Abstract
Environmental shotgun sequencing (or metagenomics) is widely used to survey the communities of microbial organisms that live in many diverse ecosystems, such as the human body. Finding the protein-coding genes within the sequences is an important step for assessing the functional capacity of a metagenome. In this work, we developed a metagenomics gene prediction system Glimmer-MG that achieves significantly greater accuracy than previous systems via novel approaches to a number of important prediction subtasks. First, we introduce the use of phylogenetic classifications of the sequences to model parameterization. We also cluster the sequences, grouping together those that likely originated from the same organism. Analogous to iterative schemes that are useful for whole genomes, we retrain our models within each cluster on the initial gene predictions before making final predictions. Finally, we model both insertion/deletion and substitution sequencing errors using a different approach than previous software, allowing Glimmer-MG to change coding frame or pass through stop codons by predicting an error. In a comparison among multiple gene finding methods, Glimmer-MG makes the most sensitive and precise predictions on simulated and real metagenomes for all read lengths and error rates tested.
INTRODUCTION
Prokaryotes inhabit a diverse array of environmental niches and account for most of the world's biomass (1–3). They play an integral role in many ecosystems, including the human body where a typical individual carries 10–100 times more prokaryotic cells than human cells (4). The DNA sequences of these microorganisms provide us with important information about their identities, capabilities and evolution. Traditional methods for obtaining these sequences have required scientists to select a single microbe of interest, isolate it in culture and sequence its genome to high coverage (5).
Because many microbes cannot be isolated and grown in culture, researchers have increasingly turned to sequencing DNA directly from environmental samples, an approach often called ‘metagenomics’ (6,7). Metagenomics is an effective tool for exploring natural environments (e.g. acid mine drainage (8), ocean water (9), and soil (10)) and environments on and within the human body (11). With the development of improved sequencing technologies from companies such as 454 Life Sciences and Illumina, DNA sequence reads can be obtained at increasingly higher throughput and lower cost. These transformative technologies have made it possible to use metagenomics to simultaneously analyze the genomes of entire communities of microbes in an ever-broadening collection of environments.
Identifying the protein-coding genes in an organism's genome is a fundamental step in any genome sequence analysis, and metagenomics is no different. Whether the assembly produces large contigs (8) or is highly fragmented (9), many new and interesting genes can be extracted (12). The goal of some metagenomic experiments is to compare microbial communities across environments (13,14). In these cases, accurate gene prediction is critical to perform a functional comparison (15,16).
It has long been known that sequences coding for proteins have statistical properties that differentiate them from non-coding sequences, which allows gene finding programs to identify open reading frames (ORFs) that represent protein-coding genes (17,18,19). Sequence composition is the most important discriminative feature, due primarily to the fact that the triplet patterns of coding DNA differ from non-coding DNA. These patterns can be captured by Markov chain models, which need to be trained on a set of ORFs from the same species or a close relative. State of the art prokaryotic gene finding softwares typically achieve >99% sensitivity and high precision on finished genomes (20).
As environmental shotgun sequencing has become more prevalent, computational gene prediction approaches have adapted to the particular challenges of these data. Since the source organisms of the sequences are unknown, the foremost challenge is training the statistical gene feature models. Following training, the gene finder must make predictions on short sequence fragments that frequently contain only part of a gene. Further complicating matters, metagenomic assemblies have many low-coverage contigs and unassembled singleton reads (21,22), in which sequencing errors are prevalent and create difficulties for gene prediction (23). In contrast, finished genomes have many fewer sequencing errors thanks to their deeper and more uniform coverage.
Despite these challenges, a number of methods for predicting genes in metagenomic sequences have been published, reporting varying degrees of success (24–28). All these previous methods incorporate sequence GC-content into their prediction algorithms to choose model parameters. GC-content is a simple way to identify training genomes that are likely to be evolutionarily related, and whose genes might have similar sequence composition. This task's goal overlaps considerably with that of computationally assigning sequences a phylogenetic classification, which implicitly identifies close relatives. Phylogenetic classification of metagenomic sequences is a well-studied problem for which much better statistics than GC-content have been developed (29–32). In this article, we recommend using a more sophisticated classification scheme, based on the Phymm system (29), to parameterize gene prediction models for metagenomic sequences and show that it works much better than GC-content. We further enhance model parameter estimation using unsupervised sequence clustering, in which the relationships between sequences are elucidated via a partition of the sequences into clusters, generally without the use of reference (or ‘training’) genomes (33–35). In previous work, sharing information between sequences within clusters during training improved the identification of translation initiation sites (36). We demonstrate that the use of a more advanced clustering method Scimm (34) to allow an unsupervised retraining step significantly boosts full gene accuracy as well.
In previous work, our group demonstrated that the Glimmer gene prediction software is highly effective, routinely identifying >99% of the genes in complete prokaryotic genomes (20). However, Glimmer was not designed for the highly fragmented, error-prone sequences that typify metagenomic sequencing projects today. In this article, we develop a metagenomics gene prediction system called Glimmer-MG based on the Glimmer framework. As described above, Glimmer-MG implements a metagenomics pipeline that incorporates classification and clustering of the sequences prior to gene prediction. In addition, we addressed the other metagenomics gene prediction challenges with novel and effective solutions. Glimmer-MG incorporates a thorough probabilistic model for gene length and start/stop codon presence to aid prediction of truncated genes on short sequence fragments. Glimmer-MG predicts insertion, deletion and stop codon-introducing substitution errors in order to more closely track coding frames in raw error-prone sequences. Our implementation of these features produces the most sensitive and precise gene predictions on realistically simulated metagenomes. On a set of real 454 reads from the human gut microbiome (37), Glimmer-MG makes many more gene predictions matching known proteins than other programs.
Glimmer-MG is freely available as open-source software from www.cbcb.umd.edu/software/glimmer-mg under the Perl Artistic License (www.perl.com/pub/a/language/misc/Artistic.html).
MATERIALS AND METHODS
Glimmer
Glimmer's salient feature is its use of interpolated Markov models (IMMs) for capturing gene composition (18). IMMs are variable-order Markov chain models that maximize the model order for each specific oligonucleotide window based on the amount of training data available. IMMs then interpolate the nucleotide distributions between the chosen order and one greater. Thus, IMMs construct the most sophisticated composition model that the training data sequences support. To segment the sequence into coding and non-coding sequence, Glimmer uses a flexible ORF-based framework that incorporates knowledge of how prokaryotic genes can overlap and upstream features of translation initiation sites (TIS) like the ribosomal binding site (RBS). Glimmer extracts every sufficiently long ORF from the sequence and scores it by the log-likelihood ratio of generating the ORF between models trained on coding versus non-coding sequence. The features included in the log-likelihood ratio are composition via the IMMs, RBS via a position weight matrix (PWM) and start codon usage. For simplicity, features are assumed to be independent so that the overall score can be computed as a sum of the individual feature log-likelihood ratios. A dynamic programming algorithm finds the set of ORFs with maximum score subject to the constraint that genes cannot overlap for more than a certain threshold, e.g. 30 bp.
Additional features
Glimmer is ineffective on metagenomic sequences because its gene composition model is trained under the assumption that the sequences all originated from a single genome. Recent approaches both relax this assumption and add new features used to discriminate between coding and non-coding sequence. One approach called MetaGeneAnnotator (MGA) uses a similar framework to Glimmer by scoring ORFs and choosing a high scoring set using dynamic programming (25). MGA incorporates additional gene features, of which we add three—ORF length, adjacent gene orientation, and adjacent gene distance—to Glimmer. Below, we describe how to compute models for these features given an annotated genome. In the sections to follow, we further explain how such genomes are obtained.
First, we seek probability distributions for the length of coding and non-coding ORFs. For the coding model, our sample data are the lengths of annotated genes in the training genome. For the non-coding model, the lengths of non-coding ORFs that meet a minimum length threshold (75 bp) and a maximum overlap threshold with a gene (30 bp) are considered. One can estimate the distributions using a non-parametric method based on the histogram of lengths or a parametric method where one assumes a well-studied probability distribution and computes the maximum likelihood parameters (38). We use both methods to obtain our estimate. Where training data are plentiful, such as for common gene sizes, a non-parametric approach (such as kernel smoothing) offers greater modeling accuracy than any parameterized distribution. But when data are sparse, such as for very long ORFs, the non-parametric approach fails. For example, we cannot assign a useful probability to an ORF larger than any in our training set though it should obviously receive a large log-likelihood ratio score. A parameterized distribution can assign meaningful probabilities to ORFs of any length. We analyzed a number of distributions and found that a Gamma distribution most accurately modeled the gene length distributions examined and produced the highest accuracy gene predictions.
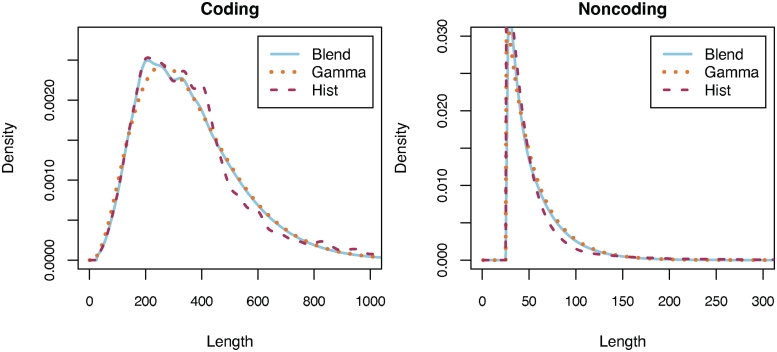
To combine the two versions, we use a histogram after kernel smoothing with a Gaussian kernel (38) for the first quartile (as determined by the raw counts), a Gamma distribution with maximum likelihood parameters for the last quartile and a linear combination of the two with a linearly changing coefficient in between (e.g. Figure 1). Performance was robust to other blending schemes and to the points at which the model changes. We score an ORF with the log-likelihood ratio that the the feature was generated by the coding versus non-coding model and add it to the ORF's overall score.
Figure 1.
Distributions for coding and non-coding ORF lengths (in amino acids) from Deinococcus radiodurans R1 estimated using the Gamma distribution (Gamma), a smoothed histogram (Hist), and a blend of the first two (Blend) that uses the histogram model for the first quartile, the Gamma model for the last quartile, and a linear combination in between. The Hist model offers greater accuracy for short and medium sized ORFs (e.g. the deviation from Gamma at 200 bp in the coding plot), but is useless for very long ORFs, which Gamma can model more effectively. The shape of the D. radiodurans length distributions are typical of the prokaryotic genomes examined, but Glimmer-MG estimates the distributions for each genome individually.
ORFs truncated by the end of their fragments require an adjustment to the length model. We know that the total length of a truncated ORF with X bp on a fragment is at least X and should therefore be scored higher than a complete X bp ORF. We accomplish this by modeling the joint distribution of the length and the presence of start and stop codons (Supplementary Methods).
Features computed on pairs of adjacent genes also capture useful information. For example, genes are frequently arranged nearby in the same orientation to form transcriptional units called operons (39). Alternatively, consecutive genes with opposing ‘head-to-head’ orientations (where the 5′-ends of the genes are adjacent) tend to be further apart to allow room for each gene's respective RBS. We added two features of adjacent genes: their orientation with respect to each other and the distance between them. Again, we need distributions for coding and non-coding ORFs to score a pair of adjacent genes by their log-likelihood ratio. The gene model uses all adjacent pairs of annotated genes. For the non-coding model, we consider pairs including non-coding ORFs that satisfy the length and overlap constraints with their adjacent annotated genes.
For adjacent gene orientation, we count the number of times each adjacent arrangement appears in the training data and normalize the counts to probabilities. The adjacent gene distance model is estimated similarly to the gene length models described above. However, common parameterized distributions were not a good fit for the distances so we rely solely on a smoothed histogram. Because one gene's start codon often overlaps another gene's stop codon due to shared nucleotides, we do not smooth the histogram for distances implying overlapping start or stop codons. We incorporate these features during Glimmer's dynamic programming algorithm for choosing ORFs by adding the log-likelihood ratios when linking an ORF to its previous adjacent ORF.
Classification
All previously published approaches to metagenomic gene prediction parameterize the gene composition models as a function of the sequence GC-content. For example, MetaGeneMark computes (offline) a logistic regression for each dicodon frequency as a function of GC-content for a large set of training genomes and sets its hidden Markov model parameters (online) according to the GC-content of the metagenomic sequence (28). For whole genomes, gene composition model training has traditionally been performed on annotated close evolutionary relatives rather than genomes with similar GC-content (40). Many methods for assigning a taxonomic classification to a metagenomic sequence are currently available (29–32). Here, we suggest using one of these methods called Phymm (29), rather than GC-content, to find evolutionary relatives of the metagenomic sequences on which to train. Phymm trains an IMM on every reference genome in GenBank (41), scores each input sequence with all IMMs and assigns a classification at each taxonomic level according to the reference genome of the highest scoring IMM. Phymm's IMMs are single-periodic and trained on all genomic sequence, in contrast to Glimmer's IMMs which are three-periodic and trained only on coding sequences.
Thus, before predicting genes, we run Phymm on the input sequences to score each sequence with each reference IMM. To train the gene prediction models, we use gene annotations for the genomes corresponding to the highest scoring IMMs. These annotations are taken from NCBI's RefSeq database (42). Though classification with Phymm is very accurate, the highest scoring IMM is rarely from the sequence's exact source genome. For this reason, we found that training over multiple genomes (e.g. 43) captured a broader signal that improved prediction accuracy. Though most of the training can be performed offline, the models over multiple genomes must be combined online for each sequence. Features such as the length, start codon and adjacent gene distributions are easy to combine across multiple training genomes by simply summing the feature counts.
IMMs cannot be combined quickly, and saving trained IMMs for all combinations of two or three genomes would require too much disk space. In practice, pairs of genomes with similar composition are far more likely to be top classification hits together and we can restrict our offline training to only these pairs (Supplementary Methods).
Glimmer-MG's RBS model trains using ELPH (http://cbcb.umd.edu/software/ELPH), a motif finder based on Gibbs sampling, to learn a 6-bp PWM from the 25-bp upstream of every gene in the training set. We train these PWMs offline for each individual reference genome, but like the other features, RBS modeling for metagenomic sequences benefits from the broader signal obtained by combining over multiple training genomes. Averaging PWMs for the top three Phymm classifications can be done quickly, but dilutes the signal. Instead, we generalized the RBS model in Glimmer-MG to score the upstream region of each start codon using a mixture of PWMs in equal proportions. Thus, a gene's RBS score is the probability that the best 6 bp motif in the 25-bp upstream of the start codon was generated by a mixture of three PWMs normalized by a null model based on GC-content to a log-likelihood ratio.
Two interesting cases warrant further discussion. First, a novel sequence may not be phylogenetically related to any known reference genome in the database. Here, Phymm's highest scoring IMMs will merely represent the reference genomes with most similar nucleotide composition. Prior work demonstrating the relationship between even simple nucleotide composition statistics and prediction model parameters supports the validity of this strategy (24–28). In addition, we did not detect a significant relationship between prediction accuracy and the divergence of a sequence from the reference genome database (Supplementary Figure S2). Second, some sequences will contain horizontally transferred genes. While single genome gene prediction typically cannot implement a model general enough to predict these genes accurately, Glimmer-MG is more robust because Phymm will likely ‘mis-classify’ the sequence containing the gene by scoring the sequence more highly with IMMs more representative of the genome from which the gene was transferred than the sequence's true source genome.
Clustering
The following prediction pipeline has been applied successfully on whole prokaryotic genomes. First, train models on a finished and annotated close evolutionary relative. Make initial predictions, but then retrain the models on them and make a final set of predictions (40). By using Phymm to find training genomes, we replicate the first step in this pipeline for application to metagenomics. However, retraining on the entire set of sequences would combine genes from many different organisms and yield a non-specific and ineffective model. If the sequences could be separated by their source genome, retraining could be applied.
We accomplish this goal using Scimm, an unsupervised clustering method for metagenomic sequences that models each cluster with a single-periodic IMM (34). After initially partitioning the sequences into a specified number of clusters, Scimm repeats the following three steps until the clusters are stable: train IMMs on the sequences assigned to their corresponding clusters, score each sequence using each cluster IMM and reassign each sequence to the cluster corresponding to its highest scoring IMM. While Scimm may not partition the sequences exactly by their source organism, the mistakes that it tends to make do not create significant problems for retraining gene prediction models. In cases where Scimm merges sequences from two organisms together, they are nearly always phylogenetically related at the family level (34). Though some families can be quite diverse, this shared phylogenetic relationship, combined with the nucleotide composition similarity that Scimm more directly identifies, is encouraging. Scimm sometimes separates sequences from a single organism into multiple clusters, but this occurs most often for highly abundant organisms, in which case there will usually still be enough training data in each cluster to be informative. The Phymm classifications that have already been obtained imply an initial clustering at a specified taxonomic level (e.g. family), which can be used as an initial partition for the iterative clustering optimization in a mode of the program referred to as PhyScimm (34). Using PhyScimm also implicitly chooses the number of clusters, removing this free variable.
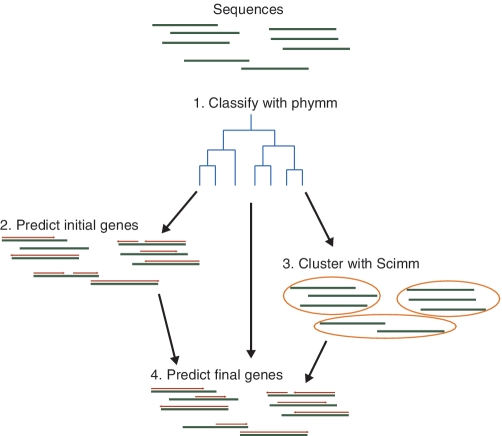
After clustering the sequences, we focus on each cluster individually to retrain the coding IMM, RBS and start codon models before making the final predictions within that cluster. The ORF length and adjacent ORF feature distributions are more difficult to estimate from short sequence fragments, so we continue to learn them using the Phymm classifications to whole annotated genomes. If the cluster is too small, retraining may not have enough data to capture the gene features, and prediction accuracy may decrease. We tested various thresholds and requiring at least 80 Kb of predicted coding sequence for retraining produced the highest accuracy predictions. For clusters with less, we do not retrain and instead finalize the gene predictions from the initial iteration. Accuracy may also decrease if the cluster is heterogeneous and does not effectively model some of its sequences. For each sequence, we compute the ratio between the likelihood that the cluster IMM versus its top scoring Phymm IMM generated the sequence. If the ratio is too low, we assume that the cluster does not represent this sequence well enough and finalize its initial predictions. The full pipeline for metagenomic gene prediction is depicted in Figure 2.
Figure 2.
Glimmer-MG pipeline. First, we classify the sequences with Phymm in order to find related reference genomes to train the feature models. We use these to make initial gene predictions. Next, we cluster the sequences with Scimm, starting at an initial partition from the Phymm classifications. Within each cluster, we retrain the models on the initial predictions before using all information to make the final set of predictions.
Sequencing errors
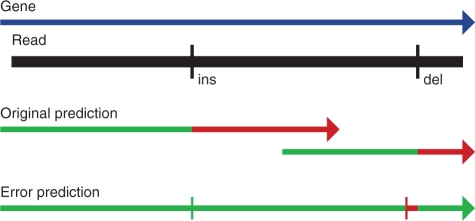
Gene prediction on raw sequencing reads or contigs with low coverage must contend with sequencing errors. The most prevalent type of error made by the 454 sequencing technology is an insertion or deletion (indel) at a homopolymer run. Indels cause major problems for gene prediction by shifting the coding frame of the true gene, making it impossible for a method without a model for these errors to predict it exactly. When Glimmer-MG encounters a shifted gene, the most frequent outcome is two predictions, each of which covers half of the gene up to the point of the indel and then beyond (Figure 3). Such predictions have limited utility.
Figure 3.
Indel errors. Depicted above is a common case where indel sequencing errors disrupt a gene prediction. This 454 simulated 526 bp read falls within a gene in the forward direction, but has an insertion at position 207 and a deletion at position 480. Without modeling sequencing errors, Glimmer-MG begins to correctly predict the gene (shown in green), but is shifted into the wrong frame by the insertion (shown in red) and soon hits a stop codon. Downstream, Glimmer-MG makes another prediction in the correct coding frame, but it too is forced into the wrong frame by the deletion. By allowing Glimmer-MG to predict frameshifts from sequencing errors, the prediction follows the coding frame nearly perfectly. The insertion site is exactly predicted and the deletion site is only off by 19 bp.
While the problems caused by sequencing errors have been known for some time (23,22), only recently has a good solution been published in the program FragGeneScan (26). FragGeneScan uses a hidden Markov model where each of the three indexes into a codon are represented by a model state, but allows irregular transitions between the codon states that imply the presence of an indel in the sequence. On simulated sequences containing errors, FragGeneScan achieves far greater accuracy than previous methods that ignore the possibility of errors.
Since Glimmer-MG uses an ORF-based approach to gene prediction, we must take a more ad hoc approach to building an error model into the algorithm. First, we address 454 indel errors. When Glimmer-MG is scoring the composition of an ORF using the coding and non-coding IMMs, we allow branching into alternative reading frames. More specifically, we traverse the sequence and identify low-quality base calls (defined below) that are strong candidates for a sequencing error. At these positions, we split the ORF into three branches. One branch scores the ORF as is. The other two switch into different frames to finish scoring, implying an insertion and deletion prediction. ORFs that change frames are penalized by the log-likelihood ratio of the predicted correction's probability to the original base call probability. A maximum of two indel predictions per ORF is used to limit the computation time. After scoring all ORFs, ORFs with the same start and stop codon (but potentially different combinations of interior indels) are clustered and only the highest scoring version is kept. All remaining ORFs are pushed to the dynamic programming stage where the set of genes with maximum score subject to overlap constraints is chosen. However, the algorithm is further constrained to disallow an indel prediction in a region of overlapping genes.
Focusing on low-quality base calls (typically <5–10% of the sequence) makes the computation feasible. If quality values are available for the sequences, either from the raw read output or the consensus stage of an assembler, Glimmer-MG uses them and designates base calls less than a quality value threshold as potential branch sites. For 454 sequences that are missing quality values, we designate the final base of homopolymer runs longer than a length threshold as potential branch sites.
In Illumina reads, indels are rare, and the primary errors are substitutions (44). Most substitutions do not affect a start or stop codon and are nearly irrelevant to the gene prediction. We focus on the most detrimental error, which is a substitution that converts a regular codon to a stop codon, thus prematurely truncating the gene. To predict such errors, we consider substitution errors to remove each stop codon in the sequence. That is, for every ORF, we consider an altered ORF where the previous stop codon did not exist, thus combining the current ORF with the previous one in the same frame. Similarly to the 454 error model, we penalize these altered ORFs with the log-likelihood ratio (based on the quality values) comparing the probability that the stop codon contains a sequencing error that changed it from a regular codon to the probability that it truly is a stop codon. All normal and altered ORFs are considered during the dynamic programming stage to choose the maximum scoring set of ORFs.
Whole genomes
Although we implemented the additional gene features with metagenomics in mind, they improve accuracy on whole genomes as well. In Glimmer3.0, the following pipeline was recommended (20). First, using a program called long-orfs, find long non-overlapping ORFs in the sequence with amino acid composition that is typical of prokaryotic genomes. Train the coding IMM on these sequences, and predict genes on the genome. On the initial predictions, train the RBS and start codon models. Finally, make a second set of gene predictions incorporating the new models.
We recommend a similar scheme for a new whole-genome pipeline, designated as Glimmer3.1. As before, we use long-orfs to train an IMM and predict an initial set of genes. Without a length model, these initial predictions tend to include many erroneous small gene predictions. We use a log-likelihood ratio threshold to filter out the lowest scoring ones. On the remaining genes, we retrain all models—IMM, RBS, start codons, length and adjacency features—before predicting again. To eliminate any remaining bias from the initial prediction and filtering, we retrain and predict one final time.
The preceding pipeline is unsupervised, but we can do slightly better on average by following Glimmer-MG and using GenBank reference genomes. In this pipeline, we first classify our new genome with Phymm to find similar reference genomes. Alternatively, a researcher may be able to specify these genomes based on prior knowledge. We train RBS, start codon, length and adjacency models from the RefSeq annotations of these similar genomes as described above. For the gene IMM, accuracy is better if we use long-orfs compared with an IMM trained on relative reference genomes. After making initial predictions, we retrain the IMM, RBS and start codon models before predicting genes a final time.
Simulated metagenomes
We constructed simulated datasets from 1206 prokaryote genomes in GenBank (41) as of November 2010. Since Glimmer-MG involves clustering the sequences, it is important to have realistic simulated metagenomes. For each metagenome, we randomly chose 50 organisms and included all chromosomes and plasmids. We sampled organism abundances from the Pareto distribution, a power law probability distribution that has previously been used for modeling metagenomes (45). Reference genomes included in the metagenome were removed from Phymm's database so that the sequences appeared novel and unknown. To simulate a single read, we selected a chromosome or plasmid with probability proportional to the product of its length and the organism's abundance and then chose a random position and orientation from that sequence. To enable comparison between experiments with different read lengths and error rates, we simulated 20 metagenomes (i.e. organisms, abundances, read positions and read orientations) and used them to derive each experiment's dataset. We labeled the reads using gene annotations that are not described as hypothetical proteins from RefSeq (42).
In experiments where we considered sequencing errors, we focused on three prevailing technologies. Two varieties of high-throughput, short-read technologies with very different characteristics have become ubiquitous tools for sequencing genomes, including metagenomics (46). The Illumina sequencing platform generates 35–150 bp length reads with sequencing errors consisting almost entirely of substitutions (44). The 454 sequencing platform generates 400–550 bp length reads where indels make up nearly all of the errors (47). Less popular in recent studies due to greater expense and lesser throughput is Sanger sequencing with read lengths of 600–1000 bp and both substitution and indel sequencing errors. We include Sanger sequencing both because previous programs were designed and tested with the technology in mind and because the reads resemble contigs assembled from the more prevalent short read technologies with respect to length and errors tending to occur at the fragments' ends.
To imitate Sanger reads, we used the lengths and quality values from real reads taken from the NCBI Trace Archive (41) as templates. That is, for each fragment simulated from a genome as described above, we randomly chose a real Sanger read from our set to determine the length and quality values of the simulated read. Then we simulated errors into the read according to the quality values and using a ratio of five substitutions per indel. To achieve a specific error rate for a dataset, we multiplied the probability of error at every base by a factor defined by the desired rate. To generate simulated reads to imitate the Illumina platform, we similarly used real 124 bp reads as templates to obtain quality values, but injected only substitution errors. For 454 reads, we used a read simulator called FlowSim, which closely replicates the 454 stochastic sequencing process to generate the sequences and their quality values (43). We conservatively quality trimmed all read ends to avoid large segments of erroneous sequence.
Accuracy
We computed accuracy a few ways to capture the multiple goals of gene prediction. Sensitivity is the ratio of true positive predictions to the number of true genes, and precision is the ratio of true positive predictions to the number of predicted genes. Since the RefSeq annotations tend to be incomplete after the removal of hypothetical proteins, which are unconfirmed computational predictions, we consider sensitivity to be the more important measure as ‘false positive’ predictions may actually be real genes. For this reason, precision values in the experiments we performed are artificially low and should be interpreted carefully. For all experiments, we computed the sensitivity and precision of the 5′-and 3′-ends of the genes separately. Since there is only a single 3′ site, 3′ prediction is generally given more attention. There are frequently many choices for the 5′-end of the gene and a paucity of sequence information to discriminate between them. Adding to the difficulty, most of the 5′ annotations in even the high-quality RefSeq database are unverified.
In experiments with sequencing errors, indels shift the gene's frame and substitutions can compromise the start and stop codons. To measure the ability of the gene prediction to follow the coding frame, we compute sensitivity and precision at the nucleotide level. That is, every nucleotide is considered a unit and a true positive prediction must annotate the nucleotide as coding in the correct frame. A gene prediction that is correct until a sequencing error indel but predicted in the wrong frame beyond gets partial credit, whereas a gene prediction that identifies the error location and shifts the frame of the prediction gets full credit.
RESULTS
Whole genomes
First, we evaluated the accuracy of the previous Glimmer3.0 iterated pipeline versus the new version with additional features, Glimmer3.1, and the metagenomics pipeline, Glimmer-MG. We predicted genes in 12 reference genomes that cover a wide range of the prokaryotic phylogeny and were previously used to compare Glimmer3.0 with Glimmer2 (20). Results for each of these genomes are displayed in Supplementary Table S1.
Glimmer3.1 maintains the high 3′ sensitivity of Glimmer3.0, but improves the precision by 1.3% on average mainly by predicting fewer short genes (42 predictions <150 bp per genome versus 68) due to the length model. Glimmer-MG increases precision another 1.0% by using additional models in the initial iteration, such as for gene length, learned accurately from close evolutionary relatives. Glimmer3.1 also significantly improves TIS prediction as 5′ sensitivity increases by 1.3% and precision by 1.8%. This improvement is attributable to its ability to assign greater scores to upstream start codons (which are longer genes) and penalize adjacent genes for unlikely arrangements like long overlaps. Glimmer-MG boosts sensitivity and precision relative to Glimmer3.1 by another 0.5 and 1.2%, respectively.
Simulated metagenomes — perfect reads
To compare Glimmer-MG to previous methods for metagenomics gene prediction, we first predicted genes on simulated metagenomes with perfect read data without sequencing errors using Glimmer-MG, MetaGeneAnnotator (25), MetaGeneMark (28), and FragGeneScan (26). MetaGeneAnnotator and MetaGeneMark runs used default parameters, and we set FragGeneScan's parameters for error-free sequences. Table 1 displays the programs' averaged accuracies over the 20 simulated metagenomes for each read technology. To best explore the programs' performances, we would ideally plot a series of sensitivity and precision data points similar to a receiver operating characteristic (ROC) curve. However, the other programs lack an accessible parameter to trade off sensitivity versus precision. Since this can easily be done for Glimmer-MG by adding a constant to every ORF's score, we display accuracy results for two versions of Glimmer-MG. The first, labeled Glimmer-MG, is our recommended mode where a constant 1.0 is added to every ORF's score. The second, labeled Glimmer-MG (Prec), reports accuracy at the point where Glimmer-MG's precision passes that of all other programs. Glimmer-MG (Prec) serves to compare Glimmer-MG to the other programs for users whose particular application requires highly precise predictions at some expense to sensitivity.
Table 1.
Accuracy on simulated metagenomes with perfect reads
| Technology | Method | 3′ Sensitivity | 3′ Precision | 5′ Sensitivity | 5′ Precision |
|---|---|---|---|---|---|
| Sanger | Glimmer-MG | 0.986 | 0.709 | 0.900 | 0.648 |
| (870 bp) | Glimmer-MG (Prec) | 0.986 | 0.709 | 0.900 | 0.648 |
| MetaGeneMark | 0.969 | 0.707 | 0.857 | 0.625 | |
| MetaGeneAnnotator | 0.969 | 0.702 | 0.846 | 0.613 | |
| FragGeneScan | 0.962 | 0.667 | 0.823 | 0.570 | |
| 454 | Glimmer-MG | 0.984 | 0.718 | 0.917 | 0.669 |
| (535 bp) | Glimmer-MG (Prec) | 0.982 | 0.721 | 0.916 | 0.673 |
| MetaGeneMark | 0.964 | 0.718 | 0.877 | 0.653 | |
| MetaGeneAnnotator | 0.966 | 0.707 | 0.853 | 0.625 | |
| FragGeneScan | 0.959 | 0.680 | 0.859 | 0.609 | |
| Illumina | Glimmer-MG | 0.945 | 0.695 | 0.919 | 0.676 |
| (120 bp) | Glimmer-MG (Prec) | 0.925 | 0.718 | 0.901 | 0.700 |
| MetaGeneMark | 0.901 | 0.717 | 0.871 | 0.693 | |
| MetaGeneAnnotator | 0.915 | 0.686 | 0.839 | 0.629 | |
| FragGeneScan | 0.932 | 0.663 | 0.904 | 0.643 |
Technology refers to the sequencing technology emulated. 3′ Sensitivity/Precision refer to sensitivity and precision based on predicted genes matching at their 3′-ends. Sensitivity is computed as the percentage of true genes that are correctly predicted, and precision is the percentage of predicted genes that are correct. 5′ Sensitivity/Precision is computed similarly using matches at the 5′-end (the start codon) of each gene. Accuracies are bolded when they are significantly greater than all other methods in that experiment using a one-sided sign test and 0.05 P-value cutoff [ignoring Glimmer-MG versus Glimmer-MG (Prec)].
Overall Glimmer-MG emerged as the clear best method, achieving the greatest sensitivity for every read length. Glimmer-MG's 3′ sensitivity was 1.3–1.8% greater than the second best method in each experiment, and its 5′ sensitivity was better by margins of 1.5% for Illumina reads up to 4.3% for Sanger reads. Though Glimmer-MG's 3′ precision was slightly less than MetaGeneMark on 454 reads, Glimmer-MG (Prec) demonstrates that our program can simultaneously exceed MetaGeneMark's 3′ precision and 3′ sensitivity. On Illumina 120 bp reads, MetaGeneMark made much fewer predictions than the other programs leading to higher precision (2.3% greater than Glimmer-MG for 3′), but much lower sensitivity (4.4% less than Glimmer-MG for 3′). Again, Glimmer-MG (Prec) shows that Glimmer-MG can achieve this level of precision while still maintaining greater sensitivity. FragGeneScan was designed for these short reads and had better sensitivity than MetaGeneMark or MetaGeneAnnotator, but ≥1.3% less accuracy than Glimmer-MG by all measures.
As an added benefit of first classifying the reads, Glimmer-MG can identify sequences that are likely to use an irregular translation code, such as Mycoplasma bacteria where TGA codes for tryptophan rather than a stop codon. On the 0.35% of the reads in our simulated datasets that used irregular codes, Glimmer-MG predicted genes on the 454 reads with 91.1% sensitivity and 55.2% precision compared with the next best MetaGeneMark's 65.1% sensitivity and 37.6% precision. This difference was similar for other read lengths.
To assess the value of clustering and retraining, we also computed accuracy for Glimmer-MG's initial predictions. For each read type, retraining increased 3′ sensitivity 0.6–1.9% while slightly decreasing 3′ precision 0.4–0.6%. Illumina 3′ sensitivity increased 1.9% because Phymm is less able to identify appropriate training genomes to aid the initial predictions; classification accuracy at the genus-level drops from 73.1% for Sanger reads to 34.3% for Illumina reads. After retraining, 5′ sensitivity increased 1.5–1.9% with a similar level of precision, an improvement expected based on prior work (36). Given that retraining increases accuracy, predictions on the few sequences placed in small or heterogenenous clusters will be slightly less accurate on average than predictions that benefited from retraining.
Simulated metagenomes - error reads
Real metagenomic sequences will inevitably contain sequencing errors, and prior work showed that current gene prediction software struggles with these errors (23). The recently published method FragGeneScan specifically models indel sequencing errors and achieves much greater accuracy than other approaches when the sequences are short and error-prone (26). To compare Glimmer-MG to FragGeneScan on reads containing errors, we simulated metagenomes as described using error rates ranging from 0% to 2%. We allowed Glimmer-MG to predict indels for Sanger and 454 reads and substitutions in stop codons for Illumina. We ran FragGeneScan using predefined model parameters meant for the sequencing technology and closest error rate. We included MetaGeneMark with default parameters due to its competitiveness on perfect reads, but excluded MetaGeneAnnotator for clarity because we found its performance lacking, similarly to previous evaluations (26). Table 2 displays the programs' averaged accuracies at the nucleotide level over the 20 simulated metagenomes for each read technology and error rate.
Table 2.
Accuracy on simulated metagenomes with error reads
| Method | Error | Sanger |
454 |
Illumina |
|||
|---|---|---|---|---|---|---|---|
| Sensitivity | Precision | Sensitivity | Precision | Sensitivity | Precision | ||
| Glimmer-MG | 0 | 0.989 | 0.756 | 0.988 | 0.752 | 0.952 | 0.686 |
| 0.005 | 0.972 | 0.741 | 0.907 | 0.677 | 0.944 | 0.675 | |
| 0.010 | 0.957 | 0.729 | 0.836 | 0.625 | 0.938 | 0.673 | |
| 0.020 | 0.928 | 0.709 | 0.732 | 0.547 | 0.927 | 0.670 | |
| Glimmer-MG | 0 | 0.989 | 0.756 | 0.988 | 0.754 | 0.921 | 0.724 |
| (Prec) | 0.005 | 0.972 | 0.744 | 0.907 | 0.677 | 0.910 | 0.717 |
| 0.010 | 0.954 | 0.739 | 0.836 | 0.625 | 0.901 | 0.716 | |
| 0.020 | 0.922 | 0.724 | 0.732 | 0.547 | 0.886 | 0.714 | |
| FragGeneScan | 0 | 0.977 | 0.740 | 0.975 | 0.735 | 0.935 | 0.663 |
| 0.005 | 0.953 | 0.699 | 0.846 | 0.621 | 0.923 | 0.640 | |
| 0.010 | 0.938 | 0.687 | 0.778 | 0.565 | 0.913 | 0.632 | |
| 0.020 | 0.914 | 0.674 | 0.678 | 0.501 | 0.900 | 0.625 | |
| MetaGeneMark | 0 | 0.983 | 0.755 | 0.979 | 0.753 | 0.903 | 0.718 |
| 0.005 | 0.584 | 0.743 | 0.605 | 0.622 | 0.889 | 0.716 | |
| 0.010 | 0.558 | 0.739 | 0.463 | 0.559 | 0.877 | 0.714 | |
| 0.020 | 0.519 | 0.723 | 0.322 | 0.483 | 0.859 | 0.710 | |
Error refers to the average rate at which errors were simulated into the sequences. Sensitivity/Precision refer to sensitivity and precision based on predicted genes matching at the nucleotide level. Sensitivity is computed as the percentage of true gene nucleotides that are correctly predicted, and precision is the percentage of predicted gene nucleotides that are correct. Accuracies are bolded when they are significantly greater than all other methods in that experiment using a one-sided sign test and 0.05 P-value cutoff [ignoring Glimmer-MG versus Glimmer-MG (Prec)].
Glimmer-MG outperformed FragGeneScan with respect to both sensitivity and precision on all read lengths and error rates. The improvement was particularly evident for 454 reads where, for example, Glimmer-MG achieved 5.8% greater sensitivity and 6.0% greater precision than FragGeneScan at a 1.0% error rate. Glimmer-MG's limit of 2 indels per gene did not hinder gene prediction at a higher rate of 2.0% as accuracy remained greater than FragGeneScan. MetaGeneMark achieves high precision on Sanger and Illumina reads, but with far lesser sensitivity. Even at MetaGeneMark's levels of precision, Glimmer-MG (Prec) demonstrates that Glimmer-MG achieves greater sensitivity.
Like prior work, our experiments demonstrate the difficulty of predicting genes on sequences with errors. For 454 reads where indel errors shift the gene frames, Glimmer-MG sensitivity plummeted 8% for even a 0.5% error rate. The decrease in accuracy for Sanger or Illumina reads, where the errors are mostly from substitutions, should be less worrisome to researchers. Glimmer-MG sensitivity dropped 1.7% for Sanger reads and 0.8% for Illumina reads when the error rate increased from 0% to 0.5%.
Modeling indel errors within Glimmer-MG significantly boosted performance for 454 reads. Without it, Glimmer-MG predicted with 41.9% sensitivity and 43.5% precision at a 2.0% error rate, compared to 73.2% and 54.7% with indel prediction. Sanger read prediction saw a meaningful 6.0% increase in sensitivity by modeling indels at a 2.0% error rate. Accuracy was not nearly as bad without modeling substitution errors for Illumina reads (91.0% sensitivity and 67.2% precision), which demonstrates the robustness of gene prediction to substitution errors. Nevertheless, sensitivity increased 1.7% at the expense of a 0.2% decrease in precision when Glimmer-MG was allowed to predict substitution errors that create false stop codons.
Comparing Glimmer-MG initial and final prediction accuracies indicated that sequencing errors increase the value of retraining. For 454 reads, sensitivity increased 0.8% after retraining without errors, and 2.8% with 2.0% errors. Since retraining occurred on initial predictions with lower precision, this result may be unintuitive. We can explain this as follows. Without sequencing errors, Glimmer-MG's predictions are very accurate so that the potential benefit of retraining and predicting again is limited. However, when there are sequencing errors, predicting coding sequence around indels is far more difficult, and the enhanced ability of Glimmer-MG's retrained models to identify coding sequence affects accuracy more significantly.
We measured Glimmer-MG and FragGeneScan's accuracy predicting indels in the 454 simulated reads to determine the degree to which it affected gene prediction accuracy. To do so, we computed a matching between the predicted and true indels in coding regions and called a pair separated by <15 bp a true positive. At a 1.0% error rate, Glimmer-MG correctly predicted 23.2% of the indels, with 63.8% precision. FragGeneScan more readily shifts the gene frame and made 1.9 times more indel predictions. However, they resulted in fewer true positives than Glimmer-MG (19.2% sensitivity) and far lower precision at 28.4%. For indels predicted correctly by both programs, Glimmer-MG's prediction was 2.3 bp away from the actual position on average compared with 5.2 bp away for FragGeneScan. Thus, by focusing on low-quality nucleotides in the sequences, Glimmer-MG identifies indel positions more effectively than FragGeneScan. Sensitivity for both methods may seem low, but note that, in some cases, the frame of the coding sequence can still be closely followed without predicting the correct error. For example, two nearby insertions will generally result in a deletion prediction, which restores the proper frame more parsimoniously than two insertion predictions.
Human gut microbiome
We evaluated Glimmer-MG's performance on two real metagenomic datasets from a human gut microbiome study of obese and lean twins (37). Sample TS28 consists of 303K reads sequenced by the 454 GS FLX Titanium with average length of 335 bp, and sample TS50 consists of 550K reads sequenced by the 454 GS FLX with average length of 204 bp. Evaluating prediction accuracy is more difficult for real metagenomes where there is no gold standard to compare against. We aligned the translated gene predictions made by Glimmer-MG, FragGeneScan and MetaGeneMark against the NCBI non-redundant protein database with BLAST (41,48), and considered a prediction to be a true positive if it matched a database protein with BLAST E <0.001.
Each methods' predictions and BLAST hits are displayed in Table 3. Glimmer-MG made the most aligning predictions, 4.5% more than FragGeneScan and 6.5% more than MetaGeneMark. Precision has the caveat that a ‘false positive’ prediction does not match anything in the database, but may still represent a novel gene. Glimmer-MG demonstrated the highest precision. In contrast to the simulated data experiments, MetaGeneMark predictions had lower precision than the other two algorithms. For genes that were predicted by both methods, the aligned portions of Glimmer-MG predictions were 1.4% longer than those from FragGeneScan and 10% longer than those from MetaGeneMark. Based on these results, Glimmer-MG is a much better option for predicting genes on this human microbiome dataset.
Table 3.
Human gut microbiome
| Method | Predictions | BLAST hits | Time (min) |
|---|---|---|---|
| Glimmer-MG | 853,293 | 669,257 (0.784) | 784 |
| FragGeneScan | 820,231 | 640,223 (0.781) | 172 |
| MetaGeneMark | 808,380 | 628,295 (0.777) | 35 |
Predictions refers to the number of gene predictions made by each method. BLAST hits refers to the number of predictions for which a BLAST alignment with E-value <0.001 was found to a non-redundant protein database. Following the count in parentheses is the proportion of predictions with such an alignment.
To compare the computational requirements of each method, we timed the programs on these datasets. MetaGeneMark does not consider the possibility of errors in the sequences, which leads to the lowest accuracy, but the fastest run time. Glimmer-MG is the most computationally demanding of the three programs, mainly because it performs two iterations of prediction and a retraining step. Though required, we did not include classification and clustering of the sequences in Glimmer-MG's run time, because these are integral steps in most metagenomic analysis pipelines that would typically be performed independent of which program were used to predict genes.
CONCLUSION
A number of exciting projects over the last few years have demonstrated the value of environmental shotgun sequencing. As sequencing technologies are refined, the technique has the potential to make an even greater impact. However, the resulting mixtures of reads from populations of usually unknown organisms are difficult to analyze. To realize the full potential, metagenomics bioinformatics, including gene prediction, must improve. For example, projects seeking to discover new organisms such as the Global Ocean Sampling Expedition (9,49) need accurate gene prediction to explore the functional repertoire of the many novel sequences obtained (12). Projects focused on more well-known environments are also typically interested in characterizing the functional capabilities of the microbial communities, often for comparison (13,14). Methods to perform such functional comparisons benefit greatly from accurate identification of genes (15,16).
In this article, we introduced a number of novel and effective techniques for metagenomics gene prediction in the software package Glimmer-MG. By modeling gene lengths and the presence of start and stop codons, Glimmer-MG successfully accounts for the truncated genes so common on metagenomic sequences. Where previous approaches parameterize prediction models using only the GC-content of the sequence, Glimmer-MG first classifies the sequences using a leading phylogenetic classifier Phymm and trains models using the results. By clustering the reads using the unsupervised method Scimm, we elegantly allow retraining of prediction models on the sequences themselves. Augmenting gene prediction with classification and clustering produces the most accurate predictions in our experiments. Furthermore, Glimmer-MG produces highly accurate predictions on whole genomes, by automatically identifying related training genomes, which boosts the accuracy of initial predictions used for retraining.
Insertion and deletion sequencing errors in real metagenomics data wreak havoc on gene sequences. In Glimmer-MG, we take a novel approach to predict indels in error-prone sequences by focusing the search for frameshifts at low-quality positions. This results in more accurate identification of simulated indel positions than a previous method FragGeneScan. Glimmer-MG is also the first gene prediction method to predict substitution errors affecting stop codons, which improves accuracy for Illumina reads. In our experiments with real gut microbiome reads and simulated metagenomes with multiple sequencing technologies, Glimmer-MG predicts genes on error-prone sequences far more accurately than all other methods.
Despite Glimmer-MG's contributions, predicting genes on error-prone sequences, particularly those with indels, remains a difficult problem. On simulated 454 reads with a 2.0% error rate, Glimmer-MG's sensitivity is 73.2% and precision is 54.7%, indicating that many genes are predicted incorrectly. While further computational improvements may appear, substantial gains are more likely to come from reducing the error rate via improvements to assembly of the metagenomic reads and the base-calling accuracy of the sequencing technologies. Highly diverged organisms where the optimal model parameters are unclear present another challenge to metagenomics gene prediction. However, our results here and previous work indicate that nucleotide composition models generalize well to these sequences. Finally, Glimmer-MG has the caveat that it is more computationally expensive than existing software. Classification and clustering of the sequences are required before prediction, but we expect most researchers will not find this disruptive given that these are common components of metagenomic analysis pipelines. The gene prediction step in Glimmer-MG is also more expensive than previous software due to the use of a more sophisticated probabilistic model, modeling of sequencing errors and multiple iterations. Users will need to evaluate the trade-off between greater accuracy and computational expense for their particular data. But because gene prediction with any of the programs can be easily parallelized and significant computational resources are increasing accessible [e.g. via cloud computing (49,50)], we expect many users will find the additional comptuation worthwhile.
Overall, Glimmer-MG represents a substantial advance in metagenomics gene prediction on a number of fronts. The software should prove useful for a variety of applications.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1 and 2, and Supplementary Methods.
FUNDING
National Institute of Health grants (R01-LM083873 and R01-HG006677). Funding for open access charge: National Institute of Health grants (R01-LM083873 and R01-HG006677).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Derrick Wood for helpful TIS discussion and manuscript comments.
REFERENCES
- 1.Curtis TP, Sloan WT, Scannell JW. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. USA. 2002;99:10494. doi: 10.1073/pnas.142680199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lozupone CA, Knight R. Global patterns in bacterial diversity. Proc. Natl. Acad. Sci. USA. 2007;104:11436. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. USA. 1998;95:6578. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett MC, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 6.Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 2004;68:669. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schloss PD, Handelsman J. Metagenomics for studying unculturable microorganisms: cutting the Gordian knot. Genome Biol. 2005;6:229. doi: 10.1186/gb-2005-6-8-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 9.Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, et al. The Sorcerer II Global Ocean Sampling Expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tringe SG, Von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 11.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K, Eisen JA, Heidelberg KB, Manning G, Li W, et al. The Sorcerer II Global Ocean Sampling Expedition: expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brulc JM, Antonopoulos DA, Berg Miller ME, Wilson MK, Yannarell AC, Dinsdale EA, Edwards RE, Frank ED, Emerson JB, Wacklin P, et al. Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc. Natl. Acad. Sci. USA. 2009;106:1948. doi: 10.1073/pnas.0806191105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dinsdale EA, Edwards RA, Hall D, Angly F, Breitbart M, Brulc JM, Furlan M, Desnues C, Haynes M, Li L, et al. Functional metagenomic profiling of nine biomes. Nature. 2008;452:629–632. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- 15.Kristiansson E, Hugenholtz P, Dalevi D. ShotgunFunctionalizeR: an R-package for functional comparison of metagenomes. Bioinformatics. 2009;25:2737. doi: 10.1093/bioinformatics/btp508. [DOI] [PubMed] [Google Scholar]
- 16.Sharon I, Bercovici S, Pinter RY, Shlomi T. Pathway-based functional analysis of metagenomes. J. Comput. Biol. 2011;18:495–505. doi: 10.1089/cmb.2010.0260. [DOI] [PubMed] [Google Scholar]
- 17.Borodovsky M, Mclninch JD, Koonin EV, Rudd KE, Médigue C, Danchin A. Detection of new genes in a bacterial genome using Markov models for three gene classes. Nucleic Acids Res. 1995;23:3554–3562. doi: 10.1093/nar/23.17.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fickett JW, Tung C. Assessment of protein coding measures. Nucleic Acids Res. 1992;20:6441–6450. doi: 10.1093/nar/20.24.6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen K, Pachter L. Bioinformatics for whole-genome shotgun sequencing of microbial communities. PLoS Comput. Biol. 2005;1:106–12. doi: 10.1371/journal.pcbi.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mavromatis K, Ivanova N, Barry K, Shapiro H, Goltsman E, McHardy AC, Rigoutsos I, Salamov A, Korzeniewski F, Land M, et al. Use of simulated data sets to evaluate the fidelity of metagenomic processing methods. Nat. Methods. 2007;4:495–500. doi: 10.1038/nmeth1043. [DOI] [PubMed] [Google Scholar]
- 23.Hoff KJ. The effect of sequencing errors on metagenomic gene prediction. BMC Genomics. 2009;10:520. doi: 10.1186/1471-2164-10-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoff KJ, Lingner T, Meinicke P, Tech M. Orphelia: predicting genes in metagenomic sequencing reads. Nucleic Acids Res. 2009;37(Suppl. 2):W101–W105. doi: 10.1093/nar/gkp327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noguchi H, Taniguchi T, Itoh T. MetaGeneAnnotator: detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes. DNA Res. 2008;15:387–396. doi: 10.1093/dnares/dsn027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rho M, Tang H, Ye Y. FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res. 2010;38:e191. doi: 10.1093/nar/gkq747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yok NG, Rosen GL. Combining gene prediction methods to improve metagenomic gene annotation. BMC Bioinformatics. 2011;12:20. doi: 10.1186/1471-2105-12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu W, Lomsadze A, Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38:e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brady A, Salzberg SL. Phymm and PhymmBL: metagenomic phylogenetic classification with interpolated Markov models. Nat. Methods. 2009;6:673–676. doi: 10.1038/nmeth.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diaz NN, Krause L, Goesmann A, Niehaus K, Nattkemper TW. TACOA – Taxonomic classification of environmental genomic fragments using a kernelized nearest neighbor approach. BMC Bioinformatics. 2009;10:56. doi: 10.1186/1471-2105-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monzoorul Haque M, Ghosh TS, Komanduri D, Mande SS. SOrt-ITEMS: sequence orthology based approach for improved taxonomic estimation of metagenomic sequences. Bioinformatics. 2009;25:1722. doi: 10.1093/bioinformatics/btp317. [DOI] [PubMed] [Google Scholar]
- 32.Patil KR, Haider P, Pope PB, Turnbaugh PJ, Morrison M, Scheffer T, McHardy AC. Taxonomic metagenome sequence assignment with structured output models. Nat. Methods. 2011;8:191–192. doi: 10.1038/nmeth0311-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chatterji S, Yamazaki I, Bai Z, Eisen J. Research in Computational Molecular Biology. Springer: 2008. CompostBin: a DNA composition-based algorithm for binning environmental shotgun reads; pp. 17–28. [Google Scholar]
- 34.Kelley DR, Salzberg SL. Clustering metagenomic sequences with interpolated Markov models. BMC Bioinformatics. 2010;11:544. doi: 10.1186/1471-2105-11-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kislyuk A, Bhatnagar S, Dushoff J, Weitz JS. Unsupervised statistical clustering of environmental shotgun sequences. BMC Bioinformatics. 2009;10:316. doi: 10.1186/1471-2105-10-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu G, Guo J, Liu Y, Zhu H. MetaTISA: Metagenomic Translation Initiation Site Annotator for improving gene start prediction. Bioinformatics. 2009;25:1843–1845. doi: 10.1093/bioinformatics/btp272. [DOI] [PubMed] [Google Scholar]
- 37.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning. New York: Springer; 2009. [Google Scholar]
- 39.Rocha EPC. The organization of the bacterial genome. Annu. Rev. Genet. 2008;42:211–233. doi: 10.1146/annurev.genet.42.110807.091653. [DOI] [PubMed] [Google Scholar]
- 40.Majoros WH. Methods for Computational Gene Prediction. Cambridge, UK: Cambridge University Press; 2007. [Google Scholar]
- 41.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2011;39:D32–D37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Balzer S, Malde K, Lanzén A, Sharma A, Jonassen I. Characteristics of 454 pyrosequencing dataenabling realistic simulation with flowsim. Bioinformatics. 2010;26:i420. doi: 10.1093/bioinformatics/btq365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Angly FE, Willner D, Prieto-Davó A, Edwards RA, Schmieder R, Vega-Thurber R, Antonopoulos DA, Barott K, Cottrell MT, Desnues C, et al. The GAAS metagenomic tool and its estimations of viral and microbial average genome size in four major biomes. PLoS Comput. Biol. 2009;5:e1000593. doi: 10.1371/journal.pcbi.1000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shendure J, Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 47.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 50.Schatz MC, Langmead B, Salzberg SL. Cloud computing and the dna data race. Nat. Biotechnol. 2010;28:691. doi: 10.1038/nbt0710-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.