Abstract
Global mechanisms defining the gene expression programs specific for hematopoiesis are still not fully understood. Here, we show that promoter DNA demethylation is associated with the activation of hematopoietic-specific genes. Using genome-wide promoter methylation arrays, we identified 694 hematopoietic-specific genes repressed by promoter DNA methylation in human embryonic stem cells and whose loss of methylation in hematopoietic can be associated with gene expression. The association between promoter methylation and gene expression was studied for many hematopoietic-specific genes including CD45, CD34, CD28, CD19, the T cell receptor (TCR), the MHC class II gene HLA-DR, perforin 1 and the phosphoinositide 3-kinase (PI3K) and results indicated that DNA demethylation was not always sufficient for gene activation. Promoter demethylation occurred either early during embryonic development or later on during hematopoietic differentiation. Analysis of the genome-wide promoter methylation status of induced pluripotent stem cells (iPSCs) generated from somatic CD34+ HSPCs and differentiated derivatives from CD34+ HSPCs confirmed the role of DNA methylation in regulating the expression of genes of the hemato-immune system, and indicated that promoter methylation of these genes may be associated to stemness. Together, these data suggest that promoter DNA demethylation might play a role in the tissue/cell-specific genome-wide gene regulation within the hematopoietic compartment.
INTRODUCTION
The orchestrated regulation of genome-wide gene expression directs the complex logistics of cell fate determination during the development/ontogeny of higher organisms. Whereas some genes are constitutively expressed throughout the different developmental stages, others are specifically expressed in a temporal (i.e. developmental stage) and spatial (i.e. lineage specific) fashion. Regulation of gene expression in blood cell development is of particular interest, given the diversity of functionally different hematopoietic cell types and the large range of hematological malignancies affecting a variety of hematopoietic cell subsets (B-cells, T-cells, NK-cells, myeloid cells, red blood cells, platelets, etc.) at distinct developmental stages [i.e. stem cell (HSC), B progenitor (pro-B), B precursor (pre-B), mature B cells, plasma cells, etc.]. Although there is considerable information available about gene expression patterns in blood cells, there is an important gap in our understanding about its regulation.
Epigenetic marks participate prominently in gene regulation (1). Genomic DNA methylation, one of the best-studied epigenetic modifications, is an important means of regulating gene expression with alterations in this process being associated with cancer and other diseases (1,2). As a means of gene inactivation, DNA methylation has been implicated in the stable silencing of undifferentiated embryonic stem cell (ESC)-associated genes such as OCT4 and NANOG (3,4). Although many genes are hypermethylated in ESCs, demethylation processes during cell differentiation have yet to be studied in depth. Previous genome-wide methylation studies in ESCs described a scenario in which most of the CpG-rich promoters were unmethylated, while CpG-poor promoters, generally associated with highly tissue-specific genes, tend to be hypermethylated (5,6). As certain genes whose expression is restricted to the hematopoietic system, such as the genes for myeloperoxidase (7), globin (8,9), c-fms (10), lysozyme (11), the granulocyte colony-stimulating factor (G-CSF) receptor (12), perforin (13), platelet glycoprotein VI (GPVI/GP6) (14) and FOXP3 (15) are nonetheless regulated by methylation in a lineage- and developmental stage-dependent manner, we hypothesized that promoter demethylation might be an important mechanism of gene regulation within the hematopoietic system/hierarchy.
We used Infinium DNA methylation arrays to study the genome-wide role of promoter DNA demethylation in the hematopoietic system. We have compared the methylation status of human ESCs, cord blood (CB), CD34+ hematopoietic stem and progenitor cells (HSPCs) and five highly represented differentiated blood white cell lineages: neutrophils, B lymphocytes, NK cells, CD8+ T lymphocytes and CD4+ T lymphocytes. Using this approach in combination with RNA/protein expression experiments, we identified several hundreds of genes that become demethylated during hematopoietic differentiation. In vitro differentiation of CB-derived CD34+ HSPCs supported the role of promoter demethylation during hematopoietic differentiation. Importantly, analyses of iPSCs derived from CD34+ HSPCs demonstrated that the promoter methylation-associated repression of hematopoietic genes is closely associated with stemness and that most but not all the promoter DNA demethylation observed during hematopoietic differentiation is reverted upon cellular reprogramming.
MATERIALS AND METHODS
Human ESC lines
Cell pellets and/or DNA/RNA were used from the following well-characterized hESC lines: SHEF-1, SHEF-4, SHEF-5, SHEF-7, H7, H14, H14S9, H7S14, HS181 and I3. Human ESCs were maintained undifferentiated in a feeder-free culture as previously described (16,17). Briefly, hESCs were cultured in Matrigel (BD Biosciences, Bedford, MA, USA)-coated T25 flasks in either mouse embryonic fibroblast (MEF), human foreskin fibroblast (HFF) or mesenchymal stromal cell (MSC)-conditioned medium (MEF-CM, HFF-CM or MSC-CM) supplemented with 8 ng/ml basic fibroblast growth factor (bFGF; Miltenyi, Madrid, Spain) Approval from the Spanish National Embryo Ethical Committee (ISCIII) was obtained to work with these hESCs lines.
Primary tissue
Fresh CB units from healthy newborns were obtained from The Málaga Cord Blood Bank upon approval by local Ethics and Biozahard Board Committee. CB samples were pooled to reduce variability between individual freshly isolated CB units (n = 5). Mononuclear cells (MNC) were isolated using Ficoll-Hypaque (GE Healthcare, Stockholm, Sweden). After erythrocyte lysis (Lysis solution, StemCell Technologies, Vancouver, Canada), CD34+ cells were purified by magnetic bead separation using the human CD34 MicroBead kit (Miltenyi, Munich, Germany) and the AutoMACS Pro separator (Miltenyi) according to manufacturer's instructions (18). Lymphocytes and neutrophils were isolated from peripheral blood (PB) from healthy adult volunteers by centrifugation, using Histopaque-1077 (Sigma, St Louis, MO, USA). CD4+ and CD8+ T cells were enriched from PB MNCs using magnetic bead isolation (Miltenyi). NK and B cells were FACS-sorted (FACSAria, Becton Dickinson, San Jose, CA, USA) based on their specific expression of the surface markers CD56 and CD19, respectively. Purity of all isolated cellular fractions was >95%. Hematological samples were pooled to reduce interindividual variability (n = 5). DNA from human normal primary tissues was obtained from Biochain (Hayward, CA, USA).
Generation of iPSC from CD34+ CB cells
CD34+ HSCs purified from CB (1 × 105 cells/ml) were pre-stimulated 48 h in Stem Span medium (Stem Cells Technologies, Grenoble, France) supplemented with SCF (100 ng/ml), FLT3L (100 ng/ml) and IL-3 (10 ng/ml) in fibronectin-coated plates (Becton Dickinson) (19). Induced PSC were generated by infection of prestimulated CD34+ HSC with high-titer viral vectors expressing the human reprogramming factors Oct4, Sox2, Klf4 and c-myc (OKSM), as previously described (20). Viral particles pseudotyped with the VSV-G protein were generated on 293T cells using a standard calcium–phosphate transfection protocol and were concentrated by ultracentrifugation (21). Cells were infected overnight; the following day, viral supernatant was removed and transduced CD34+ HSC were washed with Stem Span medium and allowed to grow for 3 days before transfer onto irradiated (4000 Gy) HFFs (16). Cultures were maintained in KO-DMEM supplemented with 20% serum replacement (Invitrogen, Carlsbad, CA, USA), 1% non-essential amino acids, 1 mM l-glutamine, 0.1 mM β–mercaptoethanol and 50 ng/ml bFGF). Emerging iPSC colonies were identified and passaged onto fresh feeders (20). iPSC from CB-derived CD34+ cells were fully characterized and proved to be bona fide iPSC, as they displayed ESC morphology, ESC-associated transcription factors and cell surface markers, silenced ectopic reprogramming factors and they differentiated into the three germ layers in vitro (embryoid bodies) and in vivo (teratomas) (22). These iPSC lines have been deposited according to Spanish Legislation in The Spanish Stem Cell Bank (http://www.isciii.es/htdocs/terapia/terapia_lineas.jsp).
DNA methylation profiling using bead arrays
Microarray-based DNA methylation profiling was performed on all samples with the HumanMethylation27 DNA Analysis BeadChip (Illumina, San Diego, CA, USA). This array allows interrogation of 27 578 highly informative CpG sites per sample at single-nucleotide resolution for more than 14 000 genes. This 12-sample BeadChip features content derived from the NCBI CCDS database (Genome Build 36) and is supplemented with more than 1000 cancer-related genes. HumanMethylation27 BeadChip content also targets the promoter regions of 110 miRNA genes. The Infinium Methylation Assay accomplishes this high multiplexing by combining bisulfite conversion of genomic DNA and whole-genome amplification (WGA) sample preparation with direct, array-based capture and enzymatic scoring of the CpG loci. Bisulfite conversion of DNA was performed using the EZ DNA Methylation Kit (Zymo Research, Orange, CA, USA) according to manufacturer's procedures, with the modifications described in the Infinium Assay Methylation Protocol Guide. Processed DNA samples were hybridized to the BeadChip (Illumina). The assay interrogates the chemically differentiated loci using two site-specific probes, one designed for the methylated locus (M bead type) and another for the unmethylated locus (U bead type). Single-base extension of the probes incorporates a labeled ddNTP, which is subsequently stained with a fluorescence reagent. The methylation level for the interrogated locus is determined by calculating the ratio of the fluorescent signals from the methylated versus unmethylated sites. The ratio of fluorescent signals was then computed from the two alleles according to the following formula:
This beta value is a quantitative measure of DNA methylation levels of specific CpG, and ranges from 0 for completely unmethylated to 1 for completely methylated.
The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE30090.
Before analyzing the methylation data, we excluded possible sources of technical and gender-specific biases. Technical bias were minimized by excluding probes with detection P-values >0.01 in ≥5% samples. Gender-specific bias was avoided by excluding probes in sex chromosomes.
Differential methylation and computational gene expression analysis
The amount of bisulfite-modified target DNA that hybridizes to each spot of the Illumina chip was quantified and standardized over a range from 0.0 to 1.0 (effectively 0% and 100% likelihood of gene promoter hypermethylation, respectively). In this work, all sequences with at least 70% likelihood of being hypermethylated (hybridization signal ≥0.7) were considered hypermethylated for each specific sample, whereas sequences whose equivalent signal was below 30% (hybridization signal <0.3) were considered unmethylated. For each differentiated tissue, a CpG site was considered demethylated when it was hypermethylated in hESC and demethylated in all the samples corresponding to that specific tissue. A CpG site was considered as tissue specifically demethylated when it was demethylated exclusively in that tissue or cell type. In in vitro CD34+ differentiation (Diff-CD34) and de-differentiation (CD34-iPSCs) experiments, we only considered CpG sites showing a change in methylation values of at least 20% relative to CD34+ progenitors. Gene ontology (GO) and database expression analyses were performed with the DAVID GO Web-based tool (23,24), comparing demethylated gene groups against a background comprising all the genes in the Illumina array.
We compared the methylation data with available gene expression data. For solid tissues and whole-PB gene expression data was taken from Amazonia (http://amazonia.transcriptome.eu), which comprises human transcriptome list annotations. Gene expression data was downloaded from GEO (http://www.ncbi.nlm.nih.gov/geo) to compare methylation and expression data in different PB cell subsets (CD8+: GSM565269, GSM565270, GSM565271, GSM565272; CD4+: GSM565273, GSM565274, GSM565275; NK: GSM565293, GSM565294, GSM565295, GSM565296; B cells: GSM565308, GSM565309, GSM565310, GSM565311, GSM565312, GSM565313, GSM565314; neutrophils: GSM565378, GSM565379, GSM565380, GSM565381, GSM565382). Box plots were used to estimate the distribution of gene expression (log scale). In each tissue or cell type, equality of densities referred to specific demethylated gene expression levels compared to the other tissue or cell types was confirmed using L1 criterion (25). We used very restrictive criteria to select genes with expression associated with specific demethylation in a given tissue or blood cell subset (Supplementary Tables S1 and S2): for each tissue or cell type, differences (significant at 5%) were calculated between average expression values of specific genes and the maximum average of the other tissues (Student's t-test). Resample methods were used to confirm or discard other complex hypotheses.
To determine the role of demethylation in priming genes for further expression, we compared our methylation data with the available expression data of activated hematopoietic cells (B cells, CD4+ cells and NK cells) obtained from the GEO database GSE22886 (B cells: GSM565315, GSM565316, GSM565317, GSM565318; CD4+: GSM565276, GSM565277; NK: GSM565302, GSM565303, GSM565304, GSM565305, GSM565306, GSM565307). We followed the same criteria mentioned above to select genes with expression associated with specific demethylation in a given blood cell subset (Supplementary Table S3).
To enable a comparison of gene expression between lymphoid/myeloid progenitors and terminally differentiated cell types, we compared our methylation data with expression data of a subset of hematologic progenitor and terminally differentiated hematological cells obtained from BioGPS (Hematopoietic Atlas, Data set_302) (26). We selected those genes whose expression associated with specific demethylation in a blood cell subset was higher than the likelihood thresholds for the non-specific ones. These thresholds were computed as the upper bound in a 95% confidence interval for the ‘non-specific’ means (Supplementary Table S4).
The clustering heatmaps and bar plots using methylation and expression data were prepared with BeadStudio software (Illumina) and Microsoft Excel tools.
Pyrosequencing
Sodium bisulfite modification of 0.5 µg genomic DNA isolated from various tissues was carried out with the EZ DNA Methylation Kit following the manufacturer's protocol. Bisulfite-treated DNA was eluted in 15 µl volumes with 2 µl for each PCR. The set of primers for PCR amplification and sequencing were designed using a specific software pack (PyroMark assay design version 2.0.01.15). Primer sequences were designed to hybridize with CpG-free sites to ensure methylation-independent amplification.
PCR was performed with primers biotinylated to convert the PCR product to single-stranded DNA templates. We used the Vacuum Prep Tool (Biotage, Sweden) to prepare single-stranded PCR products according to manufacturer's instructions. Pyrosequencing reactions and methylation quantification were performed in a PyroMark Q24 System version 2.0.6 (Qiagen). Graphic representation of methylation values shows bars identifying CpG sites that present percentage methylation values.
RNA purification and real-time RT–PCR analysis
RNA was isolated with TRIzol Reagent (Invitrogen) according to manufacturer's instructions. For RT–PCR, 1 µg total RNA was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative real-time RT–PCR was performed using SYBR green universal master mix and the ABI PRISM 7900 sequence-detection system (Applied Biosystems). Data are expressed as mean ± SD of three replicates of each experiment.
Western blot
Cell lysates for protein analysis were prepared by radio-immunoprecipitation assay buffer extraction and analyzed by western blot using anti-β-actin (Sigma) and anti-PI3Kδ (Santa Cruz).
Flow cytometry
iPSC from CD34+ cells were harvested with 0.1% collagenase IV (Invitrogen) and dissociated into single cells before staining. CD34+-derived iPSCs and CD34+ cells (2 × 105) were incubated 30 min with the following fluorochrome-conjugated monoclonal antibodies: PerCP/Cy5.5-anti-CD34 (Biolegend), PE-anti-CD4, FITC-anti-CD34, PE-anti-CD28 and PE-anti-CD31 (BD Biosciences). A fluorochrome-matched isotype control was always used. The proportion of living, dead and apoptotic cells were determined with 7AAD and the Annexin V-FITC apoptosis detection kit (Immunostep, Salamanca, Spain), as previously described (27). Data acquisition was carried out in a FACSCanto II Cytometer and data was analyzed using FACSDiva software (Becton Dickinson).
Colony forming unit assay
The CB-derived CD34+-enriched fraction (2 × 103 cells/cm2) was plated in methycellulose-based medium supplemented with SCF (50 ng/ml), GM-CSF (10 ng/ml), IL-3 (10 ng/ml) and erythropoietin (3 U/ml; Methocult GF H4434; StemCell Technologies). After 12–14 days in culture, colonies were counted and scored (28,29). Colonies were subsequently scraped from the tissue culture plate and used for genomic DNA extraction.
RESULTS
Identification of tissue-specific promoter-demethylated genes using bead arrays
We compared the DNA methylation status of 27 578 sequences in 10 independently isolated human hESC lines and 16 healthy tissues corresponding to eight normal tissue types [colon, bone, fat, muscle, liver, thyroid, brain and blood (PB white cells)], using Illumina Infinium Methylation Arrays.
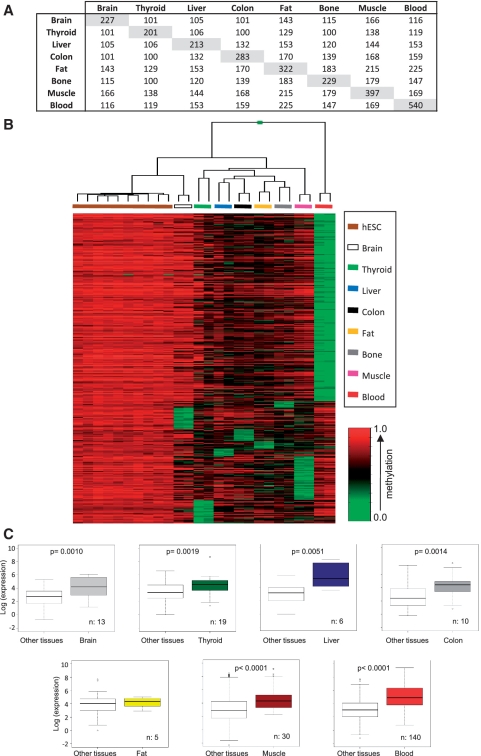
For identification of tissue-specific promoter-demethylated genes, we chose probes that showed methylation values (average_beta) <30% in each tissue type and methylation values (average_beta) ≥70% in at least 80% of the hESC, following previously published criteria (30). Using this approach, we identified groups of CpG probes that were demethylated in each tissue: 227 (corresponding to 200 genes) in brain, 201 (179 genes) in thyroid, 213 (189 genes) in liver, 283 (263 genes) in colon, 322 (289 genes) in fat, 229 (213 genes) in bone, 397 (354 genes) in muscle and 540 (463 genes) in blood (Supplementary Table S5 and Figure 1A). Of these, 23 CpG sites (corresponding to 21 genes) were demethylated specifically in brain (and in no other tissue), 25 (24 genes) in thyroid, 8 (8 genes) in liver, 13 (13 genes) in colon, 8 (8 genes) in fat, 7 (7 genes) in bone, 47 (47 genes) in muscle and 202 (182 genes) in blood (Supplementary Table S6). 40 CpG sites 110 (36 genes) were demethylated in all of these tissues, and a total of 413 CpG sites (383 genes) were demethylated in more than one tissue (Supplementary Table S7).
Figure 1.
DNA methylation profiling in human embryonic stem cell (hESC) lines and normal primary tissues. (A) Matrix showing the number of demethylated CpG sites in each tissue type and demethylated CpG sites shared between different tissue types. (B) Illumina array methylation clustering heatmap of hESC-hypermethylated genes specifically demethylated in each of the eight normal tissue samples. Methylation levels vary from fully methylated (red) to fully unmethylated (green). (C) Box plots of microarray-based gene expression data (log scale). In each primary tissue, tissue-specific demethylated genes exhibited higher expression levels compared to other tissues. P-values are shown. n = number of genes analyzed.
Supervised clustering of samples using exclusively the methylation signals of the tissue-specific demethylated probes (333; 310 genes) enabled correct classification of each sample within its corresponding group (Figure 1B), indicating that each sample group has a specific demethylated promoter signature. Two approaches were used to determine whether gene promoter demethylation enables establishment of tissue-specific expression programs; we used GO to analyze the function of tissue-specific demethylated promoters, and studied the relationship between our methylation data and published expression arrays on the tissue-specific expression of these genes (Table 1). Analysis of blood- (and less significantly in muscle- and liver-) specific demethylated promoters showed non-random distribution of GO terms in the biological process and cell component categories (Table 1). Tissue-specific demethylated genes were enriched for biological processes terms related to the tissue type in which they are demethylated (Table 1). In PB, promoter demethylation was associated predominantly to genes coding for membrane-associated proteins, whereas in muscle this was associated to genes encoding proteins located in contractile fiber. Of the 182 genes demethylated exclusively in blood, 49 (P = 4.47 × 10−18) were described by the highest ranking GO term 0002376–immune system process (Table 1).
Table 1.
GO and expression analysis of tissue-specific demethylated genes
| Gene set | GO |
Tissue-specific expression |
||
|---|---|---|---|---|
| GO Term | P-value | P-value | ||
| Blood | Biological process | UNIPROT tissue | ||
| Immune system process | 4.47E-18 | Blood | 1.74E-08 | |
| Immune response | 1.53E-12 | Spleen | 1.54E-07 | |
| Defense response | 1.41E-10 | Lymph | 4.14E-07 | |
| Signal transduction | 6.86E-07 | Thymus | 2.90E-04 | |
| Response to stimulus | 1.06E-06 | |||
| Cell activation | 2.84E-06 | |||
| Leukocyte activation | 4.35E-05 | |||
| Innate immune response | 4.38E-05 | |||
| Cell component | ||||
| Membrane | 3.30E-08 | |||
| Plasma membrane | 3.67E-08 | |||
| Integral to membrane | 1.10E-07 | |||
| Muscle | Biological process | UNIPROT tissue | ||
| Muscle contraction | 1.19E-04 | Skeletal muscle | 3.22E-03 | |
| Muscle system process | 1.85E-04 | |||
| Response to reactive oxygen species | 1.41E-03 | |||
| Catabolic process | 5.68E-03 | |||
| Cell component | ||||
| Contractile fiber | 1.55E-06 | |||
| Sarcomere | 2.28E-04 | |||
| Z disc | 3.26E-04 | |||
| Myofibril | 3.37E-04 | |||
| Contractile fiber part | 3.89E-04 | |||
| Brain | UNIPROT tissue | |||
| None | Alzheimer cortex | 2.71E-02 | ||
| Brain | 4.94E-02 | |||
| Fetal brain | 6.53E-02 | |||
| Liver | Biological process | UNIPROT tissue | ||
| Amine metabolic process | 3.09E-04 | Liver | 3.19E-02 | |
| Hyaluronan metabolic process | 3.25E-03 | |||
| Monocarboxylic acid metabolic process | 5.88E-03 | |||
| Oxoacid metabolic process | 1.84E-02 | |||
| Carboxylic acid metabolic process | 1.84E-02 | |||
| Organic acid metabolic process | 1.87E-02 | |||
| Cellular ketone metabolic process | 1.89E-02 | |||
| Glycosaminoglycan metabolic process | 2.16E-02 | |||
| Nitrogen compound metabolic process | 2.22E-02 | |||
| Aminoglycan metabolic process | 2.62E-02 | |||
| Polysaccharide metabolic process | 4.43E-02 | |||
| Metabolic process | 5.53E-02 | |||
| Fat | Biological process | |||
| Inflammatory response | 6.72E-03 | None | ||
| Response to wounding | 1.79E-02 | |||
| Response to stress | 2.00E-02 | |||
| Defense response | 2.16E-02 | |||
| Response to external stimulus | 4.92E-02 | |||
| Thyroid | Biological process | |||
| G-protein coupled receptor protein signaling pathway | 1.99E-02 | None | ||
| Negative regulation of caspase activity | 3.13E-02 | |||
| Negative regulation of peptidase activity | 3.65E-02 | |||
| Renal system process | 4.50E-02 | |||
| Hormone biosynthetic process | 4.66E-02 | |||
| Cell component | ||||
| Extracellular region | 1.61E-02 | |||
| Bone | None | None | ||
| Colon | None | None | ||
Analysis of these sample groups for tissue expression in databases (UniProt data bank tissue description) showed that genes demethylated in blood, muscle, brain and liver were enriched (P < 0.05) for expression in these tissues. To study this issue in more detail, we compared our methylation data with known expression data for these genes in blood cells [GEO, http://www.ncbi.nlm.nih.gov/geo/, see ‘Experimental Procedures’ section and Figure 1C]. Expression of these genes was restricted mainly to those cell types in which they are fully demethylated (Figure 1B). These analyses also showed that 23.1% of the genes specifically demethylated in brain (and in no other tissue), 6.7% in muscle, 10% in colon, 33.3% in liver, 5.3% in thyroid and 19.3% in blood were overexpressed in the tissue type in which they are demethylated (Supplementary Table S1), suggesting that promoter demethylation is not always associated with gene expression. These observations suggest that promoter DNA demethylation might be involved in tissue-specific gene expression; given the larger number of demethylated genes in white blood cells and their pronounced enrichment for hemato-immune system-related functions, this mechanism appears to be particularly relevant for immune system-specific genes.
Identification of hematological cell type-specific demethylated genes
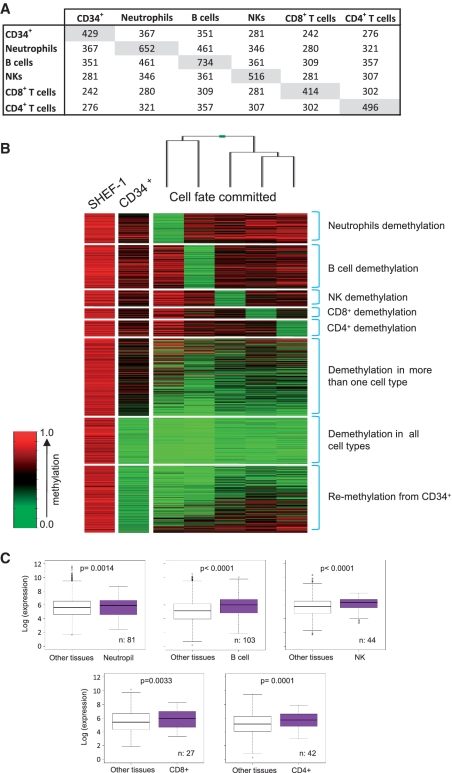
Epigenetic signaling has been implicated in establishment of the distinct hematological cell types (8,9,11–15,31–33). For a more detailed study of the cell type-dependent role of promoter demethylation in hematopoietic differentiation, we used the Infinium arrays for promoter methylation analyses in CD34+ HSPCs hematopoietic progenitor cells and five of the most abundant blood cell types: PMN (mainly neutrophils) and lymphocytes (separated as CD19+ B cells, CD56+ NK cells, CD8+ cytotoxic T cells and CD4+ helper T cells). Comparison of their methylation to that of 10 hESC lines allowed identification of 429 CpG sites (382 genes) demethylated in CD34+ hematopoietic progenitors, 652 (552 genes) in neutrophils, 734 (627 genes) in B cells, 516 (463 genes) in NK cells, 414 (366 genes) in CD8+ T cells and 496 (443 genes) in CD4+ T cells (Figure 2A and B) (Supplementary Table S8). Of these CpG sites, 25 (25 genes) were demethylated specifically in CD34+ hematopoietic progenitors, 119 (114 genes) in neutrophils, 169 (158 genes) in B lymphocytes, 68 (67 genes) in NK cells, 43 (42 genes) in CD8+ T cells and 66 (65 genes) in CD4+ T cells (Figure 2B and Supplementary Table S9).
Figure 2.
DNA methylation changes during hematopoietic differentiation. (A) Matrix showing the number of demethylated CpG sites in each hematopoietic cell subset and demethylated CpG sites shared between distinct hematopoietic cell types. (B) Illumina array methylation clustering heatmap of SHEF-1 hESC line hypermethylated genes, demethylated in at least one of the six hematopoietic cell types analyzed: CD34+ HSPCs, neutrophils, B cells, NK cells, CD8+ T cytotoxic cells (CD8+) and CD4+ T helper cells (CD4+). Methylation levels are indicated as in Figure 1B. (C) Box plots of microarray-based gene expression data (log scale). In each blood cell type, specific demethylated genes exhibited higher expression levels compared to other cell types. P-values are shown. n = number of genes analyzed.
Of the CpG sites demethylated in CD34+ HSPCs cells, 42.4% (182/429) were also hypomethylated in all terminally differentiated cells analyzed (Figure 2A). Of the 429 CpG sites demethylated in CD34+ HSPCs cells, 25 were not demethylated in terminally differentiated hematological cells types analyzed, suggesting that these genes are CD34+-specific (Supplementary Table S9). Most demethylated CpG sites in neutrophils (74.2%, 484/652) corresponded to genes previously identified as demethylated in blood (Figure 1), which can be explained by the fact neutrophils are the most abundant blood cell type.
To determine whether blood cell type-specific demethylation is restricted to the immune system, we compared the methylation status of cell type-specifically demethylated genes in the various blood cell types and in other non-hematological cell types (Supplementary Figure S1). Most of these genes (84.2%, 388/461 corresponding to 83.5%, 409/490 probes) were demethylated exclusively in the hematological system (Supplementary Table S10). Most of the CpG sites demethylated in at least one of the hematological cell types analyzed (66.44%, 796/1198; 68.64%, 694/1011 genes) were also demethylated exclusively in hematological cells (Supplementary Table S11). Collectively, these data indicate that there are at least 796 CpG sites (694 genes) that are demethylated exclusively during hematopoiesis (Supplementary Table S11), which concurs with our previous hypothesis that promoter demethylation might have a role in activation of immune system genes, and indicates that at least 694 hematologic-specific genes are regulated by promoter demethylation, more than the 182 genes identified in the whole blood sample (Supplementary Table S6). Based on the position of CpG detected in the array, the great majority (77.4%) of demethylated sites were found in low CpG density regions (Supplementary Table S11), which suggest that non-CpG island methylation plays an important role in human hematopoiesis.
To determine whether gene promoter demethylation participates in the establishment of cell type-specific expression programs in blood, we analyzed these sample groups for tissue expression in databases (UniProt data bank tissue description) and found that genes demethylated in neutrophils and B cells are enriched for expression in these cell types (P < 0.05). We compared our methylation data and earlier reports on the terminally differentiated blood cell type-specific expression of these genes obtained from the GEO database (GSE22886) (see ‘Experimental Procedures’ section and Figure 2C) (34), and found that the expression of these genes is restricted mainly to those cell types in which they are fully demethylated (Figure 2B). Moreover, these analyses showed that 17.3% of the genes specifically demethylated in neutrophils (and in no other tissue), 22.3% in B lymphocytes, 2.4% in CD4+ T cells, 3.7% in CD8+ T cells and 9.1% in NK cells were overexpressed in the tissue type in which they are demethylated (Supplementary Table S2), suggesting that, as in other tissues (Supplementary Table S1), promoter demethylation is not always associated with gene expression in blood. To study in more detail the relationship between demethylation and gene expression, we compared our methylation data with the expression data of a subset of hematologic progenitor and terminally differentiated hematological cells obtained from a different database (BioGPS, Hematopoietic Atlas, Dataset_302). These analyses showed that most (68.4%) of the demethylated genes were overexpressed in the tissue type in which they are demethylated (Supplementary Table S4). To determine whether demethylation was important in priming genes for gene expression upon cell activation, we compared the methylation data with the available expression data [GEO database (GSE22886)] of activated hematopoietic cells (B cells, CD4+ cells and NK cells). These analyses showed that 13.7% of the demethylated genes became overexpressed upon cell activation (Supplementary Table S3).
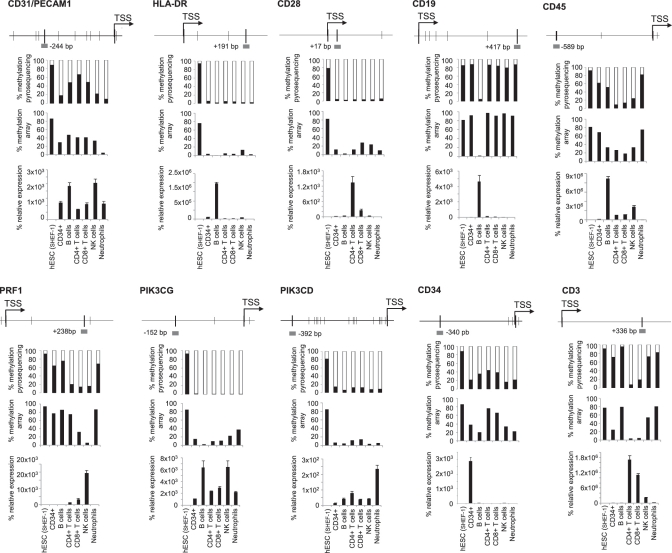
To validate the methylation array data, we selected various genes [PECAM1 (CD31), HLA-DR, CD28, CD19, CD45, PRF1, PIK3CG, PIK3CD, CD34, CD3] that are demethylated specifically in one or several blood cell types and analyzed their methylation and expression status in the same type of samples used in the arrays. In all cases, bisulfite pyrosequencing data corroborated the array results (Figure 3). We also analyzed gene expression by qPCR to determine the relationship between DNA methylation and transcription. CD19, shown by methylation arrays to be demethylated exclusively in B cells, showed a high methylation degree in all cell types analyzed except B cells. Similarly, CD3 was selectively demethylated and expressed in T cells and PRF1 in CD8+ T cells and NK cells. CD45, CD31, PIK3CG and PIK3CD (shown by methylation arrays to be demethylated in CD34+ cells and in all descendants) were demethylated and expressed at different degrees in all hematological cells. CD34, HLA-DR and CD28, although demethylated in CD34+ precursors and in hematological cells, were expressed selectively in CD34+ cells, B and T cells, respectively.
Figure 3.
Promoter methylation and expression of ten key genes of the hematopoietic system. For each gene, we depict (from top to bottom): the diagram showing the genomic region surrounding the transcriptional start site (TSS, arrow). Thin black vertical bars indicate CpG position and a grey horizontal bar indicates the location (distance in base pair from TSS) of the probe used in methylation arrays and validated by pyrosequencing in the indicated samples; percentage of promoter methylation as detected in the pyrosequencing reaction; percentage of promoter methylation as detected in the Illumina Infinium array and mRNA expression relative to GAPDH as measured by qRT–PCR in the indicated samples (n = 3, mean ± SD).
To study in greater detail the role of promoter demethylation in the establishment of specific cell lineages during hematopoietic differentiation, we examined the function of cell type-specific promoter demethylated genes by GO analyses, which showed non-random distribution of GO terms, mainly with respect to biological process (Table 2). GO terms were highly enriched for immune-related biological processes, which further supports an important role for promoter demethylation during hematopoiesis.
Table 2.
GO and expression analysis of hematopoietic lineage-specific demethylated genes
| Gene set | GO |
Tissue-specific expression |
||
|---|---|---|---|---|
| GO Term | P-value | P-value | ||
| Neutrophils | Biological process | UNIPROT Tissue | ||
| Defense response | 5.41E-06 | Neutrophil | 4.07E-04 | |
| Response to other organism | 5.60E-06 | |||
| Defense response to bacterium | 5.63E-06 | |||
| Response to biotic stimulus | 1.15E-05 | |||
| Response to stimulus | 6.40E-05 | |||
| Oxygen and reactive oxygen species metabolic process | 9.46E-05 | |||
| Multi-organism process | 9.74E-05 | |||
| Immune system process | 1.58E-04 | |||
| Response to bacterium | 3.23E-04 | |||
| Response to stress | 1.36E-03 | |||
| Cell component | ||||
| Extracellular region | 1.55E-04 | |||
| Extracellular space | 9.50E-03 | |||
| Intrinsic to membrane | 2.21E-02 | |||
| NK cells | Biological process | |||
| Defense response | 3.89E-03 | None | ||
| Cell surface receptor linked signal transduction | 2.08E-02 | |||
| Behavior | 2.27E-02 | |||
| Regulation of cytokine production | 2.38E-02 | |||
| G-protein coupled receptor protein signaling pathway | 2.96E-02 | |||
| Response to external stimulus | 3.68E-02 | |||
| Response to stimulus | 3.84E-02 | |||
| Immune system process | 4.33E-02 | |||
| Cell component | ||||
| Extracellular region | 1.37E-02 | |||
| B cells | Biological process | UNIPROT tissue | ||
| Immune response | 1.60E-10 | Spleen | 2.75E-05 | |
| Immune system process | 2.07E-09 | b-cell | 1.34E-03 | |
| Defense response | 1.28E-04 | Lymph | 1.28E-02 | |
| Response to stimulus | 7.24E-04 | Lymph node | 2.99E-02 | |
| Regulation of immune system process | 1.09E-02 | |||
| Inflammatory response | 1.12E-02 | |||
| Regulation of lymphocyte activation | 1.30E-02 | |||
| Response to other organism | 1.59E-02 | |||
| Taxis | 1.83E-02 | |||
| Chemotaxis | 1.83E-02 | |||
| Cell component | ||||
| Extracellular region | 3.43E-06 | |||
| Integral to plasma membrane | 2.55E-03 | |||
| Intrinsic to plasma membrane | 3.13E-03 | |||
| Plasma membrane part | 5.85E-03 | |||
| Plasma membrane | 9.88E-03 | |||
| CD4 T cells | Biological process | |||
| Response to stimulus | 1.66E-02 | None | ||
| Potassium ion transport | 2.71E-02 | |||
| Defense response | 2.85E-02 | |||
| Monovalent inorganic cation transport | 3.33E-02 | |||
| Immune response | 3.99E-02 | |||
| Cell component | ||||
| Extracellular region | 3.64E-04 | |||
| Extracellular region part | 1.20E-03 | |||
| Extracellular space | 1.62E-03 | |||
| CD8 T cells | Biological process | |||
| Monovalent inorganic cation transport | 5.91E-03 | None | ||
| Potassium ion transport | 6.90E-03 | |||
| Homeostatic process | 7.34E-03 | |||
| Regulation of catabolic process | 2.18E-02 | |||
| Genetic imprinting | 2.40E-02 | |||
| Ion transport | 2.48E-02 | |||
| Cell component | ||||
| Plasma membrane | 3.76E-02 | |||
| All hematologic lineages | Biological process | UNIPROT tissue | ||
| Immune system process | 4.59E-04 | Blood | 1.69E-05 | |
| Immune response | 1.27E-03 | Lymph | 1.10E-04 | |
| Leukocyte activation | 3.31E-03 | Spleen | 1.75E-03 | |
| Signal transduction | 6.17E-03 | |||
| Cell activation | 7.16E-03 | |||
| T cell activation | 1.55E-02 | |||
| Myeloid leukocyte activation | 1.68E-02 | |||
| Regulation of cellular localization | 2.49E-02 | |||
| Cell component | ||||
| Plasma membrane | 1.59E-03 | |||
| Integral to plasma membrane | 3.40E-03 | |||
| Intrinsic to plasma membrane | 3.94E-03 | |||
| Plasma membrane part | 1.19E-02 | |||
Differentiation and dedifferentiation of CD34+ HSPCs recapitulate changes in hematopoietic gene methylation
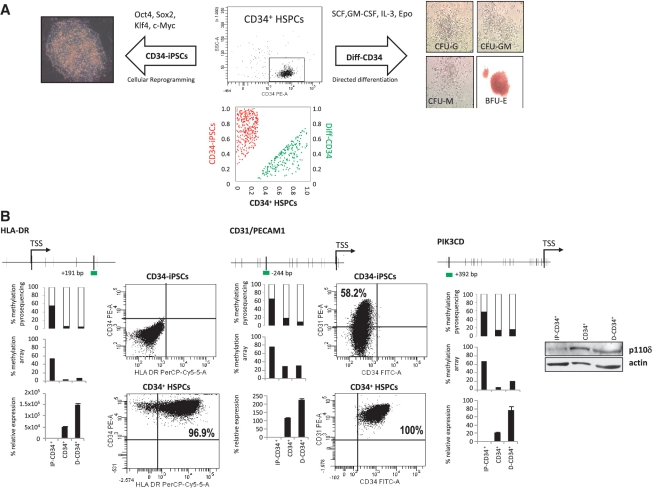
We used a dual approach to confirm the role of promoter DNA methylation in the regulation of immune system genes during ontogenic development: (i) a ‘push-forward’ in development approach in which a colony-forming unit (CFU) assay was carried out to induce in vitro myeloid differentiation of CD34+ HSPCs (Diff-CD34+) and (ii) a ‘push-back’ in development approach in which pluripotency was induced in CD34+ HSPCs (CD34-iPSCs) by viral-based ectopic expression of the reprogramming factors Oct4, Klf4, Sox2 and c-Myc (http://www.isciii.es/htdocs/terapia/terapia_lineas.jsp). Both Diff-CD34 and CD34-iPSC were analyzed by Infinium arrays in order to compare the genome-wide promoter methylation status of CD34+ HSPCs, Diff-CD34 and CD34-iPSC. We found that 29.7% of the genes (206/694; 212/767 probes) previously found to be methylated in CD34+ cells and demethylated in any of the terminally differentiated hematological cells analyzed were demethylated in the Diff-CD34 cells (Supplementary Table S12 and Figure 4). Strikingly, however, when compared to hESC, 80.1% of the genes (306/382; 341/429 probes) demethylated in CD34+ cells were hypermethylated in CD34-iPS cells, indicating a central role for promoter DNA methylation in the ontogenic development-dependent regulation of hematological genes (Supplementary Table S13 and Figure 4A).
Figure 4.
Promoter methylation and expression levels of hematopoietic genes in CD34+ HSPCs, cells differentiated from CD34+ and iPSCs generated from CD34+ HSPCs. (A) High purity sorted CB-derived CD34+ HSPCs (top middle panel) were differentiated in vitro (Diff-CD34) for 14 days in the presence of SCF, GM-CSF, IL3 and EPO. Granulocyte (CFU-G), monocyte (CFU-M), granulo-monocyte (CFU-GM) and erythroid (BFU-E) colony forming units were scored by light microscopy (right panels). Additionally, CB-derived CD34+ HSPCs were induced to travel back in development by generating iPSCs through ectopic expression of Oct4, Klf4, Sox2 and c-Myc (left panel shows a phase contrast image of a CD34-iPSC). The bottom panel shows a scatter plot of remethylated genes in CD34-iPSC (red) (Supplementary Table S13) and demethylated genes in the differentiated CD34 progeny (Diff–CD34) (green) (Supplementary Table S12). (B) Promoter methylation and expression of HLA-DR, CD31 and PIK3CD. Methylation and expression for each gene is indicated as in Figure 3. Expression by flow cytometry of each gene in CD34-iPSC (top panel) and CD34+ HSPCs (bottom panel). For PIK3CD, WB analysis was performed in CD34+ HSPCs, CD34-iPSC and Diff-CD34 (β-actin was used as loading control).
To analyze the relationship between promoter methylation and gene expression during in vitro differentiation and induction of pluripotency of CD34+ HSPCs cells, we analyzed PIK3CG, PIK3CD, HLA-DR, CD31 and CD28 by bisulfite pyrosequencing, qPCR and flow cytometry or western blotting. Pyrosequencing data corroborated array results and showed that hESC methylation affected the majority of the CpG surrounding the transcriptional start site of the genes selected (Figure 4B and Supplementary Figure S2). qPCR, flow cytometry and western blotting experiments confirmed our data showing that promoter demethylation did not necessarily involve gene upregulation. Collectively, these results support our hypothesis that immune system genes must be strictly regulated during ontogenic development (in hESC, HSC and terminally differentiated cells) and that DNA methylation might play an important role in this process.
DISCUSSION
Hematopoiesis is a highly orchestrated multi-step hierarchical process tightly regulated by the temporal gene expression regulation of transcription factors that involves the proliferation, differentiation and maturation of a very small population of self-renewing, CD34+ HSPCs into various specialized and distinct blood cell types. During this process, cellular specification is initiated by primary lineage determinants which ‘transcriptionally prime’ hematopoietic progenitors to establish a low-level expression of a mixed lineage pattern of gene expression. The primary determinants will differentiate the hematopoietic progenitors along a particular cell fate and this commitment is subsequently regulated by secondary transcription factors which function promoting a specific cell fate by repressing commitment into alternate blood lineages (35,36). In addition to transcription factors (37), cytokines (38) and miRNA (39) have long been proposed to participate in gene regulation during hematopoiesis. Nonetheless, global mechanisms involved in this process are still incompletely understood.
Promoter DNA demethylation is envisioned to be an important mechanism controlling the activation of specific genes within the hematopoietic system. Examples include the genes coding for myeloperoxidase (7), globin (8,9), c-fms (10), lysozyme (11), G-CSF receptor (12), PRF1 (13), GPVI/GP6 (14) and FOXP3 (15). As promoter DNA methylation is also implicated in the genome-wide establishment of differentiation programs in hESC (30,40), we hypothesized that promoter demethylation may contribute to genome-wide hematopoietic gene activation during hematopoiesis. We thus first evaluated the potential role of promoter DNA demethylation in a variety of somatic tissues by combining genome-wide promoter methylation and gene expression approaches in an attempt to identify promoter demethylated genes in a variety of terminally differentiated human primary tissues. Interestingly, this approach revealed that promoter demethylation-associated gene regulation is more frequent in the hematopoietic system than in non-hematopoietic tissues (463 genes demethylated in blood versus an average of 241 demethylated genes in non-hematopoietic samples. Although less frequent, promoter demethylation in non-hematopoietic tissues appears to be important for tissue differentiation, as some demethylated genes identified in non-hematopoietic tissues are also important in the identity or function of the specific tissue in which they are demethylated, in agreement with previous data (41). Examples include MYT1 (42) and RTP1 (43) in brain, OBSCN (44) and MYOT (45) in muscle, DAAO (46) and KYNU (47) in liver and TG (48) in thyroid tissue.
Upon comparing the genome-wide promoter methylation and gene expression status of a variety of hESC lines, somatic CD34+ HSPCs and five abundant white blood cell types, we identified 694 genes (Supplementary Table S11), highly enriched for hematopoietic-related functions, that undergo promoter DNA demethylation specifically during hematopoiesis. These genes were completely hypermethylated in hESCs and progressively lost promoter methylation throughout hematopoietic developmental stages in a hematopoietic cell type-specific fashion. Out of the 694 genes, 523 (75.36%) were not demethylated in CD34+ HSPCs. CD34+ HSPCs are non-cultured primary cells, suggesting that the in vitro culture is unlikely to be responsible for the hypermethylation of the genes found demethylated in a hematopoietic lineage-dependent fashion (30). Importantly, most of the genes not demethylated in CD34+ HSPCs (98.5%) become demethylated in hematopoietic terminally differentiated cell types whilst most of the genes that are demethylated in CD34+ cells (90.73%) remained demethylated in all hematopoietic terminally differentiated cell types analyzed. Although CD34+ HSPCs are commonly considered hematopoietic precursors, some studies have shown that they are composed by a heterogeneous population of cells (49) with only a minor fraction potentially differentiating into lymphocytes. We have to consider the possibility that some of the methylation differences observed in lymphoid lineages, such as CD4+ or CD8+, compared to CD34+ cells, which have been described as demethylation events might be in fact already present in lymphoid progenitor cells in the pool of CD34+ cells, and go undetected by the methylation array due to their scarcity. Further analyses of purified subpopulations of hematopoietic progenitors are thus needed to clarify this issue. Collectively, our data suggest that promoter demethylation is associated with the regulation of hematopoietic-specific genes in two developmental waves: pan-hematopoietic genes and hematopoietic lineage-specific genes.
Recent work suggest that promoter DNA demethylation is important in mouse (32,50) and human hematopoiesis (51). Ji et al. (32) analyzed hematopoietic progenitors with CHARM and, with the same cut-offs used in our study, their data show 68 genes that become demethylated in myeloid or lymphoid progenitors (Supplementary Table S14 and Supplementary Figure S3). Borgel et al. (50) used MeDip-chip to analyze different embryonic stages and they identified eight hematopoietic genes that were hypermethylated during the embryonic development and that became demethylated in hematological cells. The genes identified in the three studies (Ji's, Borgel's and ours) were quite different (Supplementary Table S14 and Supplementary Figure S3). Indeed, no genes were common to the three studies, and only eight were common to any two of the studies. In our work, we identified 686 genes that had not been identified by Ji or Borgel (Supplementary Table S14 and Supplementary Figure S3). It is possible that these genes are not regulated by promoter demethylation in mice or that our methylation arrays were more efficient in identifying these particular sequences. The fact that none of the genes identified by Borgel were identified by Ji suggests that the technology used is an important factor in determining the final set of identified genes.
Bocker et al. (51) employed the same methylation arrays used in our study and identified 770 demethylated CpG sites (change ≥15%) in myeloid cells (51). We identified 93 CpG sites in neutrophils and most (86%) of them are common to Bocker's study (Supplementary Figure S4). The discrepancy might be due to the inclusion of other types of terminally differentiated hematologic cells, non-hematologic tissues and embryonic stem cells in our study. Indeed, the inclusion of non-hematologic tissues can be an advantage of our study over the other studies as allowed for the identification of genes that become demethylated exclusively in hematologic tissues. For instance, our results indicate that 98 of the CpG sites identified by Bocker et al., can be also methylated at low levels in other non-hematologic tissues (Supplementary Figure S4).
Bocker et al. showed that promoter methylation of CD34+ cells can change over time. Since we used CD34+ cells obtained from umbilical CB, it is possible that some of our CpG sites may not have been truly hematologic genes due to the aging-dependent DNA methylation change reported by Bocker et al. To address this issue, we used our selection criteria and the data provided by Bocker et al. to identify genes demethylated in old versus young CD34+ cells (≥70% methylation in young CD34+ cells and <30% methylation in old CD34+ cells). We identified three CpG sites that became demethylated in old CD34+ (Supplementary Table S15) cells and only one of them coincided with the CpG sites identified in our study (Supplementary Table S15). Moreover, most (95.9%) of the CpG sites identified in our study remained as hematological-specific demethylated CpG sites when we used the data from adult CD34+ cells from Bocker et al. (Supplementary Figure S5). These results indicate that, in general, the CpG sites identified in our study become specifically demethylated during hematopoietic linage determination rather than during ontogenic development. Collectively, the results of the four studies suggest that promoter demethylation is an important factor in hematopoiesis but that further research is needed to fully characterize the role of promoter demethylation in mouse and human hematopoiesis.
The comparison of our methylation data with genome-wide gene expression data showed that the role of promoter demethylation during hematopoiesis is more complex. We found that regardless of the methylation status in CD34+ HSPCs, promoter demethylation did not necessarily involve gene activation/overexpression. This concurs with a previous study (52) and with a recent report showing that active demethylation in human post-mitotic hematological cells does not correlate with transcription levels (53). A possible explanation for the lack of expression of some demethylated genes is that promoter demethylation has at least, two distinct target gene-dependent effects: (i) direct mediation of gene overexpression or (ii) priming of the gene for eventual activation. This possibility is supported by the fact that many of the demethylated genes identified in this study are expressed at specific stages of the hematopoietic differentiation (i.e. hematopoietic progenitor cells or terminally differentiated cells). Among the genes we chose to analyze, examples of the first scenario include CD19, a B cell-specific receptor that is demethylated and expressed exclusively in B cells (54), and PRF1, a cytolytic protein demethylated and expressed in cytotoxic CD8+ T cells and NK cells (55). On the other hand, genes whose promoter demethylation does not necessarily result in gene activation include CD28 (56) and HLA-DR (57). Some of these genes whose demethylation does not imply immediate activation are expressed later on in response to cell transition to an active state during an immune response. These include γ-IFN (demethylated in CD8+ T cells) and FAS-L (demethylated in CD4+ T cells, CD8+ T cells and NK cells) (Supplementary Table S8), which are expressed upon T cell activation specifically at the site of an immune response and in lymphoid organs (58). This observation, together with our data, hint that promoter demethylation might be important in priming specific hematopoietic genes for activation/expression in response to external stimuli.
To study the role of promoter demethylation in hematopoiesis in more detail, we analyzed the genome-wide demethylation profiles of in vitro-differentiated CD34+ progenitors and determined the remethylation rate of CD34+ HSPCs demethylated genes in CD34+-derived iPS cells. In vitro differentiation of CD34+ progenitors resulted in demethylation of 29.7% of genes previously found to be demethylated in all blood cell lineages analyzed. CD34+-derived iPS cells restored the embryonic methylation patterns to most, but not all demethylated genes (80.1%), in line with recent works reporting incomplete epigenetic reprogramming in these cells (59) yet showing that promoter methylation-associated repression of hematopoietic genes is closely associated with stemness.
Although our data suggest that promoter demethylation might play a role in mediating genome-wide gene activation during hematopoiesis, some hematopoietic genes do not appear to use this epigenetic mechanism. One example is the cytotoxic T cell-specific antigen CD8, which is demethylated in hESC and in all hematopoietic lineages, but is expressed specifically in CD8+ T cells (60). We cannot completely rule out a role for promoter demethylation in controlling hematopoietic expression of such genes, as methylation changes might occur in a promoter region different from that included in the methylation arrays. Alternatively, it should be kept in mind that the generation of mature and functional T cells and B cells relies upon successful development and selection processes through secondary lymphoid organs such as thymus and spleen. Therefore, the expression of many hematopoietic genes and receptors being expressed during maturation in specific hematopoietic organs may be controlled by yet unknown temporal and site-specific epigenetic mechanisms. Some genes found to be demethylated during hematopoiesis were previously shown to be aberrantly hypermethylated in hematological malignancies. These include RUNX3 (61), CD79B, CD19 and TCL1 (62,63). As these genes are hypermethylated in hESCs and CD34+ HSPCs, the aberrant process in cancer could be understood as a defect in establishing an unmethylated promoter landscape during differentiation rather than as an anomalous process of de novo hypermethylation. In summary, we show that promoter demethylation might be involved in the activation of hematopoietic genes during hematopoiesis. Our data represent a genome-wide promoter demethylation landscape of human hematopoietic differentiation (Figure 5). Hematopoiesis is nonetheless a highly orchestrated multi-step hierarchical process much more complex than described here. It involves many hematopoietic organs (bone marrow, PB, spleen, thymus, liver, etc.), many intermediate stages of differentiation (64) and other less-abundant blood lineages not studied here (monocytes, basophils, eosinophils, dendritic cells, etc.). Further studies are thus needed to completely decipher the role of DNA methylation during hematopoiesis.
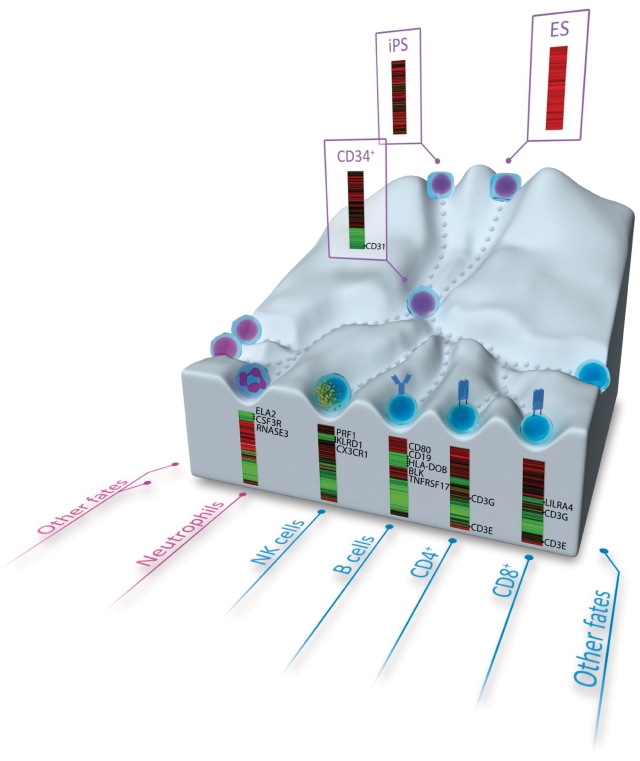
Figure 5.
Cartoon depicting the overall methylation levels of hematopoietic genes at different developmental/differentiation stages: hESC, CD34+ HSPCs and mature hematopoietic cell types (undifferentiated stages, purple nuclei; neutrophils, pink nucleus; lymphoid cells, blue nuclei). For each differentiation stage, the heatmap shows the methylation levels of a selected group of hypermethylated genes in hESCs that are demethylated in that cell type (Supplementary Table S11). Names of some key blood genes are mapped at the right of each methylation heatmap.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
P.M.'s Group is funded by The CICE (P08-CTS-3678 to P.M.) de la Junta de Andalucía, The FIS (PI070026 & PI100449) and The MICINN (PLE-2009-0111); Miguel Servet Program Fellowship (CP07/00059 to C.B.). The Marie Curie IIF (PIIF-GA-2009-236430 to V.R.-M.); ISCIII (CA10/01332 to R.M.); FPU Spanish Research Programme Fellowship (to V.C.); IUOPA (to A.F.F., R.G.U. and C.M.); Spanish Ministry of Health (PI061267; PS09/02454 to M.F.F. and PI080566 to C.L.L.); the Spanish National Research Council (CSIC; 200820I172 to M.F.F.); Community of Asturias (FYCIT IB09-106 to M.F.F.); Obra Social Cajastur, Spain (The IUOPA). Funding for open access charge: Spanish Ministry of Health (PS09/02454).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Ettore Marzocchi for three-dimensional graphics and Catherine Mark and Oficina de Investigación Biosanitaria (OIB) for editorial assistance.
REFERENCES
- 1.Fraga MF. Genetic and epigenetic regulation of aging. Curr. Opin. Immunol. 2009;21:446–453. doi: 10.1016/j.coi.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Esteller M. Epigenetics in cancer. N. Engl. J. Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 3.Hattori N, Imao Y, Nishino K, Ohgane J, Yagi S, Tanaka S, Shiota K. Epigenetic regulation of Nanog gene in embryonic stem and trophoblast stem cells. Genes Cells. 2007;12:387–396. doi: 10.1111/j.1365-2443.2007.01058.x. [DOI] [PubMed] [Google Scholar]
- 4.Hattori N, Nishino K, Ko YG, Ohgane J, Tanaka S, Shiota K. Epigenetic control of mouse Oct-4 gene expression in embryonic stem cells and trophoblast stem cells. J. Biol. Chem. 2004;279:17063–17069. doi: 10.1074/jbc.M309002200. [DOI] [PubMed] [Google Scholar]
- 5.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lubbert M, Miller CW, Koeffler HP. Changes of DNA methylation and chromatin structure in the human myeloperoxidase gene during myeloid differentiation. Blood. 1991;78:345–356. [PubMed] [Google Scholar]
- 8.Ley TJ, Chiang YL, Haidaris D, Anagnou NP, Wilson VL, Anderson WF. DNA methylation and regulation of the human beta-globin-like genes in mouse erythroleukemia cells containing human chromosome 11. Proc. Natl Acad. Sci. USA. 1984;81:6618–6622. doi: 10.1073/pnas.81.21.6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enver T, Brewer AC, Patient RK. Role for DNA replication in beta-globin gene activation. Mol. Cell. Biol. 1988;8:1301–1308. doi: 10.1128/mcb.8.3.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felgner J, Kreipe H, Heidorn K, Jaquet K, Heuss R, Zschunke F, Radzun HJ, Parwaresch MR. Lineage-specific methylation of the c-fms gene in blood cells and macrophages. Leukemia. 1992;6:420–425. [PubMed] [Google Scholar]
- 11.Klages S, Mollers B, Renkawitz R. The involvement of demethylation in the myeloid-specific function of the mouse M lysozyme gene downstream enhancer. Nucleic Acids Res. 1992;20:1925–1932. doi: 10.1093/nar/20.8.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felgner J, Heidorn K, Korbacher D, Frahm SO, Parwaresch R. Cell lineage specificity in G-CSF receptor gene methylation. Leukemia. 1999;13:530–534. doi: 10.1038/sj.leu.2401386. [DOI] [PubMed] [Google Scholar]
- 13.Lu Q, Wu A, Ray D, Deng C, Attwood J, Hanash S, Pipkin M, Lichtenheld M, Richardson B. DNA methylation and chromatin structure regulate T cell perforin gene expression. J. Immunol. 2003;170:5124–5132. doi: 10.4049/jimmunol.170.10.5124. [DOI] [PubMed] [Google Scholar]
- 14.Kanaji S, Kanaji T, Jacquelin B, Chang M, Nugent DJ, Komatsu N, Moroi M, Izuhara K, Kunicki TJ. Thrombopoietin initiates demethylation-based transcription of GP6 during megakaryocyte differentiation. Blood. 2005;105:3888–3892. doi: 10.1182/blood-2004-08-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, Schlawe K, Chang HD, Bopp T, Schmitt E, et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5:e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montes R, Ligero G, Sanchez L, Catalina P, de la Cueva T, Nieto A, Melen GJ, Rubio R, Garcia-Castro J, Bueno C, et al. Feeder-free maintenance of hESCs in mesenchymal stem cell-conditioned media: distinct requirements for TGF-beta and IGF-II. Cell Res. 2009;19:698–709. doi: 10.1038/cr.2009.35. [DOI] [PubMed] [Google Scholar]
- 17.Ramos-Mejia V, Melen GJ, Sanchez L, Gutierrez-Aranda I, Ligero G, Cortes JL, Real PJ, Bueno C, Menendez P. Nodal/Activin signaling predicts human pluripotent stem cell lines prone to differentiate toward the hematopoietic lineage. Mol. Ther. 2010;18:2173–2181. doi: 10.1038/mt.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menéndez P, Redondo O, Rodríguez A, López-Berges MC, Ercilla G, López A, Durán A, Almeida J, Pérez-Simón JA, San Miguel JF, et al. Comparison between a lyse-and-then-wash method and a lyse-non-wash technique for the enumeration of CD34+ hematopoietic progenitor cells. Cytometry. 1998;34:264–271. doi: 10.1002/(sici)1097-0320(19981215)34:6<264::aid-cyto4>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 19.Menendez P, Caballero MD, Prosper F, Del Canizo MC, Perez-Simon JA, Mateos MV, Nieto MJ, Corral M, Romero M, Garcia-Conde J, et al. The composition of leukapheresis products impacts on the hematopoietic recovery after autologous transplantation independently of the mobilization regimen. Transfusion. 2002;42:1159–1172. doi: 10.1046/j.1537-2995.2002.00190.x. [DOI] [PubMed] [Google Scholar]
- 20.Ramos-Mejia V, Munoz-Lopez M, Garcia-Perez JL, Menendez P. iPSC lines that do not silence the expression of the ectopic reprogramming factors may display enhanced propensity to genomic instability. Cell Res. 2010;20:1092–1095. doi: 10.1038/cr.2010.125. [DOI] [PubMed] [Google Scholar]
- 21.Menendez P, Catalina P, Rodriguez R, Melen GJ, Bueno C, Arriero M, Garcia-Sanchez F, Lassaletta A, Garcia-Sanz R, Garcia-Castro J. Bone marrow mesenchymal stem cells from infants with MLL-AF4+ acute leukemia harbor and express the MLL-AF4 fusion gene. J. Exp. Med. 2009;206:3131–3141. doi: 10.1084/jem.20091050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutierrez-Aranda I, Ramos-Mejia V, Bueno C, Munoz-Lopez M, Real PJ, Macia A, Sanchez L, Ligero G, Garcia-Parez JL, Menendez P. Human induced pluripotent stem cells develop teratoma more efficiently and faster than human embryonic stem cells regardless the site of injection. Stem Cells. 2010;28:1568–1570. doi: 10.1002/stem.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 24.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 25.Martínez-Camblor P, Uña-Álvarez J. Non-parametric k-sample tests: density functions vs distribution functions. Comput. Stat. Data An. 2009;53:3344–3357. [Google Scholar]
- 26.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, 3rd, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bueno C, Montes R, Menendez P, et al. The ROCK inhibitor Y-27632 negatively affects the expansion/survival of both fresh and cryopreserved cord blood-derived CD34+ hematopoietic progenitor cells: Y-27632 negatively affects the expansion/survival of CD34 + HSPCs. Stem Cell Rev. 2010;6:215–223. doi: 10.1007/s12015-010-9118-5. [DOI] [PubMed] [Google Scholar]
- 28.Catalina P, Bueno C, Montes R, Nieto A, Ligero G, Sanchez L, Jara M, Rasillo A, Orfao A, Cigudosa J, et al. Genetic stability of human embryonic stem cells: A first-step toward the development of potential hESC-based systems for modeling childhood leukemia. Leuk. Res. 2009;33:980–990. doi: 10.1016/j.leukres.2008.08.028. [DOI] [PubMed] [Google Scholar]
- 29.Bueno C, Montes R, Martin L, Prat I, Hernandez MC, Orfao A, Menendez P. NG2 antigen is expressed in CD34+ HPCs and plasmacytoid dendritic cell precursors: is NG2 expression in leukemia dependent on the target cell where leukemogenesis is triggered? Leukemia. 2008;22:1475–1478. doi: 10.1038/leu.2008.134. [DOI] [PubMed] [Google Scholar]
- 30.Calvanese V, Horrillo A, Hmadcha A, Suarez-Alvarez B, Fernandez AF, Lara E, Casado S, Menendez P, Bueno C, Garcia-Castro J, et al. Cancer genes hypermethylated in human embryonic stem cells. PLoS ONE. 2008;3:e3294. doi: 10.1371/journal.pone.0003294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lubbert M, Herrmann F, Koeffler HP. Expression and regulation of myeloid-specific genes in normal and leukemic myeloid cells. Blood. 1991;77:909–924. [PubMed] [Google Scholar]
- 32.Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, Lee H, Aryee MJ, Irizarry RA, Kim K, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Broske AM, Vockentanz L, Kharazi S, Huska MR, Mancini E, Scheller M, Kuhl C, Enns A, Prinz M, Jaenisch R, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat. Genet. 2009;41:1207–1215. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- 34.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smale ST, Fisher AG. Chromatin structure and gene regulation in the immune system. Annu. Rev. Immunol. 2002;20:427–462. doi: 10.1146/annurev.immunol.20.100301.064739. [DOI] [PubMed] [Google Scholar]
- 36.Busslinger M. Transcriptional control of early B cell development. Annu. Rev. Immunol. 2004;22:55–79. doi: 10.1146/annurev.immunol.22.012703.104807. [DOI] [PubMed] [Google Scholar]
- 37.Kehrl JH. Hematopoietic lineage commitment: role of transcription factors. Stem Cells. 1995;13:223–241. doi: 10.1002/stem.5530130304. [DOI] [PubMed] [Google Scholar]
- 38.Zhu J, Emerson SG. Hematopoietic cytokines, transcription factors and lineage commitment. Oncogene. 2002;21:3295–3313. doi: 10.1038/sj.onc.1205318. [DOI] [PubMed] [Google Scholar]
- 39.Navarro F, Lieberman J. Small RNAs guide hematopoietic cell differentiation and function. J. Immunol. 2010;184:5939–5947. doi: 10.4049/jimmunol.0902567. [DOI] [PubMed] [Google Scholar]
- 40.Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L, Jaenisch R, Fan G. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–169. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim JG, Armstrong RC, v Agoston D, Robinsky A, Wiese C, Nagle J, Hudson LD. Myelin transcription factor 1 (Myt1) of the oligodendrocyte lineage, along with a closely related CCHC zinc finger, is expressed in developing neurons in the mammalian central nervous system. J. Neurosci. Res. 1997;50:272–290. doi: 10.1002/(SICI)1097-4547(19971015)50:2<272::AID-JNR16>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 43.Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H. RTP family members induce functional expression of mammalian odorant receptors. Cell. 2004;119:679–691. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 44.Young P, Ehler E, Gautel M. Obscurin, a giant sarcomeric Rho guanine nucleotide exchange factor protein involved in sarcomere assembly. J. Cell Biol. 2001;154:123–136. doi: 10.1083/jcb.200102110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salmikangas P, Mykkanen OM, Gronholm M, Heiska L, Kere J, Carpen O. Myotilin, a novel sarcomeric protein with two Ig-like domains, is encoded by a candidate gene for limb-girdle muscular dystrophy. Hum. Mol. Genet. 1999;8:1329–1336. doi: 10.1093/hmg/8.7.1329. [DOI] [PubMed] [Google Scholar]
- 46.Khoronenkova SV, Tishkov VI. D-amino acid oxidase: physiological role and applications. Biochemistry. 2008;73:1511–1518. doi: 10.1134/s0006297908130105. [DOI] [PubMed] [Google Scholar]
- 47.Toma S, Nakamura M, Tone S, Okuno E, Kido R, Breton J, Avanzi N, Cozzi L, Speciale C, Mostardini M, et al. Cloning and recombinant expression of rat and human kynureninase. FEBS Lett. 1997;408:5–10. doi: 10.1016/s0014-5793(97)00374-8. [DOI] [PubMed] [Google Scholar]
- 48.Libert F, Vassart G, Christophe D. Methylation and expression of the human thyroglobulin gene. Biochem. Biophys. Res. Commun. 1986;134:1109–1113. doi: 10.1016/0006-291x(86)90365-7. [DOI] [PubMed] [Google Scholar]
- 49.D'Arena G, Musto P, Cascavilla N, Di Giorgio G, Zendoli F, Carotenuto M. Human umbilical cord blood: immunophenotypic heterogeneity of CD34+ hematopoietic progenitor cells. Haematologica. 1996;81:404–409. [PubMed] [Google Scholar]
- 50.Borgel J, Guibert S, Li Y, Chiba H, Schubeler D, Sasaki H, Forne T, Weber M. Targets and dynamics of promoter DNA methylation during early mouse development. Nat. Genet. 2011;42:1093–1100. doi: 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- 51.Bocker MT, Hellwig I, Breiling A, Eckstein V, Ho AD, Lyko F. Genome-wide promoter DNA methylation dynamics of human hematopoietic progenitor cells during differentiation and aging. Blood. 2011;117:e182–e189. doi: 10.1182/blood-2011-01-331926. [DOI] [PubMed] [Google Scholar]
- 52.Martin-Subero JI, Kreuz M, Bibikova M, Bentink S, Ammerpohl O, Wickham-Garcia E, Rosolowski M, Richter J, Lopez-Serra L, Ballestar E, et al. New insights into the biology and origin of mature aggressive B-cell lymphomas by combined epigenomic, genomic, and transcriptional profiling. Blood. 2009;113:2488–2497. doi: 10.1182/blood-2008-04-152900. [DOI] [PubMed] [Google Scholar]
- 53.Klug M, Heinz S, Gebhard C, Schwarzfischer L, Krause SW, Andreesen R, Rehli M. Active DNA demethylation in human postmitotic cells correlates with activating histone modifications, but not transcription levels. Genome Biol. 2010;11:R63. doi: 10.1186/gb-2010-11-6-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stamenkovic I, Seed B. CD19, the earliest differentiation antigen of the B cell lineage, bears three extracellular immunoglobulin-like domains and an Epstein-Barr virus-related cytoplasmic tail. J. Exp. Med. 1988;168:1205–1210. doi: 10.1084/jem.168.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lichtenheld MG, Olsen KJ, Lu P, Lowrey DM, Hameed A, Hengartner H, Podack ER. Structure and function of human perforin. Nature. 1988;335:448–451. doi: 10.1038/335448a0. [DOI] [PubMed] [Google Scholar]
- 56.Linsley PS, Ledbetter JA. The role of the CD28 receptor during T cell responses to antigen. Annu. Rev. Immunol. 1993;11:191–212. doi: 10.1146/annurev.iy.11.040193.001203. [DOI] [PubMed] [Google Scholar]
- 57.Das HK, Lawrance SK, Weissman SM. Structure and nucleotide sequence of the heavy chain gene of HLA-DR. Proc. Natl Acad. Sci. USA. 1983;80:3543–3547. doi: 10.1073/pnas.80.12.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Balasubramani A, Shibata Y, Crawford GE, Baldwin AS, Hatton RD, Weaver CT. Modular utilization of distal cis-regulatory elements controls Ifng gene expression in T cells activated by distinct stimuli. Immunity. 2010;33:35–47. doi: 10.1016/j.immuni.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feik N, Bilic I, Tinhofer J, Unger B, Littman DR, Ellmeier W. Functional and molecular analysis of the double-positive stage-specific CD8 enhancer E8III during thymocyte development. J. Immunol. 2005;174:1513–1524. doi: 10.4049/jimmunol.174.3.1513. [DOI] [PubMed] [Google Scholar]
- 61.Griffiths EA, Gore SD, Hooker C, McDevitt MA, Karp JE, Smith BD, Mohammad HP, Ye Y, Herman JG, Carraway HE. Acute myeloid leukemia is characterized by Wnt pathway inhibitor promoter hypermethylation. Leuk. Lymphoma. 2010;51:1711–1719. doi: 10.3109/10428194.2010.496505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ushmorov A, Leithauser F, Sakk O, Weinhausel A, Popov SW, Moller P, Wirth T. Epigenetic processes play a major role in B-cell-specific gene silencing in classical Hodgkin lymphoma. Blood. 2006;107:2493–2500. doi: 10.1182/blood-2005-09-3765. [DOI] [PubMed] [Google Scholar]
- 63.Doerr JR, Malone CS, Fike FM, Gordon MS, Soghomonian SV, Thomas RK, Tao Q, Murray PG, Diehl V, Teitell MA, et al. Patterned CpG methylation of silenced B cell gene promoters in classical Hodgkin lymphoma-derived and primary effusion lymphoma cell lines. J. Mol. Biol. 2005;350:631–640. doi: 10.1016/j.jmb.2005.05.032. [DOI] [PubMed] [Google Scholar]
- 64.Menendez P, Vargas A, Bueno C, Barrena S, Almeida J, De Santiago M, Lopez A, Roa S, San Miguel JF, Orfao A. Quantitative analysis of bcl-2 expression in normal and leukemic human B-cell differentiation. Leukemia. 2004;18:491–498. doi: 10.1038/sj.leu.2403231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.