl. Acad. Sci. U.S.A. 2008;105:11020–11025. doi: 10.1073/pnas.0800886105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lishko PV, Botchkina IL, Fedorenko A, Kirichok Y. Acid extrusion from human spermatozoa is mediated by flagellar voltage-gated proton channel. Cell. 2010;140:327–337. doi: 10.1016/j.cell.2009.12.053. [DOI] [PubMed] [Google Scholar]
- 9.Iovannisci D, Illek B, Fischer H. Function of the HVCN1 proton channel in airway epithelia and a naturally occurring mutation, M91T. J. Gen. Physiol. 2010;136:35–46. doi: 10.1085/jgp.200910379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee S-Y, Letts JA, MacKinnon R. Dimeric subunit stoichiometry of the human voltage-dependent proton channel of Hv1. Proc. Natl. Acad. Sci. U.S.A. 2008;105:7692–7695. doi: 10.1073/pnas.0803277105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tombola F, Ulbrich MH, Isacoff EY. The voltage-gated proton channel Hv1 has two pores, each controlled by one voltage sensor. Neuron. 2008;58:546–556. doi: 10.1016/j.neuron.2008.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koch HP, Kurokawa T, Okochi Y, Sasaki M, Okamura Y, Larsson HP. Multimeric nature of voltage-gated proton channels. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9111–9116. doi: 10.1073/pnas.0801553105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aggarwal SK, MacKinnon R. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 1996;16:1169–1177. doi: 10.1016/s0896-6273(00)80143-9. [DOI] [PubMed] [Google Scholar]
- 14.Schoppa NE, McCormack K, Tanouye MA, Sigworth FJ. The size of gating charge in wild-type and mutant Shaker potassium channels. Science. 1992;255:1712–1715. doi: 10.1126/science.1553560. [DOI] [PubMed] [Google Scholar]
- 15.Seoh SA, Sigg D, Papazian DM, Bezanilla F. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron. 1996;16:1159–1167. doi: 10.1016/s0896-6273(00)80142-7. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez C, Koch HP, Drum BM, Larsson HP. Strong cooperativity between subunits in voltage-gated proton channels. Nature Struct. Mol. Biol. 2010;17:51–56. doi: 10.1038/nsmb.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tombola F, Ulbrich MH, Kohout SC, I EY. The opening of the two pores of the Hv1 voltage-gated proton channel is tuned by cooperativity. Nature Struct. Mol. Biol. 2010;17:44–50. doi: 10.1038/nsmb.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larsson B, Baker OS, Dhillon DS, Isacoff EY. Transmembrane movement of the Shaker K+ channel S4. Neuron. 1996;16:387–397. doi: 10.1016/s0896-6273(00)80056-2. [DOI] [PubMed] [Google Scholar]
- 19.Mannuzzu LM, Moronne MM, Isacoff EY. Direct physical measure of conformational rearrangement underlying potassium channel gating. Science. 1996;271:213–216. doi: 10.1126/science.271.5246.213. [DOI] [PubMed] [Google Scholar]
- 20.Ruta V, Chen J, MacKinnon R. Calibrated measurement of gating-charge arginine. Cell. 2005;123:463–475. doi: 10.1016/j.cell.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 21.Pathak MM, Yarov-Yarovoy V, Agarwal G, Roux B, Barth P, Kohout S, Tombola F, Isacoff EY. Closing in on the resting state of the Shaker K+ channel. Neuron. 2007;56:124–140. doi: 10.1016/j.neuron.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 22.Alabi AR, Bahamonde MI, Jung HJ, Kin JI, Swartz KJ. Portability of paddle motif function and pharmacology in voltage sensors. Nature. 2007;450:370–376. doi: 10.1038/nature06266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Starace DM, Bezanilla F. Histidine scanning mutagenesis of basic residues of the S4 segment of the Shaker K+ channel. J. Gen. Physiol. 2001;117:469–490. doi: 10.1085/jgp.117.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Starace DM, Bezanilla F. A proton pore in a potassium channel voltage sensor reveals a focused electric field. Nature. 2004;427:548–553. doi: 10.1038/nature02270. [DOI] [PubMed] [Google Scholar]
- 25.Tombola F, Pathak MM, Gorostiza P, Isacoff EY. The twisted ion-permeation pathway of a resting voltage-sensing domain. Nature. 2007;445:546–549. doi: 10.1038/nature05396. [DOI] [PubMed] [Google Scholar]
- 26.Tombola F, Pathak MM, Isacoff EY. Voltage-sensing arginines in a potassium channel permeate and occlude cation-selective pores. Neuron. 2005;45:379–388. doi: 10.1016/j.neuron.2004.12.047. [DOI] [PubMed] [Google Scholar]
- 27.Campos FV, Chanda B, Roux B, Bezanilla F. Two atomic constraints unabiguously position the S4 segment relative to S1 and S2 segments in the closed state of Shaker K Channel. Proc. Natl. Acad. Sci. U.S.A. 2007;104:7904–7909. doi: 10.1073/pnas.0702638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sokolov S, Scheuer T, Catterall WA. Ion premeation through a voltage-sensitive gating pore in brain sodium channels having voltage sensor mutations. Neuron. 2005;47:183–189. doi: 10.1016/j.neuron.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 29.Sokolov S, Scheuer T, Caterall WA. Gating pore current in an inherited ion channelopathy. Nature. 2007;446:76–78. doi: 10.1038/nature05598. [DOI] [PubMed] [Google Scholar]
- 30.Struyk AF, Cannon SC. A Na+ channel mutation linked to hypokalemic periodic paralysis exposes a proton-selective gating pore. J. Gen. Physiol. 2007;130:11–20. doi: 10.1085/jgp.200709755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chanda B, Bezanilla F. A common pathway for charge transport through voltage-sensing domains. Neuron. 2008;57:345–351. doi: 10.1016/j.neuron.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 32.Ramsey IS, Mokrab Y, Carvacho I, Sands ZA, Sansom MSP, Clapham DE. An aqueous H+ permeation pathway in the voltage-gated proton channel Hv1. Nature Struct. Mol. Biol. 2010:1–7. doi: 10.1038/nsmb.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakata S, Kurokawa T, Nørholm MHH, Takagi M, Okochi Y, von Heijne G, Okamura Y. Functionality of the voltage-gated proton channel truncated in S4. Proc. Natl. Acad. Sci. U.S.A. 2010;107:2313–2318. doi: 10.1073/pnas.0911868107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee S-Y, Banerjee A, MacKinnon R. Two separate interfaces between the voltage sensor and pore are required for the function of voltage-dependent K+ channels. PLoS Biol. 2009;7:0676–0686. doi: 10.1371/journal.pbio.1000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato M, Pisliakov AV, Warshel A. The barrier for proton transport in aquaporins as a challenge for electrostatic models: The role of protein relaxation in mutational calculations. Proteins-Structure Function and Bioinformatics. 2006;64:829–844. doi: 10.1002/prot.21012. [DOI] [PubMed] [Google Scholar]
- 36.Burykin A, Warshel A. What really prevents proton transport through aquaporin? Charge self-energy versus proton wire proposals. Biophys. J. 2003;85:3696–3706. doi: 10.1016/S0006-3495(03)74786-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Braun-Sand S, Burykin A, Chu ZT, Warshel A. Realistic simulations of proton transport along the gramicidin channel: Demonstrating the importance of solvation effects. Journal of Physical Chemistry B. 2005;109:583–592. doi: 10.1021/jp0465783. [DOI] [PubMed] [Google Scholar]
- 38.Long SB, Tao X, Campbell EB, Mackinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 39.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 40.Phillips JC, Braun B, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacKerell AD, Jr., Bashford D, Bellott M, Dunbrack RL, Jr., Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 42.Klauda JB, Brooks BR, MacKerell AD, Jr., Venable RM, Pastor RW. An ab initio study on the torsional surface of alkanes and its effect on molecular simulations of alkanes and a DPPC bilayer. J. Phys. Chem. B. 2005;109:5300–5311. doi: 10.1021/jp0468096. [DOI] [PubMed] [Google Scholar]
- 43.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 44.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 45.Darden T, York D, Pedersen L. Particle mesh Ewald: An N•log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 46.Grubmüller H, Heller H, Windemuth A, Schulten K. Generalized Verlet algorithm for efficient molecular dynamics simulations with long-range interactions. Mol. Simul. 1991;6:121–142. [Google Scholar]
- 47.Ryckaert J-P, Ciccotti G, Berendsen HJC. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. Journal of Computational Physics. 1977;23:327–341. [Google Scholar]
- 48.Miyamoto S, Kollman P. An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 49.Martyna GJ, Tobias DJ, Klein ML. Constant-pressure molecular-dynamics algorithms. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- 50.Feller SE, Zhang Y, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 51.Humphrey W, Dalke W, Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 52.Zhu F, Tajkhorshid E, Schulten K. Collective diffusion model for water permeation through microscopic channels. Phys. Rev. Lett. 2004;93:224501–224501. doi: 10.1103/PhysRevLett.93.224501. to 224501–224504. [DOI] [PubMed] [Google Scholar]
- 53.Krepkiy D, Mihailescu M, Freites JA, Schow EV, Worcester DL, Gawrisch K, Tobias DJ, White SH, Swartz KJ. Structure and hydration of membranes embedded with voltage-sensing domains. Nature. 2009;462:473–479. doi: 10.1038/nature08542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Freites JA, Tobias DJ, White SH. A voltage-sensor water pore. Biophys. J. 2006;91:L90–L92. doi: 10.1529/biophysj.106.096065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jogini V, Roux B. Dynamics of the Kv1.2 voltage-gated K1 channel in a membrane. Biophys. J. 2007;93:3070–3082. doi: 10.1529/biophysj.107.112540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sands ZA, Sansom MSP. How does a voltage sensor interact with a lipid bilayer? Simulations of a potassium channel domain. Structure. 2007;15:235–244. doi: 10.1016/j.str.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Treptow W, Tarek M. Environment of the gating charges in the Kv1.2 Shaker potassium channel. Biophys. J. 2006;90:L64–L66. doi: 10.1529/biophysj.106.080754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Papazian DM, Shao XM, Seoh S-A, Mock AF, Huang Y, Wainstock DH. Electrostatic interactions of S4 voltage sensor in Shaker K+ channel. Neuron. 1995;14:1293–1301. doi: 10.1016/0896-6273(95)90276-7. [DOI] [PubMed] [Google Scholar]
- 59.Tiwari-Woodruff SK, Schulteis CT, Mock AF, Papazian D. Electrostatic interactions between transmembrane segments mediate folding of Shaker K+ channel subunits. Biophys. J. 1997;72:1489–1500. doi: 10.1016/S0006-3495(97)78797-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang L, Sato Y, Hessa T, von Heijne G, Lee J-K, Kodama I, Sakaguchi M, Uozumi N. Contribution of hydrophobic and electrostatic interactions to the membrane integration of the Shaker K+ channel voltage sensor domain. Proc. Natl. Acad. Sci. U.S.A. 2007;104:8263–8268. doi: 10.1073/pnas.0611007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jensen MØ, Mouritsen OG. Single-channel water permeabilities of Escherichia coli aquaporins AqpZ and GllF. Biophys J. 2006;90:2270–2284. doi: 10.1529/biophysj.105.073965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hashido M, Kidera A, Ikeguchi M. Water transport in aquaporins: osmotic permeability matrix analysis of molecular dynamics simulations. Biophys J. 2007;93:373–385. doi: 10.1529/biophysj.106.101170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hashido M, Ikeguchi M, Kidera A. Comparative simulations of aquaporin family: AQP1, AQPZ, AQP0 and GlpF. FEBS Lett. 2005;579:5549–5552. doi: 10.1016/j.febslet.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 64.Jensen MO, Dror RO, Xu HF, Borhani DW, Arkin IT, Eastwood MP, Shaw DE. Dynamic control of slow water transport by aquaporin 0: Implications for hydration and junction stability in the eye lens. Proc. Natl. Acad. Sci. U.S.A. 2008;105:14430–14435. doi: 10.1073/pnas.0802401105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tao X, Lee A, Limapichat W, Dougherty DA, MacKinnon R. A gating charge transfer center in voltage sensors. Science. 2010;328:67–73. doi: 10.1126/science.1185954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Decoursey TE. Voltage-gated proton channels and other proton transfer pathways. Physiol. Rev. 2003;83:475–579. doi: 10.1152/physrev.00028.2002. [DOI] [PubMed] [Google Scholar]
- 67.Nagle JF, Morowitz HJ. Molecular Mechanisms for Proton Transport in Membranes. Proc. Natl. Acad. Sci. U.S.A. 1978;75:298–302. doi: 10.1073/pnas.75.1.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chakrapani S, Cuello LG, Cortes DM, Perozo E. Structural dynamics of an isolated voltage-sensor domain in a lipid bilayer. Structure. 2008;16:398–409. doi: 10.1016/j.str.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilson MA, Pohorille A. Mechanism of unassisted ion transport across membrane bilayers. J. Am. Chem. Soc. 1996;118:6580–6587. doi: 10.1021/ja9540381. [DOI] [PubMed] [Google Scholar]