Abstract
In humans, mutations of a growing list of regulatory factor X (RFX) target genes have been associated with devastating genetics disease conditions including ciliopathies. However, mechanisms underlying RFX transcription factors (TFs)-mediated gene expression regulation, especially differential gene expression regulation, are largely unknown. In this study, we explore the functional significance of the co-existence of multiple X-box motifs in regulating differential gene expression in Caenorhabditis elegans. We hypothesize that the effect of multiple X-box motifs is not a simple summation of binding effect to individual X-box motifs located within a same gene. To test this hypothesis, we identified eight C. elegans genes that contain two or more X-box motifs using comparative genomics. We examined one of these genes, F25B4.2, which contains two 15-bp X-box motifs. F25B4.2 expression in ciliated neurons is driven by the proximal motif and its expression is repressed by the distal motif. Our data suggest that two X-box motifs cooperate together to regulate the expression of F25B4.2 in location and intensity. We propose that multiple X-box motifs might be required to tune specific expression level. Our identification of genes with multiple X-box motifs will also improve our understanding of RFX/DAF-19-mediated regulation in C. elegans and in other organisms including humans.
INTRODUCTION
Regulatory factor X (RFX) is an evolutionarily conserved DNA binding protein family that has been identified in organisms ranging from single cellular eukaryotes, including the budding yeast Saccharomyces cerevisiae and the fission yeast Schizosaccharomyces pombe, to humans (1). All RFX transcription factors (TFs) contain a single DNA binding domain (DBD) that is very well conserved, showing over 40% identity between yeast, nematodes and mammals and close to 100% identity in amino acid positions that are in direct contact with RFX DBDs (2,3). We have found that different organisms in the tree of life can have very different number of RFX genes. Non-metazoans except the choanoflagellates (e.g. Monosiga brevicollis) have either one RFX gene or none (2). In contrast, all metazoans have multiple RFX genes, except the nematodes which only have a single RFX gene. For example, Caenorhabditis elegans genome has a single RFX gene called daf-19, which is expressed in ciliated neurons (4). It has been proposed that nematodes could have lost some RFX genes in evolution (1,2). The first RFX TF in Drosophila melanogaster, dRFX (CG6312), was identified through a homology search for the RFX DBD and the second one, dRFX2, was identified through yeast-one-hybrid (Y1H) screening for transcription factors that bind to a putative promoter sequence (5,6). However, this gene sequence does not match any genomic region in the sequenced D. melanogaster genome. Because the D. melanogaster heterochromatin regions have not been fully sequenced due to heavy repetitive sequences (7), it cannot be ruled out that dRFX2 resides in these unsequenced regions. Because dRFX2 phylogenetically clusters with fungal RFX genes, it has been proposed that dRFX2 is in fact a fungal gene (2). Nevertheless, perfect match in any sequenced fungal species was not found (2). In a recent comparative genomics study, we have annotated the gene CG9727 in D. melanogaster as the third RFX TF dRFX1, whose DBD shows high similarity to that of human RFX5 (2). Seven RFX TFs have been uncovered in humans as well as all other mammals, including RFX6 and RFX7 that were recently identified in our laboratory (8). RFX3 in mammals has been found to be crucial for proper primary cilia development in embryo nodal cells (9), brain ependymal cells (10) and pancreatic endocrine cells (11). Recently, RFX4 was also shown to be important for cilia formation in the neural tube (12). Mutations in many of their target genes have been associated with an expanding array of devastating human disease conditions, including polycystic kidney disease (13,14) and Bardet-Biedl syndrome (BBS) (15). RFX5 regulates major histocompatibility complex class II (MHC II) gene expression in the immune system (16). RFX6, which we recently identified in the human genome, is almost exclusively expressed in the human pancreatic islets (8) and plays a role in the insulin production (17,18).
Accumulating evidence suggests that RFX genes regulate the transcription of ciliary genes in metazoans. The first critical evidence linking RFX TFs and the ciliary genes was reported by Swoboda et al. (4). The authors cloned daf-19 in the nematode C. elegans and found that it is the first and only RFX gene in C. elegans. They showed that in the absence of functional DAF-19, ciliated neurons in C. elegans lost their cilia and displayed chemosensory defects (Che), dye filling defect (Dyf) and constitutive dauer formation (Daf-c) (4). Furthermore, they demonstrated that DAF-19 regulates the expression of ciliary genes, including che-2, osm-1, osm-6 and many BBS genes through binding to a DNA element called the X-box motif, which was first identified as binding site for human RFX5 (4,19). Later, it was discovered that many ciliary genes in the fruit fly D. melanogaster are also regulated by RFX genes, including CG15161, the homolog of dyf-6 (20).
X-box motif, the binding motif of RFX DBDs, has been found to be highly conserved as well. Many validated instances of X-box motifs in yeast, C. elegans and humans are 14-bp in size. Because of their large size, X-box motifs have been used as a ciliary gene indicator in genomics and bioinformatics projects. Efimenko et al. (21) searched the C. elegans promoter regions (defined here as 1000-bp genomic region upstream of the start codon) for candidate X-box motifs that resemble an ‘average consensus X-box motif’ and identified 730 potential DAF-19 target genes in C. elegans. Independently, Blacque et al. (22) identified 53 putative DAF-19 target genes in C. elegans through searching for the presence of putative X-box motifs in promoter regions (defined as 1500-bp genomic region upstream of the start codon) and comparing relative gene expression in four different tissues. Taking advantage of the availability of two newly sequenced genomic sequences in Caenorhabditis genus, C. briggsae and C. remanei, our laboratory searched for X-box motifs in the promoter regions (defined as 2000-bp genomic region upstream of the start codon) of orthologous genes in all three species and predicted 93 candidate DAF-19 regulated genes (23), including dyf-5. The putative X-box motifs identified in these three studies all show resemblance to known X-box motifs.
Many RFX target genes have been found to contain more than one X-box motif. For example, RFX TF in S. cerevisiae negatively regulates many ribonucleotide reductase genes (e.g. RNR2, RNR3 and RNR4) through a combination of strong X-box motifs and weak X-box motifs (24). In humans, RFX1 represses MAP1A in non-neuronal cells by binding to two X-box motifs in the first exon (25). However, how multiple X-box motifs function in the same gene has never been examined in animals in vivo.
The goal of this project is to identify ciliary genes in C. elegans that may be regulated by DAF-19 through multiple X-box motifs. C. elegans was chosen as a model system for this study because of its compact genome. More importantly, C. elegans has been effectively used to identify and characterize ciliary genes. Many ciliary genes in C. elegans have readily identifiable orthologs in humans (2,4,19), which makes this study useful for understanding the function and regulatory mechanism of RFX TFs in humans.
MATERIALS AND METHODS
Strains used
Worm strains are maintained using standard procedures in 20°C unless otherwise noted (26). The following strains were used in this study: DR86 daf-19(m86), EG5003 unc-119(ed3) III; cxTi10882 IV, JT204 daf-12(sa204), JT6924 daf-12(sa204); daf-19(m86). Strains generated in this study are listed below.
| Strain name | Allele | Description |
|---|---|---|
| JNC20 | unc-119(ed3); dotSi1 [prF25B4.2::mCherry, Cb-unc-119(+)] IV | F25B4.2 Wild-type promoter |
| JNC21 | unc-119(ed3); dotSi2 [prF25B4.2::mCherry del(−140), Cb-unc-119(+)] IV | Proximal deletion |
| JNC22 | unc-119(ed3); dotSi3 [prF25B4.2::mCherry del(−190), Cb-unc-119(+)] IV | Distal deletion |
| JNC29 | unc-119(ed3); dotSi10 [prF25B4.2::mCherry del(-140) del(190), Cb-unc-119(+)] IV | Double deletion |
| JNC33 | daf-19(m86); dotSi2 | Proximal deletion in daf-19 background |
| JNC34 | daf-19(m86); dotSi10 | Double deletion in daf-19 background |
| JNC36 | daf-19(m86); dotSi1 | Wild-type promoter in daf-19 background |
| JNC37 | daf-19(m86); dotSi3 | Distal deletion in daf-19 background |
| JNC23 | unc-119(ed3); dotSi4 [prM04C9.5::mCherry, Cb-unc-119(+)] IV | dyf-5 wild-type promoter |
| JNC31 | unc-119(ed3); dotSi4 [prM04C9.5::mCherry replace −285 to −271 →gtcctcacaagtaac, Cb-unc-119(+)] IV | dyf-5 promoter replaced with distal motif |
| JNC35 | unc-119(ed3); dotSi4 [prM04C9.5::mCherry replace −285 to −271 →gtctccaatggcaac, Cb-unc-119(+)] IV | dyf-5 promoter replaced with proximal motif |
Genomic data and gene model improvement
Genomics DNA data for all four Caenorhabditis species were obtained from WS204 version of WormBase (ftp.wormbase.org/). The gene set for C. elegans was obtained from release WS204. The gene set for C. briggsae, C. remanei and C. brenneri were generated by genBlastG (27). Caenorhabditis elegans proteins (20 173) were used as input for genBlastG. These sequences represent the longest transcript if more than one alternative transcript exits. genBlastG returns 264 411 gene models for C. briggsae, 319 750 for C. remanei and 425 947 for C. brenneri. Many of these gene models are redundant gene models due to multiple orthologs (such as gene families or tandem gene duplications) in C. elegans. A filtering procedure was used so that each genomic region would contain only one gene model with the highest sequence percent identity (PID) to the query. The filtering procedure was carried out as follows: (i) all predictions are sorted by PID in decreasing order. (ii) For each two overlapping model, if the overlapping region is >5% of the length for either gene, then the model with higher PID is kept and the model with lower PID is filtered out. (iii) To ensure the quality of the gene set, we only kept gene models that show PID ≥40% to the query. The filtering procedure resulted in 16 577 gene models for C. briggsae, 18 426 for C. remanei and 23 473 for C. brenneri. In the last step, we combined these gene models with the current WormBase models to generate a hybrid gene set. In the hybrid set, genBlastG’s predicted models replace corresponding WormBase gene models if genBlastG’s prediction shows at least 2% improvement in PID. The final gene models were uploaded in GFF3 format to a MYSQL server and visualized on Generic Genome Browser (28). Mapping of orthologous relationships between genes in C. elegans and the other three species were generated by Inparanoid (29).
X-box motif search using TFMscan
We generated a position weighted matrix (PWM) for the left 6-bp and the right 6-bp based on 31 validated X-box motifs (Table 1). The PWM for each half was used by TFMscan to probe the promoter regions. A putative X-box motif was predicted by combining a left half and a right half while allowing 2–3 nt in between. TFMscan was executed with P = e−5 (30).
Table 1.
Validated X-box motifs
| Gene name | Sequence name | Distance from ATG | HMMER score | X-box sequence | Reference |
|---|---|---|---|---|---|
| che-13 | F59C6.7 | −74 | 5.33 | GTTGCTATAGCAAC | (14,21) |
| xbx-1 | F02D8.3 | −79 | 7.73 | GTTTCCATGGTAAC | (4,21,40) |
| xbx-2 | D1009.5 | −77 | 7.98 | GTTGCCATGACAAC | (21,22) |
| xbx-3 | M04D8.6 | −97 | 5.72 | GTTGTCTTGGCAAC | (21) |
| xbx-4 | C23H5.3 | −82 | 7.98 | GTTGCCATGACAAC | (21) |
| xbx-5 | T24A11.2 | −121 | 6.84 | GTCTCCATGACAAC | (21) |
| xbx-6 | F40F9.1 | −151 | 7.53 | GTTTCCATGGAAAC | (21) |
| xbx-7 | R148.1 | −69 | 4.56 | GTCACCATAGGAAC | (21) |
| ZK328.7 | −89 | 6.51 | GTTACCATGGCAAT | (22) | |
| bbs-9 | C48B6.8 | −81 | 7.53 | GTTTCCATGACAAC | (22) |
| che-11 | C27A7.4 | −85 | 7.04 | ATCTCCATGGCAAC | (21) |
| odr-4 | Y102E9.1 | −200 | 4.09 | ATCGTCATGGTAAC | (21) |
| osm-5 | Y41G9A.1 | −115 | 6.92 | GTTACTATGGCAAC | (21,36,37) |
| nhr-44 | T19A5.4 | −76 | 6.91 | GTCTTCATGGCAAC | (21) |
| nph-1 | M28.7 | −77 | 5.57 | GTTGCCAGGGGCAAC | (41) |
| nph-4 | R13H4.1 | −168 | 5.93 | ATTTCCATGACAAC | (41) |
| nud-1 | F53A2.4 | −263 | 3.81 | GTATCCATGGGAAC | (21) |
| dyf-2 | ZK520.3 | −140 | 5.84 | GTTACCAAGGCAAC | (42) |
| osm-6 | R31.3 | −100 | 6.13 | GTTACCATAGTAAC | (4,21,43) |
| dyf-3 | C04C3.5 | −88 | 4.22 | GTTTCTATGGGAAC | (44,45) |
| Y110A7A.20 | −60 | 4.26 | GTCTCTATAGCAAC | (22) | |
| che−2 | F38G1.1 | −117 | 7.05 | GTTGTCATGGTGAC | (4,21,46) |
| osm-1 | T27B1.1 | −86 | 5.27 | GCTACCATGGCAAC | (4,21,34,47) |
| bbs-1 | Y105E8A.5 | −99 | 5.45 | GTTCCCATAGCAAC | (21,48,49) |
| bbs-2 | F20D12.3 | −94 | 6.31 | GTATCCATGGCAAC | (21,48,49) |
| bbs-5 | R01H10.6 | −65 | 8.64 | GTCTCCATGGCAAC | (21,50) |
| bbs-7 | Y75B8A.12 | −94 | 6.92 | GTTGCCATAGTAAC | (21,48,49) |
| bbs-8 | T25F10.5 | −84 | 4.22 | GTACCCATGGCAAC | (21,48,49) |
| tub-1 | F10B5.4 | −183 | 5.25 | ATCTCCATGACAAC | (21,51) |
| che-12 | B0024.8 | −767 | ATCAGCTTGAAAAC | (52) | |
| dyf-5 | M04C9.5 | −285 | 5.95 | GTTACCATAGAAAC | (23,53) |
An X-box motif containing gene is considered validated when the expression of the gene is dependent on the X-box motif (mutagenesis study) or on DAF-19 (DAF-19 knock out study).
Construct generation and cloning
Deletion constructs were made by standard site-directed mutagenesis and PCR stitching methods (31). Briefly, primers were designed to contain the particular deletion. See below for a list of primers used and their sequences. A left fragment was amplified using Primer A and Primer DeletionR (either distal or proximal). A right fragment was amplified using Primer DeletionF (either distal or proximal) and Primer B. The left and right fragment is stitched together using Primer A* and B. The mCherry was amplified using Primer C and D from pCFJ190 (A gift from E.M. Jorgensen). The final stitching between the promoter fragment and mCherry was done using Primer A* and D*. Primer A* contains SbfI site and Primer D* contains SpeI site. The construct and the plasmid pCFJ178 were cut using the respective restriction enzymes in 37°C for 2.5 h. The construct was ligated into the linearized plasmid overnight at room temperature. The final ligation reaction was transformed into DH5α cells by electroporation. The transformants were plated onto lysogeny broth (LB)-Ampicillin plates. Living colonies were picked to grow in a 5 ml of LB broth and the DNA was extracted using Qiagen Mini-prep kit (Cat#:27104).
All the primers used in this study are listed below.
| Primer name | Primer sequence |
|---|---|
| F25B4.2_A | CAAAATTACCTATCGCACTACGTT |
| F25B4.2_A* | CCTGCAGGCCTGCAGGAAGCTGAAACGTCGGAGATAATAC |
| F25B4.2_B | TATCTTCTTCACCCTTTGAGACCATCATCCACGATTAATCTGAAACTCA |
| M04C9.5_A | CCTGCAGGCCTGCAGGAATTGAATTAGCCGCGGAGC |
| M04C9.5_B | TATCTTCTTCACCCTTTGAGACCATGGCTTCTTGCCCTTATATTTTCC |
| mCherry_C | ATGGTCTCAAAGGGTGAAGA |
| mCherry_D | GGCCTCTTCGCTATTACGC |
| mCherry_D* | ACGACGGCCAGTGAATTATCACTAGTACTAGT |
| F25B4.2_deletionF_distal | CACTTTTCAATTCGAAATGTCATGGGCGTTG |
| F25B4.2_deletionR_distal | CCATGACATTTCGAATTGAAAAGTGTCGAAATTCTTAGAG |
| F25B4.2_deletionF_proximal | GGCGCCACTGAAACCCGCATTTTAAACTCCAT |
| F25B4.2_deletionR_proximal | CGGGTTTCAGTGGCGCCGTGGCGACA |
| M04C9.5_replaceF_distal | GTCCTCACAAGTAACTGTCTGTTACACCCTTTTCTC |
| M04C9.5_replaceR_distal | GTTACTTGTGAGGACCAAGAGCAAACGGCGGAG |
| M04C9.5_replaceF_proximal | GTCTCCAATGGCAACTGTCTGTTACACCCTTTTCTC |
| M04C9.5_replaceR_proximal | GTTGCCATTGGAGACCAAGAGCAAACGGCGGAG |
| ChrIV-R | TGTTTACTAGACCGGGGCTC |
| mCherry-genoF | AAAACCGCACACAAAATACC |
| 178-genoF | TCCCCATTTCACCAGAGAAC |
MosSCI
DNA purified from the transformation was used directly for injection. The injection mix for MosSCI was made as suggested from the literature: pJL43.1 (50 ng/µl), purified plasmid (50 ng/µl), pGH8 (10 ng/µl), pCFJ90 (2.5 ng/µl), pCFJ104 (5 ng/µl). The mix was injected into EG5003 worms. Worms that move and show none of the mCherry markers were individually plated. To confirm an insertion, we performed PCR with primers ChrIV-F, mCherry-genoF and 178-genoR to genotype individual mothers. A worm with homozygous insertion would have a single band at ~2.2 kb; a worm with no insertion would have a single band at ~4 kb; a worm with heterozygous insertion would have both bands.
Dye filling assay
The methods for dye filling was adapted from Worm Atlas (32). Briefly, we washed one plate of mixed population using 1 ml of M9 buffer. Then, we collected worms using centrifugation at 1500 rpm for 1 min and removed the supernatant. Then, we resuspended the worms in 1 ml of M9 buffer mixed with 5 µl of 2 mg/ml DiO (Molecular Probes, Cat#:D275). To allow the worms to take up the dye, we covered the tubes in tin foil and slowly shook at room temperature for 2 h. The worms were spun down again and transferred to a fresh seeded plate to allow the dye to pass through the gut. The worms were washed and spun as before just prior to transferring worms to the glass slide.
Genetic crosses
We obtained males for each strain containing the Mos insert by heat shocking 30 L4 hermaphrodites in 33°C for 4 h. We crossed four males with Mos insert to two daf-19 (DR86) L4 hermaphrodites. In all, 15 hermaphrodite F1s were selected randomly and individually plated. The genotype of the daf-19 gene in these F1s was confirmed by Tetra-ARM PCR (33). To find homozygous Mos insertion and homozygous daf-19 mutation, we individually plated 200 F2s and screened for dauer phenotype as 85% of daf-19 worms enter the dauer stage even in favorable condition (34). Candidates were screened and confirmed by genotyping.
Microscopy visualization
Worms were immobilized using azide on 3% agarose pad. Images were captured under Zeiss spinning disc confocal microscope (Zeiss Axio Observer.Z1) equipped with Hamamatsu ImagEM camera. Image capture and visualization were performed using Volocity software (www.improvision.com).
RESULTS
Search for putative C. elegans genes containing multiple X-box motifs by comparative genomics
Our search strategy involves finding genes that have conserved X-box motifs within 500-bp of promoter region in four Caenorhabditis species. We chose the 500-bp search space because all validated X-box motif identified to date reside in this region (Table 1). In order to obtain the 500-bp promoter region, we first performed gene annotation on C. briggsae, C. remanei and C. brenneri using genBlastG (see ‘Materials and Methods’ section). The C. briggsae gene models have not been examined since its publication (35) while the genomes of C. remanei and C. brenneri have not been published.
With the revised gene set generated from genBlastG, we employed TFMscan (30) to find X-box motifs within 500-bp upstream of the start codon. We focused on searching 14-bp and 15-bp X-box motifs since all validated X-box motifs identified to date are either 14-bp or 15-bp (see ‘Materials and Methods’ section, Table 1). We tested the sensitivity of TFMscan and it was able to capture 30 out of 31 validated X-box motifs in C. elegans, which shows our search strategy is sensitive to identify bona fide X-box motifs. We searched 500-bp promoter region of every gene in C. elegans and found 91 genes that contain two or more putative X-box motifs. To further narrow down our candidate list, we asked which of the 91 genes have conserved X-box motif in C. briggsae, C. remanei and C. brenneri. Our query returned eight genes with such characteristics (Table 2).
Table 2.
Caenorhabditis elegans genes with multiple putative X-box motifs within 500-bp promoter region
| Sequence name | Gene name | Description | Putative X-box sequences | X-box position | Expression in ciliated neuron |
|---|---|---|---|---|---|
| C30F2.2 | Similar to ARD GTP-binding proteins | gtcatccaggaaaac | −27 | SAGE | |
| atcaccaatagcaac | −80 | ||||
| C53B4.7 | bre-1 | Homologous to GDP-mannose 4, 6-dehydratases | gtttccttggcaaa | −423 | GFP, SAGE |
| atttcccatagaaac | −464 | ||||
| F17A9.4 | Homologous to NADH:flavin oxidoreductase | gtttgcccaacaac | −98 | SAGE | |
| gtttctcaggcaac | −233 | ||||
| F25B4.2 | Homologous to Pellino | gtctccaatggcaac | −149 | GFP, SAGE | |
| gtcctcacaagtaac | −199 | ||||
| F32B6.9 | Homologous to Bestrophin | gtctccttgacaac | −134 | GFP, SAGE | |
| gtttctttgataac | −241 | ||||
| gttatcaaagaaac | −273 | ||||
| H01G02.2 | Homologous to CDK-Activating Kinase | gtctccatgacaac | −160 | GFP, SAGE | |
| gttggcacggtaac | −234 | ||||
| gttaccgtgccaac | −280 | ||||
| T13C2.7 | Uncharacterized | gtcatctcagataac | −80 | SAGE | |
| gtcacctaggaaac | −144 | ||||
| Y41G9A.1 | osm-5 | Part of the intraflagellar transport | atctccatgacaac | −183 | GFP, SAGE |
| gtcgtcttggagac | −270 |
Positions are listed as distance from the translational start site.
We explored whether these eight genes show expression in ciliated neurons based on existing expression data. We found five genes with anatomical expression pattern data using GFP (Table 2). One of the genes, osm-5, was already characterized as a ciliary gene (21,36,37). Existing GFP reporter construct expression showed that F25B4.2, H01G02.2, C53B4.7 and F32B6.9 have neuronal expression (38,39). SAGE data further demonstrated that these genes are expressed in many ciliated neurons, including ASER and AFD neurons. The remaining three genes, though not having anatomical expression pattern data, show ciliated neuron expression based on SAGE data.
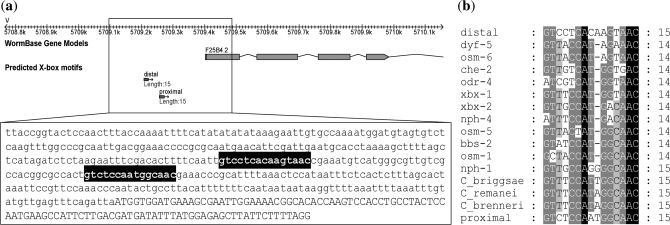
F25B4.2 is a conserved gene that harbors X-box motifs in four Caenorhabditis species but not in C. elegans
The expression pattern of F25B4.2 shown by GFP and SAGE tags indicates its potential for DAF-19 regulation. F25B4.2 contains two putative 15-bp X-box motifs in the upstream region, one located at 199-bp and the other at 149-bp upstream of the start codon (Figure 1a, Table 2). For convenience, we named the X-box motif located at −199 the distal motif and the X-box motif located at −149 the proximal motif. However, F25B4.2 was not identified as a DAF-19 target gene in any of the previous three genome-wide searches (21–23), likely due to its 15-bp X-box motifs that are different from the 14-bp consensus used. Among the two, the proximal motif displays higher conservation especially in the last six nucleotides where it is identical to many known X-box motifs (Figure 1b). However, these two motifs differ from the consensus at the third nucleotide (consensus = T) and at the eighth nucleotide (consensus = T). A single 15-bp X-box motif was also identified in three other Caenorhabditis species (Figure 1b).
Figure 1.
(a) The location and sequence of the putative X-box motifs upstream of F25B4.2. (b) The alignment of the putative proximal and distal motifs to known X-box motifs. Also include in the alignment are the putative X-box motifs in the orthologs of F25B4.2 in three other Caenorhabditis species labeled as C. briggsae, C. remanei and C. brenneri.
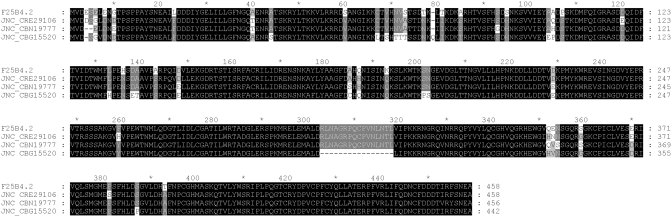
F25B4.2 is conserved in four sequenced Caenorhabdits species with >80% identity at the protein level (Figure 2). The presence of X-box in upstream region of a gene usually suggests regulation by DAF-19 (4). F25B4.2 protein sequence shows ~40% identity to human Pellino gene family. Pellino proteins are E3 ligases known to participate in balancing inflammatory response (56). Pellino proteins interact with IRAK and mediate NFkB nuclear translocation to promote activation of pro-inflammatory genes (57,58). Pellino1 is suggested to play a part in TGF-β pathways to promote anti-inflammatory response preventing hyperactivation of inflammatory response (59,60).
Figure 2.
The alignment of F25B4.2 protein sequence with its orthologs in the other four Caenorhabditis species. CBG = C. briggsae, CRE = C. remanei, CBN = C. brenneri. The sequences were aligned using ClustalW (54) and visualized using GeneDoc (55).
Functional analysis of proximal and distal motifs
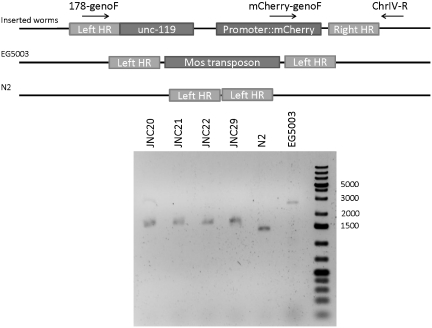
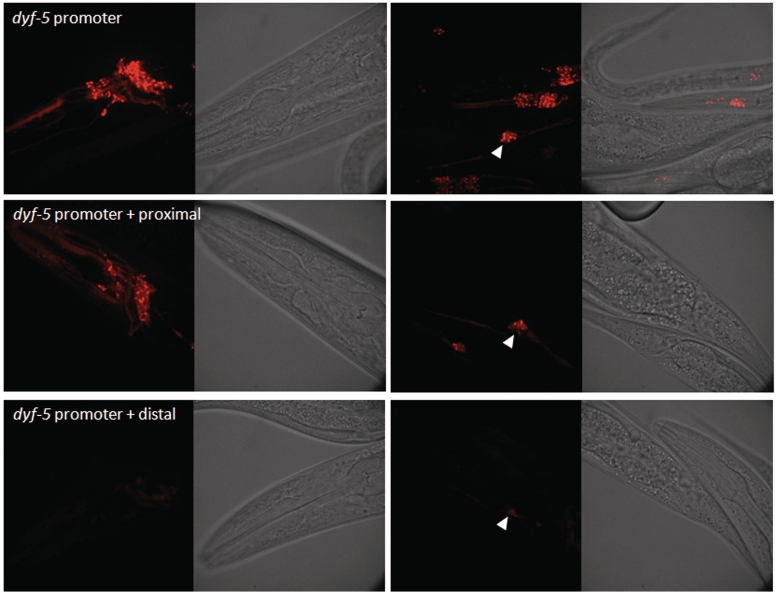
To examine whether the proximal or the distal motif is functional, we replaced the endogenous X-box motif of a known DAF-19 target gene promoter with either motif. If the motifs are functional, then it should drive ciliated neuron expression. We chose the promoter of dyf-5 for this experiment because dyf-5 was identified previously to express exclusively in ciliated neurons in a DAF-19-dependent manner (23,53). dyf-5 expression is also X-box motif dependent such that its expression is completely abolished when the 14-bp X-box motif is removed (Wang,J. and Chen,N., unpublished data). We replaced the endogenous X-box motif in dyf-5 promoter region (2000-bp upstream) with either the proximal motif or the distal motif and fused the promoter with mCherry. A single copy of the construct is integrated into the C. elegans genome at the Mos site cxTi10882 in chromosome IV using MosSCI method (61). This Mos element is located in an intergenic region with the flanking genes pointing toward each other. Hence, this location is not likely to have functional elements disrupted after reporter gene insertion. Insertion at the Mos site is confirmed by genotyping (Figure 3). The single copy-inserted construct using the wild-type dyf-5 promoter is able to reproduce the ciliated neuron expression as reported before (Figure 4: dyf-5 promoter) (23). In comparison to the endogenous X-box motif in dyf-5, the proximal element taken from F25B4.2 can drive robust expression in the amphid neurons and somewhat lower expression in the labial neurons (Figure 4: dyf-5 promoter + proximal), suggesting that the proximal motif is a functional X-box motif. Interestingly, the distal element also drives mCherry expression in ciliated neurons in C. elegans but the expression level is greatly reduced (Figure 4: dyf-5 promoter + distal). Our observations demonstrated that both motifs are functional motifs with the proximal motif able to drive much stronger expression than the distal motif.
Figure 3.
Genotyping of stably integrated strains. The insertion site on chromosome IV is depicted by the diagram on the top while an agarose gel showing the genotyping results on the bottom. The primers used for genotyping are also indicated by the arrows. Primer mCherry-genoF can only hybridize to inserted worms and not EG5003 and N2. The expected band sizes for inserted worms are 8312 bp from 178-genoF→ChrIV-R and 1564 bp from mCherry-genoF→ChrIV-R. The expected band size for EG5003 is 2700 bp [Mos1 is ~1280 bp (62–64)]. The expected band size for N2 is 1420 bp from 178-genoF→ChrIV-R. The gel image shows homozygous insertion for JNC20, 21, 22 and 29 as well as EG5003 and N2 as controls. The number on the right-hand side indicates the ladder positions.
Figure 4.
The expression pattern driven by dyf-5 promoter replacing the endogenous X-box motif with either the proximal motif or the distal motif. Proximal motif is able to drive normal expression while distal motif is unable to. The white arrows show the location of PHA and PHB neurons. Exposure time = 3 s.
F25B4.2 is expressed in ciliated neurons in a DAF-19-dependent manner
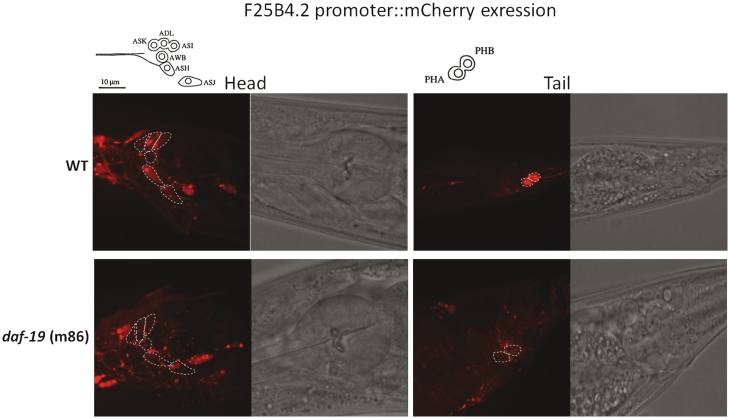
Since the proximal motif can be used to replace endogenous X-box motifs in the dyf-5 promoter to drive ciliated neuron expression, we hypothesized that F25B4.2 expression depends on DAF-19. To test this hypothesis, we examined whether the promoter of F25B4.2 drives expression in ciliated neurons and whether its expression is dependent on DAF-19. We constructed a C. elegans strain carrying a single copy mCherry transgene driven by a 3000-bp promoter region upstream of F25B4.2 using MosSCI method (61). Observation of mCherry signals indicates that F25B4.2 is expressed in ciliated neurons (Figure 5). Dye-filling method with DiO in C. elegans allows six pairs of amphid neurons and two pairs of phasmid neurons to be filled with dye. Detailed analysis of F25B4.2 expression using dye-filling shows that F25B4.2 drives gene expression in ciliated neurons, including ASK, ADL, ASI, ASH, ASJ, PHA and PHB neurons (outlined by white dash lines in Figure 5). Expression in AWB was not found. Additional expression was also observed in muscle cells during larval stages but not in adults. Similar expression pattern for this gene was observed previously in C. elegans injected with extra-chromosomal array that contained GFP reporter driven by the same putative promoter sequence (38). To confirm whether the expression pattern indicated by mCherry is dependent on DAF-19, we crossed the strain with the mCherry reporter construct to a daf-19 mutant strain (m86). In the absence of DAF-19, the expression in ciliated neurons that dye-fills in the head and tail is abolished (Figure 5). The fact that mCherry signals remain in other neurons suggests that additional cis- and trans- acting factors exist to regulate F25B4.2 expression pattern. Nevertheless, our data here shows that F25B4.2 expression in many ciliated neurons in C. elegans is dependent on DAF-19.
Figure 5.
The head and tail expression patterns of F25B4.2 3 kb upstream region fused to mCherry in either wild-type (WT) strain or daf-19(m86) strain. White dashed lines outline the ciliated neurons that dye fill. Neurons that dye fill in the head include ASK, ADL, ASI, AWB, ASH and ASJ; neurons that dye fill in the tail include PHA and PHB. Because daf-19 worms do not dye fill, the white outlines are the supposed location of these neurons. The expressions in these neurons are abolished in daf-19(m86) background. The schematic of the ciliated neurons that dye fill is shown on top. The schematic is adapted from (65). Exposure time = 3 s.
Both proximal and distal motifs function in regulating F25B4.2
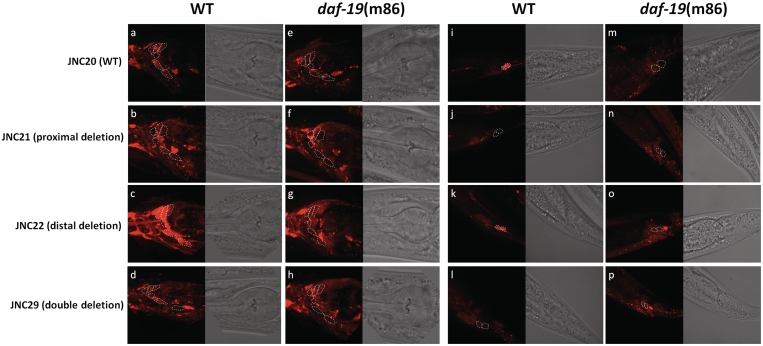
To test whether these two motifs are functional in their endogenous environment, we engineered three additional promoter fusion constructs with (i) only the proximal motif removed, (ii) only the distal motif removed and (iii) both the proximal and the distal motifs removed (Figure 6). If these motifs are functional in promoting gene expression, we would expect the expression pattern in ciliated neurons to be abolished. These constructs were also injected and integrated using the MosSCI method (61). In the strain carrying the proximal deletion construct (JNC21), we observed that many amphid neurons as well as phasmid neurons lost mCherry expression (compare Figure 6a and b; i and j). Using dye-filling with DiO, we observed specifically that ASK, ASI and ASJ neurons no longer show expression while ADL and ASH neurons retained expression. In the strain carrying the distal deletion construct (JNC22), we were surprised that it did not abolish any expression but instead enhanced expression (compare Figure 6a and c; i and k). By reducing the exposure time from 3 s to 800 ms (~4-fold), we were able to capture the expression intensity at a comparable level to that of JNC20 (the strain carrying the wild-type promoter). In the strain carrying construct with both motifs removed (JNC29), we observed similar pattern and intensity as the JNC21 where many ciliated neurons no longer show mCherry expression (Figure 6). Again, dye-filing with DiO reveals that ASK, ASI, ASJ neurons do not show mCherry expression anymore while ADL and ASH neurons retained expression. Since we used a 3-kb promoter region in our constructs, it is possible that additional X-box motifs outside of the 500-bp region are responsible for the expression remaining in ADL and ASH neurons. Searching further upstream revealed a 13-bp motif at −1092 with a weak match to X-box motifs consensus (results not shown), which can be a bona fide X-box motif. Taken together, our results suggest proximal motif but not the distal motif is responsible for driving F25B4.2 expression in ciliated neurons. However the distal motif may have a repressive role in modulating the expression level of this gene.
Figure 6.
The expression of different deletion constructs in either wild-type or daf-19(m86) backgrounds. Proximal deletion construct removes the 15 bp putative X-box at −140; distal deletion construct removes the 15-bp putative X-box at −190; double deletion construct removes both putative X-box motifs. Other than that, all sequences remain the same. White dashed lines outline the ciliated neurons that dye fill. The outlines for strains in daf-19 background are supposed locations. (a–g) show expression in the head and (i–p) show expression in the tail. Exposure time = 3 s.
In order to show whether these motifs function together with DAF-19, we have crossed JNC21, JNC22 and JNC29 to the daf-19(m86) mutant background. If the X-box motifs are functional with DAF-19, we expect these constructs in the daf-19 mutant background would show similar pattern to what was observed in Figure 5 where many ciliated neurons no longer show mCherry expression in daf-19 mutant worms. As expected, we observed nearly identical expression pattern across all constructs in daf-19 mutant strain where many ciliated neurons in both the head and tail have abolished expression (compare Figure 6e to h; m to p). The difference is especially striking for the distal deletion construct where an elevated expression level in wild-type background dropped to very low expression in daf-19 mutant (Figure 6c and g). The observation here further suggests that proximal motif is the main driving force for expression by interacting with DAF-19.
DISCUSSION
Ciliopathy is an emerging human genetic disorder caused by malformation of cilia that leads to many clinical hallmarks including obesity, polydactyly and retinal degeneration. The first link between RFX TFs and ciliary genes together with cilia development was made by examining sensory defect in C. elegans, which lead to the cloning of daf-19 (4), the only RFX TF gene in C. elegans. In the 10 years that followed this discovery, studies on Chlamydomonas reinhardtii and C. elegans greatly facilitated further ciliopathy research in mammals. Caenorhabditis elegans, in particular, has been used as a platform for identifying human BBS3, 5, 7 and 8 (48,50,66). Evidence suggests that the known 14 human BBS genes only constitute 25–50% of the ciliopathy cases (67). Therefore, additional target genes remain to be discovered. More importantly, mechanisms underlying the transcriptional regulation of these target genes by RFX TFs are not known. Such knowledge can definitely facilitate further search for RFX TF target genes.
DAF-19 target genes identified in C. elegans to date have been demonstrated to be regulated by X-box motifs. In this study, through bioinformatics and comparative genomics searches, we have found genes that have two putative X-box motifs residing in 500-bp upstream region. To ascertain the contribution of each individual X-box motif in gene expression and the possible interaction of two X-box motifs, we extensively examined the two 15-bp motifs in the genomic region upstream of F25B4.2. We show that F25B4.2 is regulated by both X-box motifs. F25B4.2 is likely orthologous to human Pellino gene. Pellino proteins are E3 ligases known to participate in balancing inflammatory response (56). Pellino proteins interact with IRAK and mediate NFkB nuclear translocation to promote activation of proinflammatory genes (57,58). Pellino1 is suggested to play a part in TGF-β pathways to promote anti-inflammatory response preventing hyperactivation of inflammatory response (59,60). However, Pellino is not currently known to have any role in cilia development or cilia maintenance in human or in any other organisms.
The proximal motif identified in this study can be seen as a ‘strong’ motif that has higher sequence conservation and drives gene expression while the distal motif can be seen as a ‘weak’ motif that do not drive gene expression as well but may function in expression level regulation. While a repressive X-box motif was never reported in C. elegans, there are ample of repressive X-box motifs in other systems. For example, RFX TF in S. cerevisiae was found to exhibit similar characteristic. S. cerevisiae RFX negatively regulates many ribonucleotide reductase genes (e.g. RNR2, RNR3 and RNR4) through a combination of strong X-box motifs and weak X-box motifs (24). Removing the weak X-box motifs only show slight expression increase (1.4–1.7 fold) and removing the strong X-box motif increase the expression level by 5-fold (24). However, simultaneous removal of all motifs elevates the expression level by 17-fold (24). Other TFs have also been found to bind to multiple regulatory elements in order to regulate gene expression to a specific level. For instance, PurR in Bacillus subtilis binds to two PurBox motifs for higher affinity (68). Drosophila gap gene hunchback (hb) is regulated by multiple bicoid (bcd) binding sites (69). Multiple NFkB binding sites work synergistically to regulate US3 gene of human cytomegalovirus (70).
In a similar fashion, human RFX1 and RFX3 repress MAP1A expression in non-neuronal cells by binding to two X-box motifs in the first exon (25). The strong and weak X-box motifs in yeast and humans may work synergistically; however, our results here suggest that X-box motifs in F25B4.2 work antagonistically. We propose that while a single strong X-box is sufficient to drive gene expression, additional weak X-box motifs could fine-tune the expression level appropriate for the gene. The molecular mechanism of the fine-tuning remains to be further explored. A simple model is that the weak motif servers as a dominant negative competitor against the strong motif. In other words, the weak site servers as a ‘transcription factor sponge’ that absorbs transcription factor in the local molecular niche so that the strong site has limited supply of the transcription factor.
In conclusion, this project represents an important step toward understanding RFX regulatory mechanism in animal genome, which will potentially help identifying functional X-box motifs as well as RFX target genes in humans.
FUNDING
Discovery Grants from the Natural Sciences and Engineering Research Council (NSERC) of Canada (to N.C. and D.L.B.). J.S.C.C. is supported by a NSERC Postgraduate Scholarship. D.L.B. is a Tier 1 Canada Research Chair (CRC). N.C. is a Michael Smith Foundation for Health Research (MSFHR) Scholar and a Canadian Institutes of Health Research (CIHR) New Investigator. Funding for open access charge: NSERC Canada.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENT
We thank Erik M. Jorgensen for plasmids and strains and Harald Hutter for discussion of results. We thank Tao Luo, Elaheh Kahrobaee Khorassani and Behnaz Kahrobaee Khorassani for technical assistance. Reagents for MosSCI were generous gifts from E.M. Jorgensen.
REFERENCES
- 1.Emery P, Durand B, Mach B, Reith W. RFX proteins, a novel family of DNA binding proteins conserved in the eukaryotic kingdom. Nucleic Acids Res. 1996;24:803–807. doi: 10.1093/nar/24.5.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chu JS, Baillie DL, Chen N. Convergent evolution of RFX transcription factors and ciliary genes predated the origin of metazoans. BMC Evol. Biol. 2010;10:130. doi: 10.1186/1471-2148-10-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gajiwala KS, Chen H, Cornille F, Roques BP, Reith W, Mach B, Burley SK. Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature. 2000;403:916–921. doi: 10.1038/35002634. [DOI] [PubMed] [Google Scholar]
- 4.Swoboda P, Adler HT, Thomas JH. The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol. Cell. 2000;5:411–421. doi: 10.1016/s1097-2765(00)80436-0. [DOI] [PubMed] [Google Scholar]
- 5.Durand B, Vandaele C, Spencer D, Pantalacci S, Couble P. Cloning and characterization of dRFX, the Drosophila member of the RFX family of transcription factors. Gene. 2000;246:285–293. doi: 10.1016/s0378-1119(00)00093-7. [DOI] [PubMed] [Google Scholar]
- 6.Otsuki K, Hayashi Y, Kato M, Yoshida H, Yamaguchi M. Characterization of dRFX2, a novel RFX family protein in Drosophila. Nucleic Acids Res. 2004;32:5636–5648. doi: 10.1093/nar/gkh895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoskins RA, Carlson JW, Kennedy C, Acevedo D, Evans-Holm M, Frise E, Wan KH, Park S, Mendez-Lago M, Rossi F, et al. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science. 2007;316:1625–1628. doi: 10.1126/science.1139816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aftab S, Semenec L, Chu JS, Chen N. Identification and characterization of novel human tissue-specific RFX transcription factors. BMC Evol. Biol. 2008;8:226. doi: 10.1186/1471-2148-8-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonnafe E, Touka M, AitLounis A, Baas D, Barras E, Ucla C, Moreau A, Flamant F, Dubruille R, Couble P, et al. The transcription factor RFX3 directs nodal cilium development and left-right asymmetry specification. Mol. Cell. Biol. 2004;24:4417–4427. doi: 10.1128/MCB.24.10.4417-4427.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baas D, Meiniel A, Benadiba C, Bonnafe E, Meiniel O, Reith W, Durand B. A deficiency in RFX3 causes hydrocephalus associated with abnormal differentiation of ependymal cells. Eur. J. Neurosci. 2006;24:1020–1030. doi: 10.1111/j.1460-9568.2006.05002.x. [DOI] [PubMed] [Google Scholar]
- 11.Ait-Lounis A, Baas D, Barras E, Benadiba C, Charollais A, Nlend Nlend R, Liegeois D, Meda P, Durand B, Reith W. Novel function of the ciliogenic transcription factor RFX3 in development of the endocrine pancreas. Diabetes. 2007;56:950–959. doi: 10.2337/db06-1187. [DOI] [PubMed] [Google Scholar]
- 12.Ashique AM, Choe Y, Karlen M, May SR, Phamluong K, Solloway MJ, Ericson J, Peterson AS. The Rfx4 transcription factor modulates Shh signaling by regional control of ciliogenesis. Sci. Signal. 2009;2:ra70. doi: 10.1126/scisignal.2000602. [DOI] [PubMed] [Google Scholar]
- 13.Praetorius HA, Spring KR. A physiological view of the primary cilium. Ann. Rev. Physiol. 2005;67:515–529. doi: 10.1146/annurev.physiol.67.040403.101353. [DOI] [PubMed] [Google Scholar]
- 14.Haycraft CJ, Schafer JC, Zhang Q, Taulman PD, Yoder BK. Identification of CHE-13, a novel intraflagellar transport protein required for cilia formation. Exp. Cell Res. 2003;284:251–263. doi: 10.1016/s0014-4827(02)00089-7. [DOI] [PubMed] [Google Scholar]
- 15.Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Ann. Rev. Genomics Hum. Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 16.Garvie CW, Stagno JR, Reid S, Singh A, Harrington E, Boss JM. Characterization of the RFX complex and the RFX5(L66A) mutant: implications for the regulation of MHC class II gene expression. Biochemistry. 2007;46:1597–1611. doi: 10.1021/bi6023868. [DOI] [PubMed] [Google Scholar]
- 17.Soyer J, Flasse L, Raffelsberger W, Beucher A, Orvain C, Peers B, Ravassard P, Vermot J, Voz ML, Mellitzer G, et al. Rfx6 is an Ngn3-dependent winged helix transcription factor required for pancreatic islet cell development. Development. 2010;137:203–212. doi: 10.1242/dev.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith SB, Qu HQ, Taleb N, Kishimoto NY, Scheel DW, Lu Y, Patch AM, Grabs R, Wang J, Lynn FC, et al. Rfx6 directs islet formation and insulin production in mice and humans. Nature. 2010;463:775–780. doi: 10.1038/nature08748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emery P, Strubin M, Hofmann K, Bucher P, Mach B, Reith W. A consensus motif in the RFX DNA binding domain and binding domain mutants with altered specificity. Mol. Cell. Biol. 1996;16:4486–4494. doi: 10.1128/mcb.16.8.4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laurencon A, Dubruille R, Efimenko E, Grenier G, Bissett R, Cortier E, Rolland V, Swoboda P, Durand B. Identification of novel regulatory factor X (RFX) target genes by comparative genomics in Drosophila species. Genome Biol. 2007;8:R195. doi: 10.1186/gb-2007-8-9-r195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Efimenko E, Bubb K, Mak HY, Holzman T, Leroux MR, Ruvkun G, Thomas JH, Swoboda P. Analysis of xbx genes in C. elegans. Development. 2005;132:1923–1934. doi: 10.1242/dev.01775. [DOI] [PubMed] [Google Scholar]
- 22.Blacque OE, Perens EA, Boroevich KA, Inglis PN, Li C, Warner A, Khattra J, Holt RA, Ou G, Mah AK, et al. Functional genomics of the cilium, a sensory organelle. Curr. Biol. 2005;15:935–941. doi: 10.1016/j.cub.2005.04.059. [DOI] [PubMed] [Google Scholar]
- 23.Chen N, Mah A, Blacque OE, Chu J, Phgora K, Bakhoum MW, Newbury CR, Khattra J, Chan S, Go A, et al. Identification of ciliary and ciliopathy genes in Caenorhabditis elegans through comparative genomics. Genome Biol. 2006;7:R126. doi: 10.1186/gb-2006-7-12-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang M, Zhou Z, Elledge SJ. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell. 1998;94:595–605. doi: 10.1016/s0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- 25.Nakayama A, Murakami H, Maeyama N, Yamashiro N, Sakakibara A, Mori N, Takahashi M. Role for RFX transcription factors in non-neuronal cell-specific inactivation of the microtubule-associated protein MAP1A promoter. J. Biol. Chem. 2003;278:233–240. doi: 10.1074/jbc.M209574200. [DOI] [PubMed] [Google Scholar]
- 26.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.She R, Chu JS, Uyar B, Wang J, Wang K. genBlastG: using BLAST searches to build homologous gene models. Bioinformatics. 2011;27:2141–2143. doi: 10.1093/bioinformatics/btr342. [DOI] [PubMed] [Google Scholar]
- 28.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 30.Liefooghe A, Touzet H, Varre J-S. Large scale matching for Position Weight Matrices. Lect. Notes Comput. Sci. 2006;4009:401–412. [Google Scholar]
- 31.Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. BioTechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- 32. Cell identification in C. elegans. 2005. Altun, Z.F. and Hall, D.H. In WormAtlas. http://www.wormatlas.org/cellID.html.
- 33.Ye S, Dhillon S, Ke X, Collins AR, Day IN. An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res. 2001;29:E88. doi: 10.1093/nar/29.17.e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- 35.Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, Chen N, Chinwalla A, Clarke L, Clee C, Coghlan A, et al. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 2003;1:E45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haycraft CJ, Swoboda P, Taulman PD, Thomas JH, Yoder BK. The C. elegans homolog of the murine cystic kidney disease gene Tg737 functions in a ciliogenic pathway and is disrupted in osm-5 mutant worms. Development. 2001;128:1493–1505. doi: 10.1242/dev.128.9.1493. [DOI] [PubMed] [Google Scholar]
- 37.Qin H, Rosenbaum JL, Barr MM. An autosomal recessive polycystic kidney disease gene homolog is involved in intraflagellar transport in C. elegans ciliated sensory neurons. Curr. Biol. 2001;11:457–461. doi: 10.1016/s0960-9822(01)00122-1. [DOI] [PubMed] [Google Scholar]
- 38.Hunt-Newbury R, Viveiros R, Johnsen R, Mah A, Anastas D, Fang L, Halfnight E, Lee D, Lin J, Lorch A, et al. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 2007;5:e237. doi: 10.1371/journal.pbio.0050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKay SJ, Johnsen R, Khattra J, Asano J, Baillie DL, Chan S, Dube N, Fang L, Goszczynski B, Ha E, et al. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb. Symp. Quant. Biol. 2003;68:159–169. doi: 10.1101/sqb.2003.68.159. [DOI] [PubMed] [Google Scholar]
- 40.Schafer JC, Haycraft CJ, Thomas JH, Yoder BK, Swoboda P. XBX-1 encodes a dynein light intermediate chain required for retrograde intraflagellar transport and cilia assembly in Caenorhabditis elegans. Mol. Biol. Cell. 2003;14:2057–2070. doi: 10.1091/mbc.E02-10-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winkelbauer ME, Schafer JC, Haycraft CJ, Swoboda P, Yoder BK. The C. elegans homologs of nephrocystin-1 and nephrocystin-4 are cilia transition zone proteins involved in chemosensory perception. J. Cell Sci. 2005;118:5575–5587. doi: 10.1242/jcs.02665. [DOI] [PubMed] [Google Scholar]
- 42.Efimenko E, Blacque OE, Ou G, Haycraft CJ, Yoder BK, Scholey JM, Leroux MR, Swoboda P. Caenorhabditis elegans DYF-2, an orthologue of human WDR19, is a component of the intraflagellar transport machinery in sensory cilia. Mol. Biol. Cell. 2006;17:4801–4811. doi: 10.1091/mbc.E06-04-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collet J, Spike CA, Lundquist EA, Shaw JE, Herman RK. Analysis of osm-6, a gene that affects sensory cilium structure and sensory neuron function in Caenorhabditis elegans. Genetics. 1998;148:187–200. doi: 10.1093/genetics/148.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ou G, Qin H, Rosenbaum JL, Scholey JM. The PKD protein qilin undergoes intraflagellar transport. Curr. Biol. 2005;15:R410–R411. doi: 10.1016/j.cub.2005.05.044. [DOI] [PubMed] [Google Scholar]
- 45.Murayama T, Toh Y, Ohshima Y, Koga M. The dyf-3 gene encodes a novel protein required for sensory cilium formation in Caenorhabditis elegans. J. Mol. Biol. 2005;346:677–687. doi: 10.1016/j.jmb.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 46.Fujiwara M, Ishihara T, Katsura I. A novel WD40 protein, CHE-2, acts cell-autonomously in the formation of C. elegans sensory cilia. Development. 1999;126:4839–4848. doi: 10.1242/dev.126.21.4839. [DOI] [PubMed] [Google Scholar]
- 47.Bell LR, Stone S, Yochem J, Shaw JE, Herman RK. The molecular identities of the Caenorhabditis elegans intraflagellar transport genes dyf-6, daf-10 and osm-1. Genetics. 2006;173:1275–1286. doi: 10.1534/genetics.106.056721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blacque OE, Reardon MJ, Li C, McCarthy J, Mahjoub MR, Ansley SJ, Badano JL, Mah AK, Beales PL, Davidson WS, et al. Loss of C. elegans BBS-7 and BBS-8 protein function results in cilia defects and compromised intraflagellar transport. Genes Dev. 2004;18:1630–1642. doi: 10.1101/gad.1194004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ansley SJ, Badano JL, Blacque OE, Hill J, Hoskins BE, Leitch CC, Kim JC, Ross AJ, Eichers ER, Teslovich TM, et al. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature. 2003;425:628–633. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- 50.Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, et al. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- 51.Mak HY, Nelson LS, Basson M, Johnson CD, Ruvkun G. Polygenic control of Caenorhabditis elegans fat storage. Nat. Genet. 2006;38:363–368. doi: 10.1038/ng1739. [DOI] [PubMed] [Google Scholar]
- 52.Bacaj T, Lu Y, Shaham S. The conserved proteins CHE-12 and DYF-11 are required for sensory cilium function in Caenorhabditis elegans. Genetics. 2008;178:989–1002. doi: 10.1534/genetics.107.082453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burghoorn J, Dekkers MP, Rademakers S, de Jong T, Willemsen R, Jansen G. Mutation of the MAP kinase DYF-5 affects docking and undocking of kinesin-2 motors and reduces their speed in the cilia of Caenorhabditis elegans. Proc. Natl Acad. Sci. USA. 2007;104:7157–7162. doi: 10.1073/pnas.0606974104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 55.Nicholas KB, Nicholas HBJ, Deerfield DWI. GeneDoc: analysis and visualization of genetic variation. EMBNET.NEWS. 1997;4 [Google Scholar]
- 56.Butler MP, Hanly JA, Moynagh PN. Kinase-active interleukin-1 receptor-associated kinases promote polyubiquitination and degradation of the Pellino family: direct evidence for PELLINO proteins being ubiquitin-protein isopeptide ligases. J. Biol. Chem. 2007;282:29729–29737. doi: 10.1074/jbc.M704558200. [DOI] [PubMed] [Google Scholar]
- 57.Rich T, Allen RL, Lucas AM, Stewart A, Trowsdale J. Pellino-related sequences from Caenorhabditis elegans and Homo sapiens. Immunogenetics. 2000;52:145–149. doi: 10.1007/s002510000249. [DOI] [PubMed] [Google Scholar]
- 58.Strelow A, Kollewe C, Wesche H. Characterization of Pellino2, a substrate of IRAK1 and IRAK4. FEBS Lett. 2003;547:157–161. doi: 10.1016/s0014-5793(03)00697-5. [DOI] [PubMed] [Google Scholar]
- 59.Chang M, Jin W, Sun SC. Peli1 facilitates TRIF-dependent Toll-like receptor signaling and proinflammatory cytokine production. Nat. Immunol. 2009;10:1089–1095. doi: 10.1038/ni.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi KC, Lee YS, Lim S, Choi HK, Lee CH, Lee EK, Hong S, Kim IH, Kim SJ, Park SH. Smad6 negatively regulates interleukin 1-receptor-Toll-like receptor signaling through direct interaction with the adaptor Pellino-1. Nat. Immunol. 2006;7:1057–1065. doi: 10.1038/ni1383. [DOI] [PubMed] [Google Scholar]
- 61.Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benjamin HW, Kleckner N. Excision of Tn10 from the donor site during transposition occurs by flush double-strand cleavages at the transposon termini. Proc. Natl Acad. Sci. USA. 1992;89:4648–4652. doi: 10.1073/pnas.89.10.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lampe DJ, Churchill ME, Robertson HM. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 1996;15:5470–5479. [PMC free article] [PubMed] [Google Scholar]
- 64.van Luenen HG, Colloms SD, Plasterk RH. The mechanism of transposition of Tc3 in C. elegans. Cell. 1994;79:293–301. doi: 10.1016/0092-8674(94)90198-8. [DOI] [PubMed] [Google Scholar]
- 65.Starich TA, Herman RK, Kari CK, Yeh WH, Schackwitz WS, Schuyler MW, Collet J, Thomas JH, Riddle DL. Mutations affecting the chemosensory neurons of Caenorhabditis elegans. Genetics. 1995;139:171–188. doi: 10.1093/genetics/139.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fan Y, Esmail MA, Ansley SJ, Blacque OE, Boroevich K, Ross AJ, Moore SJ, Badano JL, May-Simera H, Compton DS, et al. Mutations in a member of the Ras superfamily of small GTP-binding proteins causes Bardet-Biedl syndrome. Nat. Genet. 2004;36:989–993. doi: 10.1038/ng1414. [DOI] [PubMed] [Google Scholar]
- 67.Yang Z, Yang Y, Zhao P, Chen K, Chen B, Lin Y, Guo F, Chen Y, Liu X, Lu F, et al. A novel mutation in BBS7 gene causes Bardet-Biedl syndrome in a Chinese family. Mol. Vis. 2008;14:2304–2308. [PMC free article] [PubMed] [Google Scholar]
- 68.Bera AK, Zhu J, Zalkin H, Smith JL. Functional dissection of the Bacillus subtilis pur operator site. J. Bacteriol. 2003;185:4099–4109. doi: 10.1128/JB.185.14.4099-4109.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Driever W, Nusslein-Volhard C. The bicoid protein is a positive regulator of hunchback transcription in the early Drosophila embryo. Nature. 1989;337:138–143. doi: 10.1038/337138a0. [DOI] [PubMed] [Google Scholar]
- 70.Chan YJ, Tseng WP, Hayward GS. Two distinct upstream regulatory domains containing multicopy cellular transcription factor binding sites provide basal repression and inducible enhancer characteristics to the immediate-early IES (US3) promoter from human cytomegalovirus. J. Virol. 1996;70:5312–5328. doi: 10.1128/jvi.70.8.5312-5328.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]