Abstract
Cerium oxide nanoparticles (nanoceria) are versatile engineered nanoparticles (ENPs) due to their unique redox properties. We and others have previously demonstrated naked nanoceria could act as antioxidants to protect cells against oxidative damage. While the redox properties may be beneficial, the genome-wide effects of nanoceria on gene transcription and associated biological processes remain elusive. Here we applied functional genomic approach to examine the genome-wide effects of nanoceria on global gene transcription and cellular functions in mouse neuronal cells. Importantly, we demonstrated nanoceria induced chemical- and size-specific changes in the murine neuronal cell transcriptome. The nanoceria specially contributed more than 83% of uniquely altered gene population and associate with a unique spectrum of genes related to neurological disease, cell cycle control and growth. These observations suggest an in-depth assessment of potential health effects of naked nanoceria and other naked nanoparticles is both necessary and imminent.
Keywords: Cerium oxide, nanoceria, nanoparticles, neuronal cells, genomics
Background
Despite nanomaterials having been used in a broad range of industrial and consumer products, the biological effects after exposure remain largely unknown. Nano cerium oxide (CeO2) or nanoceria are popular engineered nanoparticles (ENPs) used in various applications, including catalysis, solar cells, fuel additives and chemical abrasives.1 However, its main application, in the field of catalysis, stems from its unique properties compared to other materials. Ceria, especially in nano form, is a special material for two reasons. First, it is able to undergo significant reduction of its cerium cations from a +4 to +3 oxidation state and the accompanying loss of oxygen without structural (i.e., phase) changes.2 That is, it can become increasingly non-stoichiometric while still maintaining its characteristic cubic fluorite structure.3 Second, oxygen transport through the lattice is rapid.4
In the catalysis field, nanoceria is used for its oxygen storage capacity (OSC)5 or ability to store and release oxygen. An example of this is the diffusion of oxygen out of its lattice, in an environment free of gaseous oxygen, which allows carbon monoxide (CO) to be converted to carbon dioxide.6 The OSC of ceria and its mixed oxides is helpful in facilitating a number of other reactions including steam reforming of methane,7 three-way catalysis in the automobile,8 and the water-gas shift reaction.9 However, nanoceria’s oxygen storage capabilities stem not only from the interior of the nanoparticles, but also from the surface,10 and, since catalysis is a surface science, are worthy of additional consideration. To explain the ease in reducing the surface of ceria (i.e., creating oxygen vacancies), one study suggests that surface oxygen may not be as strongly bound as bulk oxygen,11 allowing for the facile formation of oxygen vacancies. Oxygen vacancies are important surface sites, because they have been found to bind adsorbates more strongly than normal oxide sites and may assist in the dissociation of the adsorbed species.12, 13
Other than its industrial applications, ceria’s biochemical properties have been explored in recent years. Biomedical and toxicity studies of nanoceria, which focused predominantly on its redox properties, have resulted in contradictory conclusions about its effects. We, and others, have previously documented that naked nanoceria particles may act as antioxidants and protect cells against oxidative damage,14, 15 ionizing radiation16 and improve cardiac function.17 Other reports found it to be cytotoxic18, 19 and induce oxidative stress.20 These studies mainly focused on the redox capability of nanoceria reacting with reactive oxygen species (ROS) or its intermediates (ROI), and evaluated the cellular responses in oxidative stress condition. Under normal/basal conditions the genome-wide response of cells to nanoceria exposure remains largely unknown. Since nanoceria has the potential in biomedical applications, the assessment of their cellular effects and the associated molecular mechanisms are urgently needed. In this study, we report that nanoceria induced a unique set of genes related to neurological and cell cycle regulation, which are validated by expression and functional assays.
Methods
Preparation of particles
Two cerium oxide particles of different sizes (6 nm, CeO-6 and ~100 nm, CeO-M) and Al2O3 at 300 nm (AlO) were selected for experiment. CeO2 nanoparticles were prepared by combining 0.04M Ce(NO3)3• 6H2O and 0.5M hexamethylenetetramine (HMT) as described previously.21 The CeO-M was prepared by annealing similarly prepared CeO2 nanoparticles for 12 hours at 1200°C. HMT burns off in the annealing process but the particles were re-coated with HMT to be consistent with the the unannealed CeO2 nanoparticles. The size of these particles was determined by analysis of x-ray diffraction data, collected with an Inel XRG 3000 (Cu Kα1 radiation (λ=1.54056Å), by using the Debye-Scherrer equation.22 AlO was obtained from Buehler (Lake Bluff, IL).
Cell culture
All particles were sonicated for optimizing the microarray experiment. The sonicated mixture was diluted with 9 ml of FBS and added to the corresponding amount of DMEM to make 10% FBS by volume. The final particle concentration was 20 µg/ml in all experiments. Particles were freshly prepared prior to each experiment. HT22 hippocampal nerve cell line is a subclone of HT4 cells. Culture dishes were incubated for 8 hours in 5% CO2 at 37°C on an orbital shaker at constant speed in order to maintain a uniform suspension of the particles. Control cells were incubated in DMEM containing 10% sonicated FBS without particles. All experiments were done in duplicate
Microarray and data analysis
Gene expression was determined from duplicate microarray experiments using Mouse Genome 430 2.0 microarray (Affymetrix, Santa Clara, CA). Gene expression profiles were analyzed with Partek Genomic Suite 6.3 (Partek, St. Louis, MO) and used for data acquisition and normalization by robust multiarray average (RMA) algorithm. Genes specifically expressed in cells treated with CeO-6 were uploaded to the Ingenuity Pathway Analysis program (Ingenuity Systems, Mountain View, CA) to explore transcriptional profiles and to identify potential gene networks induced by the particles in the model. The Global Functional Analysis feature identified biological functions most significantly associated with the gene list. Querying the Ingenuity Pathways Knowledge Base for relationships that described a gene’s role in cellular and organismal function. The significance value associated with a function in Global Functional Analysis was expressed as p-value by right-tailed Fisher Exact Test. P-value of less than 0.01 was taken as significant.
Western blot
Whole-cell lysates were extracted using the nuclear extract kit (Active Motif, Carlsbad, CA), according to the manufacture’s protocol. Twenty micrograms of extract was mixed with Laemmli loading buffer (containing β-mercaptoethanol) and heated at 100°C for 5 minutes prior to loading onto 4–12% Tris-glycine precast gels and electrophoresed at 140 V for 90 minutes. Proteins were transferred to 0.45 µm nitrocellulose membranes (Invitrogen, Carlsbad, CA) for 2 h at 20 V at room temperature using the mini-cell Module (Invitrogen, Carlsbad, CA). Primary antibody against Htt (#2733), cyclin D1(#2922), p16(#4824), p21(#2946), and p53 (#9282) (Cell signaling technology, Danvers, MA) were diluted in 5% nonfat powdered milk in 0.5% TBS-Tween 20 in 1:200 dilution. The blot was incubated with SuperSignal West Pico substrate (Pierce, Rockford, IL) and exposed to Kodak X-OMAT film (Kodak, Rochester, NY).
Quantitative real-time RT-PCR
RNA was isolated using the RNeasy mini kit (QIAGEN Inc., Valencia, CA) according to the manufacturer's instruction. For cDNA synthesis, 500 ng total RNA was transcribed with cDNA transcription reagents using SuperScriptIII reverse transcriptase and oligo(dT)12–18 (Invitrogen) according to the manufacturer's instructions. Primers were designed using Perkin Elmer Primer Express® software. All primers were designed to be intron-spanning to preclude amplification of genomic DNA. To normalize relative levels of expression, β-actin was used as an internal reference control. Real-time RT-PCR was performed on an Applied Biosystems Gene Amp® 7900HT Sequence Detection System (Foster City, CA). Standard reaction volume was 10µl and contained 1X SYBR RT-PCR buffer, 3 mM MgCl2, 0.2 mM each of dATP, dCTP, dGTP, 0.4 mM dUTP, 0.005 U AmpliTaq Gold, 0.002 U AmpErase UNG erase enzyme, 0.35µl cDNA template and 50 nM of oligonucleotide primer. Initial steps of RT-PCR were 2 min at 50°C for UNG erase activation, a 10-min hold at 95°C and consisted of a 15-sec melt at 95°C, a 1-min annealing/extension at 60°C for a total of 30 cycles and incubated at 60°C for 1 min in the final step. The identity of the PCR products was confirmed by melting curve analysis with ABI SDS 2.0 software. The expression level of a gene in a given sample was represented as 2−ΔΔCt [ΔCT(medium)] and ΔCT = [CT(experimental)] − [CT(housekeeping)]. All reactions were performed in triplicate.
DNA flow cytometry and proliferation assay
HT22 cells were synchronized by serum deprivation for 48 h in 6-well plates at a concentration of 5 × 105 cells/well. Fresh medium with 10% FBS with or without particles was added and incubated for 8, 16 or 24 h. Both live monolayer cells and dead nonadhesive cells were collected and counted by haemocytometer with trypan blue solution (Invitrogen). For each sample, 1 × 106 cells were washed twice in buffer solution from the CycleTest Plus DNA Reagent Kit (Becton Dickinson, San Jose, CA) and resuspended in 250 µL of trypsin buffer. DNA staining for cell cycle was quantified by FACScan flow cytometer using CellQuest software (Becton Dickinson). Cell cycle and DNA degradation of cells were analyzed by ModFit LT software (Verity Software House, Topsham, MA). Sub-G1 cell populations were considered to be apoptotic. The induction of apoptosis was confirmed by examining the cells under fluorescent microscopy after double staining with propidium iodide and Annexin V-FITC according to the manufacturer’s instructions (Trevigen, Gaithersburg, MD). To measure cellular proliferation in response to particles, cells were seeded at a density of 1 × 104 per well in 24-well gelatin-coated plates and treated at 20 µg/ml as described. Cell viability was assessed using colorimetric 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay (Chemicon, Billerica, MA ) on days 0, 1, 3 and 5 according to manufacturer’s protocol. Cell viability was measured by the absorbance on an ELISA plate by setting the optical density of ELISA reader at 570 nm and a reference wavelength of 630 nm.
Results
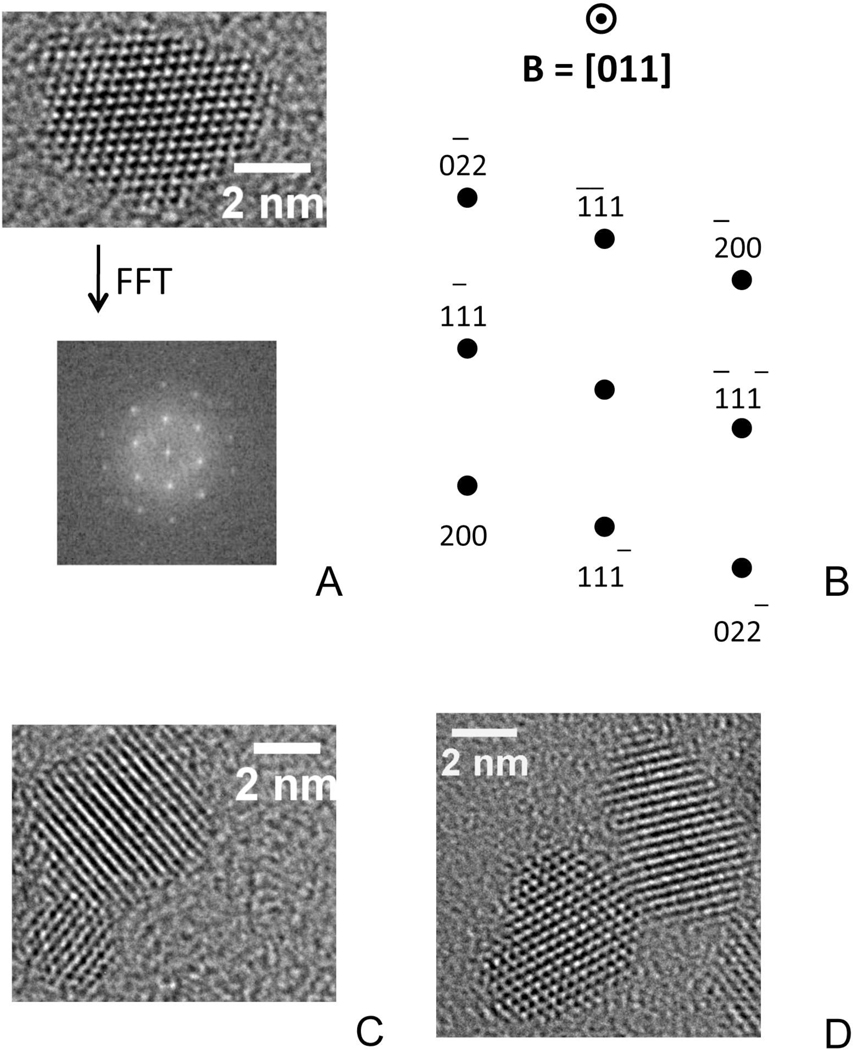
Using whole-genome expression microarray analysis we studied the transcriptional response of murine neuronal cells (HT22) following exposure to 6 nm nanoceria (CeO-6). The nanoparticles of ceria, prepared by aqueous precipitation as previously described,23,24 were 6 nm in size. Compared to micron-sized ceria, nanoceria has an expanded lattice25 due to the nature of ionic bonding in nanoparticles26 and a considerable concentration of Ce3+ cations, which are larger in size than Ce4+ cations (Figure 1).27 Moreover, the concentration of both Ce3+ ions and oxygen vacancies, which exist in a 2:1 ratio in the ceria lattice, increases as the crystallite size decreases. Therefore, 6 nm crystallites would be expected to be more easily reduced or oxidized compared to 12 nm crystallites. Note that such differences in the concentration of point defects (i.e., Ce3+ in CeO2 and oxygen vacancies) as a function of size will not be apparent in TEM images.
Figure 1.
High resolution TEM images of pure nanoceria. (A) Single crystallite, showing multiple lattice planes, subject to a Fast Fourier Transform (FFT) and the resulting diffraction pattern. (B) Schematic of the diffraction pattern including the indexed crystallographic planes and the beam direction. (C) Two crystallites of different size. (D) Two crystallites in which the left shows multiple planes while the right displays only a single set of its atomic planes
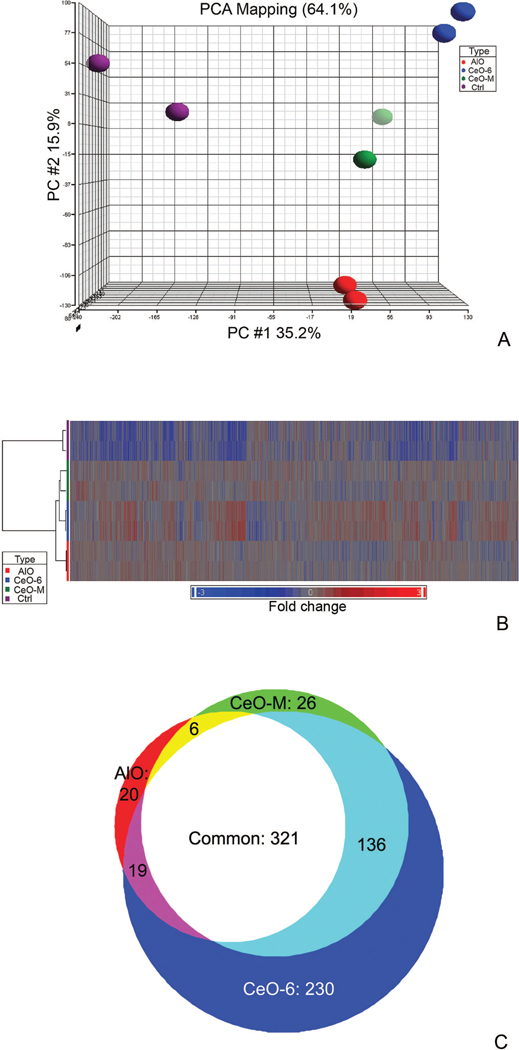
We controlled for the effect of particle size using 100 nm diameter cerium oxide (CeO-M), and for its chemical properties by using 300 nm diameter aluminum oxide particles (AlO). Nanoceria were subjected to sonication and agitation to prevent aggregation and sedimentation prior to incubation with cells to obtain consistent gene expression results (see methods). Principal Component Analysis (PCA) was applied to the microarray datasets to detect global gene-expression patterns in HT22 cells following exposure to CeO-6, CeO-M, AlO or “no particle” controls. The data revealed that each treatment produced a distinct gene expression pattern as evidenced by four spatial groups (treatment with CeO-6, CeO-M, AlO, Ctrl) - implying particle-specific responses of the cells (Figure 2, A). The datasets (represented by colored dots in the PCA diagram) suggested the expression profile of particle-treated groups were more similar as compared to the “no particle” control. The relationship between the sample groups and the gene expression signatures were highlighted by one-way hierarchical gene cluster analysis (Figure 2, B). The denogram segregated the data into two major clusters, namely particle exposure in the first branch, as contrasted to the “no-particle exposure” group; these data validated the results obtained from the PCA analysis. Further analysis on the particle exposure group observed gene signatures that were indicative of chemical and size properties of the particles. The chemical differences between cerium oxides and aluminum oxide were indicated by two cluster subgroups in the figure. In addition, we observed that the size of the cerium oxide particles affected the gene expression pattern: HT22 cells treated with nanoceria exhibited a distinct gene expression signature compared to non-nano-sized cerium oxide particles. These data indicate particle size has an active effect on cellular behavior similar to the recent report for conjugated particles.28 These results underscore the notion that nanoparticles may not be viewed as simple “carriers” for use in biomedical applications.
Figure 2.
Microarray analysis on HT22 cell exposed to particles of different sizes and chemistry. (A) Principal Component Analysis (PCA) of four HT22 groups: 1. No particle control, 2. CeO-6, 3. CeO-M and 4. AlO. 64.1% of the data were represented in the scatter plot and were divided among the three PCA component vectors: PC#1 (x-axis), PC #2 (y-axis), and PC#3 (z-axis). Biological duplicates were included in each group. (B) Unsupervised hierarchical clustering of expressed genes for HT22 cells exposed to different particles indicated two distinct clusters which segregated the particle-treated and non-treated group. Each row represents a sample and each column represents a gene. Red color represents over-expressed genes while blue color represents down-regulated genes, fold change is indicated on the bottom bar. (C) Comparison of global gene expression profiles of cells treated with different particles. One-way ANOVA analysis comparing gene expression of each treated group to control was performed, and the Venn diagram was produced by overlaying each group.
To identify genes differentially expressed following CeO-6, CeO-M and AlO exposure, one-way ANOVA with a false discovery rate (FDR) of < 0.01, and a fold change of ± 1.5 was applied. 706 differentially expressed genes were identified in the CeO-6 group (p<0.001), 489 in the CeO-M group (p<0.0001) and 350 in the AlO group (p<0.0008). To reveal unique gene signatures associated with each particle, a proportional Venn diagram was constructed by superimposing the differentially expressed genes from each group (Figure 2, C). We identified 1545 (706+489+350) genes significantly induced by particle exposure, and of these 276 (20+26+230, 17%) expressed genes were unique dependent on the particle they were exposed to. It is noteworthy that the largest number of “unique” genes occurred in the presence of nanoceria. Approximately 83% (230/276) of unique genes were derived from nanoceria group (Supplementary Materials Table S1), while CeO-M- and AlO-specific genes contributed only 9.4% (26/276) and 7.2% (19/276), respectively. One-fifth (321/1545, 20.8%,) of the total differentially expressed genes were common to all groups indicating their effect was independent of particle chemistry and size. CeO-6 and CeO-M shared more commonly expressed genes (136, light-green) than either did with AlO (19, purple and 6, yellow respectively, Figure 2, C). The expression changes of these 136 genes were reflective of differences in the chemical properties rather than the molecular size of cerium oxide and aluminum oxide. The nanoscale effect in cerium oxide was evidenced by the number of CeO-6-specific genes (230) as contrasted to CeO-M-specific genes (26).
Venn diagram analysis clearly demonstrated that exposure to nanoceria particles results in the largest number of particle-specific gene alterations. We focused on the 230 signature genes following nanoceria exposure to obtain insight into the responsible molecular events. We postulated the change in gene expression profile was coordinated through modules or gene networks that shared a common biological function. We utilized ingenuity pathway analysis to dynamically generate significant biological networks based on its biological knowledge base of relationships between genes, proteins, drugs and diseases. Initially we attempted functional analysis to identify known disease models that resembled the nanoceria gene signature. The top biological functions associated with each gene network were determined by identifying relationships between the genes in the network and their cellular and organismal function. The results were ‘weighted” based on their statistical significance and gene number of involved genes; “neurological disease” and “cancer” models were suggested [13 genes (p= 0.0004) and 57 genes (p=0.0005), respectively] as relevant potential biological responses of the cell model. Other relevant disorders that passed the statistical test included cardiovascular disease (7 genes), viral function (7 genes) and organismal injury (10 genes). These were excluded in subsequent analysis on their low probability and the lack of relevance to the cell model.
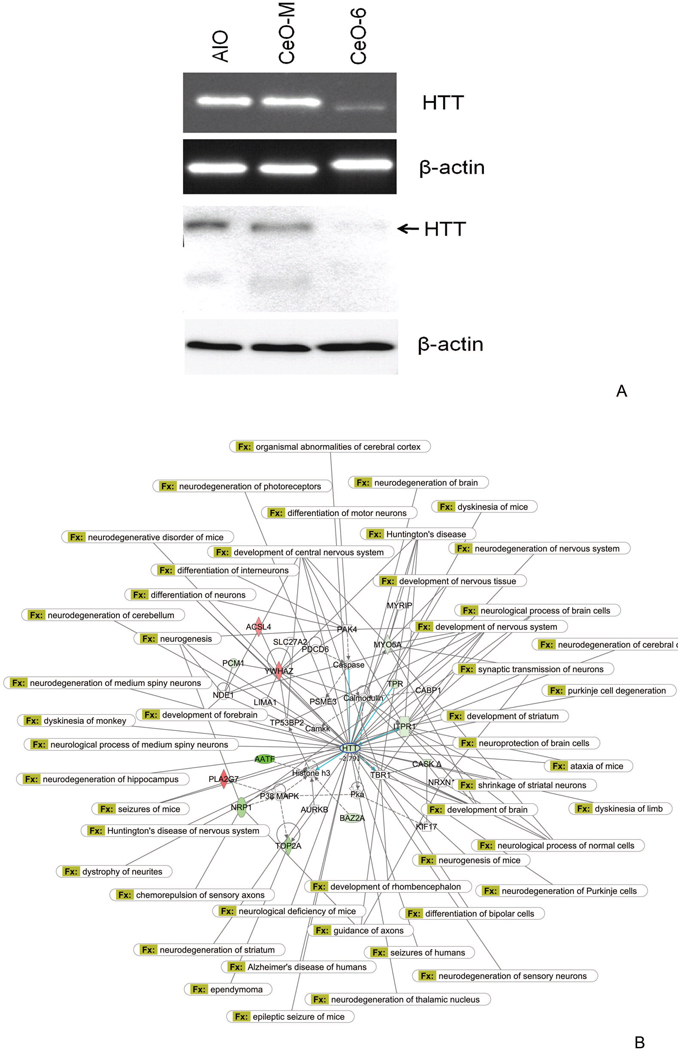
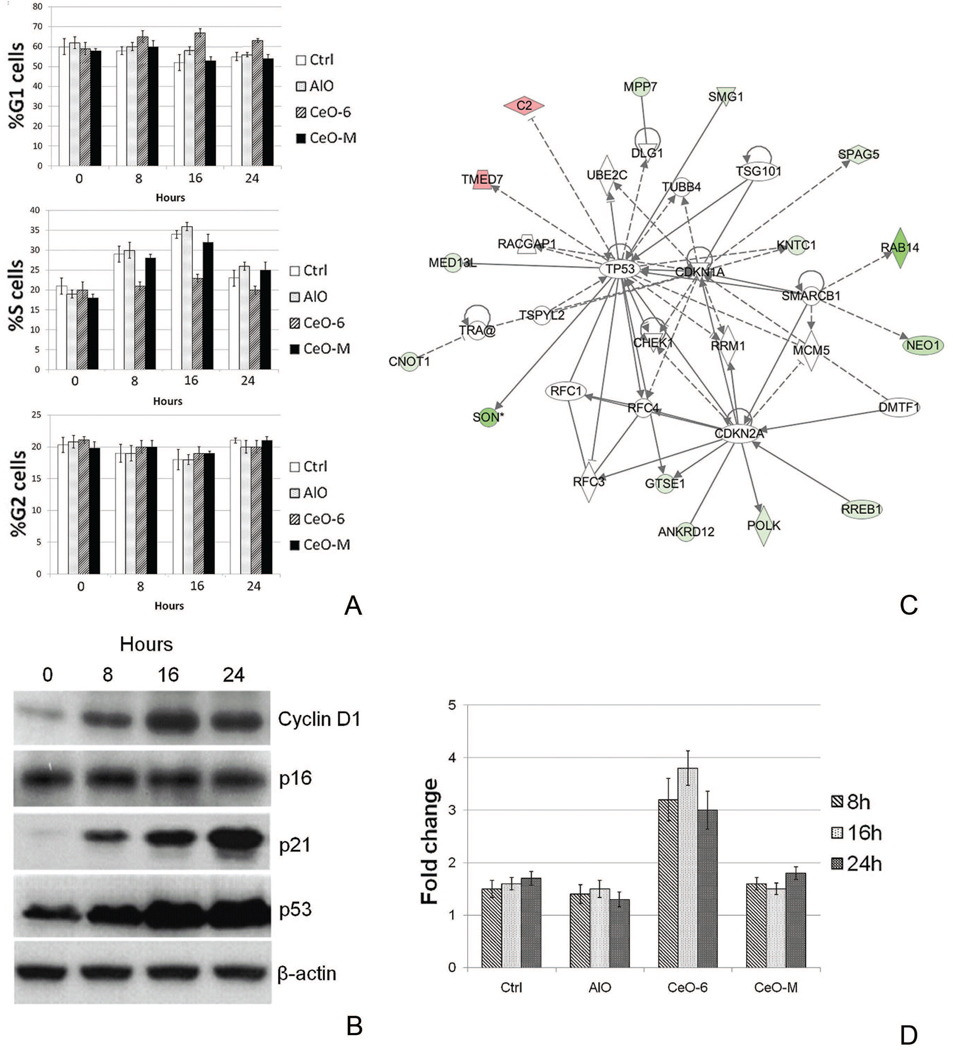
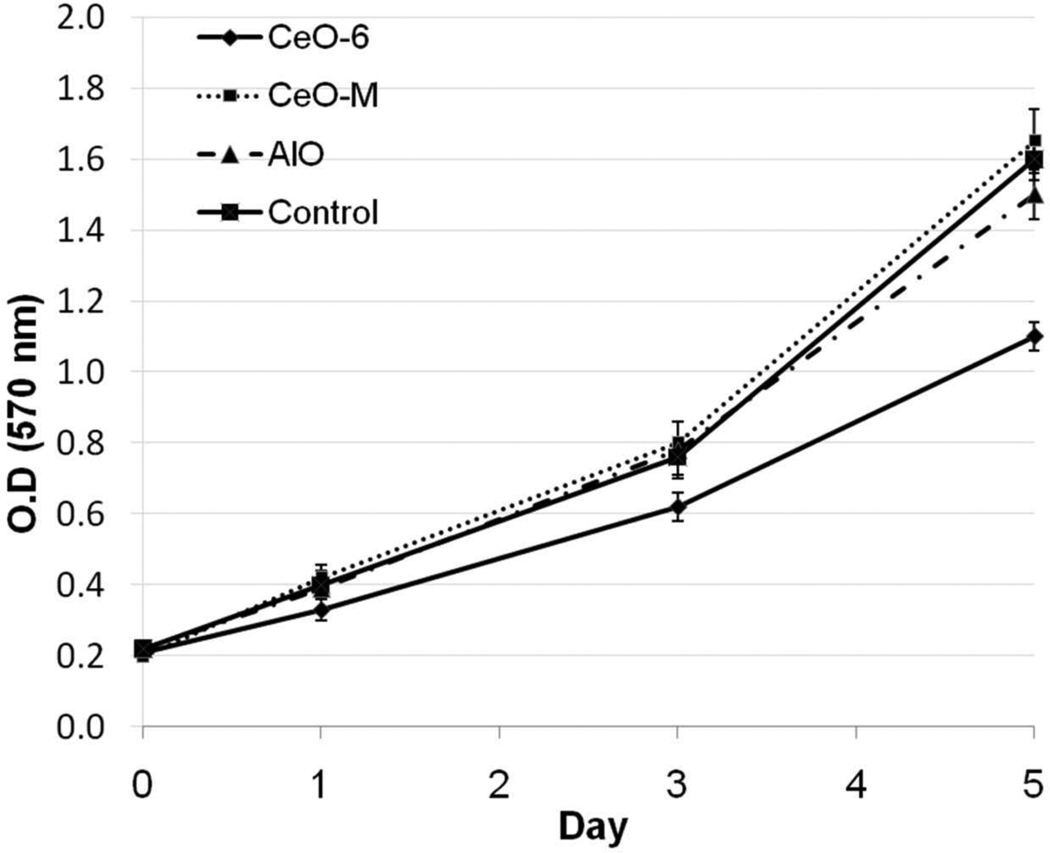
Subsequently genes were sorted by their biological function and relevance to the disease model. Interestingly, after nanoceria exposure we observed profound down-regulation (>3 fold) of the huntingtin (Htt) gene in various pathways within the neurological disease model (Figure 3, A, Supplementary Materials Table S2 and Figure 5, A). Htt is known to be important in various developmental processes; complete knockout of mouse Htt is embryonic lethal,29–31 while mice carrying less than 50% of wild-type Htt display malformations of the cortex and striatum32, 33. To obtain biological insight into the interaction of Htt and other genes we created an Htt-centric gene network (Figure 3, B) including those genes whose expression is affected by nanoceria (Figure 4, A) to highlight its involvement in neurological pathophysiology. In the cancer model, genes involved in cell cycle-related processes were over-represented (Supplementary Materials Table S3); this suggests nanoceria might perturb cell cycle control. Cell cycle analysis confirmed nanoceria inhibited cell cycle progression from G1 to S phase (c). Gene network analysis implied nanoceria inhibited the G1/S transition via regulators such as TP53, p21 (CDKN1A) and p16 (CDKN2A), which led to downstream effects on other genes (Figure 4, B, Supplementary Materials Table S3 and Figure 5, B). Induction of p21 (CDK1A) and TP53 were observed starting at 8 h, but there was no indication of altered p16 expression (Figure 4, C), suggesting inhibition of the cell cycle effect may be mediated by a p53 and p21 dependent mechanism. It was surprising to find that nanoceria increased the expression of cyclin D, since this would be expected to enhance, rather than inhibit, progression through G1. Thus, these particles may not simply act as a brake at the G1/S transition but rather, they may disrupt normal regulation of cell cycle components. In addition, we observed increased apoptosis during exposure as assessed by the sub-G1 population (Figure 4, D) that led to growth inhibition starting at 48 h (Figure 6).
Figure 3.
Htt-centric gene network in HT22 cells in response to nanoceria treatment. (A) Expression validation demonstrated Htt RNA and protein are down-regulated upon nanoceria exposure. Upper panel: semi-quantitative RT-PCR, lower panel: Western blot analysis using antibody against Htt. Beta-actin was used as control. (B) The interaction of Htt with other signature genes in response to nanoceria exposure is shown. Red indicates up-regulated genes, green represents down-regulated genes. Genes with a white background were not part of the nanoeria gene signature, but were integrated into the computationally generated network based on evidence in the knowledge base with strong biological relevance to that network. Known neurological disorders associated with Htt were indicated by connections to Htt. Top biological functions in this network include nervous system development and function, cellular assembly and organization, and cell signaling.
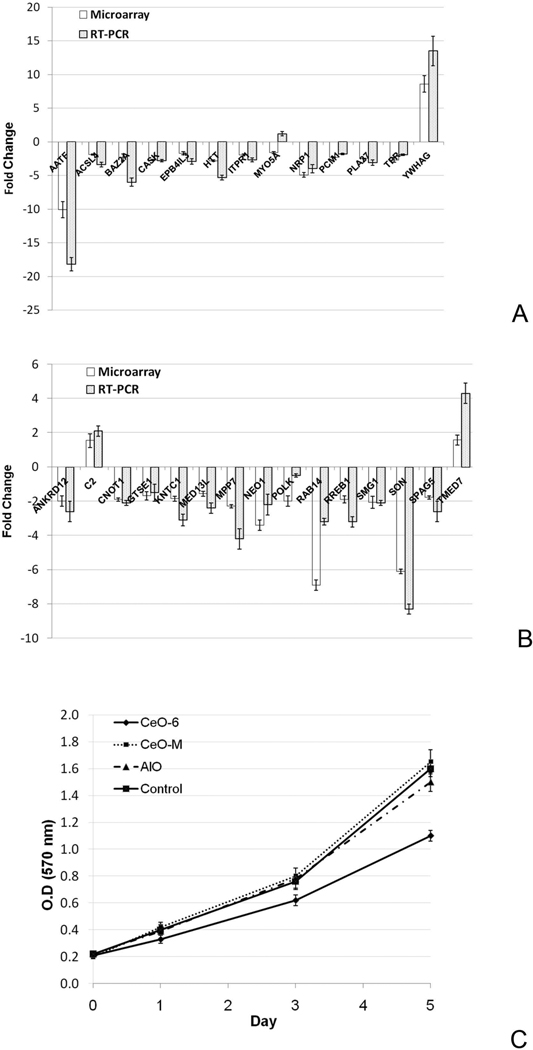
Figure 5.
Validation of microarray results in the gene networks. (A) Htt gene network. (B). The cell cycle related gene network. In both studies, genes that were specifically altered by nanoceria exposure in the corresponding gene network were selected from the microarray experiment (Black bar) and validated by real-time RT-PCR (Grey bar). The data are presented as fold change in nanoceria treated cells compared to the control HT22 cells.
Figure 4.
Cell cycle related network in HT22 cells in response to nanoceria treatment. (A) Distribution of HT22 cells at different stages of the cell cycle at 0, 8, 16 and 24 h post exposure to the particles. (B) The involvement of key cell cycle regulators and other signature genes in response to nanoceria exposure is illustrated. Red indicates up-regulated genes, green indicates down-regulated genes. Genes with a white background were not part of the nanoceria gene signature, but were integrated into the computationally generated network based on evidence in the knowledge base with strong biological relevance to that network. Top biological functions in this network include cell cycle, DNA replication, recombination and repair, and connective tissue development. (C) Immunoblot demonstrating changes in expression of several key cell cycle control proteins at specific time intervals after nanoceria exposure. D: Percentage of apoptotic HT22 cells as assessed by the sub-G1 population. The data are expressed as fold increase relative to 0 h (mean ±SD).
Figure 6.
Proliferation assay of cells exposed to different particles. Five-day proliferation of CeO-6, CeO-M, AlO and Control-exposed samples was assessed using MTT proliferation assay.
Discussion
The present study demonstrates cellular exposure to naked nanoceria particles induces changes in gene expression. Previous research on nanoceria, prepared in a similar manner, but with 0.0375M Ce(NO3)3• 6H2O rather than the 0.04M concentration used in this study,24 shows that the precipitated nanoparticles tend to be octahedrons or truncated octahedrons, as seen in typical transmission electron microscopy images of such particles (Figure 1) with crystallographic planes having {111} Miller indices primarily exposed at the nanoparticle surface. The nanoparticles, moreover, have a high surface area and few linear or planar defects. Therefore, we believe nanoceria prepared in this study better could give better evaluation. Importantly, these gene expression changes (biological responses) are related to the size and chemical/physical characteristics of nanoceria. Whether other naked ENPs have similar properties remains unclear.
The exact mechanism(s) by which nanoceria interacts with neuronal cells and causes such unique genomic alterations remains largely unknown at this moment. However, abundant research on ceria in the catalysis field has shown that the surface of nanoceria is a dynamic place, and it is here where reactive oxygen species like peroxides and superoxides have been observed to form when molecular oxygen, O2, adsorbs on a defect site.34, 35 This tendency is the basis for a recently developed hydrogen peroxide sensor that incorporates 3–5 nm ceria nanoparticles in its design.36 Some suggested these unstable oxygen species are part of reaction pathways to catalyze redox reactions on doped ceria such as CO oxidation,37 diesel soot oxidation38 and reactions in other oxides.39 These species may also help to re-oxygenate oxygen vacancies on the surface of ceria,40,41 which is an important step in the redox cycle in various biological pathways. Superoxide species, in particular, have been shown by theoretical calculations to be key entities in low temperature oxygen buffering,42 which is the ability to attenuate rapid variations in oxygen partial pressure.43 Moreover, the surface of ceria is a region of rapid oxygen transport. For instance, at 400°C, the surface oxygen diffusion coefficients are high compared to other oxides; combined with a small amount of rhodium metal, these coefficients for cerium oxide and silicon oxide are 6×10−16m/s and 3×10−20m/s, respectively—a four order of magnitude difference.44 Oxygen hopping even occurs at room temperature according to a recent atomic force microscopy study.45
Given these unique properties of nanoceria, We speculate the unique effects of nanoceria may result from size-dependent interactions with membrane receptors.28 We also postulated it could induced directly at the transcriptional level or indirectly through affecting other genomic programs, including affecting other cellular pathways (e.g. ROS related genes or pathways, which we have shown in our previous study) or factors that lead to altered gene expression regulation and protein activities (Supplementary Materials Figure S3). To answer that properly we need to apply various approaches to show interaction evidence between nanoceria and the genome. This will be addressed in our next study.
Neural cell apoptosis occurs in neurodegenerative diseases,46 and deletion or down-regulation of Htt may also lead to apoptosis.47 Due to the limitations of current in-vitro model, it is not clear whether similar apoptotic and growth effect are cell-type specific and will lead to similar neurological effects. Interestingly, the apoptotic and growth inhibition properties of naked nanoceria could be a novel way to control cancer cell proliferation. In fact, gold nanoparticle was recently shown to sensitize radiotherapy of prostate cancer cell by altering cell cycle regulation.48, 49 Since current application of nanoparticles in cancer treatment have been largely applied as a vehicle for drug delivery,50, 51 targeted delivery of un-conjugated nanoceria or other nanoparticles could be a potential alternative in cancer treatment. This simple approach might reduce the cost of treatment and would not cause side effects from the conjugated drug. Nevertheless, the mechanism and potential interaction of Htt and p53/p21 in apoptosis and growth regulation need to be further studied.
Finally, we have demonstrated the utility of novel analytical methods applied to genomic datasets. This approach helps identifying underlying biological processes and builds molecular networks to explore novel biological pathways associated with nanoceria. Exposure to these nanoparticles led to extensive and unique gene alterations in a murine neuronal cell model. Previous studies of nanoceria have predominantly focused on the potential beneficial effects of its redox capacity, whereas our study provides evidence that the biological responses to nanoceria in an oxidative stress-free state may be detrimental. The consequences of nanoceria exposure in the neuronal cell model were largely associated with down-regulation of Htt and deregulation of its related pathways, inhibition of the G1/S transition, induction of apoptosis, and growth inhibition. This preliminary study has only focused on one cell model, it is important to validate whether nanoceria would produce similar consequences with other cell types, such as skin fibroblasts and lung cells, which are the immediate targets in the exposure route to these particles. Nevertheless, the demonstration that nanoceria impacts cell regulation makes it reasonable to speculate that other naked ENPs may have corresponding unique biochemical effects. The results obtained with this in vitro cell model following exposure to nanoceria may not be extrapolated as evidence of in vivo (human) consequences, but they do identify the potential health effects of these nanoparticles, particularly since nanoparticles are known to penetrate through skin52 lung53 and the blood-brain barrier54. In addition, since these particles evoke biological responses independent of the presence of conjugated materials, our results suggest the further study on the potential biological side effects of released of naked nanoparticles from the conjugated form (e.g. linked to drugs or other active components) need to be considered. In summary, our results validate concerns about the un-regulated use of nanoparticles in consumer products, and the potential side effects of nanoparticles on cellular processes cannot be overlooked.
Supplementary Material
Acknowledgment
The authors thank editorial assistance of the NIH Fellows Editorial Board. We also thank Prof. Dave Schubert at the Salk Institute, La Jolla, CA for providing the HT22 cell line.
Funding: This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), Eunice Kennedy Shriver National Institute of Child Health and Human Development, and Department of Energy under Award Number DOE DE FG02-05ER15730, and the MRSEC Program of the National Science Foundation (# DMR-0213574).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Information on conflict of interests, disclosures and financial support
All the authors declare no conflict of interests and disclosures involved in this project.
Supplementary Materials
Details on the gene network analyses are available in the Supplementary Materials.
References
- 1.Gao F, Lu Q, Komarneni S. Fast synthesis of cerium oxide nanoparticles and nanorods. J Nanosci Nanotechnol. 2006;6:3812–3819. doi: 10.1166/jnn.2006.609. [DOI] [PubMed] [Google Scholar]
- 2.Mogensen M, Sammes NM, Tompsett GA. Physical, chemical and electrochemical properties of pure and doped ceria. Solid State Ionics. 2000;129:63–94. [Google Scholar]
- 3.Greenwood NN, Earnshaw A. Chemistry of the Elements. Oxford, England: Pergamon Press; 1989. [Google Scholar]
- 4.Fullerton DJ, Westwood AVK, Brydson R, Twigg MV, Jones JM. Deactivation and regeneration of Pt/gamma-alumina and Pt/ceria-alumina catalysts for methane combustion in the presence of H2S. Catal Today. 2003;81:659–671. [Google Scholar]
- 5.Mamontov E, Egami T, Brezny R, Koranne M, Tyagi S. Lattice defects and oxygen storage capacity of nanocrystalline ceria and ceria-zirconia. J Phys Chem B. 2000;104:11110–11116. [Google Scholar]
- 6.Boaro M, Vicario M, Leitenburg Cd, Dolcetti G, Trovarelli A. The use of temperature-programmed and dynamic/transient methods in catalysis: characterization of ceria-based, model three-way catalysts. Catal Today. 2003;77:407–417. [Google Scholar]
- 7.Sutthisripok W, Sattayanurak S, Sikong L. Effect of specific surface area on oxygen storage capacity (OSC) and methane steam reforming reactivity of CeO2. J Porous Mat. 2008;15:519–525. [Google Scholar]
- 8.Suda A, Yamamura K, Sobukawa H, Ukyo Y, Tanabe T, Nagai Y, Dong F, Sugiura M. Effect of the amount of pt loading on the oxygen storage capacity of ceria-zirconia solid solution. J Ceram Soc Jpn. 2004;112:623–627. [Google Scholar]
- 9.Gupta A, Hegde MS. Ce0.78Sn0.2Pt0.02O2-delta: A new non-deactivating catalyst for hydrogen production via water-gas shift reaction. Appl Catal B-Environ. 99:279–288. [Google Scholar]
- 10.Yao HC, Yao YFY. Ceria in Automotive Exhaust Catalysts .1. Oxygen Storage. J Catal. 1984;86:254–265. [Google Scholar]
- 11.Tschope A. Interface defect chemistry and effective conductivity in polycrystalline cerium oxide. J Electroceram. 2005;14:5–23. [Google Scholar]
- 12.Henderson MA, Epling WS, Perkins CL, Peden CHF, Diebold U. Interaction of molecular oxygen with the vacuum-annealed TiO2(110) surface: Molecular and dissociative channels. J Phys Chem B. 1999;103:5328–5337. [Google Scholar]
- 13.Campbell CT, Peden CHF. Chemistry - Oxygen vacancies and catalysis on ceria surfaces. Science. 2005;309:713–714. doi: 10.1126/science.1113955. [DOI] [PubMed] [Google Scholar]
- 14.Das M, Patil S, Bhargava N, Kang JF, Riedel LM, Seal S, Hickman JJ. Auto-catalytic ceria nanoparticles offer neuroprotection to adult rat spinal cord neurons. Biomaterials. 2007;28:1918–1925. doi: 10.1016/j.biomaterials.2006.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schubert D, Dargusch R, Raitano J, Chan S-W. Cerium and yttrium oxide nanoparticles are neuroprotective. Biochemical and Biophysical Research Communications. 2006;342:86–91. doi: 10.1016/j.bbrc.2006.01.129. [DOI] [PubMed] [Google Scholar]
- 16.Tarnuzzer RW, Colon J, Patil S, Seal S. Vacancy engineered ceria nanostructures for protection from radiation-induced cellular damage. Nano Lett. 2005;5:2573–2577. doi: 10.1021/nl052024f. [DOI] [PubMed] [Google Scholar]
- 17.Niu J, Azfer A, Rogers LM, Wang X, Kolattukudy PE. Cardioprotective effects of cerium oxide nanoparticles in a transgenic murine model of cardiomyopathy. Cardiovascular Research. 2007;73:549–559. doi: 10.1016/j.cardiores.2006.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thill A, Zeyons O, Spalla O, Chauvat F, Rose J, Auffan M, Flank AM. Cytotoxicity of CeO2 nanoparticles for Escherichia coli. Physico-chemical insight of the cytotoxicity mechanism. Environmental Science and Technology. 2006;40:6151–6156. doi: 10.1021/es060999b. [DOI] [PubMed] [Google Scholar]
- 19.Lin W, Huang YW, Zhou XD, Ma Y. Toxicity of cerium oxide nanoparticles in human lung cancer cells. Int J Toxicol. 2006;25:451–457. doi: 10.1080/10915810600959543. [DOI] [PubMed] [Google Scholar]
- 20.Park EJ, Choi J, Park YK, Park K. Oxidative stress induced by cerium oxide nanoparticles in cultured BEAS-2B cells. Toxicology. 2008;245:90–100. doi: 10.1016/j.tox.2007.12.022. [DOI] [PubMed] [Google Scholar]
- 21.Zhang F, Chan S-W, Spanier JE, Apak E, Jin Q, Robinson RD, Herman IP. Cerium oxide nanoparticles: Size-selective formation and structure analysis. Applied Physics Letters. 2002;80:127–129. [Google Scholar]
- 22.Cullity BD. Elements of X-Ray Diffraction. 2nd edn. Addison Wesley Publishing Company; 1978. [Google Scholar]
- 23.Zhang F, Chen C-H, Hanson JC, Robinson RD, Herman IP, Chan S-W. Phases in ceria-zirconia binary oxide (1-x)CeO2-xZrO2 nanoparticles: the effect of particle size. Journal of the American Ceramic Society. 2006;89:1028–1036. [Google Scholar]
- 24.Zhang F, Jin Q, Chan S-W. Ceria nanoparticles: size, size distribution, and shape. Journal of Applied Physics. 2004;95:4319–4326. [Google Scholar]
- 25.Spanier JE, Robinson RD, Zhang F, Chan S-W, Herman IP. Size-dependent properties of CeO2-y nanoparticles as studied by Raman scattering. Physical Review B. 2001;64:245407/245401–245407/245408. [Google Scholar]
- 26.Perebeinos V, Chan S-W, Zhang F. 'Madelung model' prediction for dependence of lattice parameter on nanocrystal size. Solid State Communications. 2002;123:295–297. [Google Scholar]
- 27.Zhang F, Wang P, Koberstein J, Khalid S, Chan S-W. Cerium oxidation state in ceria nanoparticles studied with X-ray photoelectron spectroscopy and absorption near edge spectroscopy. Surface Science. 2004;563:74–82. [Google Scholar]
- 28.Jiang W, Kim BY, Rutka JT, Chan CW. Nanoparticle-mediated cellular response is size-dependent. Nat Nano. 2008;3:145–150. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- 29.Duyao MP, Auerbach AB, Ryan A, Persichetti F, Barnes GT, McNeil SM, Ge P, Vonsattel JP, Gusella JF, Joyner AL, et al. Inactivation of the mouse Huntington's disease gene homolog Hdh. Science. 1995;269:407–410. doi: 10.1126/science.7618107. [DOI] [PubMed] [Google Scholar]
- 30.Zeitlin S, Liu JP, Chapman DL, Papaioannou VE, Efstratiadis A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington's disease gene homologue. Nat Genet. 1995;11:155–163. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
- 31.Nasir J, Floresco SB, O'Kusky JR, Diewert VM, Richman JM, Zeisler J, Borowski A, Marth JD, Phillips AG, Hayden MR. Targeted disruption of the Huntington's disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- 32.White JK, Auerbach W, Duyao MP, Vonsattel JP, Gusella JF, Joyner AL, MacDonald ME. Huntingtin is required for neurogenesis and is not impaired by the Huntington's disease CAG expansion. Nat Genet. 1997;17:404–410. doi: 10.1038/ng1297-404. [DOI] [PubMed] [Google Scholar]
- 33.Auerbach W, Hurlbert MS, Hilditch-Maguire P, Wadghiri YZ, Wheeler VC, Cohen SI, Joyner AL, MacDonald ME, Turnbull DH. The HD mutation causes progressive lethal neurological disease in mice expressing reduced levels of huntingtin. Hum Mol Genet. 2001;10:2515–2523. doi: 10.1093/hmg/10.22.2515. [DOI] [PubMed] [Google Scholar]
- 34.Binet C, Daturi M, Lavalley JC. IR study of polycrystalline ceria properties in oxidised and reduced states. Catal Today. 1999;50:207–225. [Google Scholar]
- 35.Can L, Domen K, Maruya K, Onishi T. Dioxygen Adsorption on Well-Outgassed and Partially Reduced Cerium Oxide Studied by Ft-Ir. J Am Chem Soc. 1989;111:7683–7687. [Google Scholar]
- 36.Mehta A, Patil S, Bang H, Cho HJ, Seal S. A novel multivalent nanomaterial based hydrogen peroxide sensor. Sens Actuator A-Phys. 2007;134:146–151. [Google Scholar]
- 37.Guzman J, Carrettin S, Fierro-Gonzalez JC, Hao YL, Gates BC, Corma A. CO oxidation catalyzed by supported gold: Cooperation between gold and nanocrystalline rare-earth supports forms reactive surface superoxide and peroxide species. Angew Chem-Int Edit. 2005;44:4778–4781. doi: 10.1002/anie.200500659. [DOI] [PubMed] [Google Scholar]
- 38.Gross MS, Ulla MA, Querini CA. Catalytic oxidation of diesel soot: New characterization and kinetic evidence related to the reaction mechanism on K/CeO2 catalyst. Appl Catal A-Gen. 2009;360:81–88. [Google Scholar]
- 39.Shimizu K, Satsuma A. Reaction mechanism of H-2-promoted selective catalytic reduction of NO with NH3 over Ag/Al2O3. J Phys Chem C. 2007;111:2259–2264. [Google Scholar]
- 40.Wu ZL, Li MJ, Howe J, Meyer HM, Overbury SH. Probing Defect Sites on CeO2 Nanocrystals with Well-Defined Surface Planes by Raman Spectroscopy and O-2 Adsorption. Langmuir. 26:16595–16606. doi: 10.1021/la101723w. [DOI] [PubMed] [Google Scholar]
- 41.Nolan M. Healing of oxygen vacancies on reduced surfaces of gold-doped ceria. J Chem Phys. 2009;130 doi: 10.1063/1.3110702. [DOI] [PubMed] [Google Scholar]
- 42.Huang M, Fabris S. Role of surface peroxo and superoxo species in the low-temperature oxygen buffering of ceria: Density functional theory calculations. Physical Review B. 2007;75 [Google Scholar]
- 43.Trovarelli A. Catalysis by Ceria and Related Materials. London: Imperial College Press; 2002. [Google Scholar]
- 44.Martin D, Duprez D. Mobility of surface species on oxides .1. Isotopic exchange O-18(2) with O-16 of SiO2, Al2O3, ZrO2, MgO, CeO2, and CeO2-Al2O3. Activation by noble metals. Correlation with oxide basicity. J Phys Chem. 1996;100:9429–9438. [Google Scholar]
- 45.Namai Y, Fukui KI, Iwasawa Y. Atom-resolved noncontact atomic force microscopic and scanning tunneling microscopic observations of the structure and dynamic behavior of CeO2(111) surfaces. Catal Today. 2003;85:79–91. [Google Scholar]
- 46.Kermer P, Liman J, Weishaupt JH, Bahr M. Neuronal apoptosis in neurodegenerative diseases: from basic research to clinical application. Neurodegener Dis. 2004;1:9–19. doi: 10.1159/000076665. [DOI] [PubMed] [Google Scholar]
- 47.Cattaneo E, Zuccato C, Tartari M. Normal huntingtin function: an alternative approach to Huntington's disease. Nat Rev Neurosci. 2005;6:919–930. doi: 10.1038/nrn1806. [DOI] [PubMed] [Google Scholar]
- 48.Roa W, Zhang X, Guo L, Shaw A, Hu X, Xiong Y, Gulavita S, Patel S, Sun X, Chen J, et al. Gold nanoparticle sensitize radiotherapy of prostate cancer cells by regulation of the cell cycle. Nanotechnology. 2009;20 doi: 10.1088/0957-4484/20/37/375101. 375101. [DOI] [PubMed] [Google Scholar]
- 49.Zhang X, Xing JZ, Chen J, Ko L, Amanie J, Gulavita S, Pervez N, Yee D, Moore R, Roa W. Enhanced radiation sensitivity in prostate cancer by gold-nanoparticles. Clin Invest Med. 2008;31:E160–E167. doi: 10.25011/cim.v31i3.3473. [DOI] [PubMed] [Google Scholar]
- 50.Huang YH, Zugates GT, Peng W, Holtz D, Dunton C, Green JJ, Hossain N, Chernick MR, Padera RF, Jr, Langer R, et al. Nanoparticle-delivered suicide gene therapy effectively reduces ovarian tumor burden in mice. Cancer Res. 2009;69:6184–6191. doi: 10.1158/0008-5472.CAN-09-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.J LA, Lopez-Viota M, Delgado AV, Ruiz MA. 5-Fluorouracil-loaded iron/ethylcellulose (core/shell) nanoparticles for active targeting of cancer. J Drug Target. 2009 doi: 10.1080/10611860903244207. [DOI] [PubMed] [Google Scholar]
- 52.Baroli B, Ennas MG, Loffredo F, Isola M, Pinna R, Lopez-Quintela MA. Penetration of metallic nanoparticles in human full-thickness skin. J Invest Dermatol. 2007;127:1701–1712. doi: 10.1038/sj.jid.5700733. [DOI] [PubMed] [Google Scholar]
- 53.Wallace W. Phospholipid lung surfactant and nanoparticle surface toxicity: Lessons from diesel soots and silicate dusts. Journal of nanoparticle research. 2007;9:23–38. [Google Scholar]
- 54.Kreuter J, Shamenkov D, Petrov V, Ramge P, Cychutek K, Koch-Brandt C, Alyautdin R. Apolipoprotein-mediated transport of nanoparticle-bound drugs across the blood-brain barrier. J Drug Target. 2002;10:317–325. doi: 10.1080/10611860290031877. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.