Abstract
The tumor suppressor p53 responds to a wide variety of cellular stress signals. Among potential regulatory pathways, post-translational modifications such as acetylation by CBP/p300 and PCAF have been suggested for modulation of p53 activity. However, exactly how p53 acetylation is modulated remains poorly understood. Here, we found that SET/TAF-Iβ inhibited p300- and PCAF-mediated p53 acetylation in an INHAT (inhibitor of histone acetyltransferase) domain-dependent manner. SET/TAF-Iβ interacted with p53 and repressed transcription of p53 target genes. Consequently, SET/TAF-Iβ blocked both p53-mediated cell cycle arrest and apoptosis in response to cellular stress. Using different apoptosis analyses, including FACS, TUNEL and BrdU incorporation assays, we also found that SET/TAF-Iβ induced cellular proliferation via inhibition of p53 acetylation. Furthermore, we observed that apoptotic Drosophila eye phenotype induced by either dp53 overexpression or UV irradiation was rescued by expression of dSet. Inhibition of dp53 acetylation by dSet was observed in both cases. Our findings provide new insights into the regulation of stress-induced p53 activation by HAT-inhibiting histone chaperone SET/TAF-Iβ.
INTRODUCTION
The tumor suppressor protein (p53) is induced in response to a wide variety of stress signals and regulates the transcription of genes responsible for many cellular processes, including cell cycle regulation and apoptosis. A series of post-translational modifications are involved in p53 responses to different stimuli, and some of these modifications are known to influence regulation of p53 activity. Among the many post-translational modifications of p53, acetylation has been one of the most extensively studied (1).
The histone acetyltransferases p300/CBP (CREB-binding protein) and PCAF (p300/CBP-associated factor) acetylate p53 and enhance its transcriptional activity (2–6). The acetylation of p53 is further expanded by other acetyltransferases such as hMOF and TIP60 at lysine 120 (K120) in response to DNA damage (7). p53 can be acetylated by p300/CBP at multiple lysine residues (K164, 370, 372, 373, 381, 382 and 386) and by PCAF at K320. Earlier studies using mice with seven (7KR) or six C-terminal lysines changed to arginine (6KR) displayed only minor effects in p53-mediated activity (8–10). However, loss of acetylation at all eight lysines (8KR) completely abolished p53-mediated stress response, suggesting an indispensable role for acetylation in p53 activation (11).
We previously identified SET/TAF-Iβ and pp32 as subunits of the INHAT (inhibitor of histone acetyltransferase) complex with ‘histone masking’ activity; that is binding of these proteins to histones prevents acetylation by p300/CBP and PCAF (12). Additional studies revealed that INHAT binds the N-termini of histone tails, and modifications within histone tails affect INHAT binding (13). SET/TAF-Iβ specifically binds to unacetylated, hypo-acetylated histones and not to hyper-acetylated ones, which implies a novel in vivo function in transcriptional repression (14).
INHAT is a multiprotein complex composed of highly acidic domain-containing proteins, SET/TAF-Iβ, TAF-Iα and pp32 (12). Initial biochemical studies revealed that SET/TAF-Iβ can promote adenoviral DNA replication, nucleosome assembly and transcription (15). Both the nuclear and cytoplasmic localization of SET/TAF-Iβ indicate that it has the potential to regulate and integrate cytoplasmic and nuclear signaling pathways, including mRNA transport and stability (16). As multitasking proteins, SET/TAF-Iβ and pp32 have been reported to be negative and positive regulators of caspase-independent and -dependent apoptotic signaling, respectively (17–19). In fact, SET/TAF-Iβ was originally identified as a translocated gene in acute undifferentiated leukemia, which further supports its oncogenic activity (15,20,21).
Here, we show that SET/TAF-Iβ inhibits p53 acetylation and modulates its key effects, including cell cycle arrest and apoptosis induction. In our in vivo analysis using UAS-dSet and dp53 in Drosophila, we provide evidence that dSet inhibits acetylation of Drosophila dp53 and negatively regulates dp53-mediated apoptosis.
MATERIALS AND METHODS
Plasmids
The CMX-SET/TAF-Iβ plasmid was used as described previously (12). p53 and p53 mutants were inserted into pGEX-4T1 bacterial expression vector (Amersham Biosciences) to construct glutathione S-transferase (GST) fusion proteins. In order to construct the mammalian expression vectors, we employed modified pcDNA6-HA-myc-his (Invitrogen) and used pGEX-4T1-p53 to create the HA, myc and his-tagged p53 and p53 mutants. sh-RNA against human SET/TAF-Iβ (RHS4533) was purchased from Openbiosystems.
Antibodies
Antibodies against p53 (DO-1) (Santa Cruz Biotechnology), acetyl-p53 (K320) (Millipore), acetyl-p53 (K373/382) (Millipore), acetyl lysine (Ac-K) (Santa Cruz Biotechnology), SET/TAF-Iβ (Santa Cruz Biotechnology), anti-myc (Santa Cruz Biotechnology) and β-actin (Santa Cruz Biotechnology) were employed for immunoblot, immunoprecipitation and chromatin immunoprecipitation (ChIP) analyses.
In vitro INHAT assay
INHAT assays were performed by incubating 20–30 pmol of purified GST-SET/TAF-Iβ with 1 µg of GST-p53 in HAT buffer (12) for 15 min on ice. Following pre-incubation, 1 pmol of PCAF or 1 µg of p300 along with 14[C]-acetyl CoA (50 µCi/µl, Perkin Elmer) or 100 µM acetyl coenzyme A were added for 2 h at 30°C. Reaction products were separated by SDS–PAGE and analyzed by a phosphorimager. For scintillation counting, p53-K320 peptides [PQPKKKPLDG] and p53-K383 peptides [SRRKKLMFKT] were synthesized based on the N-terminal amino acid sequences of histone H3 (Peptron). Peptides were filtered using p81 filter paper (Upstate Biotechnology) and washed three times with cold 10% TCA and 70% ethanol for 5 min at RT. The filters were then allowed to air dry, followed by the addition of 1 ml of Ultima Gold (Perkin Elmer). 14[C]-acetyl CoA was quantified using a scintillation counter.
Liquid chromatography–mass spectrometry
Synthetic peptides (p53-K320 or p53-K383) (100 µM) were used as substrates in the INHAT assay with SET/TAF-Iβ and PCAF or p300. The reaction was stopped with 10% TCA precipitation for 10 min at 4°C. After removing the precipitates by centrifugation, the supernatants were retrieved and acetylated peptides in the supernatants analyzed by liquid chromatography–mass spectrometry (LC–MS) at the Korea Basic Science Institute. The eluted peptides were separated on a Luna column (C18 PepMap 100, 150 × 1 mm 5 micron) with a linear gradient (A: 5% ACN, 0.1% formic acid; B: 95% ACN, 0.1% formic acid) at a flow rate of 50 µl/min. Typically, 5 μl of sample was injected. Mass spectrometry was performed on a linear ion trap mass spectrometer (LCQ DECA XP, Thermo Finnigan) coupled to a nano-LC system (NANOSPACE SI-2, Shiseido). The MS scan range was 160–2000 m/z.
Immunoprecipitation
For the p53 and SET/TAF-Iβ interaction assays, transfected cells were lysed in RIPA lysis buffer and immunoprecipitated with anti-p53, anti-SET/TAF-Iβ and anti-c-myc antibodies overnight at 4°C. Protein A/G agarose beads (GenDEPOT) were added for 1 h with rotation at 4°C. The bound proteins were analyzed by immunoblotting with anti-p53 and anti-SET/TAF-Iβ antibodies.
In vitro transcription and translation reactions
For in vitro transcription and translation, the [35S]-labeled SET/TAF-Iβ proteins were constructed with a coupled transcription and translation (TNT) system (Promega). In brief, 1 µg of DNA was directly added to TNT rabbit reticulocyte lysates and then allowed to react for 2 h at 30°C. The reaction mixtures were utilized in a GST pull-down assay using GST-p53 and GST-p53 deletion mutants. The precipitated proteins were separated via 10% SDS–PAGE and visualized by autoradiography.
Transfection assay
Transfection assay was conducted using the p21 promoter reporter along with CMX-SET/TAF-Iβ, HA-p53, flag-PCAF, SET/TAF-IβΔ3 and sh-SET/TAF-Iβs. The quantity of DNA in each transfection was kept constant via the addition of CMX. U2OS, 293 and H1299 cells were transfected with the p21 promoter reporter (100 ng), HA-p53, flag-PCAF (100 ng), CMX-SET/TAF-Iβ (100, 200 ng), SET/TAF-IβΔ3 (200 ng), sh-SET/TAF-Iβs (200 ng), NAP-1 (200 ng) and si-HDAC1 (200 nM). At 24 h after transfection, the cells were treated with 330 nM TSA or 5 mM nicotinamide (Sigma). Cells were then harvested and assayed for luciferase activity using a luciferase assay system (Promega). Each value is expressed as the mean of six replicates of a single assay, and the results were confirmed by performing at least three repetitions.
RT–PCR
U2OS and H1299 cells were transfected with p53, SET/TAF-Iβ and sh-SET/TAF-Iβ. Total RNAs were then isolated using RNAiso Plus (TaKaRa). cDNA was generated with 1 µg of RNA using oligodTs and reverse transcriptase (RTase) (Enzynomics). Primers and PCR conditions have been described previously (22).
Chromatin immunoprecipitation
U2OS and H1299 cells were transfected with 4 µg of DNA and harvested after 48 h. Cells were then cross-linked with 1% formaldehyde in medium for 10 min at 37°C, followed by the addition of 125 mM glycine for 5 min at RT, after which they were scraped into SDS lysis buffer. The cells were further sonicated and diluted for immunoprecipitation with antibodies as indicated. The resulting immunoprecipitates were eluted and reverse cross-linked. DNA fragments were purified and PCR amplified for quantification. Anti-p53, anti-p300 and anti-PCAF were employed for immunoprecipitation. The primer sequences were as follows: p21 (forward, 5′-GTG GCT CTG ATT GGC TTT CTG-3′; reverse, 5′-CTG AAA ACA GGC AGC CCA AG-3′) and PUMA (forward, 5′-GTA CAT CCT CTG GGC TCT GC-3′; reverse, 5′-GGA CAG TCG GAC ACA CAC-3′).
MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay
Two thousand cells were seeded (48-well plates) 24 h prior to transfection with p53, SET/TAF-Iβ and sh-SET/TAF-Iβ. After 0–4 days of incubation at 37°C, 20 µl of MTT (1 mg/ml) was added and incubated for 4 h at 37°C, followed by aspiration of the medium and addition of 200 µl of dimethyl sulfoxide (DMSO). The OD was determined using a spectrophotometer at a wavelength of 570 nm.
Flow cytometry
Forty-eight hours after transfection, cells were fixed in 70% ethanol for 24 h, treated with RNase (20 mg/ml) and stained with propidium iodide. Cell cycle profiles were analyzed by FACS Calibur (Becton Dickinson).
BrdU incorporation assay
The assay was performed according to the manufacturer’s instructions (Roche Diagnostics). U2OS and H1299 cells were cultured on 48-well plates and transfected with p53, SET/TAF-Iβ and sh-SET/TAF-Iβ. After 72 h, cells were incubated in medium containing BrdU for 2 h, after which the absorbance was measured at 370 nm (reference wavelength : 475 nm).
Apoptosis by TUNEL assay
Apoptosis was detected by TUNEL (TdT-mediated dUTP nick end labeling) assay (In Situ Cell Death Detection Kit, Roche). Briefly, H1299 cells were seeded in chamber slides and transfected with p53, SET/TAF-Iβ SET/TAF-IβΔ3 and sh-SET/TAF-Iβ. The cells were then fixed with 4% paraformaldehyde for 1 h at RT, followed by incubation for 1 h at 37°C with terminal deoxynucleotidyl transferase enzyme (TdT) in reaction buffer. The cells were then incubated with AP (alkaline phosphatase)-conjugated antibody for 1 h at 37°C, and the reaction was developed with NBT/BCIP substrate for 2 h at RT.
Drosophila culture and stocks
Drosophila melanogaster was cultured at 25°C by following the standard method. Wild-type Oregon-R, w-, GMR-gal4 and UAS-dp53 were obtained from the Bloomington Stock Center (Bloomington, USA). Drosophila Set (dSet) 810 bp coding sequences were subcloned into pUAST vector to generate the pUAS-dSet construct. UAS-dSet transgenic fly was obtained by P-element-mediated germ-line transformation (23). To express these UAS lines, the UAS/gal4 system was used (24). Detailed information is provided in the Supplementary Data.
RESULTS
Inhibition of p53 acetylation by SET/TAF-Iβ
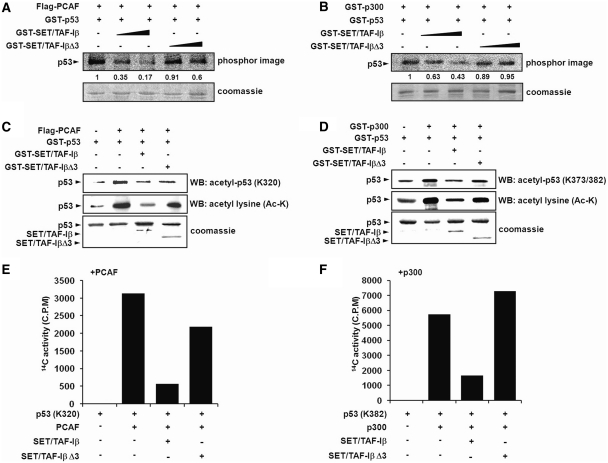
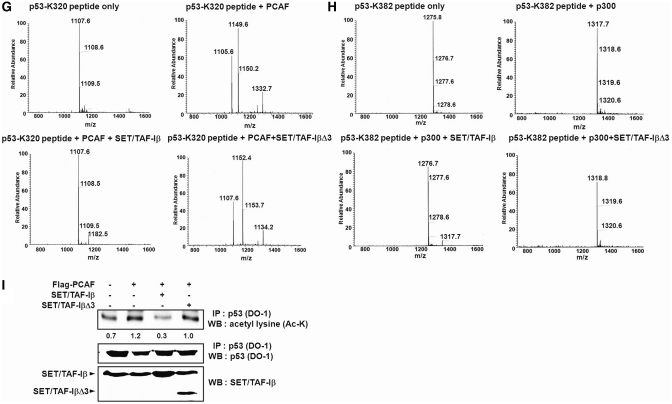
To understand the regulation of p53 acetylation through the HAT inhibitory activity of SET/TAF-Iβ, we first tested whether or not SET/TAF-Iβ can inhibit p300- and PCAF-mediated p53 acetylation using HAT assays both in vitro and in vivo. Both recombinant PCAF- and p300-acetylated p53, but the addition of increasing amounts of SET/TAF-Iβ inhibited this (Figure 1A and B). Using C-terminal INHAT domain-deleted mutant SET/TAF-IβΔ3, we verified that INHAT domain was responsible for the inhibition of p300- and PCAF-mediated p53 acetylation (Figure 1A and B). Coomassie staining revealed consistent p53 levels in the INHAT assay. Immunoblotting performed on the proteins after in vitro HAT assay showed strong inhibition of both p300- and PCAF-mediated p53 acetylation by the INHAT domain of SET/TAF-Iβ, but not by SET/TAF-IβΔ3 (Figure 1C and D). Recent studies have shown that p53 can be acetylated by p300/CBP at multiple C-terminal lysine residues, including K373 and K382 (2,25). In addition, p53 can be acetylated by PCAF at K320 (4). To further elucidate whether or not SET/TAF-Iβ can inhibit specific p53 lysine targets of p300 and PCAF, we used antibodies specific for acetyl-p53 (K320) and acetyl-p53 (K373/382) residues. Addition of SET/TAF-Iβ clearly decreased the acetylation of p53-K320 and p53-K373/382 residues (Figure 1C and D). As expected, the effects of SET/TAF-βΔ3 were minimal (Figure 1C and D). To further confirm the p53 acetylation inhibitory activity of SET/TAF-Iβ, we conducted HAT assay using p53 peptides containing two independent acetylation target lysine residues and measured acetylation levels by scintillation counting. HAT activities of PCAF and p300 as well as INHAT activity of SET/TAF-Iβ were tested first using core histones (Supplementary Figure S1A and S1B). Incubation of p53 (K320) and p53 (K382) peptides with PCAF and p300, respectively, induced strong acetylation. Addition of SET/TAF-Iβ but not SET/TAF-IβΔ3 significantly reduced acetylation levels, clearly confirming the INHAT activity of SET/TAF-Iβ toward non-histone protein p53 (Figure 1E and F). Mass spectrometry analysis demonstrated strong inhibitory activity of SET/TAF-Iβ but not SET/TAF-IβΔ3 toward both PCAF- and p300-mediated p53 acetylation (Figure 1G and H). The inhibition of endogenous p53 acetylation by SET/TAF-Iβ was further confirmed in vivo by immunoprecipitation (IP) of p53. In transient co-transfection assays in p53-positive HCT116 cells, a high level of p53 acetylation was observed (Figure 1I). As expected, overexpression of SET/TAF-Iβ induced a 4-fold decrease in p53 acetylation. The observation that overexpression of SET/TAF-IβΔ3 decreased the acetylation level of p53 could suggest that inhibition of p53 acetylation is mostly dependent on the acidic C-terminus of SET/TAF-Iβ (Figure 1I).
Figure 1.
SET/TAF-Iβ inhibits PCAF- and p300-mediated p53 acetylation. (A and B) Acetylation assays of p53 with PCAF and p300 were performed with increasing concentrations of SET/TAF-Iβ or SET/TAF-Iβ-Δ3. Autoradiogram of INHAT assay using recombinant PCAF (A) or p300 (B) with SET/TAF-Iβ and SET/TAF-IβΔ3 on GST-p53 (top) followed by coomassie staining (bottom). Numbers below phosphorimage represent quantification of p53 acetylation. (C and D) Western blot analysis of in vitro p53-acetylation assay with PCAF (C) or p300 (D), GST-p53, GST-SET/TAF-Iβ or GST-SET/TAF-IβΔ3 using anti-acetyl-p53 (K320) or anti-acetyl-p53 (K373/382) and anti-acetyl lysine. The levels of GST-SET/TAF-Iβ and SET/TAF-Iβ-Δ3 are shown in the bottom panel by coomassie staining. (E and F) p53 peptides (p53-K320 and p53-K382) were used as substrates in the INHAT assay with PCAF (E) or p300 (F) with GST-SET/TAF-Iβ or SET/TAF-Iβ-Δ3. Acetylation levels were quantified via filter binding assay and represented as raw counts per minute (cpm) incorporated. (G and H) p53 peptides were incubated with PCAF (G) or p300 (H) with GST-SET/TAF-Iβ, and the modified peptides were analyzed via LC–MS spectrometry. (I) HCT116 cells were transfected with various plasmids, as indicated, and p53 immunoprecipitates were subjected to western blotting using anti-acetyl lysine. Quantification of acetylated p53 band intensities is shown below. The amounts of immunoprecipitated p53 and transfected SET/TAF-Iβ proteins were determined by western blotting and shown in the lower panels.
SET/TAF-Iβ inhibits stress-induced p53 acetylation
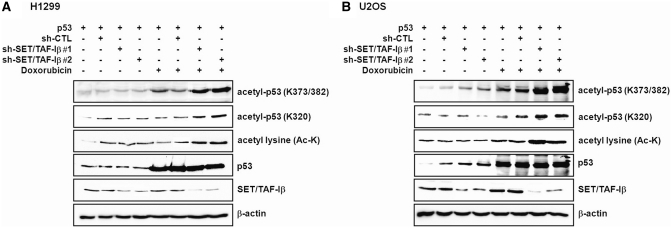
Cellular stresses, such as treatment with DNA-damaging agents, induce p53 acetylation and activity. To study the effects of SET/TAF-Iβ on stress-induced p53 acetylation in vivo, the levels of acetylated p53 were monitored using an acetylated lysine antibody and specific acetyl-p53 antibodies. Doxorubicin, a DNA-damaging agent was used to impose cellular stress. Importantly, we verified that SET/TAF-Iβ protein levels were not altered under these conditions (Figure 2A and B; Supplementary Figure S3B). Additionally, p53 protein levels were not changed by either SET/TAF-Iβ or SET/TAF-Iβ-Δ3 in the presence or absence of stress induction (Supplementary Figure S3A). In exogenous p53-expressing H1299, U2OS and 293 cells, treatment with doxorubicin resulted in increased p53 acetylation. When SET/TAF-Iβ was knocked down by two sh-SET/TAF-Iβ RNAs, further increases in p53 acetylation were induced, which indicates that endogenous SET/TAF-Iβ inhibited stress-induced p53 acetylation (Figure 2A and B; Supplementary Figure S3B). These two independent sh-SET/TAF-Iβ RNAs also successfully downregulated SET/TAF-Iβ, as confirmed by RT–PCR and western blotting (Supplementary Figure S2A and S2B).
Figure 2.
SET/TAF-Iβ inhibits p53 acetylation under stress conditions. (A and B) Western blot analysis of whole cell extracts of H1299 (A) and U2OS cells (B) transfected with p53 alone or co-transfected with sh-CTL and sh-SET/TAF-Iβ #1 or #2, followed by immunoblotting against anti-acetyl-p53(K373/382), anti-acetyl-p53(K320), anti-acetyl lysine, anti-p53, anti-SET/TAF-Iβ or anti-β-actin antibodies. Cells were either untreated or treated with 1 µM doxorubicin.
SET/TAF-Iβ interacts with p53 via INHAT domain
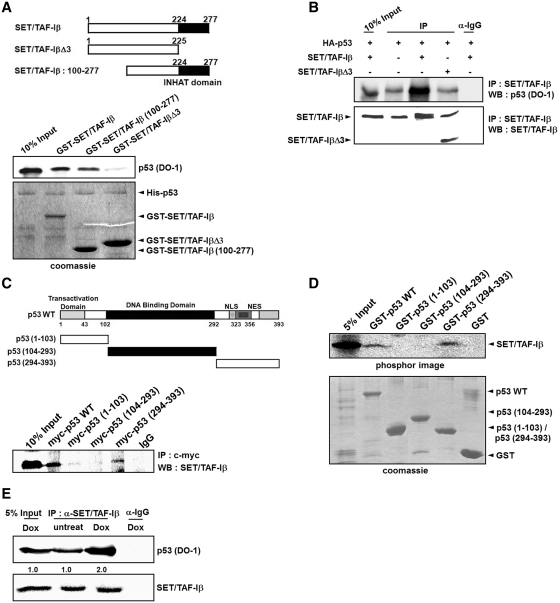
To determine whether or not p53 directly interacts with SET/TAF-Iβ, we conducted in vitro binding assays with purified GST-SET/TAF-Iβ and His-p53 protein. Compared to control GST, p53 protein strongly bound to immobilized GST-SET/TAF-Iβ, and this interaction was strictly dependent on the INHAT domain of SET/TAF-Iβ (Figure 3A). In vivo interactions between p53 and SET/TAF-Iβ but not SET/TAF-IβΔ3 were confirmed by IP with transiently transfected cell extracts using both p53 and SET/TAF-Iβ antibodies (Figure 3B). To identify the p53 domain involved in interaction, we performed an in vivo IP interaction assay using p53 deletion mutants. The results indicate that the C-terminus region of p53, which contains important acetylation target residues, was responsible for the interaction with SET/TAF-Iβ, even though the interaction was less strong than that of wild-type p53 (Figure 3C). Additionally, in vitro transcription and translation of SET/TAF-Iβ were retained on a GST-p53 and p53 (294–393) glutathione sepharose affinity matrix similar to the pattern in Figure 3C, which suggests the participation of an additional domain besides the p53 C-terminal domain in the full interaction between p53 and SET/TAF-Iβ (Figure 3D). When we compared the SET/TAF-Iβ and p53 interaction under stress and non-stress conditions, an apparent increase in the interaction under stress condition was observed (Figure 3E). It may be possible that a minor fraction of p53 associates with SET/TAF-Iβ before stress and full interaction is induced upon stress induction.
Figure 3.
SET/TAF-Iβ interacts with p53 both in vitro and in vivo. (A) Interaction assay of input and eluates from indicated GST-SET/TAF-Iβ and deletion mutants incubated with p53 and immunoblotted with anti-p53. INHAT domain of SET/TAF-Iβ is shown as a black box. The amounts of His-p53, GST-SET/TAF-Iβ and deletion mutants used in the assay were determined by coomassie staining. (B) Western blot analysis of SET/TAF-Iβ immunoprecipitates from 293 cells expressing HA-p53, SET/TAF-Iβ and SET/TAF-IβΔ3 was performed with anti-p53 antibody. The amounts of immunoprecipitated SET/TAF-Iβ and SET/TAF-Iβ-Δ3 were determined by western blotting (lower panel). (C) 293 cells were transfected with indicated myc-p53 and myc-p53 deletion mutants, immunoprecipitated with anti-myc and immunoblotted with anti-SET/TAF-Iβ antibodies. The expression levels of myc-p53 protein were determined by western blotting (lower panel). (D) In vitro transcribed and translated SET/TAF-Iβ were incubated with GST, GST-p53 or GST-p53 deletion mutants. The levels of full-length p53 and p53 deletion mutants were determined by coomassie staining. (E) Western blot analysis of the input and p53 immunoprecipitates from U2OS cells with anti-SET/TAF-Iβ antibodies. Cells were either untreated or treated with 1 µM doxorubicin. The amounts of immunoprecipitated p53 proteins were determined by western blotting (bottom). Quantification of acetylated p53 band intensities is shown below.
From these results, we can conclude that SET/TAF-Iβ targets p53 by specifically associating with its hypo-acetylated state, inhibits p53 acetylation via substrate masking from HATs, and possibly induces a transcriptional repressive state in p53 target genes.
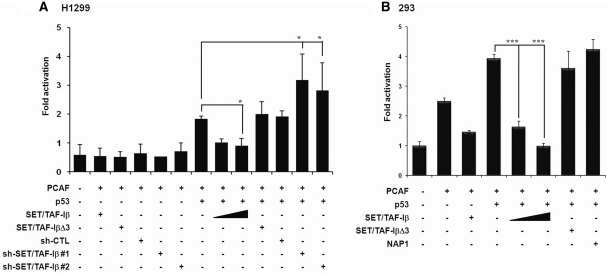
Inhibition of p53-mediated transcriptional activation by SET/TAF-Iβ
To evaluate the effect of SET/TAF-Iβ on p53-mediated transcriptional activation, we co-transfected p53, PCAF and SET/TAF-Iβ expression vectors along with the synthetic minimal p21 reporter into different cell lines. In reporter assays performed in exogenous p53-expressing H1299 cell lines, SET/TAF-Iβ reduced the activation of p21 reporter mediated by exogenous p53, showing that the regulatory activity of SET/TAF-Iβ was p53 dependent (Figure 4A). SET/TAF-Iβ-mediated repression was effective with endogenous PCAF and further evidenced with exogenous PCAF (Supplementary Figure S4A). Knockdown of SET/TAF-Iβ by two different sh-RNAs slightly upregulated endogenous p53-dependent p21 transcription and significantly activated exogenous p53-mediated p21 transcription (Figure 4A and Supplementary Figure S4B–D). Endogenous p53-mediated transactivation was significantly repressed by the addition of exogenous SET/TAF-Iβ (Supplementary Figure S4E). Again, SET/TAF-IβΔ3 had almost no effect on the transactivational activities of p53, which indicates that the physical interaction of p53 with the INHAT domain was crucial for SET/TAF-Iβ-mediated repression (Figure 4A and B; Supplementary Figure S4E). As a member of the histone chaperone nucleosome assembly protein (NAP) family, SET/TAF-Iβ shares sequence homology including a highly acidic domain with NAP-1 protein. To rule out the possibility that the observed effect of SET/TAF-Iβ on p53 was due to the presence of an acidic domain, we performed the same p21 reporter assays with NAP-1. As expected and previously reported, NAP-1 slightly upregulated p21 reporter activity mediated by p53 (Figure 4B). Indeed, it is known that NAP-1 activates p53-mediated transcription through direct interaction with p53 and p300 (26–28).
Figure 4.
SET/TAF-Iβ inhibits p53-mediated transcriptional activation. (A) H1299 cells were transfected with p53, SET/TAF-Iβ, SET/TAF-IβΔ3, sh-CTL, sh-SET/TAF-Iβs and PCAF. (B) 293 cells were transfected with the p21 promoter-luc construct along with p53, SET/TAF-Iβ, SET/TAF-IβΔ3 and NAP-1 in the presence or absence of PCAF. Data in A and B are presented as mean ± SD; n = 4. *P < 0.05 and ***P < 0.001.
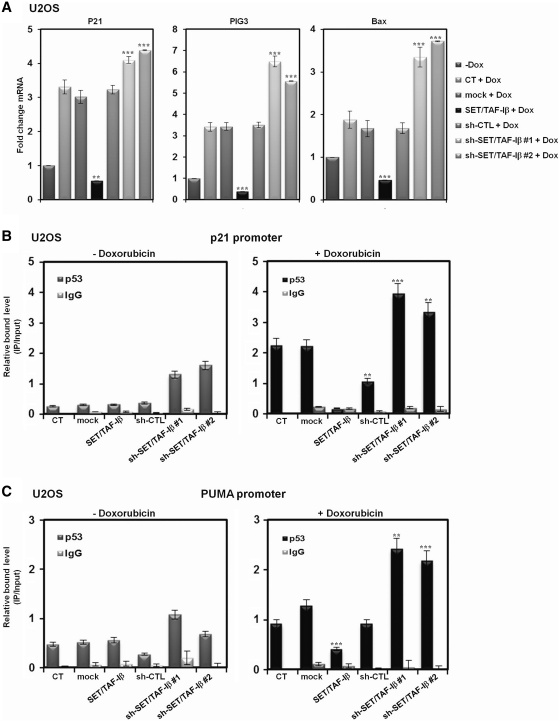
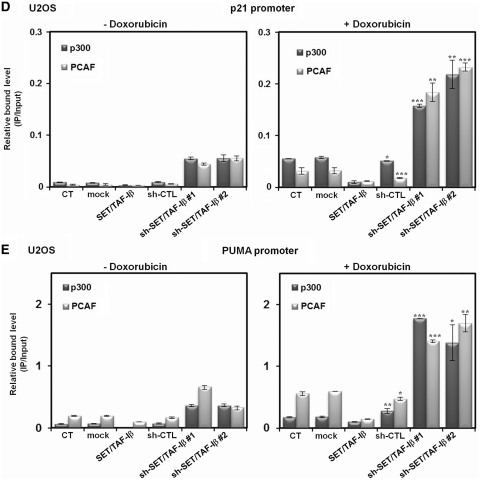
SET/TAF-Iβ represses transcription of p53 target genes
To understand the functional effects of SET/TAF-Iβ-mediated inhibition on p53 acetylation, we analyzed the effects of SET/TAF-Iβ expression on endogenous p53 target genes in U2OS cells. Real-time PCR analyses showed that overexpression of SET/TAF-Iβ significantly downregulated stress-induced expression of p53 target genes, including p21, Bax and PIG3 (Figure 5A). More importantly, knockdown of endogenous SET/TAF-Iβ by two different sh-RNAs further induced expression of the same target genes, which confirms the effect of endogenous SET/TAF-Iβ on p53 target genes (Figure 5A). To determine whether or not the regulation of p53 target genes by SET/TAF-Iβ is p53 dependent, we performed the same real-time PCR analysis in exogenous p53-expressing H1299 cells. Again, the expression of p21, Bax and PIG3 was reduced by SET/TAF-Iβ and induced by different sh-SET/TAF-Iβs only in the presence of exogenous p53, which demonstrates the p53-dependent regulatory effects of SET/TAF-Iβ (Supplementary Figure S5A and data not shown). Having established the regulation of p53 target genes by SET/TAF-Iβ, we next determined whether or not SET/TAF-Iβ modulates the recruitment of p53 to target chromatins in vivo by ChIP assays combined with real-time PCR. The recruitment of p53 to p21 promoter was significantly reduced by SET/TAF-Iβ in doxorubicin-treated U2OS cells and exogenous p53-expressing H1299 cells (Figure 5B and Supplementary Figure S5B). Notably, further increases in p53 recruitment were detected upon SET/TAF-Iβ knockdown, demonstrating that endogenous SET/TAF-Iβ inhibited the association of p53 with the p21 promoter (Figure 5B and Supplementary Figure S5B). The effects of SET/TAF-Iβ on p53 recruitment to the PUMA promoter were further investigated in U2OS cells and exogenous p53-expressing H1299 cells (Figure 5C and Supplementary Figure S5C). Again, ChIP analysis combined with real-time PCR showed that SET/TAF-Iβ significantly downregulated doxorubicin-induced p53 recruitment to the PUMA promoter, and different sh-SET/TAFIβs resulted in elevated levels of recruitment (Figure 5C and Supplementary Figure S5C). These results suggest that SET/TAF-Iβ functioned effectively in cellular stress-induced p53 activation by regulating the recruitment of p53 to target promoters. In contrast, transfection of SET/TAF-Iβ into exogenous p53-expressing H1299 cells resulted in no detectable changes in p53 recruitment to the p21 and PUMA promoters (Supplementary Figure S5D and S5E). We further analyzed p300 and PCAF recruitment to the p21 and PUMA promoters using real-time PCR and ChIP analyses. There were no noticeable changes in p300 and PCAF recruitment to either promoter under both stress and non-stress conditions. However, sh-SET/TAF-Iβ significantly increased p300/PCAF recruitment to both promoters in the presence of doxorubicin, confirming repression of transcription by SET/TAF-Iβ (Figure 5D and E).
Figure 5.
SET/TAF-Iβ represses transcription of p53-target genes. (A) Real-time PCR analysis of p53-target genes p21, Bax and PIG3 in the presence of SET/TAF-Iβ, sh-CTL and sh-SET/TAF-Iβ #1 or #2 upon doxorubicin treatment in U2OS cells. (B and C) ChIP assay of SET/TAF-Iβ and sh-SET/TAF-Iβ #1 or #2-transfected U2OS cells for measurement of p53 recruitment to the p21 (B) and PUMA promoters (C) upon doxorubicin treatment. Recruitment of p53 to the p21 or PUMA promoter was normalized to input. The data are averages of three independent experiments, and the error bars represent ± SD. **P < 0.01 and ***P < 0.001 compared with untreated control. (D and E) Recruitment of p300 and PCAF to the p21 promoter region was analyzed via ChIP assay and real-time PCR using SET/TAF-Iβ and sh-SET/TAF-Iβ #1 or #2-transfected U2OS cells upon doxorubicin treatment.
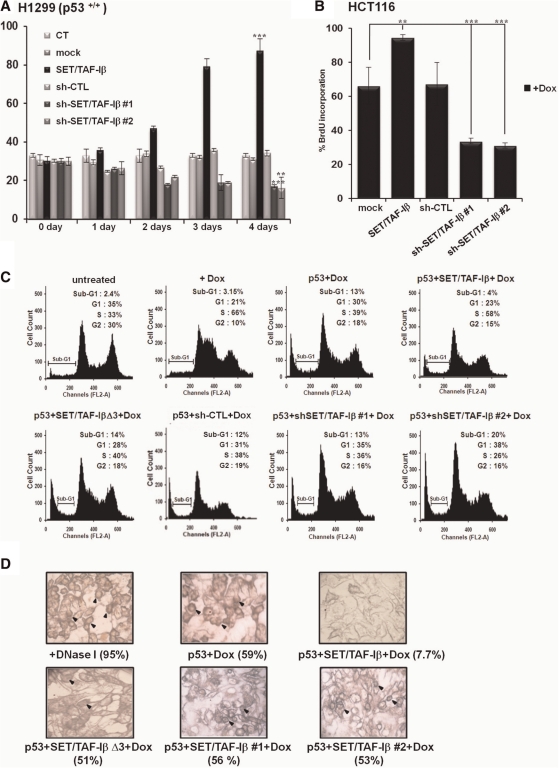
Negative regulation of p53-mediated apoptosis by SET/TAF-Iβ
To further elucidate the biological role of SET/TAF-Iβ in the cellular functions of p53, we performed an MTT assay to measure cell viability. The results of the MTT assay indicate that the strong cell death or apoptosis-inducing activity of p53 was effectively abrogated by co-transfection with SET/TAF-Iβ in exogenous p53-expressing H1299 (Figure 6A and Supplementary Figure S6A). Again, the two sh-SET/TAF-Iβ RNAs efficiently downregulated the effects of SET/TAF-Iβ on p53 and further decreased cell survival (Figure 6A). BrdU incorporation assay for measurement of cellular proliferation indicated that the growth inhibitory activity of p53 was impeded by overexpression of SET/TAF-Iβ and restored by sh-SET/TAF-Iβ, supporting SET/TAF-Iβ-mediated regulation of p53 activity in both HCT116 and H1299 cells (Figure 6B and Supplementary Figure S6B). Exogenous p53-expressing H1299 cells showed no indication of BrdU incorporation when SET/TAF-Iβ was overexpressed (Supplementary Figure S6B). FACS analysis revealed that p53-dependent G1 arrest following doxorubicin treatment was rescued by SET/TAF-Iβ expression but not by the two sh-SET/TAF-Iβ RNAs (Figure 6C). Consistent with the MTT assay results, SET/TAF-Iβ decreased the apoptotic cell population (sub-G1) induced by p53 following doxorubicin treatment (13–4%), and both sh-SET/TAF-Iβ RNAs increased the number of cells entering apoptosis from 4% to 13% and 20%, respectively (Figure 6C). The effects of endogenous SET/TAF-Iβ were confirmed by sh-SET/TAF-Iβ RNA #2 treatment, which increased the apoptotic cell population up to 20% compared to that of doxorubicin-treated cells with p53 (13%). Again, SET/TAF-IβΔ3 did not influence either G1 arrest or apoptosis induced by doxorubicin. Negative regulation of p53-mediated apoptosis by SET/TAF-Iβ was also investigated by TUNEL assay. The apoptotic nuclei in DNase I-treated control and p53 + doxorubicin-treated cells were not found in SET/TAF-Iβ-expressing H1299 cells. However, the number of dark brown-stained apoptotic cells increased upon expression of SET/TAF-IβΔ3 and both sh-SET/TAF-Iβ RNAs, which supports the negative regulation of p53-mediated apoptosis by SET/TAF-Iβ (Figure 6D).
Figure 6.
SET/TAF-Iβ represses p53-mediated cell cycle arrest and apoptosis and induces proliferation. (A) Doxorubicin-treated H1299 cells were transfected with the indicated constructs for MTT assay. Results are shown as means ± SD; n = 3. **P < 0.01 and ***P < 0.001 compared with Day 0. (B) HCT116 cells were transfected with p53, SET/TAF-Iβ, sh-CTL and sh-SET/TAF-Iβ #1 or #2 as indicated upon doxorubicin treatment. At 72-h post-transfection, cells were fixed and BrdU assays performed. Results are shown as means ± SD. n = 3. **P < 0.01 and ***P < 0.001. (C) FACS analysis of H1299 cells transfected with p53 alone or co-transfected with SET/TAF-Iβ, SET/TAF-IβΔ3, sh-CTL and sh-SET/TAF-Iβ #1 or #2, followed by doxorubicin treatment. (D) H1299 cells transiently transfected with the indicated constructs upon doxorubicin treatment were subjected to TUNEL assay. DNase I-treated cells were shown as a positive control. The numbers of TUNEL-positive cells are shown in the numbers below the pictures.
SET/TAF-Iβ inhibits UV-mediated apoptosis in Drosophila
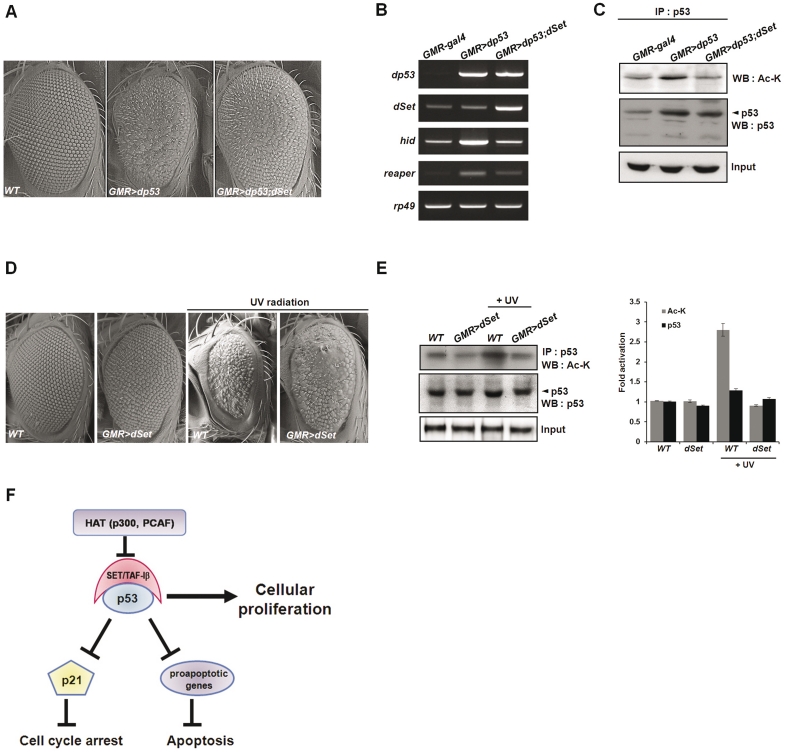
To further investigate the physiological significance of SET/TAF-Iβ in acetylated p53-mediated apoptosis, we used the developing Drosophila eye as an in vivo model system. We examined the effect of Drosophila p53 (dp53) and dSet overexpression using the eye-specific GMR-gal4 driver. The GMR driver induces target gene expression in the posterior region of the eye disc. Similar to mammalian p53, dp53 functions in the cellular response to stress, and overexpression of dp53 in the fly eye results in severe apoptosis and a small eye phenotype (29–31). Consistent with previous studies, expression of dp53 in the fly eye under direct control of the eye-specific GMR driver (GMR > dp53) resulted in a significant reduction in eye size as well as disruption of the ommatidia structure (Figure 7A). Next, we generated transgenic flies overexpressing both dp53 and dSet in the eye. The defective eye phenotype of GMR > dp53 was significantly rescued by co-expression of dSet, which suggests the negative regulation of dp53-mediated apoptosis by SET/TAF-Iβ (Figure 7A). Next, we determined whether or not dSet expression can suppress the expression of dp53-target genes. In response to ionizing radiation, dp53 targets the pro-apoptotic genes reaper and hid (30,32,33). Expression of reaper and hid was markedly reduced by co-expression of dSet, suggesting negative regulation of dp53-mediated apoptosis by SET/TAF-Iβ in the stress response pathway (Figure 7B). To examine the effect of dSet on dp53 acetylation status, we first immunoprecipitated dp53 using antibodies and then confirmed acetylation of dp53 using anti-acetyl lysine antibodies. In accordance with the SET/TAF-Iβ-mediated inhibition of p53 acetylation in this study, the acetylation level of dp53 was significantly reduced by co-expression of dp53 and dSet, suggesting that SET/TAF-Iβ has a significant effect on pd53 (Figure 7C). To extend the effects of dSet under cellular stress conditions, we examined the fly eye phenotype under UV irradiation. Compared with wild-type eye, expression of dSet alone only slightly disrupted the arrangement of ommatidia (Figure 7D). Upon UV irradiation during pupariation, the wild-type fly eye showed reduced size and a severe apoptotic phenotype (Figure 7D). On the contrary, expression of dSet produced a less severe apoptotic eye phenotype even in the presence of UV irradiation (Figure 7D). The acetylation level of dp53 induced in UV-irradiated wild-type fly significantly decreased upon expression of dSet (Figure 7E). Taken together, these results suggest that dSet inhibited acetylation of Drosophila dp53 and negatively regulated dp53-mediated apoptosis under cellular stress conditions.
Figure 7.
SET/TAF-Iβ inhibits UV-mediated apoptosis in Drosophila. (A) Overexpression of dp53 or dSet in the eye of Drosophila induced by the eye-specific GMR-gal4 driver. Adult eyes from transgenic flies were viewed by scanning electron microscopy (SEM). The eye phenotypes of wild-type fly (left), dp53-overexpresssing fly (GMR > dp53, middle) and dp53- and dSet-overexpresssing fly (GMR > dp53; dSet, right). (B) Eye-specific dp53 or dSet overexpression along with expression of the dp53-target genes hid and reaper were confirmed by RT–PCR. Equal loading of samples was confirmed using an rp49-specific primer. (C) Western blot analysis of the anti-dp53 immunoprecipitates from dp53- and dSet-overexpresssing flies with anti-acetyl lysine (top) and anti-dp53 antibodies (middle). Equal loading of samples was confirmed using anti-β-actin antibody. (D) UV irradiation-mediated apoptotic phenotypes. UV irradiation (40 000 mJ/cm2) led to a change in eye morphology when applied during pupal development. A non-irradiated adult eye (WT and GMR > dSet, left) and UV-irradiated adult eye (right) after 2 h. (E) Western blot analysis of the anti-dp53 immunoprecipitates from dp53- and dSet-overexpresssing flies and UV-irradiated flies with anti-acetyl lysine (top) and anti-dp53 antibodies (middle). Western blot images were quantitatively analyzed in the bar graph using the Image J program (right panels). (F) Model of the mechanism of SET/TAF-Iβ-mediated repression of p53 activity.
DISCUSSION
The linkage between the acetylation of histone lysine residues and transcriptional regulation has been well established (34,35). Since then, a large number of non-histone proteins have been found to be acetylated as well.
p53 protein can be activated by cellular stresses such as DNA damage, hypoxia or viral and cellular oncogenes. Ever since the first observation that non-histone protein p53 is regulated by HAT-mediated acetylation, numerous studies have been carried out to understand the post-translational modifications of p53. The level of p53 acetylation is correlated with p53 activation and acetylation by different HATs, resulting in subtle differences in the biological properties of p53.
In this study, we demonstrated that the proto-oncogene protein SET/TAF-Iβ inhibits p300- and PCAF-mediated p53 acetylation and negatively regulates p53 activity. We determined that the acidic INHAT domain of SET/TAF-Iβ located in the C-terminus is important for both inhibition of acetylation and direct interaction with p53. Moreover, stress-induced p53 acetylation was efficiently inhibited by SET/TAF-Iβ. We also observed that knockdown of SET/TAF-Iβ by different sh-RNAs significantly activates p53-mediated p21 transcription. In addition, SET/TAF-Iβ negatively regulates stress-induced p53 activation by repressing the recruitment of p53 to target promoters. By targeting p53 acetylation, SET/TAF-Iβ inhibits p53-mediated cell cycle arrest and apoptosis, which may result in cellular proliferation. In our other results, the Drosophila dp53-induced apoptotic phenotype was rescued by co-expression with dSet. Drosophila dSet inhibited dp53 acetylation as well as expression of the apoptosis target genes reaper and hid. Under stress conditions, dSet still inhibited dp53 acetylation. It has been reported that GMR driver-mediated dp53 expression in the eye disc cannot completely prevent cell death in response to whole fly body UV irradiation (36). Nonetheless, partial rescue of UV-mediated apoptotic phenotype by dSet leaves open the possibility of another apoptotic pathway besides UV-induced dp53 acetylation-mediated cell death. SET/TAF-Iβ containing the INHAT complex is a group of multifunctional proteins with acidic domains that are involved in various nuclear and cytoplasmic signaling pathways. Recent studies have indicated that INHAT complex proteins are involved in the regulation of apoptotic cell death, with SET/TAF-Iβ as an inhibitor (17,18). Since SET/TAF-Iβ is an oncoprotein, its antiapoptotic functions may contribute to the accumulation of damaged cells under stress circumstances.
Negative regulation of p53 activity by NIR (novel INHAT repressor) has been suggested as a regulatory mechanism of p53 activity (22). In this study, we expanded the role of INHAT regulator SET/TAF-Iβ to the important non-histone protein p53. We showed that INHAT subunit SET/TAF-Iβ plays a novel repressive role in p53 activity due to the inhibition of p53 acetylation. Considering that SET/TAF-Iβ is frequently upregulated in a number of cancers, we propose that SET/TAF-Iβ inhibits both p300- and PCAF-mediated p53 acetylation, which leads to inhibition of cell cycle arrest or apoptosis in the presence of different cellular stresses and guides cellular proliferation toward carcinogenesis (Figure 7F).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Education, Science and Technology (2009-0073484), Basic Science Research program through the National Research Foundation of Korea Grant (NRF); Chung-Ang University Excellent Researcher Grant (2009) (to S.S.B.). Funding for open access charge: The NRF of Korea (2009-0073484).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Prives C, Manley JL. Why is p53 acetylated? Cell. 2001;107:815–818. doi: 10.1016/s0092-8674(01)00619-5. [DOI] [PubMed] [Google Scholar]
- 2.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 3.Gu W, Shi XL, Roeder RG. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 4.Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A, Anderson CW, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avantaggiati ML, Ogryzko V, Gardner K, Giordano A, Levine AS, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 6.Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, Berger SL. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat. Rev. Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 9.Krummel KA, Lee CJ, Toledo F, Wahl GM. The C-terminal lysines fine-tune P53 stress responses in a mouse model but are not required for stability control or transactivation. Proc. Natl Acad. Sci. USA. 2005;102:10188–10193. doi: 10.1073/pnas.0503068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng L, Lin T, Uranishi H, Gu W, Xu Y. Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol. Cell. Biol. 2005;25:5389–5395. doi: 10.1128/MCB.25.13.5389-5395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seo SB, McNamara P, Heo S, Turner A, Lane WS, Chakravarti D. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell. 2001;104:119–130. doi: 10.1016/s0092-8674(01)00196-9. [DOI] [PubMed] [Google Scholar]
- 13.Schneider R, Bannister AJ, Weise C, Kouzarides T. Direct binding of INHAT to H3 tails disrupted by modifications. J. Biol. Chem. 2004;279:23859–23862. doi: 10.1074/jbc.C400151200. [DOI] [PubMed] [Google Scholar]
- 14.Kutney SN, Hong R, Macfarlan T, Chakravarti D. A signaling role of histone-binding proteins and INHAT subunits pp32 and Set/TAF-Ibeta in integrating chromatin hypoacetylation and transcriptional repression. J. Biol. Chem. 2004;279:30850–30855. doi: 10.1074/jbc.M404969200. [DOI] [PubMed] [Google Scholar]
- 15.Nagata K, Kawase H, Handa H, Yano K, Yamasaki M, Ishimi Y, Okuda A, Kikuchi A, Matsumoto K. Replication factor encoded by a putative oncogene, set, associated with myeloid leukemogenesis. Proc. Natl Acad. Sci. USA. 1995;92:4279–4283. doi: 10.1073/pnas.92.10.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brennan CM, Gallouzi IE, Steitz JA. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell. Biol. 2000;151:1–14. doi: 10.1083/jcb.151.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakravarti D, Hong R. SET-ting the stage for life and death. Cell. 2003;112:589–591. doi: 10.1016/s0092-8674(03)00151-x. [DOI] [PubMed] [Google Scholar]
- 18.Fan Z, Beresford PJ, Oh DY, Zhang D, Lieberman J. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell. 2003;112:659–672. doi: 10.1016/s0092-8674(03)00150-8. [DOI] [PubMed] [Google Scholar]
- 19.Jiang X, Kim HE, Shu H, Zhao Y, Zhang H, Kofron J, Donnelly J, Burns D, Ng SC, Rosenberg S, et al. Distinctive roles of PHAP proteins and prothymosin-alpha in a death regulatory pathway. Science. 2003;299:223–226. doi: 10.1126/science.1076807. [DOI] [PubMed] [Google Scholar]
- 20.von Lindern M, van Baal S, Wiegant J, Raap A, Hagemeijer A, Grosveld G. Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3' half to different genes: characterization of the set gene. Mol. Cell. Biol. 1992;12:3346–3355. doi: 10.1128/mcb.12.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adachi Y, Pavlakis GN, Copeland TD. Identification and characterization of SET, a nuclear phosphoprotein encoded by the translocation break point in acute undifferentiated leukemia. J. Biol. Chem. 1994;269:2258–2262. [PubMed] [Google Scholar]
- 22.Hublitz P, Kunowska N, Mayer UP, Muller JM, Heyne K, Yin N, Fritzsche C, Poli C, Miguet L, Schupp IW, et al. NIR is a novel INHAT repressor that modulates the transcriptional activity of p53. Genes Dev. 2005;19:2912–2924. doi: 10.1101/gad.351205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 24.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 25.Luo J, Li M, Tang Y, Laszkowska M, Roeder RG, Gu W. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc. Natl Acad. Sci. USA. 2004;101:2259–2264. doi: 10.1073/pnas.0308762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shikama N, Chan HM, Krstic-Demonacos M, Smith L, Lee CW, Cairns W, La Thangue NB. Functional interaction between nucleosome assembly proteins and p300/CREB-binding protein family coactivators. Mol. Cell. Biol. 2000;20:8933–8943. doi: 10.1128/mcb.20.23.8933-8943.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asahara H, Tartare-Deckert S, Nakagawa T, Ikehara T, Hirose F, Hunter T, Ito T, Montminy M. Dual roles of p300 in chromatin assembly and transcriptional activation in cooperation with nucleosome assembly protein 1 in vitro. Mol. Cell. Biol. 2002;22:2974–2983. doi: 10.1128/MCB.22.9.2974-2983.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rehtanz M, Schmidt HM, Warthorst U, Steger G. Direct interaction between nucleosome assembly protein 1 and the papillomavirus E2 proteins involved in activation of transcription. Mol. Cell. Biol. 2004;24:2153–2168. doi: 10.1128/MCB.24.5.2153-2168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ollmann M, Young LM, Di Como CJ, Karim F, Belvin M, Robertson S, Whittaker K, Demsky M, Fisher WW, Buchman A, et al. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell. 2000;101:91–101. doi: 10.1016/S0092-8674(00)80626-1. [DOI] [PubMed] [Google Scholar]
- 30.Brodsky MH, Nordstrom W, Tsang G, Kwan E, Rubin GM, Abrams JM. Drosophila p53 binds a damage response element at the reaper locus. Cell. 2000;101:103–113. doi: 10.1016/S0092-8674(00)80627-3. [DOI] [PubMed] [Google Scholar]
- 31.Jin S, Martinek S, Joo WS, Wortman JR, Mirkovic N, Sali A, Yandell MD, Pavletich NP, Young MW, Levine AJ. Identification and characterization of a p53 homologue in Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 2000;97:7301–7306. doi: 10.1073/pnas.97.13.7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brodsky MH, Weinert BT, Tsang G, Rong YS, McGinnis NM, Golic KG, Rio DC, Rubin GM. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol. Cell. Biol. 2004;24:1219–1231. doi: 10.1128/MCB.24.3.1219-1231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou L, Steller H. Distinct pathways mediate UV-induced apoptosis in Drosophila embryos. Dev. Cell. 2003;4:599–605. doi: 10.1016/s1534-5807(03)00085-6. [DOI] [PubMed] [Google Scholar]
- 34.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 35.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 36.Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.