Abstract
Thiostrepton, a macrocyclic thiopeptide antibiotic, inhibits prokaryotic translation by interfering with the function of elongation factor G (EF-G). Here, we have used 70S ribosome binding and GTP hydrolysis assays to study the effects of thiostrepton on EF-G and a newly described translation factor, elongation factor 4 (EF4). In the presence of thiostrepton, ribosome-dependent GTP hydrolysis is inhibited for both EF-G and EF4, with IC(50) values equivalent to the 70S ribosome concentration (0.15 µM). Further studies indicate the mode of thiostrepton inhibition is to abrogate the stable binding of EF-G and EF4 to the 70S ribosome. In support of this model, an EF-G truncation variant that does not possess domains IV and V was shown to possess ribosome-dependent GTP hydrolysis activity that was not affected by the presence of thiostrepton (>100 µM). Lastly, chemical footprinting was employed to examine the nature of ribosome interaction and tRNA movements associated with EF4. In the presence of non-hydrolyzable GTP, EF4 showed chemical protections similar to EF-G and stabilized a ratcheted state of the 70S ribosome. These data support the model that thiostrepton inhibits stable GTPase binding to 70S ribosomal complexes, and a model for the first step of EF4-catalyzed reverse-translocation is presented.
INTRODUCTION
The bacterial ribosome is a major target for several classes of natural antibiotics, which inhibit nearly all steps of translation (1–3). In the past decade, several atomic resolution X-ray crystal structures of various antibiotics bound to the ribosome have been elucidated and have assisted in demystifying the mode of action of many of these translational inhibitors (4–9). Not surprisingly, it has been revealed that many antibiotics inhibit translation by binding to functionally important regions of ribosomal RNA (rRNA) such as the peptidyltransferase center (5,8), the decoding center (4) and the exit tunnel (6,9). Another target for several antibiotics is the translation factor binding site on the large (50S) ribosomal subunit, which includes the ribosomal components responsible for stimulating the GTPase activity of several translation factors (10,11). The factor binding site consists of the universally conserved sarcin-ricin loop (SRL) of 23S rRNA as well as the ‘GTPase associated center’, which in Escherichia coli consists of a conserved region of 23S rRNA bound to ribosomal protein L11 and the pentameric complex L10•(L7/12)4 (7,12,13).
Thiostrepton is a well-studied antibiotic belonging to the thiopeptide family of highly modified macrocyclic peptides produced as secondary metabolites by actinomycetes within the genus Streptomyces (14,15). Thiostrepton exerts its inhibitory function by binding to the ribosome within the GTPase-associated center, in a cleft formed between the N-terminal domain (NTD) of L11 and 23S rRNA loops H44 and H45 (7,16–19). Due to the low aqueous solubility of thiostrepton (20,21), there has traditionally been low interest in its clinical use. However, the observation that thiostrepton inhibits the growth of the malarial parasite Plasmodium falciparum (22–25) and induces apoptosis in breast cancer cells (26,27) has greatly renewed interest in the therapeutic potential of thiostrepton. Additionally, the total synthesis of thiostrepton has been completed (28–31), and it has been discovered that fragments of thiostrepton retain biological activity (32).
Although there is evidence that thiostrepton inhibits steps in the initiation (20,33) and termination (34) phases of translation, the most studied effect of thiostrepton is its inhibition of elongation factor G (EF-G) function on the ribosome (20,35–42). The mechanism by which thiostrepton inhibits EF-G function is actively debated. Results from early investigations led to the formation of the ‘classical’ model of thiostrepton action, which holds that thiostrepton prevents ribosome-dependent GTP hydrolysis by EF-G via inhibition of stable binding of EF-G to the ribosome (35–37,43). This was the predominant model for the mode of action of thiostrepton until the results of rapid kinetic experiments suggested that thiostrepton allows binding and GTP hydrolysis by EF-G, but inhibits the subsequent steps of phosphate release and factor turnover (41). Support for this model has been provided by similar time-resolved kinetic experiments (42,44) as well as a cryo-electron microscopy (cryo-EM) structure purporting to show EF-G bound to the 70S•thiostrepton complex (45). However, a subsequent report provided renewed support for the notion that thiostrepton prevents ribosome binding and GTP hydrolysis by EF-G altogether (20), in agreement with recent structural evidence which suggested that the presence of thiostrepton is incompatible with stable binding of EF-G (7).
In this report, we demonstrate that thiostrepton is a potent inhibitor of GTPase activity and stable ribosome binding by EF-G as well as by a recently characterized elongation factor with high homology to EF-G, elongation factor 4 (EF4), also known as LepA (46). Interestingly, an EF-G mutant lacking domains 4 and 5 is insensitive to thiostrepton in both ribosome binding and GTPase activity. Implications of the presented results for the mode of action of thiostrepton are discussed, and attempts are made to reconcile our results with seemingly contradictory non-equilibrium studies. Additionally, results of chemical footprinting analyses of the EF4•ribosome complex is described, which suggest that EF4 stabilizes a novel conformational state of tRNA bound to the ribosome. Based on these data and previously reported observations, a model for the first step of EF4-catalyzed reverse translocation is presented.
MATERIALS AND METHODS
Reagents
Guanosine-5′-triphosphate (GTP) and guanosine-5′-(β,γ-imino)triphosphate (GDPNP) were purchased from Sigma; guanosine-5′-[γ-32P]-triphosphate was purchased from American Radiolabeled Chemicals, Inc.; deoxythymidine-5′-[α-32P]-triphosphate was purchased from Perkin Elmer; thiostrepton, Sephacryl S-300 HR resin and Ni-NTA His-Bind resin were purchased from Fisher Scientific.
Cloning and Mutagenesis
The open reading frames for EF-G and EF4 (LepA) were amplified by PCR from the genomic DNA of E. coli MRE600. The primers used for cloning EF-G were 5′-GGTGGTGGATCCATCGCTCGTACAACACCCATCGC-3′ and 5′-GGTGGTGGTCTCGAGTTAATGCCGGTGAGCAACGTTACTCGG-3′ for the N- and C-termini, respectively. The primers used for cloning EF4 were 5′-GGTGGTGGATCCATGAAGAATATACGTAACTTTTCGATCATAGC-3′ and 5′-GGTGGTGGTCTCGAGTTATTTGTTGTCTTTGCCGACGTGCAGAATGGC-3′ for the N- and C-termini, respectively. Each open reading frame was cloned into a pET27 derivative (pSV281) that contained a TEV protease-cleavable His6 N-terminal tag using BamHI (5′) and XhoI (3′) restriction sites. The plasmid for the EF-G(Δ4,5) C-terminal truncation mutant was constructed by introducing a stop codon following domain 3 (Gly 484) using the primer 5′-GGTGGTGGTGAGCTCGCTTACCGTGAAACTATCCGCCAG-3′ and then inserting the resultant reading frame into the pSV281 vector in the same manner as EF-G and EF4.
Preparation of GTPases and ribosomes
All GTPases (EF-G, EF4, and EF-GΔ4,5) were expressed in E. coli BL21(DE3) cells by the addition of 0.5 mM IPTG and grown at 15°C for 16–18 h with shaking. Cells were pelleted at 7000 rpm in a GS3 rotor at 4°C, resuspended in GTPase lysis buffer [50 mM Tris–HCl (pH 7.5), 60 mM NH4Cl, 7 mM MgCl2, 15 mM imidazole, 25% glycerol, 6 mM β-mercaptoethanol], and lysed by sonication. The soluble fraction was clarified by centrifugation at 18 000 rpm at 4°C in an SS34 rotor. Each GTPase was subsequently purified by chelated nickel affinity chromatography as described previously (47,48). Purified fractions of each GTPase were subsequently dialyzed into GTPase storage buffer [50 mM Tris–HCl (pH 7.5), 60 mM NH4Cl, 7 mM MgCl2, 50% glycerol, 1 mM dithiothreitol].
Affinity-tagged 70S ribosomes were produced and purified from the E. coli JE28 cell line as described previously (49). Briefly, JE28 cells were grown at 37°C with shaking to mid-log phase and subsequently placed on ice and cooled to 4°C. The cells were harvested and resuspended in JE28 lysis buffer [20 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 150 mM KCl, 30 mM NH4Cl, 5 mM imidazole, 1 mM DTT]. Resuspended cells were subsequently lysed by sonication, and the soluble fraction was isolated by centrifugation at 18 000 rpm at 4°C in an SS34 rotor. The supernatant was filtered through a 0.2 µm pore size syringe filter and subsequently purified by chelated nickel affinity chromatography [wash buffer:20 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 150 mM KCl, 500 mM NH4Cl, 5 mM imidazole, 1 mM DTT; elution buffer: 20 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 150 mM KCl, 30 mM NH4Cl, 150 mM imidazole, 1 mM DTT]. Following purification, ribosomes were dialyzed into JE28 salt wash buffer [20 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 150 mM KCl, 500 mM NH4Cl, 1 mM DTT], pelleted twice at 150 000g (60Ti rotor, 57 400 rpm, 2 h, 4°C), and resuspended in ribosome storage buffer [50 mM Tris–HCl (pH 7.5), 30 mM NH4Cl, 7 mM MgCl2, 25% glycerol]. Purified, concentrated ribosomes were flash frozen in liquid nitrogen and stored at −80°C.
GTP hydrolysis assays
The rates of GTP hydrolysis by various GTPases were assayed using γ-32P-labeled GTP according to previous studies (50). Ribosome functional complexes were assembled in reaction buffer [90 mM K-HEPES (pH 7.5), 100 mM NH4Cl, 20 mM Mg(OAc)2] with the following components: 70S ribosomes (0.2 µM), GTPase (0.5 µM) and [γ-32P]-GTP (10 µM). For time course assays, thiostrepton (10 µM) was pre-incubated with 70S at 37°C for 10 min. In single time-point dose–response assays, the thiostrepton concentration was varied while all other reaction components remained constant as listed above. Following GTP hydrolysis, the reaction was quenched by adding 20 µl of the reaction mixture to 380 µl of 5% (w/v) activated charcoal resuspended in 50 mM NaH2PO4. Inorganic phosphate was isolated from the reaction mixture by centrifugation at 7000 g for 10 min, and 50 µl of the supernatant was added to 200 µl Optima Gold scintillation fluid. The amount of 32P-labeled phosphate was quantified with a Wallac Trilux 1450 Microbeta scintillation counter.
Ribosome binding assays
The binding of GTPases to 70S ribosomes in the GTP state was assayed using a size exclusion/centrifugation protocol. Ribosome functional complexes were constructed in GTPase reaction buffer with 1.0 µM 70S, 4.0 µM GTPase, 1 mM GDPNP and 10 µM thiostrepton, and the reactions were incubated at 37°C for 20 min. For each reaction, 60 µl was added to 500 µl Sephacryl-300 HR resin that had been pre-equilibrated in GTPase binding buffer with 1 mM GDPNP in a microfuge spin column. The column was immediately centrifuged at 2000 rpm for 2 min. Unbound GTPase was retained within the resin while 70S ribosomal complexes completely eluted through the column matrix. The flow-through was collected, precipitated with cold acetone, and subsequently analyzed with 10% (v/v) Tris–glycine SDS–PAGE gels.
Chemical footprinting
Ribosome functional complexes were formed as previously described (48,51) and were initially formed by the addition of 70S ribosomes (0.5 µM), mRNA 32 (5′-GGCAAGGAGGUA AAAAUGUUUAAAGGUAAAUCUACU-3′, 1.0 µM) and N-Ac-Phe-tRNAPhe (1.0 µM) in reaction buffer (80 mM HEPES·KOH, pH 7.8; 100 mM NH4Cl; 20 mM MgCl2; 1 mM dithiothreitol) and was incubated for 20 min at 37°C. The P site-bound tRNA was then deacylated by the addition of puromycin (1 mM), which was incubated for an additional 20 min at 37°C. The resulting complex was then incubated with either EF-G or EF4 in the presence of either GDP or GDPNP for 20 min at 37°C before chemical modification. Chemical footprinting procedures were performed as previously described (52). Ribosome functional complexes were reacted with dimethyl sulfate for 8 min at 37°C, the modified rRNA was purified by phenol/chloroform extraction, and resultant rRNA template was used for subsequent primer extension analysis.
RESULTS
Thiostrepton inhibits the ribosome-stimulated GTPase activity of EF-G and EF4
The effect of thiostrepton on the ribosome-dependent GTPase activity of translation factors EF-G and EF4 was evaluated using purified 70S ribosomes and translation factors from E. coli. In vitro GTPase reactions were carried out employing [γ-32P]GTP, and GTPase activity was measured by quantifying the amount of hydrolyzed 32Pi that remained in solution subsequent to activated charcoal extraction, as a function of time (see ‘Materials and Methods’ section). Experimental conditions were designed to mimic those of previous thiostrepton investigations (20,41,42), in order to facilitate a comparison of our results with those of similar studies. A noteworthy discrepancy is that we found thiostrepton to have poor aqueous solubility when using conditions employed in a previous report (100 µM thiostrepton in the presence of 2% (v/v) DMSO) (41). Therefore, with the exception of dose–response experiments in which the concentration of thiostrepton was varied widely, thiostrepton was utilized at a concentration of 10 µM in 2% (v/v) DMSO.
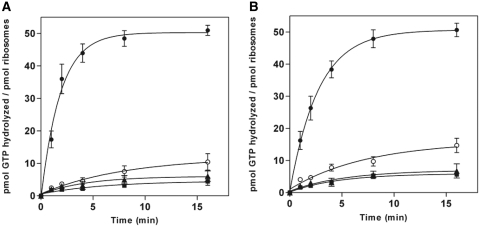
In agreement with the conventional view of translational GTPases, the intrinsic GTPase activity of EF-G and EF4 alone was very low, and the activity of these factors was strongly stimulated by the presence of 70S ribosomes, in which case the majority of available [γ-32P]GTP was hydrolyzed within the first 2 min of the reaction (Figure 1). However, when the GTPase activity of EF-G and EF4 was examined upon reacting with 70S ribosomes that were pre-incubated with 10 µM thiostrepton, a striking reduction in the release of hydrolyzed 32Pi was observed. In the presence of thiostrepton-treated ribosomes, the extent of GTP hydrolysis observed was comparable to that of the intrinsic activity of the factors alone, suggesting that thiostrepton is a strong inhibitor of multiple-round, ribosome-dependent GTP hydrolysis by both EF-G and EF4. For EF-G, this result differs from that of a previous study (41) in which thiostrepton caused a notable decrease in the initial rate of GTP hydrolysis, yet still allowed for the full extent of the reaction to occur.
Figure 1.
Thiostrepton inhibition of ribosome-dependent multiple-turnover GTP hydrolysis. (A) EF-G and (B) EF4. Samples consisted of GTPase + ribosomes (filled circle), GTPase + ribosomes + thiostrepton (open circle), GTPase alone (filled square) or ribosomes alone (filled triangle). Reactions contained 0.5 μM GTPase and/or 0.2 μM ribosomes, as well as 10 μM [γ-32P]GTP, 2% DMSO, and, where indicated, 10 μM thiostrepton. Error bars represent the standard deviation from the mean for triplicate experiments.
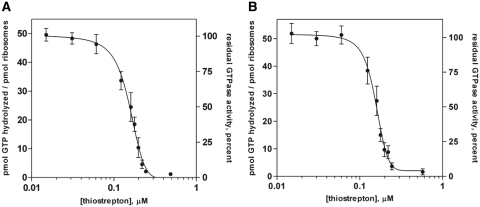
To assess the relative potency of thiostrepton in the inhibition of GTPase activity of both EF-G and EF4, dose–response experiments were performed. In these experiments, the concentrations of 70S ribosomes (0.2 µM) and translation factors (0.5 µM) were held constant while the concentration of thiostrepton was varied, and the relative extent of the GTPase reaction was measured after allowing the GTPase reaction to proceed for 10 min. For reporting relative activities, the amount of 32Pi remaining in solution in the absence of thiostrepton was assigned a value of 1.0, and all subsequent data points were normalized to this value. Titration curves are shown in Figure 2, and were virtually identical for both EF-G and EF4, resulting in an IC50 of ~0.15 µM for both factors under these reaction conditions. When the concentration of ribosomes greatly exceeded that of thiostrepton, the extent of the multiple-turnover GTPase reaction appeared unhindered by the presence of the antibiotic. However, as the concentration of thiostrepton approached equimolarity with ribosomes (0.2 µM), GTPase activity is rapidly suppressed, and when the thiostrepton concentration greatly exceeds that of ribosomes, the reaction is completely inhibited. These results suggest that a 1:1 molar ratio of thiostrepton to ribosomes is sufficient for complete inactivation of EF-G and EF4 GTP hydrolysis activity, which is consistent with previous reports (18,20).
Figure 2.
Titration of thiostrepton concentration and its effect on ribosome-dependent GTP hydrolysis activity. (A) EF-G and (B) EF4. Plots indicate the amount of GTP hydrolyzed (left ordinate axes) as well as percent residual GTPase activity (right ordinate axes) after 10 min in the presence of ribosomes and thiostrepton. Hundred percent activity corresponds to the amount of hydrolyzed GTP measured after 10 min in the absence of thiostrepton. Reactions contained 0.5 μM GTPase, 0.2 μM ribosomes, 10 μM [γ-32P]GTP, 2% DMSO and thiostrepton (at concentrations indicated on the abscissa). Error bars represent the standard deviation from the mean for triplicate experiments.
Thiostrepton inhibits stable binding of EF-G•GDPNP and EF4•GDPNP to the 70S ribosome
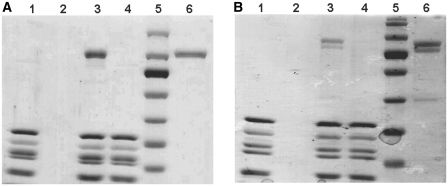
To investigate the mode of inhibition of thiostrepton, its effect on the stable binding of both EF-G and EF4 in the presence of a non-hydrolyzable analog of GTP (GDPNP) to 70S ribosomes was assessed. Since it has been suggested that equilibrium ribosome-binding experiments employing ultracentrifugation may cause the premature dissociation of weakly bound factors (53), we utilized an alternative binding assay reliant on gel filtration, which allows for a direct observation of binding without artifacts associated with ultracentrifugation. Briefly, 70S ribosomes were incubated with translation factors and the non-hydrolyzable GTP analog, GDPNP, in the presence or absence of thiostrepton. Ribosomal complexes were then applied to a gel filtration resin, eluted from the resin by centrifugation, and analyzed by SDS–PAGE. Ribosomal complexes eluted as expected, while unbound factors were retained within the resin. Results of ribosome-binding experiments using EF-G and EF4 are shown in Figure 3. Control samples consisting solely of ribosomes or translation factors were also subjected to this assay. Ribosomes in the absence of EF-G or EF4 (Figure 3, lane 1) indicated that there was no band present for the GTPase molecular weight, while low molecular weight bands corresponding to ribosomal proteins associated with the 70S were present. When the gel filtration assay was performed with free GTPase in the absence of ribosomes (Figure 3, lane 2), a band for the GTPase molecular weight did not appear in the SDS–PAGE gel, indicating that it was retained in the resin. In the absence of thiostrepton, EF-G and EF4 clearly form stable complexes with the ribosome, as evidenced from the presence of the correct molecular weight bands in the lanes containing 70S ribosomes and GDPNP. In contrast to previously published reports based on fast kinetic experiments that indicate transient binding of EF-G to 70S•thiostrepton complexes (41,42,45), the presence of thiostrepton resulted in the inability of EF-G•GDPNP to bind stably to the 70S ribosome, as indicated by the absence of a band on the gel for EF-G. Similarly, the binding of EF4•GDPNP to 70S ribosomes was inhibited by thiostrepton and did not result in the elution of EF4 with 70S ribosomes in the GTP-state. These data do not detect transient interactions and are therefore not contradictory to previous kinetic data (41,42,45).
Figure 3.
Effect of thiostrepton on stable binding of GTPases to the 70S ribosome. (A) EF-G and (B) EF4. Lanes: 1, ribosomes only; 2, GTPase only; 3, ribosomes, GTPase and GDPNP; 4, ribosomes, thiostrepton, GTPase and GDPNP; 5, MW standards; 6, GTPase positive control. Concentrations of components were: 1.0 μM ribosomes, 4.0 μM GTPase, 10 μM thiostrepton and 1 mM GDPNP.
An EF-G mutant lacking domains 4 and 5 is insensitive to the effects of thiostrepton on both GTPase activity and ribosome binding
To evaluate the role of domains 4 and 5 of EF-G in the mechanism of inhibition by thiostrepton, a mutant EF-G construct lacking these domains (EF-GΔ4,5) was cloned, expressed, purified and subjected to the GTPase activity assays and ribosome binding experiments as described above for wild-type EF-G and EF4.
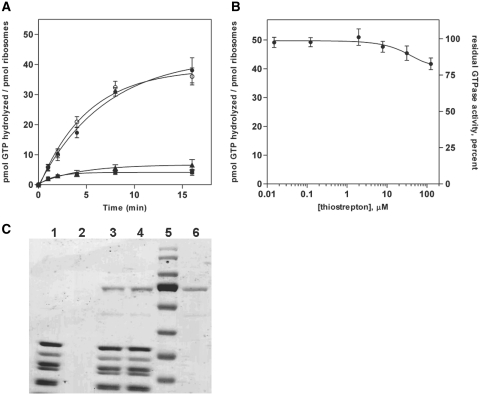
Results of time-based multiple-turnover GTP hydrolysis experiments for EF-GΔ4,5 are shown in Figure 4. Similar to full-length EF-G, the EF-GΔ4,5 construct shows low intrinsic GTPase activity, but the presence of 70S ribosomes causes significant stimulation of GTP hydrolysis (although the ribosome-stimulated GTP hydrolysis activity of EF-GΔ4,5 is ~2.5-fold slower than that of full-length EF-G). Interestingly, in the presence of thiostrepton, EF-GΔ4,5 shows no reduction in its ribosome-dependent GTPase activity, as measured in the time-based GTP hydrolysis assay (Figure 4A). Dose–response experiments (Figure 4B) also indicate that this insensitivity to thiostrepton by EF-GΔ4,5 persists even when the concentration of the antibiotic is 500-fold greater than the ribosome concentration (0.2 µM ribosomes, 100 µM thiostrepton). As mentioned above, thiostrepton was found to have poor aqueous solubility at 100 µM and was insoluble at concentrations above 100 µM, so it is possible that insoluble thiostrepton aggregates may have contributed to the observed decrease in GTPase activity by EF-GΔ4,5 in a physiologically irrelevant manner when thiostrepton was present at concentrations above 100 µM (data not shown).
Figure 4.
Deletion of domains 4 and 5 from EF-G results in insensitivity to thiostrepton. (A) Multiple turnover GTP hydrolysis. Samples consisted of GTPase + ribosomes (filled circle), GTPase + ribosomes + thiostrepton (open circle), GTPase alone (filled square) or ribosomes alone (filled triangle) (conditions as in Figure 1). (B) Effect of thiostrepton concentration (conditions as in Figure 2). (C) Effect of thiostrepton on EF-GΔ4,5 ribosome binding ability. Lanes: 1, ribosomes only; 2, GTPase only; 3, ribosomes, GTPase and GDPNP; 4, ribosomes, thiostrepton, GTPase and GDPNP; 5, MW standards; 6, GTPase positive control (conditions as in Figure 3). Error bars represent the standard deviation from the mean for triplicate experiments.
To assess whether the observed insensitivity of the EF-GΔ4,5 deletion construct to thiostrepton is correlated with a restored ability to bind ribosomes in the presence of the antibiotic, gel filtration/SDS–PAGE binding experiments were performed, as described above for full-length EF-G and EF4. The results of these experiments (Figure 4C) indicate that EF-GΔ4,5•GDPNP binds to thiostrepton-treated 70S ribosomes to an extent which is indistinguishable from the binding of EF-GΔ4,5•GDPNP to untreated 70S ribosomes.
EF-G and EF4 bind to conserved bases in 23S rRNA and stabilize ribosomal ratcheting
In order to assess the binding interface between EF4 and the 70S ribosome, ribosome•tRNA•mRNA•GTPase complexes were formed with a single, deacylated tRNA bound to the 30S P site. Ribosomes containing a defined mRNA sequence and N-acetyl-Phe-tRNAPhe bound to the 30S P site were reacted with the antibiotic puromycin, which deacylates P site-bound tRNA, which allows for the determination of tRNA binding sites in the 23S rRNA (A, P or E) by chemical footprinting. The remaining complex contains a single, deacylated tRNAPhe bound to the 30S P site. The resultant complex was then reacted with either EF-G or EF4 in the presence of GDPNP to form stable complexes for chemical probing with dimethyl sulfate.
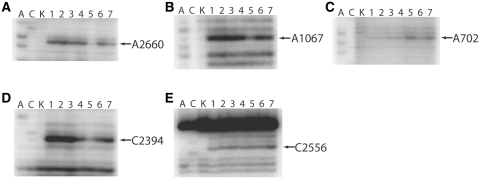
Functional complexes were constructed in a reaction buffer containing 20 mM Mg2+ in the absence of polyamines in order to maintain the P site-bound tRNAPhe in the classical P/P state (48). As reported previously, addition of EF-G•GDPNP to this complex stabilizes the hybrid P/E state (48). After reacting the 70S ribosomal complex with EF-G and EF4, strong protections were observed at A2660 of the SRL in the presence of GDPNP (Figure 5A, lanes 5 and 7, respectively). In contrast to the strong SRL protections, the GAC was differentially protected for EF-G and EF4, with EF-G displaying a much stronger protection at A1067 and EF4 only displaying a modest protection (Figure 5B, lanes 5 and 7, respectively). These protection patterns suggest that both EF-G and EF4 bind to the same conserved regions of the 23S rRNA and likely have similar binding interfaces between the 70S ribosome and each GTPase. Another striking similarity in the chemical protection patterns of EF-G and EF4 are observed at A702 of the intersubunit bridge, B7a. In the presence of GDPNP, both EF-G and EF4 cause an enhancement of chemical modification at A702 (Figure 5C, lanes 5 and 7, respectively), a result that was previously shown to be indicative of hybrid P/E state formation (47,48). In contrast to this similarity, the presence of EF4•GDPNP does not result in a protection of C2394, a 23S base that is strongly protected in the presence of hybrid P/E state tRNA (Figure 5D, lane 7). EF-G, by comparison, strongly protects C2394 for the identical 70S ribosome functional complex, as shown previously (Figure 5D, lane 5) (48). Lastly, the presence of EF4•GDPNP causes an enhancement of modification at C2556, a conserved 23S rRNA base in the 50S A-loop (Figure 5E, lane 7).
Figure 5.
Chemical footprinting of 70S ribosome·tRNAPhe ·GTPase·GDPNP complexes. (A and C), sequencing lanes; (K), unmodified rRNA; (1) 70S ribosomes and mRNA32; (2) 70S ribosomes, mRNA32 and N-Ac-Phe-tRNAPhe; (3) 70S ribosomes, mRNA32, N-Ac-Phe-tRNAPhe and puromycin; (4) 70S ribosomes, mRNA32, N-Ac-Phe-tRNAPhe, puromycin and EF-G·GDP; (5) 70S ribosomes, mRNA32, N-Ac-Phe-tRNAPhe, puromycin and EF-G·GDPNP; (6) 70S ribosomes, mRNA32, N-Ac-Phe-tRNAPhe, puromycin and EF4·GDP; (7) 70S ribosomes, mRNA32, N-Ac-Phe-tRNAPhe, puromycin and EF4·GDPNP. (A) 23S rRNA sarcin-ricin loop (SRL, GTPase binding site); (B) 23S rRNA GTPase-associated center (GAC); (C) 16S rRNA intersubunit bridge B7a; (D) 23S rRNA E site; (E) 23S rRNA A-loop.
DISCUSSION
The effect of thiostrepton on stable GTPase•ribosome interactions
In an attempt to reconcile seemingly conflicting proposed mechanisms of the mode of translational inhibition by the thiopeptide antibiotic thiostrepton, we analyzed its effect on multiple-turnover GTP hydrolysis and stable ribosome binding by EF-G, an EF-G mutant lacking domains 4 and 5 (EF-GΔ4,5) and EF4, a recently discovered GTPase elongation factor which bears strong structural and functional similarities with EF-G (46). Our results provide renewed support for the classical model of translational inhibition by thiostrepton, which proposes that the presence of the antibiotic prevents the stable binding of GTPase elongation factor EF-G, thus inhibiting ribosome-dependent GTP hydrolysis and rendering the factor non-functional. Additionally, we show that the EF-G homolog EF4 is also strongly inhibited by thiostrepton, and propose that this is likely through a mechanism similar to that of EF-G.
The current predominantly cited model for the mechanism of inhibition of EF-G by thiostrepton, based primarily on rapid kinetic analyses, holds that thiostrepton stabilizes a conformation of the ribosome which is compatible with EF-G binding and GTPase activation (41,42,44). However, the results of GTP hydrolysis assays presented here indicate that thiostrepton is a potent inhibitor of ribosome-dependent GTP hydrolysis by EF-G and EF4, which is in agreement with several studies that demonstrate similar inhibition of ribosome-dependent multiple-turnover EF-G GTP hydrolysis by thiostrepton (20,35,39,40). Curiously, some investigations have shown that, while thiostrepton slows GTP hydrolysis, the presence of the antibiotic still allows the reaction to proceed to a significant extent, with reports of 50–100% of available GTP hydrolyzed within a 10 min GTP hydrolysis experiment, depending on reaction conditions (41,54). Despite our attempts to replicate these results, we have only observed strong inhibition of GTP hydrolysis by thiostrepton, which persisted throughout the duration of the experiments. Notably, when we attempted to replicate experiments that showed the most dramatic level of GTP hydrolysis in the presence of thiostrepton (41), we found the antibiotic to have very poor solubility in the conditions utilized in that study (100 uM thiostrepton in 2% DMSO). Similar complications arose in other studies that have attempted to replicate those results (20).
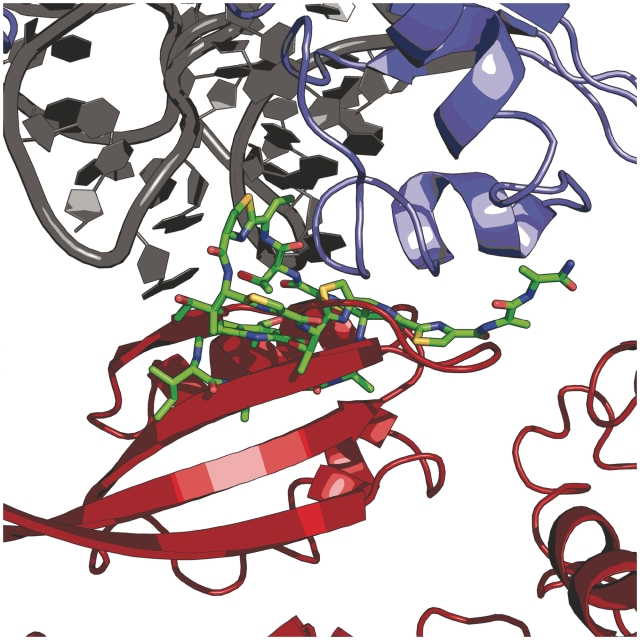
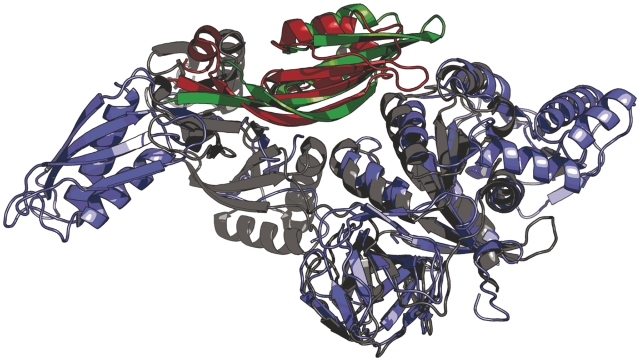
One implication of the commonly purported model for thiostrepton inhibition of EF-G is that thiostrepton inhibits turnover of the factor from the ribosome (41). Therefore, this proposed EF-G•ribosome•thiostrepton complex should be sufficiently stable to be observable in solution equilibrium binding experiments. However, we demonstrate that pre-incubation of ribosomes with thiostrepton results in the inability of EF-G•GDPNP and EF4•GDPNP to form stable complexes with the ribosome. This is in line with previously reported ultracentrifugation-based equilibrium binding experiments, which also failed to show any detectable stable binding of EF-G•GTP to the ribosome•thiostrepton complex (20). However, those results were challenged based on the argument that while the ribosome•thiostrepton complex may allow for EF-G binding, its overall affinity for EF-G is likely reduced, which could account for the dissociation of EF-G from the ribosome•thiostrepton complex upon ultracentrifugation (53). Importantly, the gel filtration/SDS–PAGE binding assay we employed did not require the use of ultracentrifugation, in order to address these concerns. Additionally, our results are consistent with an alignment of EF-G [from a recent X-ray crystal structure of EF-G bound to the 70S ribosome (53)] with the X-ray crystal structure of the thiostrepton-bound 50S ribosomal subunit (Figure 6) (7,53). This alignment suggests that the presence of thiostrepton is incompatible with the formation of a stable EF-G•ribosome complex due to significant steric clashing between thiostrepton and domain 5 of EF-G. A model based on a similar alignment had been previously presented, but employed lower resolution cryo-EM data for modeling ribosome-bound EF-G coordinates into the ribosome•thiostrepton structure (7). Regardless, a similar clash between thiostrepton and domain 5 was also observed in the previous model. We propose that this interaction is a key determinant of thiostrepton inhibition of EF-G. Our observation that deletion of domains 4 and 5 from EF-G enables the binding and GTP hydrolysis by the factor is consistent with this notion and also implies that the binding of thiostrepton to the ribosome does not cause significant peripheral conformational changes within the GTPase factor-binding region at large. This is further substantiated by the observation that release factor 3, a GTPase that does not possess domain 5, does not show any inhibition of ribosome-dependent GTPase activity using the time-dependent conditions described herein (Supplementary Data). Previous structural data indicate EF4 contains a domain that is homologous to domain 5 of EF-G (Figure 7) (55,56). Therefore, it is likely that a similar clash between thiostrepton and domain 5 of EF4 is responsible for the inhibition of ribosome binding and GTP hydrolysis by that factor.
Figure 6.
Alignment of the 50S•thiostrepton and 70S•EF-G X-ray crystal structures. Structural alignment of X-ray crystal structures (PDB entries 3CF5 and 2WRI) indicated significant steric clash between domain V of EF-G (red) and ribosome-bound thiostrepton (green). 23S rRNA is shown in gray and ribosomal protein L11 is colored blue (7,53).
Figure 7.
Superposition of EF-G and EF4. Structural alignment reveals strong structural homology between domain V of EF-G (red, PDB entry 1DAR) and the homologous domain in EF4 (green, PDB entry 3CB4). The remainder of EF-G and EF4 is colored blue and gray, respectively (55,56).
Although our results do not confirm the conclusions of time-resolved experiments, these ostensibly disparate proposed scenarios for thiostrepton inhibition of translation need not be mutually exclusive. Kinetic studies have suggested that the binding of EF-G to the ribosome is a two-step reaction, with thiostrepton allowing the initial step but inhibiting the second, causing an estimated 10-fold overall reduction in EF-G•ribosome binding affinity (42,44). Our results are compatible with the observation that EF-G is able to transiently adopt this proposed initial binding step in the presence of thiostrepton. However, the results presented here, which fail to demonstrate a stable EF-G•ribosome•thiostrepton complex, suggest that this interaction is not stable under physiological conditions. Interestingly, a structural model based on cryo-EM has been reported which purports to show EF-G bound to the ribosome•thiostrepton complex (45). In that model, EF-G was found to adopt a novel conformation in which domain 5 is significantly displaced from its position as observed in the fusidic acid-stabilized, post-translocation state, in which domain 5 makes contacts with the GTPase-associated center of the ribosomal translation factor binding site (13,53). It is possible that this cryo-EM model represents the initial labile state of the proposed two-step binding model for EF-G and that the conditions used in the cryo-EM sample preparation allowed for the observation of this otherwise unstable conformation. Unfortunately, we are unable to reconcile differences between our GTP hydrolysis experiments and those of others, which show significant multiple-turnover GTP hydrolysis activity in the presence of thiostrepton. Therefore, further investigations will be necessary to resolve these discrepancies. However, based on the accumulated evidence of thiostrepton•ribosome•translation factor interactions, we propose that it is implausible that thiostrepton inhibits factor turnover on the ribosome.
EF4•ribosome interactions during reverse translocation
To further examine the nature of interactions between EF4 and 70S ribosome in the GTP state, we employed chemical footprinting to characterize the mode of binding. The similarity in chemical protections for both EF4 and EF-G in the presence of GDPNP suggests that both GTPases bind to the same conserved rRNA regions in their GTP state. Notably, EF4 appears to bind more weakly to the 70S ribosome in comparison with EF-G as evidenced by weaker chemical protections at the GAC (Figure 5B) and due to the presence of a weaker band for EF4 in the gel filtration assay (Figure 3B). This is consistent with previously estimated KDvalues for EF4•GDPNP binding to various ribosomal complexes of ~0.25–1 μM (57), which are considerably weaker than the KD of ~26 nM determined for EF-G•GTP binding to a similar ribosomal complex (58).
Further chemical footprinting revealed that EF4•GDPNP enhances the DMS modification of 16S A702 identically to the same complexes with EF-G•GDPNP. This modification enhancement has previously been shown to be associated with the hybrid P/E state of ribosome-bound tRNA and ribosomal ratcheting (47,48,59,60). Surprisingly, EF4•GDPNP did not protect C2394 as is the case with EF-G. C2394 makes a Watson–Crick basepair with the 3′-CCA end of E site-bound tRNA (61), and this result suggests that EF4•GDPNP does not support hybrid P/E state tRNA conformation when a single tRNA is bound to the 30S P site. Taken together, these chemical footprinting data support a model for the first step in EF4-catalyzed reverse translocation. In this model, EF4•GDPNP binding stabilizes a ratcheted conformation of the 70S ribosome that is not associated with movement of tRNA from the classical P/P state to the hybrid P/E state.
These data illustrate the significance of the E site to the directionality of translocation. It was shown previously that the presence of E-site tRNA is required for both spontaneous (62,63) and EF4-catalyzed reverse translocation (46), likely because a tRNA must be bound to the 30S P site in all translocation intermediate states. In contrast to the E-site tRNA requirement for EF4, EF-G may stabilize forward translocation by inducing E-site tRNA release following GTP hydrolysis (48). In the absence of the translocated E-site tRNA, reverse translocation would not be thermodynamically favorable, and the resultant complex would remain in the post-translocation state.
Previous studies indicate that the ratcheted state stabilized by EF-G destabilizes the 30S P-site tRNA•mRNA interaction and increases the propensity of various functional complexes to mRNA back-slippage (48), a mechanism that may also occur with other GTPases that stabilize a ratcheted 70S conformation, such as EF4. This decrease in P-site tRNA interaction is a likely requirement for rapid, reverse translocation, as the interaction between the 30S P site and the anticodon stem loop of tRNA is the strongest interaction on the small subunit and must be destabilized for movement of tRNA in the reverse direction (64), which would also likely require the presence of an E-site tRNA for reverse translocation to the P site. We hypothesize that the ratcheted state associated with EF4•GDPNP stabilizes the hybrid ‘A/P’ and ‘P/E’ states that represent a ‘pre-reverse-translocation’ conformation, where one tRNA is bound to the 30S A site/50S P site (A/P) and a second tRNA is bound to the 30S P site/50S E site (P/E) simultaneously. Lastly, EF4•GDPNP enhances the DMS modification of C2556, a conserved base in the 50S A-loop, which is an essential component of the peptidyltransferase center and makes direct contacts with A-site tRNA. According to a previous cryo-EM study of the EF4•GDPNP complex, EF4 interacts directly with an A-site tRNA during the process of reverse translocation and stabilizes a newly characterized ‘A/L’ state of ribosome-bound tRNA (57). The ‘A/L’ state tRNA interacts directly with 23S helix 92, which consists of the A-loop and C2556, and is proposed to represent an intermediate during the reverse translocation mechanism (57). Further data are needed to resolve whether the structural results presented herein are coincident with the ‘A/L’ state or resemble a structural complex that precedes ‘A/L’ formation.
In light of these observations, along with recent studies which implicate EF4 as a GTPase that rescues stalled ribosomes (46,57,65), we hypothesize that EF4 recognizes a 70S state which harbors an aberrant E-site tRNA that did not dissociate following EF-G-catalyzed forward translocation, due to unfavorable conditions such as high ionic strength or low temperature. Indeed, under conditions of high magnesium concentration, the presence of EF4 has been shown to increase the rate of protein synthesis by 5-fold (65). Since high magnesium concentrations stabilize ribosome-bound tRNAs in the classical states (A/A, P/P and E/E) (48), it seems plausible that dissociation of a newly translocated E-site tRNA may be inhibited under such conditions, causing the adoption of irregular E-site tRNA conformations which may then be recognized by EF4.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Research Corporation Cottrell College Science Award (grant number 10786). Funding for open access charge: Western Washington University faculty mini-grant; Research Corporation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge Marion Brodhagen for use of her phosphorimager and AMGEN for their donation of a scintillation counter.
REFERENCES
- 1.Poehlsgaard J, Douthwaite S. The bacterial ribosome as a target for antibiotics. Nat. Rev. Microbiol. 2005;3:870–881. doi: 10.1038/nrmicro1265. [DOI] [PubMed] [Google Scholar]
- 2.Sohmen D, Harms JM, Schlunzen F, Wilson DN. SnapShot: antibiotic inhibition of protein synthesis I. Cell. 2009;138:1248 e1241. doi: 10.1016/j.cell.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Spahn CM, Prescott CD. Throwing a spanner in the works: antibiotics and the translation apparatus. J. Mol. Med. 1996;74:423–439. doi: 10.1007/BF00217518. [DOI] [PubMed] [Google Scholar]
- 4.Brodersen DE, Clemons WM, Jr, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- 5.Dunkle JA, Xiong L, Mankin AS, Cate JH. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc. Natl Acad. Sci. USA. 2010;107:17152–17157. doi: 10.1073/pnas.1007988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen JL, Ippolito JA, Ban N, Nissen P, Moore PB, Steitz TA. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell. 2002;10:117–128. doi: 10.1016/s1097-2765(02)00570-1. [DOI] [PubMed] [Google Scholar]
- 7.Harms JM, Wilson DN, Schluenzen F, Connell SR, Stachelhaus T, Zaborowska Z, Spahn CM, Fucini P. Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol. Cell. 2008;30:26–38. doi: 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Ippolito JA, Kanyo ZF, Wang D, Franceschi FJ, Moore PB, Steitz TA, Duffy EM. Crystal structure of the oxazolidinone antibiotic linezolid bound to the 50S ribosomal subunit. J. Med. Chem. 2008;51:3353–3356. doi: 10.1021/jm800379d. [DOI] [PubMed] [Google Scholar]
- 9.Tu D, Blaha G, Moore PB, Steitz TA. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121:257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Mohr D, Wintermeyer W, Rodnina MV. GTPase activation of elongation factors Tu and G on the ribosome. Biochemistry. 2002;41:12520–12528. doi: 10.1021/bi026301y. [DOI] [PubMed] [Google Scholar]
- 11.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330:835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diaconu M, Kothe U, Schlunzen F, Fischer N, Harms JM, Tonevitsky AG, Stark H, Rodnina MV, Wahl MC. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell. 2005;121:991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 13.Moazed D, Robertson JM, Noller HF. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature. 1988;334:362–364. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]
- 14.Bagley MC, Dale JW, Merritt EA, Xiong X. Thiopeptide antibiotics. Chem. Rev. 2005;105:685–714. doi: 10.1021/cr0300441. [DOI] [PubMed] [Google Scholar]
- 15.Donovick R, Pagano JF, Stout HA, Weinstein MJ. Thiostrepton, a new antibiotic. I. In vitro studies. Antibiot. Annu. 1955;3:554–559. [PubMed] [Google Scholar]
- 16.Bowen WS, Van Dyke N, Murgola EJ, Lodmell JS, Hill WE. Interaction of thiostrepton and elongation factor-G with the ribosomal protein L11-binding domain. J. Biol. Chem. 2005;280:2934–2943. doi: 10.1074/jbc.M407008200. [DOI] [PubMed] [Google Scholar]
- 17.Jonker HR, Ilin S, Grimm SK, Wohnert J, Schwalbe H. L11 domain rearrangement upon binding to RNA and thiostrepton studied by NMR spectroscopy. Nucleic Acids Res. 2007;35:441–454. doi: 10.1093/nar/gkl1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson J, Cundliffe E, Stark M. Binding of thiostrepton to a complex of 23-S rRNA with ribosomal protein L11. Eur. J. Biochem. 1979;98:261–265. doi: 10.1111/j.1432-1033.1979.tb13184.x. [DOI] [PubMed] [Google Scholar]
- 19.Xing Y, Draper DE. Cooperative interactions of RNA and thiostrepton antibiotic with two domains of ribosomal protein L11. Biochemistry. 1996;35:1581–1588. doi: 10.1021/bi952132o. [DOI] [PubMed] [Google Scholar]
- 20.Cameron DM, Thompson J, March PE, Dahlberg AE. Initiation factor IF2, thiostrepton and micrococcin prevent the binding of elongation factor G to the Escherichia coli ribosome. J. Mol. Biol. 2002;319:27–35. doi: 10.1016/S0022-2836(02)00235-8. [DOI] [PubMed] [Google Scholar]
- 21.Weisblum B, Demohn V. Thiostrepton, an inhibitor of 5OS ribosome subunit function. J. Bacteriol. 1970;101:1073–1075. doi: 10.1128/jb.101.3.1073-1075.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clough B, Strath M, Preiser P, Denny P, Wilson IR. Thiostrepton binds to malarial plastid rRNA. FEBS Lett. 1997;406:123–125. doi: 10.1016/s0014-5793(97)00241-x. [DOI] [PubMed] [Google Scholar]
- 23.Goodman CD, Su V, McFadden GI. The effects of anti-bacterials on the malaria parasite Plasmodium falciparum. Mol. Biochem. Parasitol. 2007;152:181–191. doi: 10.1016/j.molbiopara.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 24.McConkey GA, Rogers MJ, McCutchan TF. Inhibition of Plasmodium falciparum protein synthesis. Targeting the plastid-like organelle with thiostrepton. J. Biol. Chem. 1997;272:2046–2049. doi: 10.1074/jbc.272.4.2046. [DOI] [PubMed] [Google Scholar]
- 25.Schoof S, Pradel G, Aminake MN, Ellinger B, Baumann S, Potowski M, Najajreh Y, Kirschner M, Arndt HD. Antiplasmodial thiostrepton derivatives: proteasome inhibitors with a dual mode of action. Angew Chem. Int. Ed. Engl. 2010;49:3317–3321. doi: 10.1002/anie.200906988. [DOI] [PubMed] [Google Scholar]
- 26.Bhat UG, Halasi M, Gartel AL. Thiazole antibiotics target FoxM1 and induce apoptosis in human cancer cells. PLoS One. 2009;4:e5592. doi: 10.1371/journal.pone.0005592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwok JM, Myatt SS, Marson CM, Coombes RC, Constantinidou D, Lam EW. Thiostrepton selectively targets breast cancer cells through inhibition of forkhead box M1 expression. Mol. Cancer Ther. 2008;7:2022–2032. doi: 10.1158/1535-7163.MCT-08-0188. [DOI] [PubMed] [Google Scholar]
- 28.Nicolaou KC, Safina BS, Zak M, Estrada AA, Lee SH. Total synthesis of thiostrepton, part 1: construction of the dehydropiperidine/thiazoline-containing macrocycle. Angew Chem. Int. Ed. Engl. 2004;43:5087–5092. doi: 10.1002/anie.200461340. [DOI] [PubMed] [Google Scholar]
- 29.Nicolaou KC, Safina BS, Zak M, Lee SH, Nevalainen M, Bella M, Estrada AA, Funke C, Zecri FJ, Bulat S. Total synthesis of thiostrepton. Retrosynthetic analysis and construction of key building blocks. J. Am. Chem. Soc. 2005;127:11159–11175. doi: 10.1021/ja0529337. [DOI] [PubMed] [Google Scholar]
- 30.Nicolaou KC, Zak M, Safina BS, Estrada AA, Lee SH, Nevalainen M. Total synthesis of thiostrepton. Assembly of key building blocks and completion of the synthesis. J. Am. Chem. Soc. 2005;127:11176–11183. doi: 10.1021/ja052934z. [DOI] [PubMed] [Google Scholar]
- 31.Nicolaou KC, Zak M, Safina BS, Lee SH, Estrada AA. Total synthesis of thiostrepton, part 2: construction of the quinaldic acid macrocycle and final stages of the synthesis. Angew Chem. Int. Ed. Engl. 2004;43:5092–5097. doi: 10.1002/anie.200461341. [DOI] [PubMed] [Google Scholar]
- 32.Nicolaou KC, Zak M, Rahimipour S, Estrada AA, Lee SH, O'Brate A, Giannakakou P, Ghadiri MR. Discovery of a biologically active thiostrepton fragment. J. Am. Chem. Soc. 2005;127:15042–15044. doi: 10.1021/ja0552803. [DOI] [PubMed] [Google Scholar]
- 33.Brandi L, Marzi S, Fabbretti A, Fleischer C, Hill WE, Gualerzi CO, Stephen Lodmell J. The translation initiation functions of IF2: targets for thiostrepton inhibition. J. Mol. Biol. 2004;335:881–894. doi: 10.1016/j.jmb.2003.10.067. [DOI] [PubMed] [Google Scholar]
- 34.Brot N, Tate WP, Caskey CT, Weissbach H. The requirement for ribosomal proteins L7 and L12 in peptide-chain termination. Proc. Natl Acad. Sci. USA. 1974;71:89–92. doi: 10.1073/pnas.71.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bodley JW, Lin L, Highland JH. Studies on translocation. VI. Thiostrepton prevents the formation of a ribosome-G factor-guanine nucleotide complex. Biochem. Biophys. Res. Commun. 1970;41:1406–1411. doi: 10.1016/0006-291x(70)90543-7. [DOI] [PubMed] [Google Scholar]
- 36.Egebjerg J, Douthwaite S, Garrett RA. Antibiotic interactions at the GTPase-associated centre within Escherichia coli 23S rRNA. EMBO J. 1989;8:607–611. doi: 10.1002/j.1460-2075.1989.tb03415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Highland JH, Lin L, Bodley JW. Protection of ribosomes from thiostrepton inactivation by the binding of G factor and guanosine diphosphate. Biochemistry. 1971;10:4404–4409. doi: 10.1021/bi00800a009. [DOI] [PubMed] [Google Scholar]
- 38.Hummel H, Bock A. Thiostrepton resistance mutations in the gene for 23S ribosomal RNA of halobacteria. Biochimie. 1987;69:857–861. doi: 10.1016/0300-9084(87)90212-4. [DOI] [PubMed] [Google Scholar]
- 39.Modolell J, Cabrer B, Parmeggiani A, Vazquez D. Inhibition by siomycin and thiostrepton of both aminoacyl-tRNA and factor G binding to ribosomes. Proc. Natl Acad. Sci. USA. 1971;68:1796–1800. doi: 10.1073/pnas.68.8.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pestka S. Thiostrepton: a ribosomal inhibitor of translocation. Biochem. Biophys. Res. Commun. 1970;40:667–674. doi: 10.1016/0006-291x(70)90956-3. [DOI] [PubMed] [Google Scholar]
- 41.Rodnina MV, Savelsbergh A, Matassova NB, Katunin VI, Semenkov YP, Wintermeyer W. Thiostrepton inhibits the turnover but not the GTPase of elongation factor G on the ribosome. Proc. Natl Acad. Sci. USA. 1999;96:9586–9590. doi: 10.1073/pnas.96.17.9586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seo HS, Kiel M, Pan D, Raj VS, Kaji A, Cooperman BS. Kinetics and thermodynamics of RRF, EF-G, and thiostrepton interaction on the Escherichia coli ribosome. Biochemistry. 2004;43:12728–12740. doi: 10.1021/bi048927p. [DOI] [PubMed] [Google Scholar]
- 43.Skold SE. Chemical crosslinking of elongation factor G to the 23S RNA in 70S ribosomes from Escherichia coli. Nucleic Acids Res. 1983;11:4923–4932. doi: 10.1093/nar/11.14.4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan D, Kirillov SV, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Mol. Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stark H, Rodnina MV, Wieden HJ, van Heel M, Wintermeyer W. Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell. 2000;100:301–309. doi: 10.1016/s0092-8674(00)80666-2. [DOI] [PubMed] [Google Scholar]
- 46.Qin Y, Polacek N, Vesper O, Staub E, Einfeldt E, Wilson DN, Nierhaus KH. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell. 2006;127:721–733. doi: 10.1016/j.cell.2006.09.037. [DOI] [PubMed] [Google Scholar]
- 47.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 48.Spiegel PC, Ermolenko DN, Noller HF. Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. RNA. 2007;13:1473–1482. doi: 10.1261/rna.601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ederth J, Mandava CS, Dasgupta S, Sanyal S. A single-step method for purification of active His-tagged ribosomes from a genetically engineered Escherichia coli. Nucleic Acids Res. 2009;37:e15. doi: 10.1093/nar/gkn992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frolova L, Le Goff X, Zhouravleva G, Davydova E, Philippe M, Kisselev L. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA. 1996;2:334–341. [PMC free article] [PubMed] [Google Scholar]
- 51.Ermolenko DN, Spiegel PC, Majumdar ZK, Hickerson RP, Clegg RM, Noller HF. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat. Struct. Mol. Biol. 2007;14:493–497. doi: 10.1038/nsmb1243. [DOI] [PubMed] [Google Scholar]
- 52.Stern S, Moazed D, Noller HF. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 53.Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lentzen G, Klinck R, Matassova N, Aboul-ela F, Murchie AI. Structural basis for contrasting activities of ribosome binding thiazole antibiotics. Chem. Biol. 2003;10:769–778. doi: 10.1016/s1074-5521(03)00173-x. [DOI] [PubMed] [Google Scholar]
- 55.al-Karadaghi S, Aevarsson A, Garber M, Zheltonosova J, Liljas A. The structure of elongation factor G in complex with GDP: conformational flexibility and nucleotide exchange. Structure. 1996;4:555–565. doi: 10.1016/s0969-2126(96)00061-5. [DOI] [PubMed] [Google Scholar]
- 56.Evans RN, Blaha G, Bailey S, Steitz TA. The structure of LepA, the ribosomal back translocase. Proc. Natl Acad. Sci. USA. 2008;105:4673–4678. doi: 10.1073/pnas.0801308105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Connell SR, Topf M, Qin Y, Wilson DN, Mielke T, Fucini P, Nierhaus KH, Spahn CM. A new tRNA intermediate revealed on the ribosome during EF4-mediated back-translocation. Nat. Struct. Mol. Biol. 2008;15:910–915. doi: 10.1038/nsmb.1469. [DOI] [PubMed] [Google Scholar]
- 58.Lancaster L, Lambert NJ, Maklan EJ, Horan LH, Noller HF. The sarcin-ricin loop of 23S rRNA is essential for assembly of the functional core of the 50S ribosomal subunit. RNA. 2008;14:1999–2012. doi: 10.1261/rna.1202108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang W, Dunkle JA, Cate JH. Structures of the ribosome in intermediate states of ratcheting. Science. 2009;325:1014–1017. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmeing TM, Moore PB, Steitz TA. Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. RNA. 2003;9:1345–1352. doi: 10.1261/rna.5120503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Konevega AL, Fischer N, Semenkov YP, Stark H, Wintermeyer W, Rodnina MV. Spontaneous reverse movement of mRNA-bound tRNA through the ribosome. Nat. Struct. Mol. Biol. 2007;14:318–324. doi: 10.1038/nsmb1221. [DOI] [PubMed] [Google Scholar]
- 63.Shoji S, Walker SE, Fredrick K. Reverse translocation of tRNA in the ribosome. Mol. Cell. 2006;24:931–942. doi: 10.1016/j.molcel.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lill R, Robertson JM, Wintermeyer W. Affinities of tRNA binding sites of ribosomes from Escherichia coli. Biochemistry. 1986;25:3245–3255. doi: 10.1021/bi00359a025. [DOI] [PubMed] [Google Scholar]
- 65.Pech M, Karim Z, Yamamoto H, Kitakawa M, Qin Y, Nierhaus KH. Elongation factor 4 (EF4/LepA) accelerates protein synthesis at increased Mg2+ concentrations. Proc. Natl Acad. Sci. USA. 2011;108:3199–3203. doi: 10.1073/pnas.1012994108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.