Abstract
Non-coding RNAs (ncRNAs) play key roles in diverse cellular activities, and efficient ncRNA function requires extensive posttranscriptional nucleotide modifications. Small nucleolar RNAs (snoRNAs) are a group of ncRNAs that guide the modification of specific nucleotides in ribosomal RNAs (rRNAs) and small nuclear RNAs. To investigate the physiological relevance of rRNA modification in vertebrates, we suppressed the expression of three snoRNAs (U26, U44 and U78), either by disrupting the host gene splicing or by inhibiting the snoRNA precursor processing, and analyzed the consequences of snoRNA loss-of-function in zebrafish. Using a highly sensitive mass spectrometric analysis, we found that decreased snoRNA expression reduces the snoRNA-guided methylation of the target nucleotides. Impaired rRNA modification, even at a single site, led to severe morphological defects and embryonic lethality in zebrafish, which suggests that rRNA modifications play an essential role in vertebrate development. This study highlights the importance of posttranscriptional modifications and their role in ncRNA function in higher eukaryotes.
INTRODUCTION
A majority of non-coding RNAs (ncRNAs) undergo posttranscriptional modifications. To date, more than 100 types of modifications that are thought to be crucial for RNA function have been identified in various RNA species (1,2). For example, a tRNA molecule contains 5–10 modified sites, and functional studies in Escherichia coli have shown that these modifications are essential for codon recognition (3). In plants, all microRNAs and small interfering RNAs undergo 2′-O-methylation at their 3′ termini, which protects the RNA from exonucleotic degradation (4–6). Similarly, piwi-interacting RNAs, which are expressed only in germ cells, are 2′-O-methylated at their 3′-ends (7–10); however, the function of this modification is currently unknown.
Ribosomal RNAs (rRNAs), which are the most abundant ncRNAs in the cell, also undergo several modifications. There are three types of modifications in eukaryotic rRNAs: (i) methylation of 2′-hydroxyls (Nm), (ii) conversion of uridine to pseudouridine (ψ) and (iii) methylation of bases (mN) (11). In humans, there are 103 Nm, 96 ψ and 9 mN modification sites (12). Analyses of 3D modification maps for the yeast and E. coli ribosomes revealed that most of the rRNA modifications occur in the functionally important areas of ribosomes (~60% in yeast and 95% in E. coli) (11). Loss of rRNA modification at multiple sites within the ribosome-decoding center in yeast affects cell growth and ribosome activity (13–15). In eukaryotes, the Nm and ψ modifications are catalyzed by an assemblage of small RNAs and proteins termed the small nucleolar ribonucleoprotein (snoRNP) particle. The small nucleolar RNAs (snoRNAs), which are a component of the snoRNP, guide these modifications (16). There are primarily two types of snoRNA, the box C/D type and box H/ACA type, which are classified on the basis of their box elements and 2D structure. Box C/D snoRNAs guide 2′-O-methylation and box H/ACA snoRNAs guide pseudouridylation (17). In vertebrates, almost all snoRNA genes are located within the introns of genes (intronic) that code for proteins. However, some snoRNA host genes do not code for proteins. On the other hand, in plants and yeast, most of the snoRNAs are encoded as clusters (polycistronic) or as independent genes (monocistronic) (18–20). Although the type, gene organization and copy number of snoRNAs can vary among species, the mechanism of snoRNA-guided rRNA modification is evolutionarily conserved (21).
Mutations in snoRNA genes have been associated with several human diseases, such as congenital disorders and cancer. Prader-Willi syndrome (PWS) is a neurogenetic disorder that is caused by the loss of paternally-expressed imprinted genes within chromosome 15q11-q13, which includes large clusters of HBII-52 snoRNAs and HBII-85 snoRNAs (22–24). Decreased U50 snoRNA expression was seen in patients diagnosed with B-cell lymphoma who exhibited a chromosomal translocation between the U50HG and BCL6 genes (25). A mutation in the U50 snoRNA gene (2-bp deletion) was also observed in prostate cancer cell lines (26) and primary breast cancer tumors (27). Moreover, several snoRNAs were overexpressed in non-small-cell lung cancer (NSCLC) patients, which suggest that snoRNAs may serve as biomarkers for NSCLC (28).
Thus, it is becoming increasingly clear that snoRNAs may be associated with human disease. Systematic studies of snoRNA function are crucial for understanding the physiological relevance of rRNA modification in vertebrates. Here, we describe the development of snoRNA-deficient zebrafish, through blocking the synthesis of snoRNAs with morpholino antisense oligonucleotides (MOs). For the first time, we show that loss of snoRNA expression impairs rRNA modification at one location on the 28S rRNA, which leads to profound developmental defects in this vertebrate model.
MATERIALS AND METHODS
Morpholino oligonucleotide injections
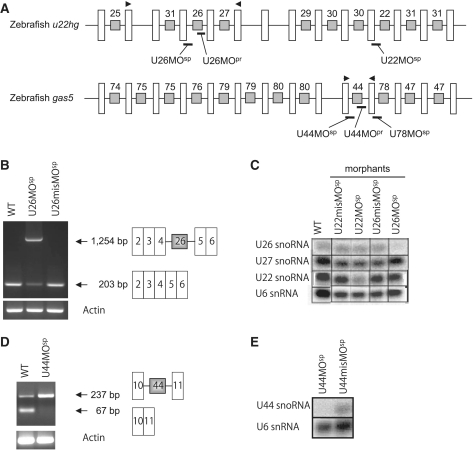
The MOs were obtained from Gene Tools, LLC (USA). For the U26 snoRNA, the splice site-targeted MO (MOsp) was designed at the exon 4/intron 4 boundary region of u22hg (Figure 1A). The U44 snoRNA and U78 snoRNA MOsps were designed within the exon10/intron 10 and exon 11/intron11 boundary regions of gas5, respectively (Figure 1A). For the precursor-MOs (MOpr), the 3′-terminal regions of the snoRNA precursor sequences within the introns (the fourth intron of u22hg for U26 snoRNA and the 10th intron of gas5 for U44 snoRNA) were targeted (Figure 1A). As a control, mismatch morpholinos (control MOs) with five mispaired bases were used. The sequences of the MOs are listed in Supplementary Table S1. Using our previous methods (29), a constant volume of MOs at the following concentrations (1.5–6 ng/embryo) was injected into one-cell stage embryos: U26MOsp at 5 µg/µl; U44MOsp and U44MOpr at 7.5 µg/µl; and U26MOpr at 20 µg/µl. The control MOs were injected using the same volume.
Figure 1.
The snoRNA-deficient zebrafish have reduced mature snoRNA expression. (A) The genomic structure of u22hg and gas5 in zebrafish. The white bars represent the exons and the black lines connecting the white bars represent the introns. The gray boxes within the introns indicate the snoRNA genes, which are numbered according to their human orthologs. The morpholinos were designed to target either the splicing (MOsp) or maturation (MOpr) of the snoRNAs, and the morpholino binding sites are shown in thick black lines. The arrowheads indicate the primer binding sites for RT–PCR. The u22hg and gas5 genomic sequences were obtained from the database under the accession numbers NW003334572.1 and NW001879345.1, respectively. (B) sqRT–PCR indicating that the improperly spliced transcript (1254 bp including intron 4) in the U26 morphants (middle lane) is increased compared with the normal u22hg transcript (203 bp without intron 4) in wild-type and control embryos. (C) Northern blotting of total RNA from morphants (U26MOsp and U22MOsp) and control embryos (U26misMOsp and U22misMOsp) using radiolabeled snoRNA probes. The U26 morphants have decreased expression of mature U26 snoRNA, and the expression of other snoRNAs transcribed from the same host gene was not affected. (D and E) sqRT–PCR and northern blotting showing the accumulation of unspliced precursor transcript (237 bp including intron 10) and a decrease in mature U44 snoRNA in the U44MOsp morphants. The U6 snRNA probe was used as loading control for the northern blotting.
Northern blot analysis
The total RNA was extracted using a TRIzol Reagent (Invitrogen, USA) according to the manufacturer’s instructions. For each sample, 10 µg of total RNA was separated on a 1% denaturing agarose gel and blotted according to standard procedures (25). The blots were hybridized overnight at 42°C in hybridization buffer (5× SSPE, 1× Denhardt's solution, 0.5% SDS, 50% formamide, 25 µg/ml salmon DNA and 100 µg/ml tRNA) containing 1000 cpm LNA (locked nucleic acid) probes labeled with [γ-33P] ATP by T4 polynucleotide kinase (Takara, Japan). The probe sequences are listed in Supplementary Table S2.
Semi-quantitative RT–PCR
The total RNA was isolated from 30 h postfertilization (hpf) embryos using a TRIzol Reagent (Invitrogen, USA), and sqRT–PCR was performed with a one-step RT–PCR kit (Qiagen, Germany). The reaction conditions were as described previously (30), except for a change in template concentration (0.5 µg total RNA in a 20 µl reaction mixture). The primers used were as follows: U26-forward, 5′-CAACGATGACTACTGCGACTC-3′; U26-reverse, 5′-CATAAACCCATCCTCTGCAGC-3′; U44-forward, 5′-TCTTCATGACTGCCATCCTT-3′; U44-reverse, 5′-CCAAGTAACATTCTTCATATTGCAC-3′; actin-forward, 5′-GCCCATCTATGAGGGTTACG-3′; and actin-reverse, 5′-GCAAGATTCCATACCCAGGA-3′.
Mass spectrometry
The total RNA was separated on a 4% polyacrylamide gel containing 7 M urea. The 18 S and 28S rRNAs were excised from the gel, eluted in buffer (400 mM sodium acetate pH 5.3, 1 mM EDTA, 0.1% SDS), and subsequently digested with RNase A or RNase T1. The RNase-digested fragments (250 fmol) were then subjected to capillary liquid chromatography/nano electrospray ionization-mass spectrometry according to a previously described protocol (31).
RESULTS
Zebrafish u22hg and gas5 encode a number of snoRNAs
The human U22 host gene (U22HG) is a non-protein coding gene that encodes nine snoRNAs (eight different types) in its introns (32). Our analysis of the zebrafish genome revealed a similar cluster of snoRNA genes in the introns of the zebrafish ortholog u22hg (Figure 1A). In addition, a comparison of zebrafish u22hg with orthologous genes in humans, frog and puffer fish revealed the following features: (i) seven snoRNAs are conserved between zebrafish and humans, although the encoding intron positions are not identical; (ii) unlike humans, zebrafish u22hg contains two copies of U30 and three copies of U31 snoRNA gene; and (iii) U28 snoRNA is absent in zebrafish and puffer fish, although it is conserved in humans and Xenopus (Supplementary Figure S1A). The 5′-terminal oligopyrimidine (5′ TOP) tract, which is a characteristic feature of the transcription start site in human U22HG, is also present in zebrafish u22hg. Similar to the human gene, zebrafish u22hg is likely a non-protein coding gene because the exons are small (<50 nt), contain only short ORFs (<49 amino acids), and have no predicted significant protein homology.
Similarly, the human growth arrest-specific 5 gene (GAS5) is a non-protein coding gene that encodes 10 different types of snoRNAs (33). We found that the zebrafish ortholog contains eight of these snoRNAs, except for U77 and U81. However, four snoRNAs (U75, U79, U80 and U47) are present in duplicate (Figure 1A and Supplementary Figure S1B). In this study, we targeted three snoRNAs (U26, U44 and U78) that are present as a single copy in the zebrafish genome to achieve a specific loss-of-function effect. The U26 and U78 snoRNAs guide ribose methylation at positions 398 (Am398) and 3745 (Gm3745) in the 28S rRNA, respectively, while the U44 snoRNA guides ribose methylation at position 163 (Am163) in the 18 S rRNA.
MOs effectively inhibit snoRNA expression in zebrafish
To inhibit snoRNA expression in zebrafish, we employed two types of MOs: splice-MO (MOsp), which disrupts the splicing of the host gene, and precursor-MO (MOpr), which inhibits snoRNA precursor processing. The splice MO for U26 snoRNA (U26MOsp) was designed to target the exon 4/intron 4 boundary region of u22hg and disrupt U26 snoRNA synthesis (Figure 1A). Similarly, a splice MO targeting the exon 10/intron 10 boundary region of gas5 was designed to inhibit U44 snoRNA synthesis (Figure 1A). For the precursor MOs, the precursor sequence of the snoRNAs (U26 and U44) within the introns was targeted, in contrast to the splicing region (Figure 1A).
Loss of snoRNA expression was confirmed by semi-quantitative RT–PCR (sqRT–PCR) and northern blot analysis of total RNA that was extracted from MO-injected embryos (morphants). As is shown in Figure 1B, sqRT–PCR revealed that the U26 snoRNA precursor accumulated in the U26MOsp morphants, which indicates that the MO disrupted host gene splicing. Northern blot analysis showed decreased mature U26 snoRNA expression, but unaltered mature U22 and U27 expression in these morphants, which indicates that the U26MOsp specifically inhibited U26 snoRNA synthesis (Figure 1C). Similarly, zebrafish embryos injected with U44MOsp showed an accumulation of the U44 precursor transcript and a decrease in mature U44 snoRNA expression (Figure 1D and E).
rRNA methylation is decreased in snoRNA-deficient zebrafish
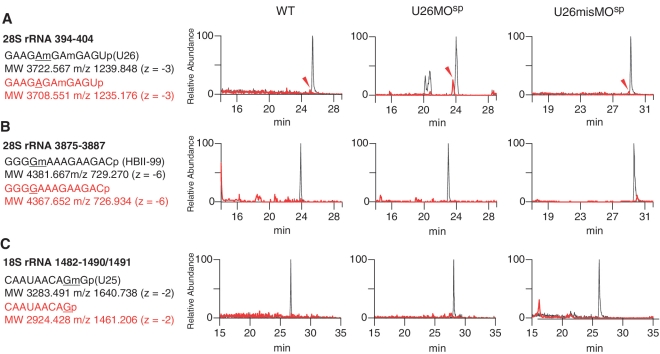
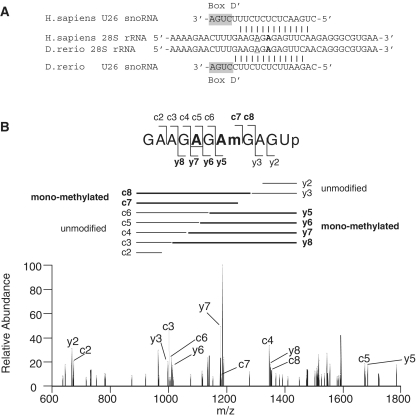
To determine whether rRNA modification was altered in the morphants, we used a highly sensitive detection method of RNA mass spectrometry (liquid chromatography/nano electrospray ionization mass spectrometry; LC/MS). Specifically, we analyzed complex mixtures of 28S and 18S rRNA fragments that were isolated from the morphants. Among the three morphants (U26MO, U44MO and U78MO), we could analyze only the rRNA fragments from the U26MO morphants, because the fragment that contains the U26 snoRNA target site (Am398) has a unique molecular mass and could be discriminated from the other 28S rRNA fragments. The 28S rRNA isolated from the wild-type and U26MOsp morphants was digested by RNase A and subjected to LC/MS analysis. In the wild-type embryos, the 11-mer RNA fragment (positions 394–404) that contains two ribose methylations at positions 398 and 400 was detected (Figure 2A). We sequenced the dimethylated 11-mer fragment by MS/MS using collision-induced dissociation (CID) and confirmed that positions 398 and 400 were methylated as reported (34) (Supplementary Figure S2). When the U26 snoRNA was inhibited by U26MOsp or U26MOpr, the same 11-mer fragment lacking a single methylation was clearly detected (Figure 2A and Supplementary Figure S3A). To determine the nucleotide that was not methylated in these morphants, the 11-mer fragment with monomethylation was analyzed by CID, which indicated that there was deficient methylation at position 398 (Figure 3A, B and Supplementary Table S3).
Figure 2.
28S rRNA methylation is decreased in the U26 morphants. LC/MS analyses of RNase A-digested 28S rRNA fragments and RNase T1-digested 18S rRNA fragments from wild-type (WT, left panels), U26 morphants (U26MOsp, middle panels) and control embryos (U26misMOsp, right panels). (A–C) Mass chromatograms of RNase A-digested 28S rRNA fragments showing the accumulation of a mono-methylated fragment containing the U26 snoRNA-specific modification site (arrowhead in A) in U26 morphants. The other snoRNA-specific modification sites in the 28S rRNA (HBII-99 snoRNA-guided guanosine at position 3878; B) and the 18S rRNA (U25 snoRNA-guided guanosine at position 1490; C) show no detectable accumulation of unmethylated fragments in the U26 morphants. The spectra for the methylated and unmethylated (mono-methylated in A) fragments are shown in black and red, respectively. The sequence and molecular weight of these fragments and their corresponding m/z values are indicated. The snoRNA-specific target nucleotide in each rRNA fragment is underlined.
Figure 3.
A398 methylation of the 28S rRNA is decreased in the U26 morphants. (A) The 28S rRNA region that contains the methylated adenosine (Am, guided by U26 snoRNA) at position 398 (underlined) is highly conserved between humans and zebrafish. The boxD signature motif of the U26 snoRNA and the base-pairing interactions with 28S rRNA are shown. Another proximal adenosine (indicated in bold), which is methylated by U81 snoRNA, is also highlighted. (B) The collision-induced dissociation (CID) spectrum of the mono-methylated fragment (precursor ion m/z 1235.0) obtained from the U26 morphants (as in Figure 2A). The A398 site is underlined in the fragment sequence (upper panel). The delineated fragment pattern (middle panel) corresponds to the dissociated fragments obtained after CID analysis of the mono-methylated fragment as in Figure 2A. The assignments of the product ions are indicated in the CID spectrum (lower panel), and the nomenclature for the product ions of the nucleic acids are as described by McLuckey et al. (49). The mono-methylated product ions are shown in bold. The observed and calculated m/z values of each product ion are listed in Supplementary Table S3.
There was no difference in the degree of methylation at sites guided by the other snoRNAs (e.g. Gm 3878 in 28S rRNA, which is guided by HBII-99 snoRNA, or Gm1490 in 18S rRNA, which is guided by U25 snoRNA) in both the U26MOsp (Figure 2B and C) and U26MOpr morphants (Supplementary Figure S3B and C). Thus, the U26 snoRNA-guided modification was specifically inhibited in the U26MO morphants.
Impaired rRNA modification leads to developmental abnormalities in zebrafish
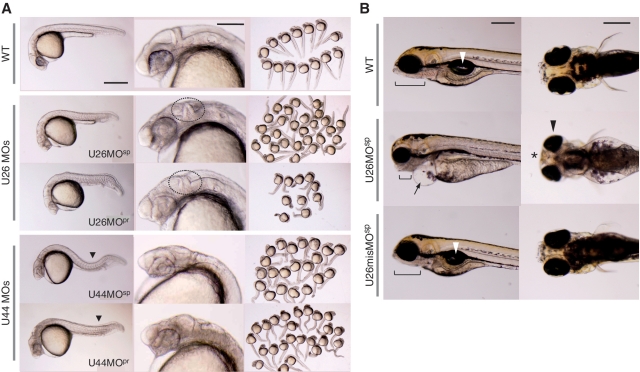
To investigate the role of rRNA modification in zebrafish embryogenesis, we performed a phenotypic analysis of snoRNA-deficient embryos at various stages of development. Loss of snoRNA expression resulted in growth mpairment and developmental delay with specific abnormalities in various organs that depended upon the type of snoRNA inhibited. At 27 hpf, both the U26MOsp and U26MOpr morphants displayed an overall decreased body size with specific deformities in the head region, such as an indistinct midbrain–hindbrain boundary (mhb) and delayed pigmentation of the eyes (Figure 4A). At 5 days postfertilization (dpf), the morphants showed an abnormal jaw structure, pericardial edema, underdeveloped internal organs and malformed eyes and mouth (Figure 4B). These embryos died by 7 dpf. The embryos injected with a control MOs (U26misMOsp and U26misMOpr) did not display any of these phenotypes (Figure 4B and Supplementary Figure S4).
Figure 4.
Developmental defects in the snoRNA-deficient zebrafish. (A) A lateral view of wild-type embryos and morphants (left column), close-up images of the head region (middle column), and an overview of embryos (right column) at 27 hpf. Both the U26 and U44 morphants display deformities in the brain region and reduced eye pigmentation (middle column). The mhb is not clearly delineated in the U26 morphants (dotted circle). The U44 morphants display ventrally or laterally bent trunks (solid black triangle) and an incomplete yolk sac extension (solid line). Scale bars: 500 µm (left column), 200 µm (middle column). (B) Lateral (left column) and ventral (right column) views of wild-type embryos and U26MOsp morphants at 5 dpf. The U26 morphants display an underdeveloped jaw structure (solid line) and pericardial edema (arrow), as well as malformed eyes (black arrowhead) and mouth (asterisk). The internal organs, including the swim bladder (white arrowhead), were only observed in the wild-type and U26misMO-injected embryos. Scale bars: 200 µm.
Injection of MOsp and MOpr to inhibit U44 snoRNA resulted in severe hypoplasia of the brain and delayed pigmentation of the eyes (Figure 4A). In addition, these morphants showed an incomplete yolk sac extension and ventrally or laterally bent trunks. These embryos died by 7 dpf.
Similar effects on development were evident when expression of U78 snoRNA was inhibited with a splice MO. U78 snoRNA-deficient zebrafish displayed a decreased body size and an incomplete yolk sac extension. Interestingly, brain defects in the U78MOsp morphants were restricted to the hindbrain, and there were no obvious defects in any other regions (Supplementary Figure S5). These embryos died by 8 dpf.
Collectively, these results show that impaired rRNA modification owing to loss of snoRNA expression causes severe developmental defects and leads to embryonic lethality in zebrafish. Our data indicate that RNA modifications mediated by snoRNAs play a crucial role in vertebrate development. In addition, we observed snoRNA-dependent phenotypes, such as an indistinct mhb in the U26 morphants, characteristic bent trunks in the U44 morphants, or a hindbrain-specific malformation in the U78 morphants; these data suggest that site-specific rRNA modifications are important for specific organ development.
DISCUSSION
Over the past decade, functional analyses of RNA modifications in bacteria and lower eukaryotes have shown that nucleotide modifications are important for stabilization, maturation, turnover and localization of ncRNAs (5,35–37). However, similar studies in higher eukaryotes have been poorly described. In yeast, loss of rRNA modification at a single site in the ribosome-decoding center has no apparent effect on cell growth, but modification loss at multiple sites within this region affects cell growth and ribosome activity (13–15). In this study, we have demonstrated that the loss of rRNA modification, even at a single site, can have deleterious effects on early development in zebrafish.
Ribose methylation at the 2′-hydroxy group can be detected with a primer extension assay, where the extension stops at the methylated site depending on the dNTP concentration (38). However, it is difficult to quantify the frequency of methylation, especially for partial methylation, using this method because the signal intensity can vary at the target sites, depending upon th structural conformation of the RNA and dNTP calibration. In addition, this technique does not allow for absolute quantification of the modified nucleotide. On the other hand, direct analysis of RNA fragments with mass spectrometry allows for an accurate quantification of any type of modification with high precision and reproducibility (31,39). Using LC/MS analysis, we were able to detect the absence of methylation at position 398 in U26 snoRNA-deficient zebrafish.
According to the mass chromatogram (Figure 2A), the 11-mer fragment lacking Am398 constitutes ~20% of the total RNA fragments. This limited fraction of the ribosome was affected by U26MO treatment, which prevented methylation at position 398. Because the embryo also contains maternal ribosomes, a large part of the methylated fragment may have originated from the maternal pool. Thus, we hypothesize that the unmethylated position 398 in 28S rRNA was from de novo RNA synthesized during zygotic transcription, which indicates that the developing tissue in an embryo may contain high concentration of unmethylated ribosomes. Interestingly, we observed that the U26 snoRNA-deficient zebrafish displayed defective morphogenesis and embryonic lethality. The results indicate that partial loss of methylation may significantly interfere with the ribosomal activity, leading to severe developmental phenotypes. It is known that expression of mutant ribosomes carrying point mutations at specific residues in the rRNA in a wild-type background confers a dominant lethal phenotype in E. coli (40,41). Thus, methylation of adenosine at position 398 in 28S rRNA may conceivably play a crucial role in vertebrate development.
It can be argued that the loss of host gene expression, rather than the individual snoRNAs, may be responsible for the observed phenotypes in the snoRNA-deficient zebrafish. Although we have not confirmed whether depletion of u22hg and gas5 host genes has any effects on zebrafish development, we believe that the deformities in the brain and the other associated abnormalities are not an off-target effect of host gene depletion for several reasons. First, suppression of both U26 and U44 snoRNA expression by two different types of MOs (splice inhibitory and precursor binding) resulted in similar phenotypes. Second, properly spliced gas5 mRNA transcript was detected in both U44MOpr and U44misMOpr injected embryos (Supplementary Figure S6), but phenotypes were observed only in U44MOpr morphants, indicating that the loss of snoRNA expression, rather than the host gene defect, caused these phenotypes. Third, specific phenotypes were found that were associated with the type of snoRNA inhibited. For example, the mhb was deformed in the U26, but not the U44 morphants. Fourth, it is known that the human U26 snoRNA host gene U22HG mRNA is rapidly degraded most likely by nonsense-mediated mRNA decay (32), and a similar pathway may exist in zebrafish.
Defects in ribosome biogenesis have been linked to many human diseases called ribosomopathies, a rare collection of genetic disorders that are associated with increased cancer susceptibility (42,43). Diamond-Blackfan anemia (DBA) represents the first and the most extensively studied human disease caused by defects in ribosomal proteins (RPs) (44). RPS19 is most commonly mutated in DBA, although some patients show mutations in several other RP genes (45,46). In Treacher Collins syndrome, Shwachman-Diamond syndrome and X-linked dyskeratosis congenita (X-DC), mutations have been found in genes that are essential for rRNA processing and maturation (47). In X-DC, the mutated gene encodes dyskerin, a protein component of H/ACA snoRNP that catalyzes pseudouridylation of RNAs. Hypomorphic Dkc1 mutant mice (Dkc1m) recapitulate many clinical features of X-DC and display impaired rRNA modification (48). Because snoRNAs guide rRNA modification and because rRNA modifications appear to be associated with human disease, systematic studies of RNA modifications through the manipulation of snoRNA expression in vertebrate models are crucial for understanding the importance of ncRNAs in fundamental biological processes. The snoRNA-deficient zebrafish developed in this study may prove to be useful tools for such studies in the future.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology and the Japan Society for the Promotion of Science (20200070, 22370065, 22659186 and 22790989). Funding for open access charge: Ministry of Education, Culture, Sports, Science and Technology.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs Maki Yoshihama and Hidetsugu Torihara for technical support and useful discussions. The authors also thank Drs Hideyuki Yamamoto and Noriko Maeda (University of the Ryukyus, Japan) for their advice and suggestions.
REFERENCES
- 1.Limbach PA, Crain PF, McCloskey JA. Summary: the modified nucleosides of RNA. Nucleic Acids Res. 1994;22:2183–2196. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunin-Horkawicz S, Czerwoniec A, Gajda MJ, Feder M, Grosjean H, Bujnicki JM. MODOMICS: a database of RNA modification pathways. Nucleic Acids Res. 2006;34:D145–D149. doi: 10.1093/nar/gkj084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki T. Biosynthesis and function of tRNA wobble modifications. In: Grosjean H, editor. Fine-Tuning of RNA Functions by Modification and Editing. Berlin Heidelberg: Springer-Verlag; 2005. pp. 23–69. [Google Scholar]
- 4.Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′ end uridylation activity in Arabidopsis. Curr. Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ebhardt HA, Thi EP, Wang MB, Unrau PJ. Extensive 3′ modification of plant small RNAs is modulated by helper component-proteinase expression. Proc. Natl Acad. Sci. USA. 2005;102:13398–13403. doi: 10.1073/pnas.0506597102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 8.Kirino Y, Mourelatos Z. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat. Struct. Mol. Biol. 2007;14:347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- 9.Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K, Suzuki T. The 3′-termini of mouse piwi-interacting RNAs are 2′-O-methylated. Nat. Struct. Mol. Biol. 2007;14:349–350. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- 10.Saito K, Sakaguchi Y, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of PIWI-interacting RNAs at their 3′ ends. Genes Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 12.Maden BE. The numerous modified nucleotides in eukaryotic ribosomal RNA Prog. Nucleic Acid Res. Mol. Biol. 1990;39:241–303. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- 13.King TH, Liu B, McCully RR, Fournier MJ. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell. 2003;11:425–435. doi: 10.1016/s1097-2765(03)00040-6. [DOI] [PubMed] [Google Scholar]
- 14.Liang XH, Liu Q, Fournier MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell. 2007;28:965–977. doi: 10.1016/j.molcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 15.Liang XH, Liu Q, Fournier MJ. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA. 2009;15:1716–1728. doi: 10.1261/rna.1724409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;3:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 17.Balakin AG, Smith L, Fournier MJ. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 18.Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu. Rev. Biochem. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 19.Lowe TM, Eddy SR. A computational screen for methylation guide snoRNAs in yeast. Science. 1999;283:1168–1171. doi: 10.1126/science.283.5405.1168. [DOI] [PubMed] [Google Scholar]
- 20.Hüttenhofer A, Kiefmann M, Meier-Ewert S, O'Brien J, Lehrach H, Bachellerie JP, Brosius J. RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J. 2001;20:2943–2953. doi: 10.1093/emboj/20.11.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertrand E, Fournier MJ. The snoRNPs and related machines: ancient devices that mediate maturation of rRNA and other RNAs. In: Olson MOJ, editor. The Nucleolus. Georgetown, TX: Landes Bioscience Publishing; 2004. pp. 223–257. [Google Scholar]
- 22.Cavaillé J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, Bachellerie JP, Brosius J, Hüttenhofer A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl Acad. Sci. USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inshore S, Stem S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 24.Sahoo T, del Gaudio D, German JR, Shinawi M, Peters SU, Person RE, Garnica A, Cheung SW, Beaudet AL. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat. Genet. 2008;40:719–721. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka R, Satoh H, Moriyama M, Satoh K, Morishita Y, Yoshida S, Watanabe T, Nakamura Y, Mori S. Intronic U50 small-nucleolar-RNA (snoRNA) host gene of no protein-coding potential is mapped at the chromosome breakpoint t(3;6)(q27;q15) of human B-cell lymphoma. Genes Cells. 2000;5:277–287. doi: 10.1046/j.1365-2443.2000.00325.x. [DOI] [PubMed] [Google Scholar]
- 26.Dong XY, Rodriguez C, Guo P, Sun X, Talbot JT, Zhou W, Petros J, Li Q, Vessella RL, Kibel AS, et al. SnoRNA U50 is a candidate tumor suppressor gene at 6q14.3 with a mutation associated with clinically significant prostate cancer. Hum. Mol. Genet. 2008;17:1031–1042. doi: 10.1093/hmg/ddm375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong XY, Guo P, Boyd J, Sun X, Li Q, Zhou W, Dong JT. Implication of snoRNA U50 in human breast cancer. J. Genet. Genomics. 2009;36:447–454. doi: 10.1016/S1673-8527(08)60134-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao J, Yu L, Mei Y, Guarnera M, Shen J, Li R, Liu Z, Jiang F. Small nucleolar RNA signatures as biomarkers for non-small-cell lung cancer. Mol. Cancer. 2010;9:198. doi: 10.1186/1476-4598-9-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uechi T, Nakajima Y, Nakao A, Torihara H, Chakraborty A, Inoue K, Kenmochi N. Ribosomal protein gene knockdown causes developmental defects in zebrafish. PLoS ONE. 2006;1:e37. doi: 10.1371/journal.pone.0000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chakraborty A, Uechi T, Higa S, Torihara H, Kenmochi N. Loss of ribosomal protein L11 affects zebrafish embryonic development through a p53-dependent apoptotic response. PLoS ONE. 2009;4:e4152. doi: 10.1371/journal.pone.0004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimura S, Suzuki T. Fine-tuning of the ribosomal decoding center by conserved methyl-modifications in the Escherichia coli 16S rRNA. Nucleic Acids Res. 2010;38:1341–1352. doi: 10.1093/nar/gkp1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tycowski KT, Shu MD, Steitz JA. A mammalian gene with introns instead of exons generating stable RNA products. Nature. 1996;379:464–466. doi: 10.1038/379464a0. [DOI] [PubMed] [Google Scholar]
- 33.Smith CM, Seitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol. Cell. Biol. 1998;18:6897–6909. doi: 10.1128/mcb.18.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lustrate L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006;34:D158–D162. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaneko T, Suzuki T, Kapushoc ST, Rubio MA, Ghazvini J, Watanabe K, Simpson L, Suzuki T. Wobble modification differences and subcellular localization of tRNAs in Leishmania tarentolae: implication for tRNA sorting mechanism. EMBO J. 2003;22:657–667. doi: 10.1093/emboj/cdg066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321:1490–1492. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49:341–351. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 38.Motorin Y, Muller S, Behm-Ansmant I, Branlant C. Identification of modified residues in RNAs by reverse transcription-based methods. Methods Enzymol. 2007;425:21–53. doi: 10.1016/S0076-6879(07)25002-5. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki T, Ikeuchi Y, Noma A, Suzuki T, Sakaguchi Y. Mass spectrometric identification and characterization of RNA-modifying enzymes. Methods Enzymol. 2007;425:211–229. doi: 10.1016/S0076-6879(07)25009-8. [DOI] [PubMed] [Google Scholar]
- 40.Powers T, Noller HF. Dominant lethal mutations in a conserved loop in 16S rRNA. Proc. Natl Acad. Sci. USA. 1990;87:1042–1046. doi: 10.1073/pnas.87.3.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson J, Kim DF, O'Connor M, Lieberman KR, Bayfield MA, Gregory ST, Green R, Noller HF, Dahlberg AE. Analysis of mutations at residues A2451 and G2447 of 23S rRNA in the peptidyltransferase active site of the 50S ribosomal subunit. Proc. Natl Acad. Sci. USA. 2001;98:9002–9007. doi: 10.1073/pnas.151257098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chakraborty A, Uechi T, Kenmochi N. Guarding the ‘translation apparatus’: defective ribosome biogenesis and the p53 signaling pathway. WIREs RNA. 2011;2:507–522. doi: 10.1002/wrna.73. [DOI] [PubMed] [Google Scholar]
- 43.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat. Genet. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- 45.Lipton JM, Ellis SR. Diamond Blackfan anemia 2008-2009: broadening the scope of ribosome biogenesis disorders. Curr. Opin. Pediatr. 2010;22:12–19. doi: 10.1097/MOP.0b013e328334573b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flygare J, Aspesi A, Bailey JC, Miyake K, Caffrey JM, Karlsson S, Ellis SR. Human RPS19, the gene mutated in Diamond-Blackfan anemia, encodes a ribosomal protein required for the maturation of 40S ribosomal subunits. Blood. 2007;109:980–986. doi: 10.1182/blood-2006-07-038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freed EF, Bleichert F, Dutca LM, Baserga SJ. When ribosomes go bad: diseases of ribosome biogenesis. Mol. Bio. Syst. 2010;6:481–493. doi: 10.1039/b919670f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruggero D, Grisendi S, Piazza F, Rego E, Mari F, Rao PH, Cordon-Cardo C, Pandolfi PP. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science. 2003;299:259–262. doi: 10.1126/science.1079447. [DOI] [PubMed] [Google Scholar]
- 49.McLuckey SA, Vanberkel GJ, Glish GL. Tandem Mass-Spectrometry of Small Multiply Charged Oligonucleotides. J. Am. Soc. Mass Spectrom. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.