Abstract
Double-stranded RNA (dsRNA) can enter different pathways in mammalian cells, including sequence-specific RNA interference (RNAi), sequence-independent interferon (IFN) response and editing by adenosine deaminases. To study the routing of dsRNA to these pathways in vivo, we used transgenic mice ubiquitously expressing from a strong promoter, an mRNA with a long hairpin in its 3′-UTR. The expressed dsRNA neither caused any developmental defects nor activated the IFN response, which was inducible only at high expression levels in cultured cells. The dsRNA was poorly processed into siRNAs in somatic cells, whereas, robust RNAi effects were found in oocytes, suggesting that somatic cells lack some factor(s) facilitating siRNA biogenesis. Expressed dsRNA did not cause transcriptional silencing in trans. Analysis of RNA editing revealed that a small fraction of long dsRNA is edited. RNA editing neither prevented the cytoplasmic localization nor processing into siRNAs. Thus, a long dsRNA structure is well tolerated in mammalian cells and is mainly causing a robust RNAi response in oocytes.
INTRODUCTION
Double-stranded RNA (dsRNA), a double helix formed by two antiparallel RNA strands, is a unique structure whose recognition is important in host defense and regulation of gene expression. The recognition and effects of dsRNA are mediated by a diverse set of proteins harboring dsRNA binding domains (dsRBD), [reviewed in Ref. (1)].
One of the evolutionarily conserved effects of dsRNA is represented by RNA interference (RNAi), sequence-specific degradation of RNAs complementary to the sequence of the dsRNA [reviewed in Ref, e.g. (2,3)]. RNAi is initiated by the RNase III enzyme Dicer, which is cleaving dsRNA at ~21 nt intervals, generating short interfering RNA (siRNA) duplexes with two nucleotide 3′ overhangs. One of the siRNA strands is loaded onto the RNA-induced silencing complex (RISC), where it serves as a guide for cleaving perfectly complementary mRNAs. In mammalian cells, RNAi effects can also be induced experimentally by siRNAs (4) or by microRNAs (5). Endogenous microRNAs (miRNAs) are genome-encoded small RNAs produced by cleavage of pre-processed short hairpin precursors by Dicer and are also loaded on a RISC-like complex [reviewed in Ref. (6)]. For the purpose of this study, we use the term RNAi for the pathway that is induced by long dsRNA, i.e. as it was originally described by Fire et al. (7). RNAi operates in most eukaryotes and often serves as a defense mechanism against viruses and repetitive sequences. Hence, it is often viewed as a form of innate immunity (8). However, RNAi in mammals does not appear to play an antiviral role (9) and its endogenous function has only been clearly documented for oocytes and embryonic stem cells where it targets repetitive elements and regulates endogenous genes [reviewed in Ref (10)]. Nevertheless, experimental induction of RNAi with ectopically expressed long dsRNA in different somatic cells (11–14) suggests capacity for siRNA generation from dsRNA also in other cell types.
Mammalian somatic cells can respond to dsRNA in a sequence-independent manner. A pioneering work by Hunter et al. (15) showed that different types of dsRNA, including the poly I:C duplex, can block translation in reticulocyte lysates. Analysis of this phenomenon identified protein kinase R (PKR), an enzyme activated upon binding to dsRNA that blocks translation by phosphorylating the α-subunit of eukaryotic initiation factor 2 (eIF-2α) (16). Activation of PKR represents part of a complex response to foreign molecules known as the interferon (IFN) response [reviewed in Ref (17)], which includes activation of the NFκB transcription factor and a large number of IFN-stimulated genes (ISGs) (18). In addition to PKR, several other proteins recognizing dsRNA are integrated into the IFN response. DDX58 (RIG-I) and MDA5 sense cytoplasmic dsRNA and activate IFN expression. The 2′,5′-oligoadenylate synthetase produces 2′,5′-linked oligoadenylates that induce a general degradation of RNAs by activating latent RNase L [reviewed in Ref, e.g. (19,20)].
Another dsRNA-associated mammalian pathway is represented by RNA editing, which is mediated by adenosine deaminases acting on RNA (ADARs). These dsRNA recognizing enzymes convert adenosines to inosines, thus affect stability and coding potential of modified RNAs [reviewed in Ref (21,22)]. The effects of dsRNA editing are complex and the degree of editing may affect the subsequent fate of the edited RNA, leading to its nuclear retention (23) or degradation (24). Nevertheless, mRNAs carrying edited dsRNA hairpins in their 3′-UTR can be transported to the cytoplasm and translated, as evidenced by the presence of such mRNAs on polysomes (25). Editing of long dsRNA in Caenorhabditis elegans antagonizes the transgene-induced RNAi in somatic cells by retaining edited dsRNA in the nucleus (26). However, the reported role of the cytoplasmic RISC component Tudor-SN in the degradation of hyperedited RNAs (>50% conversion) indicates more complex interplay between editing and RNAi (24). Mammalian ADARs can reduce efficiency of RNAi in two ways. First, they can erode dsRNA to the point where it is either no longer a suitable substrate for Dicer processing or, in the case of successful Dicer cleavage, the resulting siRNAs have changed specificity to base pair with target mRNAs. Second, they can reduce availability of siRNAs by directly binding to them, an effect that seems to be independent of ADAR editing activity (27,28).
While the three main mammalian dsRNA-responding pathways mentioned above have been individually characterized in substantial detail, the interactions between them are still poorly understood. Co-existence of these pathways certainly involves recognition of different types of dsRNA substrates and their possible sequestration in different cellular compartments or cell types [reviewed in Ref (19,29)]. The latter phenomenon underlies the common simplistic view that cytoplasmic dsRNA is toxic to somatic cells because it activates the IFN response, while nuclear dsRNA is edited and thus prevented to enter the cytoplasm. However, such interpretation is challenged by the growing list of reports showing induction of RNAi by intracellular expression of long dsRNA in transformed and primary somatic cells (4,11–13,30,31).
To obtain new insights into the effects of dsRNA in various types of somatic cells, we produced a transgenic mouse model ubiquitously expressing long dsRNA. We have previously developed a transgene that generates dsRNA within the 3′-UTR of a protein-coding transcript. This dsRNA takes the form of a long hairpin with a perfect ~0.5 kb stem, which is flanked by long single-stranded 5′ and 3′ overhangs. Using a transgene with the Mos gene sequence in the hairpin and oocyte-specific ZP3 promoter, we induced an efficient and highly specific RNAi effect in mouse oocytes (32,33). Physiologically, the Mos gene encodes for a dormant maternal mRNA, which is stored in the oocyte until the resumption of meiosis (34). Elimination of the Mos maternal mRNA by transgenic RNAi phenocopies the null mutation (32), which manifests as parthenogenetic activation of ovulated eggs and ovarian cysts. Otherwise, Mos−/− animals appear normal (35,36).
Here, we report an adaptation of the Mos hairpin transgene (for simplicity referred to as MosIR) for ubiquitous, constitutive expression of dsRNA in transgenic mice. We show that in somatic cells of transgenic animals, dsRNA does not induce the IFN response, is inefficiently processed by Dicer and its editing is barely detectable. This suggests that a long dsRNA structure embedded in a transcript produced by RNA polymerase II in the nucleus of somatic cells is not a potent trigger of any of the three common pathways responding to dsRNA. When MosIR RNA levels were increased in cell culture experiments, we observed more frequent editing while IFN pathway activation and RNAi effects were still negligible. The IFN response was induced only with high levels of expressed dsRNA in somatic cells. In contrast to somatic cells, the MosIR induced a robust RNAi effect in oocytes suggesting that female germ cells represent a tissue adapted to directing dsRNA into the RNAi pathway.
MATERIALS AND METHODS
Plasmids and transgenes
Schematic structures of the relevant parts of plasmid constructs used in the project are shown in the Supplementary Figure S1. pCAGEGFP-MosIR (for simplicity, referred to as MosIR) was produced by transferring the EagI fragment carrying the Mos inverted repeat inserted in the pCR II plasmid (37) into the SspBI site downstream of the enhanced green fluorescent protein (EGFP) coding sequence in the pCAGEGFP plasmid (38). pCAGEGFP-Mos3 (for simplicity, referred to as Mos3) was produced by inserting a PCR-amplified 973 bp fragment of the Mos transcript (corresponding to nucleotides 114–1089 of the Mos cDNA sequence NM020021) into the SspBI site downstream of the EGFP coding sequence in the pCAGEGFP plasmid. pCAGEGFP-MosP (for simplicity, referred to as MosP) was produced by inserting the same PCR-amplified 973 bp fragment of the Mos transcript into the SnaBI site between the cytomegalovirus (CMV) enhancer and β-actin promoter. Insertion into the pCAGEGFP was verified by restriction digest and sequencing. The Mos sequence in Mos3 and MosP fragments was inserted in the sense orientation relative to the EGFP transcription and a KpnI site in the Mos was eliminated by blunt-ending and re-ligation, allowing for distinguishable reporter sequences from the endogenous Mos sequences. EGFP expression from MosP and Mos3 plasmids were compared with the original EGFP plasmid by flow cytometry (FACS) analysis of HEK293 cells (for simplicity, referred to as 293) transfected with equimolar amounts of each plasmid. This analysis showed that, at similar transfection efficiency, Mos3 and MosP EGFP expression reached ~25 and 80% of pCAGEGFP, respectively (data not shown). The lower Mos3 EGFP expression was probably caused by lower stability or reduced translatability of the EGFP mRNA that was expanded in its 3′-UTR by an additional 1 kb of sequence. For each MosIR, Mos3 and MosP plasmid, SalI and HindIII were used to release the transgenic cassette for producing transgenic animals. phRL-TK-Mos3 (for simplicity, referred to as RL-Mos3) production was described previously (39). Firefly luciferase (for simplicity, referred to as FF) served as a non-targeted control.
Transgenic mice
Transgenic mice were generated at the transgenic facility of the Friedrich Miescher Institute by injecting linearized DNA into male pronuclei of C57BL/6 × BALB/c 1-cell embryos. Transgene-positive mice were identified by PCR (primer sequences are listed in the Supplementary Table SIII). For the MosIR transgene, four different founder animals (one female and three males) were obtained and examined further. One female founder animal was sterile and showed the Mos null phenotype [parthenogenetic activation of unfertilized oocytes (35,36)]. F1 progeny of founder males was examined for the expression of EGFP and down-regulation of Mos mRNA in oocytes. The line with the strongest Mos null phenotype in female animals was expanded and used for further analysis. For Mos3 and MosP reporter lines, founder animals were crossed with C57BL/6 × BALB/c animals and the line with the strongest EGFP expression for each reporter was used further.
Oocyte isolation and culture
Adult transgenic or wild-type female siblings were superovulated using 5 U of pregnant mare serum gonadotropin (PMSG, Intervet). Oocytes were isolated as described previously (40). Images of oocytes and organ samples were obtained using a digital camera mounted to a stereomicroscope SZX16 equipped with a 100 W mercury lamp and GFPA filter (Olympus).
Cell culture and transfection
Human 293 cells were maintained in Dulbecco's Modified Eagle Medium (DMEM, Invitrogen) supplemented with 10% FCS (Invitrogen), penicillin (100 U/ml, Invitrogen) and streptomycin (100 µg/ml, Invitrogen) at 37°C and 5% CO2 atmosphere.
For transfection, cells were typically plated on a 24-well plate, grown to 70% density and transfected using Turbofect in vitro Tranfection Reagent (Fermentas) according to the manufacturer's protocol. Cells were transfected with 100 ng of both untargeted (FF) and targeted (RL-Mos3) reporter plasmids and various amount of MosIR plasmid (50–500 ng per well). The total amount of transfected DNA was kept constant by adding pCAGEGFP. After 48 h, cells were washed with PBS and lysed with the Passive Lysis Buffer (Promega). Luciferase reporter expression was assessed using the Dual-Luciferase Reporter Assay (Promega) and luminiscence intensity was measured by Modulus Microplate Multimode Reader (Turner Biosystems).
The 293 cells stably expressing both RL-Mos3 and FF were established by co-transfecting reporter plasmids with the pPuro selection plasmid carrying puromycin resistance (500 ng of each plasmid per well in a 6-well plate). To select positive clones, cells were passaged in the presence of 1.5 µg/ml of puromycin. The positively selected individual clones were tested for Renilla and firefly luciferase expression using the Dual-Luciferase Reporter Assay. Clones yielding luciferase expression similar to that of the transiently transfected cells were used for further studies.
Isolation of mouse embryonic fibroblasts
Mouse embryos were isolated at embryonic day 12.5. The head and internal organs were removed, the torso was homogenized using a needle under sterile conditions and subsequently cultured in DMEM supplemented with 10% FCS, penicillin (100 U/ml, Invitrogen), and streptomycin (100 µg/ml, Invitrogen) at 37°C and 5% CO2 atmosphere. Mouse embryonic fibroblasts (MEFs) within the first three passages were used for experiments.
Detection of RNase T1-resistant MosIR RNA
The fraction of MosIR RNA, which folded into dsRNA, was approximated from the comparison of non-treated RNA (input) and RNase T1-treated (T1) samples. MosIR-positive MEFs or 293 cells transfected with 2 µg of MosIR per well in a 6-well plate were harvested 48 h post-transfection. The cells were washed twice with PBS, liberated with a rubber scraper into a 1.5 ml tube, and hypotonically lysed with three volumes of water [experiments where cells were permeabilized with 0.1% Triton X-100 yielded the same results (data not shown)]. Cell lysates were incubated with RNase T1 (5000 U) for 30 min at 37°C. Negative-control samples were denaturated at 85°C for 5 min and immediately placed on ice to prevent perfect pairing of complementary strands prior to addition of RNase T1. RNase T1 was inactivated by phenol–chloroform extraction using the RNA Blue reagent (Top Bio). DNA was removed by Turbo DNase (Ambion) treatment. All RNA samples were produced in the same volume (30 µl). If needed, concentration of RNA recovered after treatment was equalized using Escherichia coli rRNA (Roche). From each sample, 1 µg of RNA was then reverse-transcribed using RevertAid M-MuLV Reverse Transcriptase (Fermentas) and a MosIR-specific primer or using Superscript II (Invitrogen) and random hexamer primers with similar results. Reverse transcriptase was omitted in ‘–RT’ samples. cDNA was amplified using Maxima SYBR Green/ROX qPCR Master Mix (Fermentas) and PCR products were resolved on 1.5% agarose gel stained with ethidium bromide (Sigma Aldrich).
Microarray analysis
The 293 and HeLa cell lines were analyzed in duplicates 24 h after transfection with 500 ng/well of the MosIR plasmid in a 6-well plate. Controls were transfected with equal amounts of the parental pCAGEGFP plasmid. Microarray analysis of 5 µg of total RNA per sample was performed as described previously (41). Principal component analysis and hierarchical clustering of the GeneChip Robust Multiarray Averaging (GC-RMA) normalized data were performed by Partek software (Partek Inc., USA). The bioconductor Limma package (42) was used to identify differentially expressed genes and generate MA plots. The heatmap for IFN-related gene expression was created using TM4 microarray software suite (43). Microarray data were deposited in the NCBI GEO database (GSE27316).
Next generation sequencing
Brain and kidney tissues were obtained from a freshly sacrificed 17-week old transgenic female. HeLa and 293 cells were transfected with 500 ng/well of MosIR plasmid in a 6-well plate and cells were collected 48 h post-transfection. Small RNA fractions were isolated using mirPremier microRNA Isolation Kit (Sigma) according to the manufacturer's instructions. RNA concentration was determined using a Nanodrop 1000 spectrophotometer, and the quality of RNA was checked by 8% polyacrylamide gel electrophoresis.
Library construction and deep sequencing of small RNAs were performed by Seqomics (Szeged, Hungary) using SOLiD (version 3.0) sequencing platform. For the bioinformatic analysis, the bar code and 3′ adaptor sequences were removed from raw sequence reads using an algorithm requiring at least 3-nt exact matches between 35-nt reads and the adaptor sequence. Sequencing quality and depth of all samples was comparable (Supplementary Figure S3, the complete Seqomics Sequencing Report is available on request). Reads were mapped onto the following sequences: miRNA [MirBase release 15; http://www.mirbase.org/, (44)], tRNA [Genomic tRNA database; http://lowelab.ucsc.edu/GtRNAdb/, (45)], rRNA [http://www.arb-silva.de/, (46)] and MosIR plasmid. Only perfect matches were considered for the first round of analysis. The annotation order was miRNA, tRNA, rRNA and plasmid sequence. To avoid mapping of some reads to multiple categories, we removed each read from the dataset once it was annotated to a particular sequence category. The remaining reads not matching any RNA sequences were marked as ‘other’. For RNA (A > I) editing analysis, ‘other’ reads were remapped to the above mentioned selected sequences allowing for one or more A/G mismatches. All mapping software used in the analysis were programmed using the Visual Basic 2010 platform (Microsoft). Lists of generated 35-nt reads in color-coded format were deposited in the GEO database (GSE26577).
RNA isolation and quantitative real-time RT–PCR
Oocytes for real-time PCR analysis were washed 3-times in PBS, placed individually in pure water with 10 U of RiboLock RNase inhibitor (Fermentas) while 0.14 pg/µl of rabbit globin mRNA (Sigma Aldrich) and 0.2 µg/µl of E. coli rRNA (Roche) was added to act as an external standard and to inhibit sample RNA degradation by RNases. Samples were stored at −80°C prior to reverse transcription. Total RNA from organs and cultured cells was isolated by phenol–chloroform extraction using the RNA Blue reagent (Top-Bio) according to the manufacturer's instructions. Expression of specific mRNA was analyzed by quantitative real-time PCR (qPCR) as described previously (39). Briefly, 1 µg of total RNA was reverse transcribed using RevertAid M-MuLV reverse transcriptase (Fermentas) and random hexanucleotides (Fermentas). Reverse transcriptase was omitted in control (−RT) samples. The resulting cDNA was diluted three times with water and a 3 µl aliquot was used as a template for a 10 µl qPCR reaction. qPCR was performed on the Mx3000P (Stratagene) or LC480 (Roche) machines using Maxima SYBR Green qPCR Master Mix (Fermentas). Hypoxanthine–guanine phosphoribosyltransferase (Hprt1) was used as an internal housekeeping gene control (all primers are listed in the Supplementary Table SIII). Values of crossing points (CPs) were evaluated and corrected according to PCR efficiency for each reaction. The statistical significance of relative expression changes of target mRNA levels normalized to a housekeeping gene was analyzed by the pair-wise fixed reallocation randomization test using the REST 2008 software (47).
RESULTS AND DISCUSSION
Transgenic mice ubiquitously expressing long dsRNA are viable
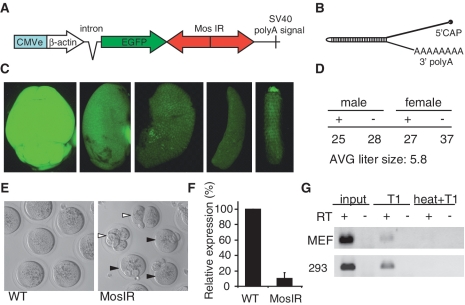
To achieve ubiquitous long dsRNA expression in the mouse, we employed the same general design (Figure 1A) that was used for transgenic RNAi in mouse oocytes (32). To control dsRNA expression, we chose a strong chimeric promoter composed of the CMV enhancer and β-actin core promoter (CAG) that provided efficient, ubiquitous expression in transgenic mice (48). The MosIR transgene produced an mRNA that carried an EGFP coding sequence and 3′-UTR containing a long dsRNA hairpin of the Mos sequence (Figure 1B).
Figure 1.
Analysis of MosIR expression and phenotype. (A) Schematic composition of the MosIR transgene. (B) A schematic structure of the MosIR transcript folded into a long dsRNA hairpin. The length of the stem is 520 bp. (C) MosIR transgene produces EGFP in somatic tissues. From left to right: brain, kidney, liver, spleen, tail. All images were taken with the same settings. No autofluorescence was visible under these conditions in internal organs of wild-type mice. (D) MosIR transgene (+) shows normal segregation into male and female progeny in crosses of MosIR males and wild-type female mice. (E) MosIR positive females phenocopy the Mos null phenotype: parthenogenetic activation of ovulated eggs. The left panel shows normal metaphase II-arrested eggs. The right panel shows transgenic parthenogenotes with two extruded polar bodies (black arrowheads) and cleaving parthenogenotes (white arrowheads). (F) qPCR of Mos mRNA in fully-grown oocytes obtained from three transgenic animals reveals strong reduction of Mos mRNA level compared to wild-type oocytes. Error bars = SEM. (G) RNase T1-resistant MosIR RNA can be detected in MEFs isolated from MosIR embryos and in 293 cells transfected with the MosIR plasmid. Hypotonically lysed cells were treated with RNase T1 for 30 min at 37°C before RNA extraction (T1). The efficiency of RNase T1 digestion was tested by disrupting secondary RNA structures by heat before RNase T1 digestion (heat+T1). RT indicates reverse-transcribed RNA (+) and controls without reverse transcriptase (−). cDNA was amplified with MosIR-specific primers for 31 cycles (MEF) and 30 cycles (293) of PCR. No amplification was observed with primers located in the single-stranded EGFP coding sequence (data not shown).
The MosIR transgene offered several advantages. First, the EGFP coding sequence upstream of the Mos hairpin yields a sufficient amount of green fluorescence to monitor transgene expression. Second, this RNA efficiently folds into dsRNA upon transcription in vitro (37) and it induces an efficient and highly specific RNAi effect in the oocyte (32,33). Third, the Mos null phenotype manifests as a parthenogenetic activation of unfertilized eggs (35,36), providing an easily assayed positive control for RNAi effects. Fourth, the loss of Mos has no phenotype in somatic cells (35,36) and Mos mRNA levels are negligible in somatic cells (49). Therefore, while RNAi effects can be conveniently monitored with an appropriate reporter target, the MosIR is also suitable for studying sequence-independent effects of dsRNA expression in either the presence or absence of the target mRNA.
We obtained four MosIR transgenic lines upon pro-nuclear injection of the MosIR transgene. All of them showed green fluorescence in all tissues examined, documenting that the transgene was ubiquitously expressed (Figure 1C; data not shown). The line derived from the male founder 317.3 showed the strongest Mos phenotype in the female progeny and was selected for detailed analysis of effects caused by MosIR expression. Transgenic females had reduced fertility. Mating of five different females for several weeks resulted in only one litter of two animals. This was an expected consequence of parthenogenetic activation caused by RNAi-mediated down-regulation of Mos mRNA in oocytes (Figure 1E and F). This effect indirectly demonstrated formation of MosIR-derived dsRNA in oocytes and was identical to the results obtained with the oocyte-specific version of the MosIR transgene (32,33).
Transmission of the MosIR transgene via the male germline showed normal Mendelian distribution and an average litter size of 5.8 (Figure 1D), which did not significantly differ from litters of this strain in our mouse facility. This indicated that the MosIR did not cause embryonic lethality. Apart from the reduced female fertility, transgenic mice showed no other apparent phenotype suggesting the MosIR transcript has no effect on cell growth or viability. At the same time, observable green fluorescence in different organs indicated that the MosIR expression was ubiquitous. The amount of EGFP fluorescence varied across different organs (Figure 1C). The brightest signal was found in the brain and testis and the lowest in the liver and spleen. A similar pattern of EGFP signal was also observed in organs of transgenic mice with other EGFP-carrying transgenes controlled by the same promoter but devoid of the inverted repeat (38) (Figure 2B). Notably, MosIR transgene generated less EGFP than other transgenes driven by the CAG promoter (Supplementary Figure S1C). Transfection of equal amounts of pCAGEGFP and MosIR plasmids into 293 cells showed discrepancy between mRNA and EGFP fluorescence in MosIR transfected cells (Supplementary Figure S2A and B) and thus, suggested that lower EGFP fluorescence in MosIR mice was caused by lower translatability of the MosIR transcript. The MosIR transcript was readily detectable by qPCR in somatic tissues and its relative abundance correlated with relative amounts of EGFP fluorescence among tissues (Supplementary Figure S2C).
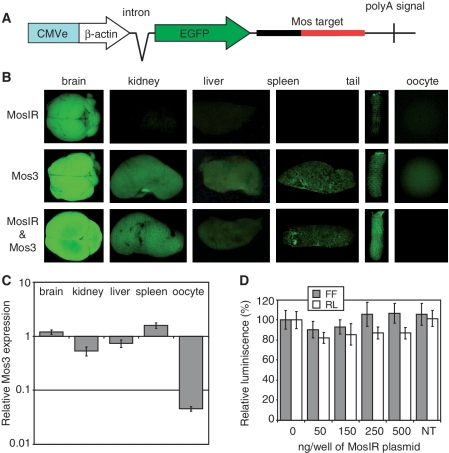
Figure 2.
Absence of RNAi in somatic cells. (A) A schematic composition of the Mos3 reporter transgene. (B) EGFP signal in different organs of MosIR, Mos3 and Mos3 & MosIR mice. Organs were collected from siblings of one litter. All images were taken with the same settings. The exposure of was shorter than in Figure 1C in order to obtain non-saturated brain EGFP signal. (C) qPCR analysis of different tissues shows little, if any, RNAi in somatic tissues and a strong RNAi effect in oocytes. Mos3 reporter expression in Mos3 & MosIR mice is shown relative to its expression in Mos3 mice. Samples were collected from two Mos3 and two Mos3 & MosIR animals and analyzed by qPCR in triplicates. Hprt was used as an internal normalization standard. Tissue heterogeneity could contribute to the lower level of Mos3 in the kidney. Error bars = SEM. (D) MosIR does not induce RNAi when transfected to somatic cells. The 293 cells stably transfected with a non-targeted firefly luciferase (FF) reporter and a targeted Renilla luciferase (RL) reporter (carrying Mos sequence in its 3′-UTR, Supplementary Figure S1B) were grown in 24-well plates and transfected with increasing amounts of the MosIR plasmid. pCAGEGFP was added to transfection mixtures to balance different amounts of the MosIR plasmid and to maintain the total amount of transfected DNA constant. Cells were harvested 48 h after transfection and luciferase activity was analyzed. Both luciferase activities are shown relative to cells transfected with 0 ng of the MosIR plasmid. NT = non-transfected cells. Transfection efficiency estimated by microscopic examination of EGFP fluorescence was >90%. Error bars = SEM.
Although the Mos null phenotype indicated expression of dsRNA in the female germline, further evidence that the MosIR transcripts formed dsRNA in vivo in somatic cells was necessary. To assess formation of MosIR dsRNA in intact cells, we used RNase T1 to degrade single-stranded RNA prior to RNA extraction from MEFs derived from the MosIR mice since the extraction can cause formation of dsRNA that does not exist in vivo (50). RNase T1-resistant MosIR RNA was analyzed by qPCR. We successfully amplified the Mos fragment from RNase T1-treated samples but not from samples where secondary RNA structure was disrupted by heat before RNase T1 digestion (Figure 1G). These results provide qualitative evidence for the presence of MosIR dsRNA in intact cells. It remains unclear whether the low yield RNase T1-resistant RNA (~5–10% of the input when estimated by qPCR) is caused by instability of dsRNA during T1 treatment or whether it reflects reduced formation of dsRNA from the MosIR transcript. The latter would suggest that somatic cells actively reduce levels of dsRNA originating from endogenous transcripts, either at the level of RNA binding proteins, which prevent dsRNA formation and/or by a duplex unwinding activity. In any case, our data show that the MosIR transgene is ubiquitously transcribed, triggers RNAi in oocytes and generates dsRNA in MEFs derived from transgenic mice. This conclusion is also supported by evidence provided by experiments presented further below, which show that the MosIR RNA is edited and processed into siRNAs in somatic cells.
MosIR transcript efficiently enters the RNAi pathway only in oocytes
The MosIR transcript efficiently entered the RNAi pathway in oocytes as evidenced by the reduction of Mos mRNA and presence of the Mos null phenotype in MosIR mice (Figure 1E and F). As Mos is not expressed in somatic tissues (49), we produced a transgenic EGFP reporter carrying the Mos sequence in the 3′-UTR (Figure 2A), allowing us to monitor RNAi effects in mouse tissues by microscopy. The Mos3 transgene produced bright EGFP fluorescence, which was stronger than that of the MosIR (Figure 2B), allowing for rapid identification of RNAi effects. In any case, the fact that both transgenes produced EGFP was not an obstacle for studying RNAi effects since the expression of MosIR and Mos3 transgenes could be distinguished by qPCR.
With the exception of oocytes, where EGFP fluorescence was reduced, examination of EGFP fluorescence in whole organs or in organ cryosections did not reveal any obvious RNAi effect when samples from mice carrying both MosIR and Mos3 transgenes were compared to samples from mice carrying the Mos3 transgene alone (Figure 2B; data not shown). Likewise, analysis of the Mos3 transcript level by qPCR did not suggest the occurrence of robust somatic RNAi (Figure 2C). Although we did observe relative reduction of the Mos3 RNA in kidney and liver samples, this was much less profound compared with oocytes and may have been a result of tissue heterogeneity.
To further address the potential of the MosIR dsRNA to induce RNAi in somatic cells, we performed experiments in cultured cells. 293 cells carrying stably integrated non-targeted FF and targeted Renilla luciferase (RL-Mos3) reporters were transfected with increasing doses of the MosIR transgene that lead to proportionally increasing levels of MosIR RNA (Supplementary Figure S2A). However, even in cells with the highest expression of MosIR, no statistically significant RNAi effect was found (Figure 2D). Furthermore, we did not observe significant RNAi effects upon transient transfection of the MosIR transgene and both luciferase reporters into 293, HeLa, or MCF-7 cells (data not shown).
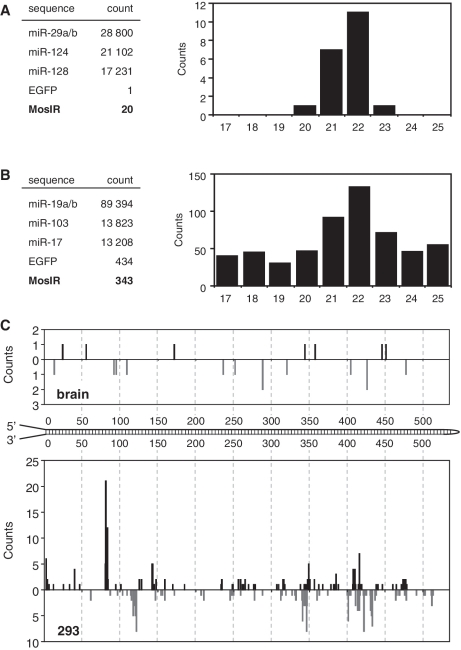
To directly address whether the MosIR RNA enters the RNAi pathway, we analyzed kidney and brain for the presence of MosIR-derived siRNAs using high throughput sequencing by the SOLiD technology (sequencing depth ~4 × 106 reads). Kidney and brain samples were selected because both tissues showed high MosIR expression and kidney samples also showed somewhat reduced levels of the Mos3 reporter. SOLiD sequencing of tissue samples produced a distinct peak of 21–23 nt small RNAs, the size expected for Dicer products (Supplementary Figure S3A). The number of putative Mos siRNAs was minimal (20 in the brain and 4 in the kidney). This contrasted with hundreds of thousands of potential Dicer cleavage products, including ~70 000 sequences derived from the three most abundant miRNAs (Figure 3A). Notably, only one putative siRNA sequence derived from the single-stranded EGFP region of the MosIR transcript was found in each organ sample. Since virtually all Mos sequences in the brain and kidney samples were in the range of Dicer products (Figure 3A), we concluded that a small fraction of MosIR RNA was processed by Dicer into siRNAs in somatic tissues but their abundance was too low to cause a robust RNAi effect.
Figure 3.
Deep sequencing of MosIR-derived small RNAs. (A) Deep sequencing of small RNAs from brain using SOLiD technology. Absolute counts of the most abundant miRNAs and 20–23 nt RNAs derived from the EGFP coding sequence (EGFP) and from the Mos inverted repeat (MosIR) are shown on the left. The graph shows distribution of 17–25 nt small RNAs derived from the Mos inverted repeat. (B) Deep sequencing of small RNAs from 293 cells transfected with MosIR plasmid using SOLiD technology. Data are organized as in panel A. (C) Positions of putative MosIR siRNAs along the MosIR hairpin. Graph depicts cumulative counts of 5′-ends of putative MosIR-derived siRNAs (20–24 nt long reads) found in the transgenic brain and 293 cells transiently transfected with the MosIR transgene.
To further address MosIR dsRNA conversion into siRNA, we performed SOLiD sequencing of small RNA from 293 cells transiently transfected with high dose of MosIR plasmid. SOLiD sequencing yielded 343 putative Mos siRNAs (20–23 nt long), which localized along the predicted MosIR dsRNA and were enriched among small RNAs of 17–25 nt in length (Figure 3B and C). Interestingly, mapping of small RNAs onto the transfected plasmid revealed a complex picture of RNA expression, including RNA fragments derived from both strands of EGFP, from the antisense strand of the intron and the plasmid backbone (Supplementary Figure S3B). This spurious transcription likely accounts for higher level of EGFP-derived sequences (434) when compared with the number of MosIR-derived sequences (343) (Figure 3B). However, RNAs of the size expected for Dicer products were not enriched among small RNAs derived from the EGFP coding sequence (Supplementary Figure S3C) arguing that the peak of MosIR-derived 21–23 nt sequences includes Dicer products. Taking into account the background of other Mos small RNAs, it appears that Mos siRNAs could be up to 10 times more abundant in MosIR-transfected 293 cells than in transgenic tissues, which likely reflects MosIR overexpression in transfected cell. However, the amount of Mos siRNAs estimated by SOLiD sequencing in transfected 293 cells was two orders of magnitude below abundant endogenous miRNAs (Figure 3B).
Detailed examination of SOLiD results from 293 cells revealed several thousand EGFP and MosIR sequences longer than 30 nt, whereas, only one was found in the brain (Supplementary Figure S3C). Since fragments of similar length originating from endogenous mRNAs were not detected (for example, we found only one fragment from Hprt, a transcript used as an internal standard for qPCR), the MosIR fragments longer than 30 nt likely represent a snapshot of degradation intermediates of overexpressed MosIR RNA, rather than artifacts of sample preparation. Although RNA degradation intermediates would be expected to be much less stable than siRNAs, we found that putative Mos siRNAs in transfected 293 cells were an order of magnitude less abundant than the degradation intermediates (Supplementary Figure S3C). This result is consistent with the hypothesis that Dicer does not efficiently process MosIR dsRNA into siRNAs in somatic cells.
Since the MosIR transcript caused robust RNAi effects in oocytes but not in somatic cells, we propose that oocytes are a privileged tissue for endogenous RNAi in which dsRNA is efficiently presented to Dicer. This model is consistent with studies reporting abundant endogenous siRNAs in oocytes (51,52) and few, if any, endogenous siRNAs in somatic cells (53–56). Nonetheless, our results do not rule out that other types of long dsRNA molecules can be processed by Dicer in somatic cells. There are several reports of RNAi induction with long dsRNA in somatic cells where long dsRNA was formed by transcripts, which differ from the MosIR RNA (11–14). The MosIR transcript is a capped and polyadenylated translatable mRNA, which contains long 3′-UTR dsRNA stem and long single-stranded overhangs. These features may help the dsRNA to be tolerated in somatic cells and negatively affect processing by Dicer. It was shown that long single-stranded overhangs can contribute to the reduced efficiency of Dicer processing in vitro (57). Nuclear retention of edited MosIR dsRNA probably does not significantly affect MosIR processing by Dicer. As it will be discussed further below, RNA editing observed in transgenic tissues was below the reported threshold for nuclear retention. In addition, we have found adenosine-deaminated MosIR RNA in the polysomal fraction in transgenic MEFs.
The MosIR expression does not induce transcriptional silencing
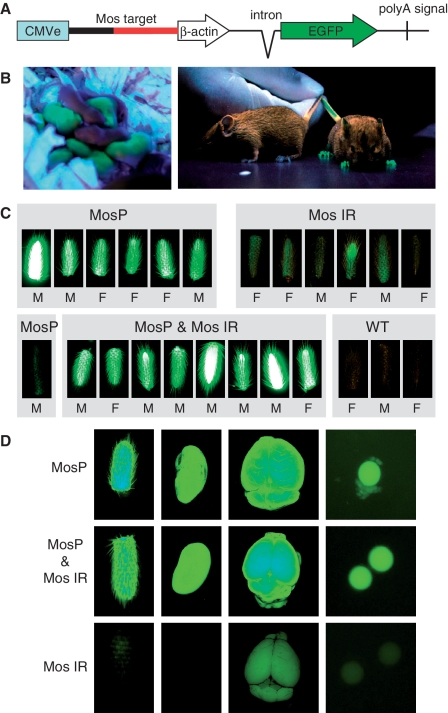
Short RNAs produced by Dicer in some model species also cause transcriptional silencing by inducing formation of transcriptionally repressive chromatin [reviewed in Ref (58,59)]. Although induction of transcriptional silencing by siRNAs has been reported for mammals (e.g. 60–62), the nature of the mechanism is still debated. We have previously shown that the MosIR transcript does not induce DNA methylation in the oocyte (63). However, that experiment was based on oocyte-specific expression of dsRNA targeting the endogenous Mos gene coding sequence and not a promoter, which would be a better suited target for studying transcriptional silencing. To address this caveat, we generated another reporter, named MosP, where the cognate sequence was inserted into the chimeric promoter, between the CMV enhancer and the β-actin core promoter (Figure 4A). The inserted sequence had a minimal effect on the activity of the promoter as estimated by FACS analysis of cultured cells transfected by equimolar amounts of MosP and the parental pCAGEGFP plasmid devoid of Mos insertion (data not shown).
Figure 4.
MosIR has no effect on transcriptional silencing of cognate sequences. (A) A schematic composition of the MosP reporter transgene. (B) EGFP signal in MosP-positive and WT newborn and adult mice. (C) EGFP fluorescence in tails of mice carrying MosP and MosIR transgenes is not reduced. Bright signal in some MosP-positive tails (e.g. first on the left in the top row) is caused by the lack of pigmentation of the tails. The MosP tail on the left in the lower row belongs to an F1 animal where we observed a spontaneous transcriptional silencing accompanied by DNA methylation of the MosP promoter (Supplementary Figure S4). All images were obtained using the same settings. (D) EGFP signal in tail, kidney, brain and oocytes of MosP, MosIR and MosP & MosIR mice. Organs were collected from siblings of one litter and their images were taken with the same camera settings.
Transgenic mice carrying the MosP transgene displayed strong EGFP expression (Figure 4B). In the F1 progeny of the MosP founder male and a wild-type female, we observed one case of spontaneous silencing of the reporter, which was accompanied by DNA methylation of the Mos sequence in the promoter (Figure 4C and Supplementary Figure S4). This event suggested that the transgene is prone to silencing involving epigenetic changes to the Mos sequence in the promoter. However, when MosP animals were crossed with MosIR animals, the MosP reporter was never silenced in the presence of the MosIR transgene (Figure 4C and D). These results showed that the MosIR transgene did not induce transcriptional silencing of homologous sequences in somatic cells. Likewise, siRNAs generated in oocytes and early embryos did not induce transcriptionally repressive chromatin at the MosP promoter, which would be propagated into the soma. These results expand the list of observations that question the nature of a putative mammalian RNA silencing pathway, which would employ siRNAs to induce transcriptionally silent chromatin in trans, a process commonly observed in plants (64–66).
MosIR expression does not induce the IFN response in somatic cells
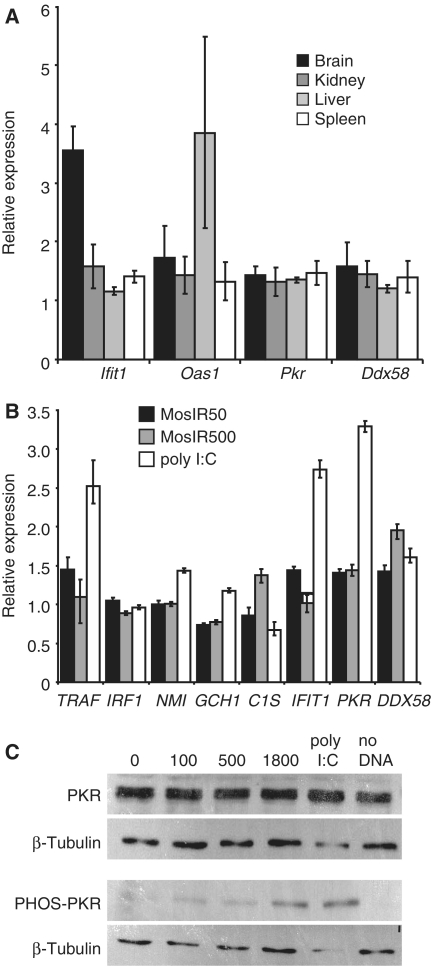
It is generally believed that long dsRNA causes sequence-independent effects in somatic cells due to the activation of a complex set of pathways generally referred to as the IFN response. However, it is unlikely that the MosIR transgene induced the typical IFN response because the MosIR mice developed normally and showed no obvious phenotype. We used qPCR to analyze transcriptional activation of several genes (Ifit1, Oas1, Pkr, Ddx58) associated with the IFN response in the brain, kidney, liver and spleen (Figure 5A). Except for Ifit1 in the brain (3.6-fold up) and Oas1 in the liver (increased in one of the three animals), we did not find increased expression of ISGs. Together, these data suggested that the MosIR transcript did not elicit a general IFN response in somatic cells of the MosIR mice.
Figure 5.
Analysis of the IFN response genes in mice and cell lines. (A) Analysis of expression of IFN stimulated genes (ISGs) Ifit1, Oas1, Pkr and Ddx58 in different tissues of MosIR mice. The expression was estimated by qPCR and is shown relative to wild-type siblings. For each gene, samples from three MosIR animals were analyzed in duplicates. Hprt was used as an internal standard. Error bars = SEM. (B) Analysis of expression of ISGs in 293 cells transfected with 50 and 500 ng (per well in a 24-well plate, the total amount of the transfected DNA was kept constant by adding pCAGEGFP DNA) of the MosIR plasmid 48 h after transfection. The expression was estimated by qPCR and is shown relative to 293 cells transfected with 100 ng/well of the pCAGEGFP plasmid. For comparison, ISGs were stimulated by adding poly I:C to the media. HPRT1 was used as an internal standard. The experiment was performed in triplicates. Error bars = SEM. (C) Western blot analysis of induction of PKR phosphorylation by the MosIR plasmid. Cells in 6-well plates were transfected with 100, 500 or 1800 ng of the MosIR plasmid and harvested for western blot analysis 24 h after transfection. The total amount of the transfected DNA was adjusted to 2 µg in all samples by adding corresponding amount of pCAGEGFP DNA. The majority of transfected cells showed green fluorescence (data not shown), toxicity of the MosIR to the cells was not apparent. Poly I:C was added to the media to a final concentration of 1 µg/ml and it was apparently toxic to the cells.
To further examine IFN activation by the MosIR transgene, we performed a series of transfections of the MosIR plasmid into 293 cells and analyzed several common markers of the IFN response. When 293 cells were examined 48 h post-transfection (this time point was chosen to emulate the situation in transgenic mice where MosIR was continuously expressed), there was little, if any, induction of ISGs even at very high doses of MosIR. In control experiments, we could detect up-regulation of several ISGs in response to poly I:C, a commonly used ISG inducer (Figure 5B). Likewise, phosphorylation of PKR, a common marker of response to dsRNA, was seen only at very high doses of the transfected MosIR vector (Figure 5C).
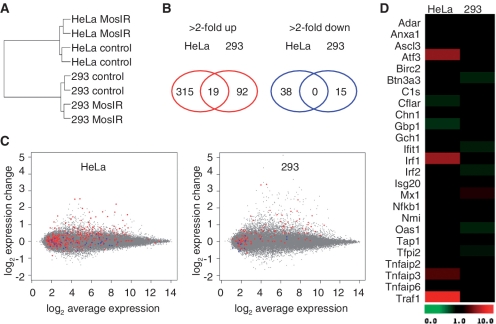
To assess in more detail the potential of the MosIR transgene to induce ISGs, we performed microarray analysis of 293 and HeLa cells transfected with the same amount of the MosIR plasmid as in the earlier described SOLiD analysis (Figure 6). Clustering analysis based on differentially expressed genes suggested that there was no common transcriptome signature in cells expressing dsRNA (Figure 6A). Overall, the number of genes with altered expression upon transfection of the MosIR plasmid was rather small and only 19 probe sets (corresponding to 17 genes) were changed more than 2-fold in both cell lines (Figure 6B and C and Supplementary Table SI). Furthermore, detailed examination of differentially expressed genes in 293 and HeLa cells revealed little, if any, evidence for the activation of the IFN pathway (Figure 6D and Supplementary Table SII). This contrasted with our earlier results where siRNA transfection into 293 cells caused a strong up-regulation of many ISGs (67). It should be noted that we have observed up-regulation of a limited set of genes related to IFN activation in HeLa cells (IRF1, IRF7, NFKB1, cytokines IL1A, CCL20, CXCL3 and TNF/related genes TRAF1 and TNFAIP3, Supplementary Table SII). However, the transcriptome changes in HeLa cells were small and did not resemble typical transcriptome changes induced either by dsRNA (18) or caused by siRNA transfection in 293 cells (67) (Figure 6C and D).
Figure 6.
Microarray analysis of HeLa and 293 cells expressing MosIR. Microarray analysis of HeLa and 293 cells transfected with MosIR plasmid (MosIR) or pCAGEGFP plasmid (control). (A) A hierarchical clustering of microarray samples using probe sets differentially expressed (>2-fold change) between MosIR and control samples in both lines shows good concordance of duplicates and clustering according to the cell type. This suggests that there is not a common set of genes induced by MosIR in these two cell lines. (B) A minimal overlap of differentially expressed genes in HeLa and 293 cells. Genes corresponding to commonly up-regulated probe sets are listed in Supplementary Table SI. (C) MA plots of HeLa and 293 transcriptome change upon MosIR expression document small and distinct changes in hybridization intensities of probe sets. The X axis shows Affymetrix probe hybridization signal intensities (log2 average expression); the Y axis, shows relative changes in probe intensity between MosIR and control samples. Each point represents one probe set. Red and blue color highlight probe sets significantly up-regulated and down-regulated in the other cell line. (D) Absence of dsRNA-induced transcriptome signature in HeLa and 293 cells expressing long dsRNA. Genes previously identified as genes induced by dsRNA treatment are shown (18).
Taken together, our data show that somatic cells tolerated MosIR expression unexpectedly well. It appears that the IFN response is only elicited when the amount of expressed dsRNA reaches a certain threshold. The tolerance to expressed dsRNA seems to be significant, considering the amounts of MosIR plasmid used in our experiments.
Adenosine deamination of MosIR RNA
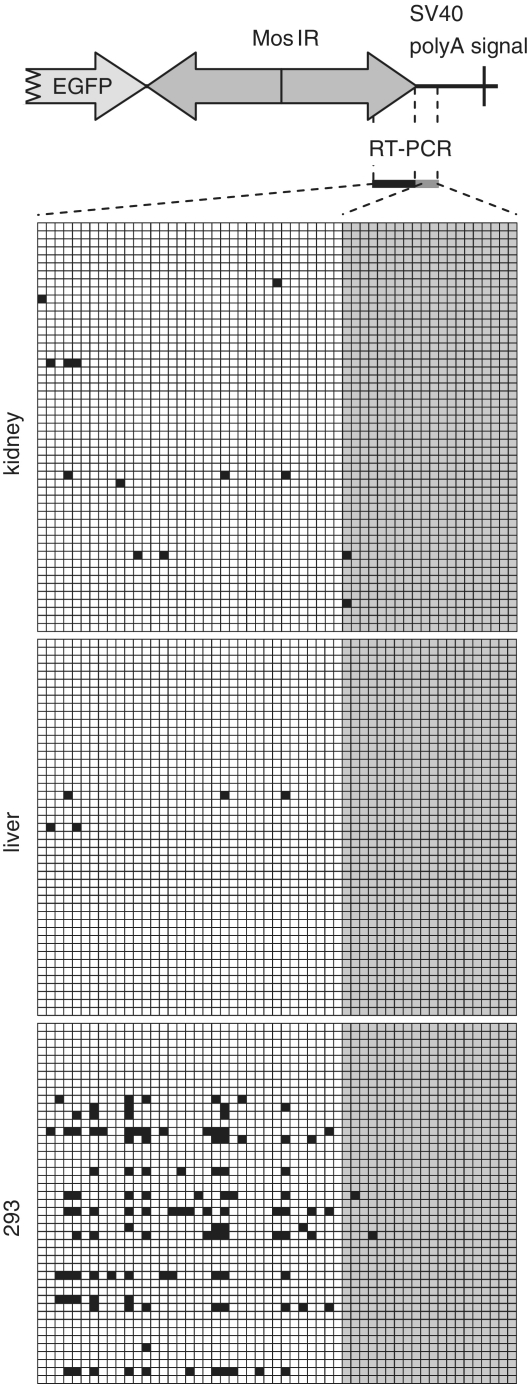
Adenosine deamination has been proposed to target nuclear dsRNA and to oppose the RNAi pathway [reviewed in Ref (68,69)]. To estimate levels of adenosine deamination of MosIR transcripts in different cell types, we analyzed the 3′-end of the MosIR hairpin region and neighboring 3′ single-stranded overhang by cloning and sequencing RT–PCR products (Figure 7). This procedure identified edited adenosines as A/G conversions in the sequence. We found a minimal number of A/G conversions in MosIR RNA obtained from kidney, liver and brain of transgenic mice (Figure 7; data not shown). These A/G conversions were mostly found in the region of the predicted RNA duplex indicating that the MosIR dsRNA was likely to be edited (Figure 7).
Figure 7.
Analysis of adenosine deamination of MosIR RNA. Adenosine deamination was analyzed by cloning RT–PCR products from the MosIR region indicated on the top. Results of editing analysis of MosIR transcripts from transgenic kidney and liver and MosIR-transfected 293 cells are shown below. Each row of squares represents one sequenced clone. Each square represents an individual adenosine in the analyzed sequence. A part of the PCR product is derived from the stem (dsRNA) sequence (white squares) and a part from single-stranded sequence (gray squares). A/G conversions are indicated by black squares.
It has been reported that editing induces nuclear retention. However, the editing frequency observed in our experiments was much lower than editing frequencies implicated in nuclear retention of edited dsRNA in Xenopus oocytes and mammalian cells (70). In addition, editing analysis in nuclear and cytoplasmic fractions from MosIR MEFs did not reveal a strong evidence for nuclear retention. We have found only 3 of 24 clones and 1 of 23 clones containing A/G conversions in nuclear and cytoplasmic fractions, respectively (Supplementary Figure S5A). Although the low frequency of editing does not allow for precise estimation of enrichment of MosIR in the nuclear fraction, these data suggest that nuclear retention is not strong. We also examined the degree of editing in MosIR transcripts found in the polysomal fraction of MEF cells derived from the MosIR mice. We found that 5 of 23 clones from the polysomal fraction contained A/G conversions (Supplementary Figure S5B–D). Edited clones contained on average 14% of edited adenosines (the most edited sequence had converted 24% of adenosines). These data suggested that editing did not prevent nuclear export and translation. Furthermore, consistent with our results, endogenous human mRNAs with edited 3′-UTRs were found previously to associate with polysomes (25).
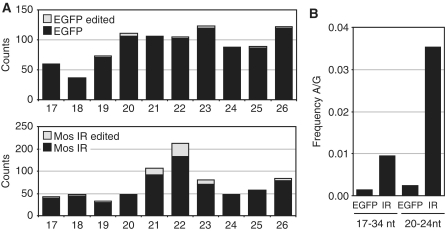
Relatively abundant editing of MosIR RNA was observed in 293 cells transfected with the MosIR plasmid. It was evident in about one-third of sequenced clones where, on average, 21% of adenosines were edited (Figure 7). Consistent with previous studies on adenosine deamination, A/G conversion was not randomly distributed along the sequence, but showed specific sequence preferences (71,72). These data further strengthen our conclusion that MosIR transcript forms dsRNA in vivo. However, editing apparently affects only a fraction of MosIR dsRNA formed in somatic cells. First, there was no enrichment of editing in RNase T1-resistant MosIR RNA from transgenic MEFs (data not shown). Second, the frequency of A/G conversions in putative MosIR siRNAs was comparable with the editing observed in longer MosIR transcripts (Figures 7 and 8). Analysis of SOLiD data from 293 cells showed that 15% of MosIR-derived small RNAs of 21–23 nt in length contained A/G conversions, while the conversion rate was negligible in small RNAs of other lengths or EGFP sequence-derived ones (Figure 8).
Figure 8.
Analysis of adenosine deamination of MosIR-derived siRNAs. (A) Distribution of putatively edited SOLiD-identified small RNAs originating from EGFP and Mos inverted repeat (MosIR) sequences in 293 cells. Small RNAs are sorted along the X-axis according to their length (17–26 nt).The Y-axis shows the number of clones with EGFP- (upper graph) or MosIR-derived sequences (lower graph). The gray portion of each column indicates the fraction of siRNAs carrying up to four A/G sequence changes. (B) Putative editing is most pronounced in SOLiD-identified siRNA-like RNAs derived from the MosIR sequence. The graph shows A/G mismatch frequency found in the total population of small RNAs and 20–24 nt long RNAs derived from EGFP or MosIR sequences in 293 cells.
The low A/G conversion rate observed in MosIR-derived small RNAs of other lengths than putative Dicer products is presumably due to increased stability of edited, Dicer-produced, siRNAs. Although we cannot exclude that editing of siRNAs takes place after Dicer processing, this scenario seems less likely than editing of the Dicer substrate. Dicer-mediated cleavage is coupled with RISC loading (73) and there is no biochemical evidence suggesting the association of ADAR with the RISC (74).
In summary, our data suggest that the MosIR forms dsRNA in vivo and that a fraction of dsRNA is edited. Since editing frequency is similar in MosIR transcripts and siRNAs derived from them, we conclude that Dicer processes a small fraction of the MosIR dsRNA hairpin regardless of whether they are edited or not. Therefore, the low amount of MosIR siRNAs in somatic cells is likely to be caused by inefficient processing of dsRNA by Dicer with editing having, at best, a secondary role as a factor responsible for the ineffective formation of endogenous siRNAs. While editing of MosIR dsRNA apparently does not appear to interfere with Dicer processing, it could still negatively influence efficiency of RNAi, since editing changes the siRNA substrate specificity.
CONCLUSIONS
We have established a transgenic mouse model that ubiquitously expresses a long RNA hairpin. There are several independent lines of evidence suggesting that expression of the MosIR transgene in mammalian cells produces dsRNA in vivo. At the same time, it appears that somatic cells are able to reduce dsRNA formation, either by preventing dsRNA duplex formation or by unwinding it. In contrast to the common view that dsRNA in mammalian somatic cells imparts a detrimental effect, we did not observe any typical sequence-independent response associated with activation of the IFN pathway. Our study suggests that long dsRNA structures in cellular mRNAs are well tolerated and activate the IFN system only at very high concentrations. Under normal conditions, such dsRNA-containing mRNAs are partially edited and poorly processed by Dicer in somatic cells, perhaps because somatic cells lack a factor facilitating siRNA biogenesis. Effective RNAi has been observed only in oocytes, providing yet more evidence that the female germline is a privileged tissue in terms of directing dsRNA into the RNAi pathway. Altogether, our data support a model that the effects of long dsRNA in mammalian cells are influenced by additional factors, which determine how cells respond to such dsRNA. These factors certainly involve the cellular history of dsRNA and structural features, such as 5′-end modifications and length of sequences flanking dsRNA stem.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Czech Science Foundation (grant GACR 204/09/0085); EMBO SDIG (program #1488, partial); Purkynje Fellowship (to P.S., partial); European Community Framework Seven Program Integrated Project “Sirocco” (Research in the laboratory to W.F.); Novartis Research Foundation (to Friedrich Miescher Institute). Funding for the open access charge: GACR 204/09/0085.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jean-Francois Spetz for production of transgenic mice, Edward Oakeley for microarray analysis, Michael Jantsch for antibodies, Susan Wagner for help with polysome profiling and Radek Sindelka for help with single-cell qPCR.
REFERENCES
- 1.Tian B, Bevilacqua PC, Diegelman-Parente A, Mathews MB. The double-stranded-RNA-binding motif: interference and much more. Nat. Rev. Mol. Cell Biol. 2004;5:1013–1023. doi: 10.1038/nrm1528. [DOI] [PubMed] [Google Scholar]
- 2.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 5.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 6.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 7.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 8.Obbard DJ, Gordon KH, Buck AH, Jiggins FM. The evolution of RNAi as a defence against viruses and transposable elements. Philos. Trans. R Soc. Lond. B Biol. Sci. 2009;364:99–115. doi: 10.1098/rstb.2008.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Umbach JL, Cullen BR. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009;23:1151–1164. doi: 10.1101/gad.1793309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Svoboda P, Flemr M. The role of miRNAs and endogenous siRNAs in maternal-to-zygotic reprogramming and the establishment of pluripotency. EMBO Rep. 2010;11:590–597. doi: 10.1038/embor.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shinagawa T, Ishii S. Generation of Ski-knockdown mice by expressing a long double-strand RNA from an RNA polymerase II promoter. Genes Dev. 2003;17:1340–1345. doi: 10.1101/gad.1073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diallo M, Arenz C, Schmitz K, Sandhoff K, Schepers U. Long endogenous dsRNAs can induce complete gene silencing in mammalian cells and primary cultures. Oligonucleotides. 2003;13:381–392. doi: 10.1089/154545703322617069. [DOI] [PubMed] [Google Scholar]
- 13.Yi CE, Bekker JM, Miller G, Hill KL, Crosbie RH. Specific and potent RNA interference in terminally differentiated myotubes. J. Biol. Chem. 2003;278:934–939. doi: 10.1074/jbc.M205946200. [DOI] [PubMed] [Google Scholar]
- 14.Bhargava A, Dallman MF, Pearce D, Choi S. Long double-stranded RNA-mediated RNA interference as a tool to achieve site-specific silencing of hypothalamic neuropeptides. Brain Res. Brain Res. Protoc. 2004;13:115–125. doi: 10.1016/j.brainresprot.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Hunter T, Hunt T, Jackson RJ, Robertson HD. The characteristics of inhibition of protein synthesis by double-stranded ribonucleic acid in reticulocyte lysates. J. Biol. Chem. 1975;250:409–417. [PubMed] [Google Scholar]
- 16.Meurs E, Chong K, Galabru J, Thomas NS, Kerr IM, Williams BR, Hovanessian AG. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- 17.Sadler AJ, Williams BR. Structure and function of the protein kinase R. Curr. Top. Microbiol. Immunol. 2007;316:253–292. doi: 10.1007/978-3-540-71329-6_13. [DOI] [PubMed] [Google Scholar]
- 18.Geiss G, Jin G, Guo J, Bumgarner R, Katze MG, Sen GC. A comprehensive view of regulation of gene expression by double-stranded RNA-mediated cell signaling. J. Biol. Chem. 2001;276:30178–30182. doi: 10.1074/jbc.c100137200. [DOI] [PubMed] [Google Scholar]
- 19.Gantier MP, Williams BR. The response of mammalian cells to double-stranded RNA. Cytokine Growth Factor Rev. 2007;18:363–371. doi: 10.1016/j.cytogfr.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeCerbo J, Carmichael GG. Retention and repression: fates of hyperedited RNAs in the nucleus. Curr. Opin. Cell Biol. 2005;17:302–308. doi: 10.1016/j.ceb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 22.Bass BL. How does RNA editing affect dsRNA-mediated gene silencing? Cold Spring Harb. Symp. Quant. Biol. 2006;71:285–292. doi: 10.1101/sqb.2006.71.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Z, Carmichael GG. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106:465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
- 24.Scadden AD. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat. Struct. Mol. Biol. 2005;12:489–496. doi: 10.1038/nsmb936. [DOI] [PubMed] [Google Scholar]
- 25.Hundley HA, Krauchuk AA, Bass BL. C. elegans and H. sapiens mRNAs with edited 3' UTRs are present on polysomes. RNA. 2008;14:2050–2060. doi: 10.1261/rna.1165008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knight SW, Bass BL. The role of RNA editing by ADARs in RNAi. Mol. Cell. 2002;10:809–817. doi: 10.1016/s1097-2765(02)00649-4. [DOI] [PubMed] [Google Scholar]
- 27.Yang W, Wang Q, Howell KL, Lee JT, Cho DS, Murray JM, Nishikura K. ADAR1 RNA deaminase limits short interfering RNA efficacy in mammalian cells. J. Biol. Chem. 2005;280:3946–3953. doi: 10.1074/jbc.M407876200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heale BS, Keegan LP, McGurk L, Michlewski G, Brindle J, Stanton CM, Caceres JF, O'Connell MA. Editing independent effects of ADARs on the miRNA/siRNA pathways. EMBO J. 2009;28:3145–3156. doi: 10.1038/emboj.2009.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Q, Carmichael GG. Effects of length and location on the cellular response to double-stranded RNA. Microbiol. Mol. Biol. Rev. 2004;68:432–452. doi: 10.1128/MMBR.68.3.432-452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gan L, Anton KE, Masterson BA, Vincent VA, Ye S, Gonzalez-Zulueta M. Specific interference with gene expression and gene function mediated by long dsRNA in neural cells. J. Neurosci. Methods. 2002;121:151–157. doi: 10.1016/s0165-0270(02)00230-3. [DOI] [PubMed] [Google Scholar]
- 31.Tran N, Raponi M, Dawes IW, Arndt GM. Control of specific gene expression in mammalian cells by co-expression of long complementary RNAs. FEBS Lett. 2004;573:127–134. doi: 10.1016/j.febslet.2004.07.075. [DOI] [PubMed] [Google Scholar]
- 32.Stein P, Svoboda P, Schultz RM. Transgenic RNAi in mouse oocytes: a simple and fast approach to study gene function. Dev. Biol. 2003;256:187–193. doi: 10.1016/s0012-1606(02)00122-7. [DOI] [PubMed] [Google Scholar]
- 33.Stein P, Zeng F, Pan H, Schultz RM. Absence of non-specific effects of RNA interference triggered by long double-stranded RNA in mouse oocytes. Dev. Biol. 2005;286:464–471. doi: 10.1016/j.ydbio.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 34.Gebauer F, Richter JD. Synthesis and function of Mos: the control switch of vertebrate oocyte meiosis. Bioessays. 1997;19:23–28. doi: 10.1002/bies.950190106. [DOI] [PubMed] [Google Scholar]
- 35.Colledge WH, Carlton MB, Udy GB, Evans MJ. Disruption of c-mos causes parthenogenetic development of unfertilized mouse eggs. Nature. 1994;370:65–68. doi: 10.1038/370065a0. [DOI] [PubMed] [Google Scholar]
- 36.Hashimoto N, Watanabe N, Furuta Y, Tamemoto H, Sagata N, Yokoyama M, Okazaki K, Nagayoshi M, Takeda N, Ikawa Y, et al. Parthenogenetic activation of oocytes in c-mos-deficient mice (published erratum appears in Nature 1994, 370, 391) Nature. 1994;370:68–71. doi: 10.1038/370068a0. [DOI] [PubMed] [Google Scholar]
- 37.Svoboda P, Stein P, Schultz RM. RNAi in mouse oocytes and preimplantation embryos: effectiveness of hairpin dsRNA. Biochem. Biophys. Res. Commun. 2001;287:1099–1104. doi: 10.1006/bbrc.2001.5707. [DOI] [PubMed] [Google Scholar]
- 38.Kaname T, Huxley C. Simple and efficient vectors for retrofitting BACs and PACs with mammalian neoR and EGFP marker genes. Gene. 2001;266:147–153. doi: 10.1016/s0378-1119(01)00375-4. [DOI] [PubMed] [Google Scholar]
- 39.Sarnova L, Malik R, Sedlacek R, Svoboda P. Shortcomings of short hairpin RNA-based transgenic RNA interference in mouse oocytes. J. Negat. Results Biomed. 2010;9:8. doi: 10.1186/1477-5751-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Svoboda P, Stein P, Hayashi H, Schultz RM. Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development. 2000;127:4147–4156. doi: 10.1242/dev.127.19.4147. [DOI] [PubMed] [Google Scholar]
- 41.Schmitter D, Filkowski J, Sewer A, Pillai RS, Oakeley EJ, Zavolan M, Svoboda P, Filipowicz W. Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Res. 2006;34:4801–4815. doi: 10.1093/nar/gkl646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 43.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 44.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. ‘Green mice' as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 49.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, 3rd, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feix G, Slor H, Weissmann C. Replication of viral RNA. 13. The early product of phage RNA synthesis in vitro. Proc. Natl Acad. Sci. USA. 1967;57:1401–1408. doi: 10.1073/pnas.57.5.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 53.Mineno J, Okamoto S, Ando T, Sato M, Chono H, Izu H, Takayama M, Asada K, Mirochnitchenko O, Inouye M, et al. The expression profile of microRNAs in mouse embryos. Nucleic Acids Res. 2006;34:1765–1771. doi: 10.1093/nar/gkl096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, et al. The colorectal microRNAome. Proc. Natl Acad. Sci. USA. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yi R, Pasolli HA, Landthaler M, Hafner M, Ojo T, Sheridan R, Sander C, O'Carroll D, Stoffel M, Tuschl T, et al. DGCR8-dependent microRNA biogenesis is essential for skin development. Proc. Natl Acad. Sci. USA. 2009;106:498–502. doi: 10.1073/pnas.0810766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smalheiser NR, Lugli G, Thimmapuram J, Cook EH, Larson J. Endogenous siRNAs and noncoding RNA-derived small RNAs are expressed in adult mouse hippocampus and are up-regulated in olfactory discrimination training. RNA. 2011;17:166–181. doi: 10.1261/rna.2123811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 58.Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 61.Ting AH, Schuebel KE, Herman JG, Baylin SB. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat. Genet. 2005;37:906–910. doi: 10.1038/ng1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Janowski BA, Huffman KE, Schwartz JC, Ram R, Hardy D, Shames DS, Minna JD, Corey DR. Inhibiting gene expression at transcription start sites in chromosomal DNA with antigene RNAs. Nat. Chem. Biol. 2005;1:216–222. doi: 10.1038/nchembio725. [DOI] [PubMed] [Google Scholar]
- 63.Svoboda P, Stein P, Filipowicz W, Schultz RM. Lack of homologous sequence-specific DNA methylation in response to stable dsRNA expression in mouse oocytes. Nucleic Acids Res. 2004;32:3601–3606. doi: 10.1093/nar/gkh697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moses J, Goodchild A, Rivory LP. Intended transcriptional silencing with siRNA results in gene repression through sequence-specific off-targeting. RNA. 2010;16:430–441. doi: 10.1261/rna.1808510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haraguchi T, Mizutani T, Yamamichi N, Ito T, Minoguchi S, Iba H. SiRNAs do not induce RNA-dependent transcriptional silencing of retrovirus in human cells. FEBS Lett. 2007;581:4949–4954. doi: 10.1016/j.febslet.2007.09.028. [DOI] [PubMed] [Google Scholar]
- 66.Wang F, Koyama N, Nishida H, Haraguchi T, Reith W, Tsukamoto T. The assembly and maintenance of heterochromatin initiated by transgene repeats are independent of the RNA interference pathway in mammalian cells. Mol. Cell Biol. 2006;26:4028–4040. doi: 10.1128/MCB.02189-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Svoboda P. Off-targeting and other non-specific effects of RNAi experiments in mammalian cells. Curr. Opin. Mol. Ther. 2007;9:248–257. [PubMed] [Google Scholar]
- 68.Nishikura K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat. Rev. Mol. Cell Biol. 2006;7:919–931. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hundley HA, Bass BL. ADAR editing in double-stranded UTRs and other noncoding RNA sequences. Trends Biochem. Sci. 2010;35:377–383. doi: 10.1016/j.tibs.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kumar M, Carmichael GG. Nuclear antisense RNA induces extensive adenosine modifications and nuclear retention of target transcripts. Proc. Natl Acad. Sci. USA. 1997;94:3542–3547. doi: 10.1073/pnas.94.8.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Polson AG, Bass BL. Preferential selection of adenosines for modification by double-stranded RNA adenosine deaminase. EMBO J. 1994;13:5701–5711. doi: 10.1002/j.1460-2075.1994.tb06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stefl R, Oberstrass FC, Hood JL, Jourdan M, Zimmermann M, Skrisovska L, Maris C, Peng L, Hofr C, Emeson RB, et al. The solution structure of the ADAR2 dsRBM-RNA complex reveals a sequence-specific readout of the minor groove. Cell. 2010;143:225–237. doi: 10.1016/j.cell.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC-loading complex. Proc. Natl Acad. Sci. USA. 2008;105:512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hock J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.