Abstract
Endogenous DNA damage is removed mainly via base excision repair (BER), however, whether there is preferential strand repair of endogenous DNA damage is still under intense debate. We developed a highly sensitive primer-anchored DNA damage detection assay (PADDA) to map and quantify in vivo endogenous DNA damage. Using PADDA, we documented significantly higher levels of endogenous damage in Saccharomyces cerevisiae cells in stationary phase than in exponential phase. We also documented that yeast BER-defective cells have significantly higher levels of endogenous DNA damage than isogenic wild-type cells at any phase of growth. PADDA provided detailed fingerprint analysis at the single-nucleotide level, documenting for the first time that persistent endogenous nucleotide damage in CAN1 co-localizes with previously reported spontaneous CAN1 mutations. To quickly and reliably quantify endogenous strand-specific DNA damage in the constitutively expressed CAN1 gene, we used PADDA on a real-time PCR setting. We demonstrate that wild-type cells repair endogenous damage preferentially on the CAN1 transcribed strand. In contrast, yeast BER-defective cells accumulate endogenous damage preferentially on the CAN1 transcribed strand. These data provide the first direct evidence for preferential strand repair of endogenous DNA damage and documents the major role of BER in this process.
INTRODUCTION
Endogenous DNA damage, especially that associated with reactive oxygen species (ROS), likely contributes to a large fraction of human cancers (1) and plays a role in the pathogenesis of aging and many degenerative diseases (2,3). In addition, it is well established that defects in genes required for the repair of DNA damage can result in a genetic predisposition to cancer and aging syndromes (4). Endogenous DNA damage is typically processed by base excision repair (BER), which processes primarily small, helix non-distorting base lesions and abasic sites (5). However, there is some overlap with other DNA repair pathways, including nucleotide excision repair (NER) which has been shown to repair specific types of oxidative DNA damage (6). There are more than 20 different oxidative DNA base lesions (7) and it is grossly estimated that 10 000 oxidative hits occur per cell per day in the mammalian genome (8).
Despite the profound implications of endogenous DNA damage in human diseases, the most commonly used assays for the detection of induced oxidative DNA damage, Southern blot analysis, high performance liquid chromatography with electrochemical detection (HPLC–ECD) and enzymic assays have limited applications for the study of in vivo endogenous DNA damage (9,10). Southern blot analysis for DNA damage detection is a multi-step procedure that requires large amounts of DNA and allows only a semi-quantitative analysis of DNA strand breaks (11). HPLC–ECD can accurately measure induced oxidative DNA damage and is valuable to measure specific DNA damage lesions in body fluids, but suffers from high variable estimates of the background level of DNA oxidation and requires several days to complete depending on the number of samples (10). Enzymic methods, such as the comet assay (single cell alkaline gel electrophoresis), allow the detection of single and double strand breaks as well as alkali-labile DNA sites under alkaline conditions (10). These methods have high sensitivity and low background and are widely used for the detection of induced oxidative DNA damage (10,12), but they need standardization and inter-laboratory validation (10). Although Southern blot, chromatographic and enzymic methods can detect and quantify some specific oxidative DNA lesions, they are tedious and have inadequate sensitivity for the study of in vivo endogenous DNA damage (10). Also, they reflect only a sub-fraction of induced oxidative DNA lesions and cannot map lesion distribution, an important player in repair efficiency and cell fate. PCR-based assays take advantage of polymerase elongation properties as a sensor for damage on the template DNA (13–16) and are currently one of the most reliable strategies to map and quantify chemical or radiation induced DNA damage. However, they are quite time-consuming and require a high degree of optimization for reliable damage quantification. Additionally, their relatively low sensitivity prevents their use for the detection of overall levels of endogenous DNA damage (14–16). Therefore, due to technical limitations, the precise levels of endogenous DNA damage in different cell systems and how they impact cell fate and human health are still largely unknown (7,10).
Transcription of DNA is critical for cell function and survival, and thus unrepaired or unrecognized DNA damage in the transcribed strand can be deleterious for the cell. Transcription coupled repair (TCR) of bulky DNA adducts is well characterized in eukaryote cells and results in more rapid repair of the transcribed strands compared to the non-transcribed strands (NTS) of expressed genes. Deficient TCR has been implicated or linked to xeroderma pigmentosum, Cockayne syndrome (CS), trichothiodystrophy (TTD) and UV-sensitive syndrome (UVS), although in vivo TCR observations have not been fully validated with eukaryotic cell-free systems (17). TCR was originally documented for DNA damage induced by UV light and believed to operate through NER pathways, but later reports suggested that oxidative damage is also preferentially repaired in a transcription-dependent manner (18,19). Nonetheless, several key papers supporting transcription-coupled repair of oxidative damage have been retracted and this subject is a matter of intense debate (20). Importantly, due to current technical limitations there is no direct evidence for transcription-coupled repair of oxidative or endogenous DNA damage.
To map and quantify in vivo strand-specific endogenous DNA damage, we have developed a novel rapid, reliable and highly sensitive primer-anchored DNA damage detection assay (PADDA). Our damage detection assay relies on the principle that the presence of DNA damage on the template can change the polymerase replicative rate leading to lesion bypass with or without misincorporation, or to lesion-dependent replicative arrest before or opposite to the damaged base. PADDA (Figure 1A) screens a specific DNA region for endogenous base damage by performing a single non-cycled primer-extension using nanogram amounts of DNA, a 5′-biotin-tagged primer and Vent exo– DNA polymerase. The non-cycled primer-extension generates a pool of highly specific biotin-tagged extended products, each of them derived from a single DNA molecule and strand. Each extended product has a stop, which represents replicative arrest by a damaged nucleotide. Therefore, when derived from genomic DNA, each extended product should represent DNA damage at the single-nucleotide, single-strand, single-gene and single-cell level. After the primer-extension, extended products are purified, captured on streptavidin-coated paramagnetic beads and released from genomic DNA. This highly purified pool of extended products can be used directly for real-time PCR analysis, allowing high-throughput reliable strand-specific DNA damage quantification (q-PADDA) within 5 h. Alternatively, the purified pool of extended products can be ligated to a chemically modified oligonucleotide-adapter using a highly efficient single-strand DNA ligase. This recognition sequence at the end of the extended product allows for fingerprinting analysis (f-PADDA) of damage and repair at the single-nucleotide level by revealing the places on sequence template where polymerase extension stopped.
Figure 1.
Schematic representation of PADDA and identified DNA damage. (A) A single non-cycled primer extension performed with a 5′-biotin-tagged primer and Vent exo– DNA polymerase identifies damaged nucleotides (inverted triangles), and generates a pool of highly specific biotin-tagged extended products (EP), each of them derived from a single DNA molecule and strand. Each extended product has a stop (indicated by a red star), which represents replicative arrest by a damaged nucleotide, a nick or random polymerase stalling. Some extended products will contain misincorporations (indicated by a solid red circle) that represent polymerase lesion by-pass with misincorporation. After several purification steps that include the use of biotin-binding, streptavidin-coated, paramagnetic beads, the strand-specific, highly purified, biotin-bound extended products can be used for damage fingerprinting or quantification. (B) For strand specific fingerprinting analysis (f-PADDA) of damage and repair at the single-nucleotide level, the purified pool of EPs is ligated to a chemically modified oligonucleotide-adapter using a highly efficient single-strand DNA ligase. The pool of adapter-linked EPs can be diluted to one EP per well and each EP is directly amplified and sequenced, or the EPs can be amplified as a pool, cloned and sequenced. In either case high fidelity amplification is performed using an oligonucleotide complementary to the adapter–primer and a nested gene-specific primer. Data is obtained in <2 days. (C) For strand-specific DNA damage quantification (q-PADDA) the purified pool of EPs is used directly for real-time PCR analysis. For each strand, two sets of primers, yielding a short (~70 bases) and a long (~600 bases) fragment provide data representative of total amount of template (an internal normalization control) and undamaged (2−ΔΔCT) DNA, respectively. High-throughput reliable DNA damage quantification can be obtained within 5 h. (D) Representative DNA sequence chromatographs identifying DNA damage position. Shown is the positional identification by f-PADDA of 8-oxoG as: Stop, SM and LBM. The damaged nucleotide is indicated either by the adapter's complementary sequence (arrow) or by the presence of a misincorporated nucleotide (base change). (E) Vent exo– behavior at abasic, 8-oxoG, 8-oxoA and thymine glycol (Tg) engineered DNA lesions detected by f-PADDA. (F) Quantification via q-PADDA of induced DNA damage after in vitro exposure to a dose escalation of H2O2. DNA lesion frequency was estimated using the Poisson equation. Data represents mean ± SEM.
Here, we documented for the first time that persistent endogenous nucleotide damage in the CAN1 gene co-localizes with literature reported spontaneous gene mutations, corroborating the theory that persistent endogenous DNA damage leads to mutation fixation. The use of PADDA, on a real-time PCR setting, to quantify in vivo endogenous base damage in wild-type (WT) and BER-defective (BER−) cells demonstrated that the levels of DNA damage vary significantly between yeast strains, stages of cell growth, and DNA strands. Furthermore, it allowed us to demonstrate for the first time that BER repairs preferentially endogenous DNA damage localized in the transcribed strand.
MATERIAL AND METHODS
Yeast strains
The yeast Saccharomyces cerevisiae strains used in this study were previously reported (11): SJR751 (MATα ade2-101oc his3Δ200 ura3ΔNco lys2ΔBgl leu2-R) and its isogenic derivative SJR867 (MATα ade2-101oc his3Δ200 ura3ΔNco lys2ΔBgl leu2-R ntglΔ::LEU2 ntg2Δ::hisG apn1Δ1::HIS3). SJR751 and SJR867 are referred to as WT and BER− strains, respectively. For DNA damage assessment, yeast cells were grown, in the dark at 30°C, in liquid YPAD media for ~16 h (to ~2 × 107 cells/ml, exponential phase) or for 72 h (to ~5 × 108, stationary phase).
DNA extraction and quantification
To reduce levels of artifactual DNA damage, samples were kept in low light conditions, handled with gentle pipetting, and never vortexed throughout DNA extraction procedures. A total of 2 × 109 yeast cells were harvested by centrifugation, washed in H2O, resuspended in 1 ml spheroblast buffer with 200 U Zymolyase (Zymoresearch, Orange, CA, USA) and incubated at 37°C for 2 h. Genomic DNA was then extracted with OXItek DNA isolation kit (ZeptoMetrix Corp., Buffalo, NY, USA) to minimize artifactual oxidative DNA damage. DNA was then resuspended in TE buffer and stored at 4°C. To assure precise DNA measurement, DNA concentration was measured using the Qubit Fluorometer (Invitrogen, Carlsbad, CA, USA) and Quant-iT dsDNA HS reagents.
Primer-anchored DNA damage detection assay
To screen for DNA damage, we performed a single non-cycled primer-extension in the region of interest. The oligonucleotides used in this study are described in Supplementary Table S1. For damage detection in synthetic DNA, we prepared a 22.8 μl primer extension reaction containing 1× Thermopol buffer (NEB, Ipswich, MA, USA), 0.2 mM each dNTP (Fermentas, Glen Burie, MD), 20 pmol of a 5′-biotin-tagged primer, and 20 pmol of a 64-base template containing either a synthetic abasic, 8-oxodeoxyguanine (8-oxoG), 8-oxodeoxyadenine (8-oxoA), or thymine glycol lesion, along with a control (undamaged oligo). To generate a pool of biotin-tagged extended products (Figure 1A), the mix was incubated at 94°C for 3 min, held at 85°C to add 0.08 U of Vent exo– (NEB), and then incubated at 58°C for 2 min (annealing) and 75°C for 2 min (extension). For damage detection in the CAN1 gene, primer extension reactions contained 0.8 U of Vent exo–, 2 pmol of a 5′-biotin-tagged gene specific primer, and either 50 ng of plasmid DNA, or 50–500 ng of genomic DNA. A sample containing all reagents except Vent exo– was used as a negative control for all experiments. The non-cycled primer-extension contrasts with the multiple-cycle primer-extension reaction of previous assays, and generates a pool of highly specific biotin-tagged extended products, each of them derived from a single DNA molecule and strand (Figure 1A). The use of a 5′-biotin-tagged sequence specific primer allows for subsequent steps aimed at reducing the background of the technique. Vent exo–, a truncated form of a native DNA polymerase (Vent, NEB), was chosen for this assay instead of the currently used Sequenase (13) or Taq DNA polymerase (14), because of its reported high sensitivity to detect DNA damage (21) and primer-extension efficiency (22–24). To optimize primer-extension performance, Vent exo– units were adjusted to the complexity of the DNA being screened (22).
After the primer-extension and in order to degrade unused primers, the mix was incubated with Exonuclease I (NEB) as per manufacturer's recommendation. Next, the reaction was held for 10 min at 94°C to degrade Exonuclease I and denature the DNA, facilitating the release of desired extension products from genomic DNA. The reaction was further purified using the QIAquick Nucleotide Removal Kit (Qiagen, Valencia, CA, USA) to eliminate biotin-mononucleotides and products smaller than 17 bases generated by Exonuclease I digestion.
The purified extended products were captured with biotin-binding, streptavidin-coated paramagnetic beads (0.2 mg Dynabeads® M-270 Streptavidin, Invitrogen) and washed in B&W buffer as per manufacturer's recommendation. To further remove possibly contaminating genomic DNA, samples were incubated for 5 min with 1 ml of 0.5 M NaOH, spun briefly and exposed to a magnetic field to facilitate supernatant removal. Each sample was then washed twice with 200 μl of 2× B&W buffer to assure no traces of NaOH remained. After this step, extended products were either processed for fingerprinting (mapping) nucleotide damage (f-PADDA) or for high-throughput damage quantification (q-PADDA).
Fingerprinting strand-specific DNA damage (f-PADDA)
For the purpose of mapping strand-specific nucleotide damage (f-PADDA), the 3′-end of the highly purified extended products was attached to the 5′-end of a chemically modified adapter–primer (Figure 1B) using ThermoPhage™ single-stranded DNA ligase (Prokaria, Reykjavik, Iceland). ThermoPhage was chosen for this reaction because its efficiency for single-strand ligation is higher than that of T4 RNA ligase (25) used in some PCR-based assays. The adapter–primer was phosphorylated at the 5′-end to facilitate ligation, possessed a dideoxy-C at the 3′-end to prevent self-ligation, and contained phosphorothioate bonds between the last four nucleotides at the 3′-end to prevent exonucleolytic digestion. Purified extended-products were first washed with 20 μl of 1× Thermophage ssDNA ligase buffer and then resuspended in 3.7 μl of 1× Thermophage ssDNA ligase buffer. The ligation was performed as recommended by Prokaria except 1.5 µM adapter and 10 U ssDNA ligase in a final 10 µl volume were used under the following conditions: 55°C for 1 h, 65°C for 15 min and 70°C for 15 min. Then, samples were spun briefly and the supernatant removed as before in the presence of a magnet. The adapter-ligated extended-products were washed with 1 ml of 2× B&W buffer, and then incubated with 1 ml of 0.5 M NaOH for 5 min to completely remove free adapters and non-specifically bound DNA. Finally, the supernatant was discarded and the bound products were washed with 200 µl of 2× B&W buffer. The adapter-linked extended-products pool was then washed with 20 µl of 1× Phusion™ HF PCR buffer and amplified for 35-cycles using Phusion™Hot-Start DNA Polymerase (Finnzymes), an oligonucleotide complementary to the adapter–primer and a nested gene-specific primer according to the manufacturer's instructions. Phusion™ Hot Start DNA Polymerase has the highest available accuracy (error rate = 4.4 × 10−7) and generates blunt-end products. After PCR amplification, the supernatant was collected under a magnetic field, cloned into pJET1.2 (Clone JET PCR cloning kit) and used for transformation following standard procedures. This cloning strategy yields 99% of clones positive for a cloned insert. For amplification of single clones, individual bacterial colonies were randomly selected and directly used for colony PCR using primers flanking the plasmid insert under standard conditions. PCR products were purified with Qiagen PCR purification kit, sequenced on an ABI 3730 48-capillary sequencer and analyzed using Sequencher® (Gene Codes Co.). We assayed at least two DNA samples for each unrepaired genotype and time point and analyzed a total of 222 clones from 12 independent experiments. A minimum of 20 randomly selected clones were analyzed for each genotype, phase of growth (exponential versus stationary) and repair status (repaired versus unrepaired). Primer extension after in vitro DNA repair (Supplementary Material and Methods) was assessed as for unrepaired genomic DNA by sequencing random clones.
To assess PADDA's sensitivity in the presence of different types of damage, 20 pmol total of abasic and 8-oxodA lesion oligos were combined in a pre-set ratio of 1:1 and 1:10 ratio, and used as substrates for f-PADDA. Samples were processed as above, except that instead of being amplified as a pool, the adapter-ligated extended-products were diluted (~1 × 10−7) with the aim of obtaining one single molecule per well of a 96-well PCR plate and individually amplified by Real-Time PCR. Reactions were carried out in a 25 µl final volume with Phusion HF PCR buffer, 1:75 000 dilution SYBR Green I (Invitrogen), 0.8 mM total dNTPs (Fermentas), 0.7 µM each oligo and 0.4 U Phusion™ Hot Start II DNA Polymerase under the following conditions: 98°C, 30 s; (98°C, 10 s; 64°C, 15 s; 72°C, 10 s). Amplified products, as determined by the level of fluorescent signal per well, were purified using a PCR purification kit (Qiagen) and sequenced (Figure 1B). At least 22 sequences per ratio were analyzed.
Quantifying strand-specific DNA damage by real-time PCR (q-PADDA)
For high-throughput damage quantification, the highly purified streptavidin bound extended products were re-suspended in 50 µl TE and amplified by Real-Time PCR as follows. Reactions were carried out in an IQ5 Real-Time PCR system in a 25 µl final volume with PCR buffer [40 mM Tricine–KOH (pH 8.0), 16 mM KCl, 3.5 mM MgCl2, 3.75 µg/ml bovine serum albumin], 1:75 000 dilution SYBR Green I (Invitrogen), 0.8 mM total dNTPs (Fermentas), 0.7 U Jumpstart Taq polymerase (Sigma), 0.32 µM each oligo and 1/32 of total extended product pool. Amplification was monitored and analyzed by measuring the intercalation of the fluorescent dye to double-stranded DNA according to the manufacturer's instructions. To quantify damage on each strand of the yeast CAN1 gene, two fragments of different lengths located in the same genomic region were amplified (Figure 1C). The long fragment (513–628 bases) was used to assess damage quantification. The short fragment (67–68 bases), representing undamaged DNA, was used as an internal normalization control. The PCR conditions for the different fragments were optimized to achieve similar amplification efficiencies required to compare different amplicons. All products were amplified under the following conditions: 95°C, 3 min; 40 cycles (95°C, 15 s; 63°C, 40 s). Each sample was assayed in at least three independent experiments, each with at least four wells per sample. In vitro and in vivo induction of DNA damage was performed using standard procedures (Supplementary ‘Material and Methods’ section). Quantification of in vitro induction of DNA damage was performed on a sequence verified CAN1 fragment cloned into the pJET1.2 vector (Fermentas). For endogenous or induced in vivo DNA damage quantification studies, we assayed three separate yeast cultures and DNA extractions. Fluorescence was continuously monitored versus cycle numbers as per IQ5 Real-Time PCR detection system protocol. The relative amount of undamaged template was derived using the 2−ΔΔCT method (26). To assess the relative accumulation of damage per strand, data was normalized to the respective strand in the unrepaired DNA obtained from WT cells in exponential phase. To compare relative damage between DNA strands, data was normalized to the respective strand of in vitro repaired DNA obtained from WT cells grown in exponential phase. Lesion frequency was estimated using the Poisson equation n = –ln(o), where ‘o’ is the fraction of full length fragments and ‘n’ is the average DNA lesions per fragment (11).
Statistical analysis
Data were compiled in Excel® (Microsoft) files and statistical analyses were performed using SAS® STAT Version 9.1 (SAS Institute Inc.). Proportions were compared using Fisher's Exact Tests. Independent means were compared using t-tests whose degrees of freedom were corrected, when appropriate, for inequality of variance.
Each length of extended product equals the length of the extension performed by Vent exo–, and was determined as the distance (expressed in bases) between the 3′-end of the gene-specific primer 1 and the stop position. We compared the mean length of the extended products in an analysis of variance that simultaneously modeled extended products on genotype, growth phase, and on whether the DNA samples were repaired, or not, in vitro before processing. When examination of residuals from the original model indicated that distributions of extended products were skewed, we performed the analysis of variance on log-transformed values of extended products. This modification ensured that hypothesis tests and estimates of confidence intervals were valid and trustworthy.
The rate of misincorporation per base was calculated for each genotype as the total number of lesion bypass with misincorporation (LBMs) and stops with misincorporation (SMs) divided by the total length of the extended products for that genotype. We compared rates of misincorporation using a logistic regression model. The model estimated 95% confidence intervals on genotype-specific and growth phase-specific misincorporation rates, the lower limits of which we compared to the published Vent exo– error rate (27) of 1.9 × 10−4.
To determine whether damage was detected non-randomly in a region of interest on the CAN1 gene, we calculated for each base in the region the conditional probability that the polymerase found damage at that base, given that the polymerase had not previously ‘stopped’ and was, therefore, available to detect damage at that base. A Chi-square goodness of fit test compared this set of conditional probabilities with the uniform probabilities that are expected if damage detection was a purely random process. Rejection of the test's null hypothesis suggested that damage detection was non-random. After verifying that primers detected damage non-randomly, we focused our analysis to the CAN1 ORF (CAN1, X03784) nucleotides 1180–1215, a region where frequent mutations were reported (28,29). We used Fisher's Exact Test to assess whether the damaged nucleotides detected by f-PADDA were associated with those previously reported to be the sites of mutations (28,29).
RESULTS
Validation of PADDA on artificial templates containing oxidative DNA lesions
To validate our novel methodology (Figure 1), we first determined the efficiency of PADDA to detect and map within synthetic oligonucleotides a single abasic site, 8-oxoguanine (8-oxoG), 8-oxoadenine (8-oxoA), or thymine glycol lesion. In conformity with established concepts that interpret the behavior of DNA polymerases when they encounter DNA damage on the template strand, we observed three types of data (Figure 1D): Stop (S), corresponding to a lesion-dependent replicative arrest immediately opposite or one base before the damaged nucleotide; SM, corresponding to a lesion-dependent replicative arrest with nucleotide misincorporation opposite to the damaged base; and LBM, corresponding to a lesion-dependent misincorporation opposite to the damaged base without replicative arrest. For the engineered abasic site, the designation of misincorporation was attributed every time the polymerase incorporated a nucleotide opposite to the lesion. We analyzed at least 20 independent events for each oxidative lesion. Results are summarized in Figure 1E. Abasic sites were detected in 100% of the primer extensions, either as Vent exo– blocking lesions (95%) or as LBM (5%). As a blocking lesion the abasic site resulted in 45% Stops and 50% SMs. The 8-oxoG lesion was detected in 85% of the primer extensions. The 8-oxoG lesion acted as a blocking lesion in 23% of all primer extensions (15% SMs and 8% Stops) and originated LBM's in 62% of primer extensions. Bypass of 8-oxoG by Vent exo– without misincorporation occurred occasionally (15%). The 8-oxoA lesion blocked Vent exo– 100% of the time, resulting in 86% Stops and 14% SMs. The thymine glycol lesion stopped Vent exo– 67% of the time leading exclusively to Stops. These data demonstrate that both the stop position of the extended product and the position of the nucleotide misincorporation pinpoint the location of the damaged nucleotides. Therefore, the length of the extended product and the rate of misincorporations should provide two independent measures of damage frequency. The sensitivity of the PADDA was 100% for the abasic site, 85% for the 8-oxoG lesion, 100% for the 8-oxoA lesion and 67% for the thymine glycol lesion. When we tested PADDA's ability to detect engineered lesions at pre-defined ratios, we observed both an approximately equivalent damage detection rate compared to the pre-defined ratio as well as a linear detection capability with each dilution (Supplementary Figure S1). This high sensitivity to detect, at pre-defined ratios, lesions that are relatively frequent and well established biomarkers of oxidative stress suggested that PADDA could be used for the mapping and quantification of endogenous DNA damage.
PADDA detects a broad spectrum of DNA lesions and a genotoxic dose response
To determine the sensitivity and dynamic range of q-PADDA to different spectra of DNA lesions, we quantified induced DNA damage after in vitro exposure to H2O2, methyl methanosulfonate (mms) and sodium bisulfite. To reliably quantify strand-specific DNA damage using a real-time PCR setting, we amplified two DNA fragments of different lengths: the longest fragment represents undamaged DNA and is used to estimate the number of lesions, while the shortest fragment, assumed to represent total amount of template, serves as an internal normalization control. This principle is standard in real-time PCR analysis of gene expression (26,30) and has been previously demonstrated to accurately and reliably quantify DNA damage in synthetic oligonucleotides (31). However, it has not been routinely applied for quantification of DNA damage. DNA damage was quantified for each CAN1 strand in a targeted area of ~600 bp. To ensure an unbiased comparison, we documented that amplification efficiencies are similar for all PCR fragments (Supplementary Table S2). We also demonstrated that there is a linear relationship between the Cp value and the logarithmic dilution values of total DNA (R2 ranging from 0.989 to 0.999; Supplementary Figure S2). These values enable consistent and precise determination of DNA damage with all tested DNA template quantities and support the accuracy and reliability of the following DNA damage data.
H2O2 causes oxidative damage, one of the most frequent types of endogenous damage present in vivo. Compared to the control, q-PADDA detected a significant increase in total CAN1 DNA damage for each H2O2 dose escalation, from the lowest dose tested (1 µM) to the highest (500 mM) (Figure 1F). When analyzed by DNA strand, q-PADDA detected a significant (P < 0.005) increase in damage in the NTS and TS for as little as 1 and 10 µM H2O2, respectively. The sensitivity of other DNA damage detection assays to quantify DNA damage induced in vitro by low concentrations of H2O2 has not been reported. Nonetheless, exposure of naked DNA for 1 h to 5 mM H2O2 or human cells for 1 h to 50 mM H2O2 yields a nearly identical damage pattern (32) and an equivalent global damage frequency (33). Assuming that this 10-fold difference is maintained across H2O2 concentrations, q-PADDA is at least 10- to 100-fold more sensitive than the most sensitive assays currently used to quantify induced oxidative damage: the comet assay (34), SL-RT PCR (35) and Q-PCR (35). Plus, in contrast with these assays, PADDA does not require enzymic hydrolysis of the lesions and can discriminate damage between each DNA strand.
Mms treatment causes mainly 3-methyl-adenine (3meA) and 7-methyl-guanine (7meG) (36). 3meA, but not 7meG, blocks polymerases leading to a stop 1-nucleotide before the lesion (25). 7meG is ~11 times more frequent than 3meA (37). Therefore, most of the mms induced DNA damage is not detectable by polymerase based assays unless the lesions are enzymatically removed, as occurs in vivo (38). Even so, q-PADDA is able to detect a significant strand-specific increase in DNA damage after in vitro exposure to mms (Supplementary Figure S3A). As expected, PADDA has a significantly higher sensitivity to detect in vivo than in vitro mms induced DNA damage (Supplementary Figure S3B and C).
In vitro exposure of naked DNA to sodium bisulfite leads to deamination of unmethylated cytosines to uracil, one of the most promutagenic DNA changes in vivo (39). We used this property to document that q-PADDA is able to quantify this important type of DNA alteration (Supplementary Figure S4). Overall, these data document PADDA's ability to identify a broad spectrum of DNA lesions. Furthermore, it demonstrates that q-PADDA has very high sensitivity to oxidative DNA damage and is able to discriminate a dose response over a wide range of H2O2 concentrations.
BER defective cells have high levels of endogenous DNA damage
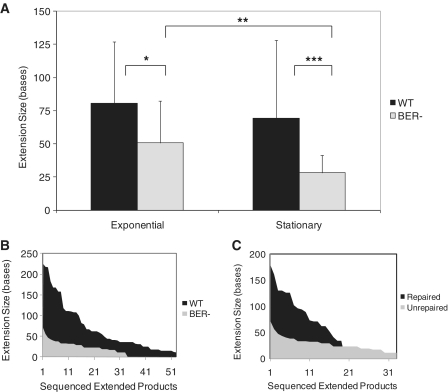
To validate f-PADDA for the detection of in vivo endogenous DNA damage, we characterized the frequency and distribution of DNA damage in the NTS of the actively transcribed CAN1 gene (40,41) in isogenic repair-proficient (WT) and repair-deficient (BER−) S. cerevisiae strains. The two most prevalent classes of endogenous DNA lesions are thought to be depurination, which results in non-coding abasic sites, and oxidative damage, which results in strand breaks, base modifications, abasic sites and DNA-protein cross-links (42). The BER-deficient triple mutant, ntg1, ntg2 and apn1, is defective in the repair of abasic sites, as well as a wide variety of oxidized bases (43,44), including thymine glycol and 8-oxoG (45). The CAN1 locus was chosen for DNA damage mapping and quantification because this same region has been used to quantify specific types of oxidative DNA damage and to obtain spontaneous mutation frequencies in these strains (11). Consistent with BER− cells being defective in the repair of endogenous DNA damage, DNA extracted from BER− cells yielded significantly shorter extended products than DNA extracted from WT cells (Figure 2A and B and Supplementary Table S3) for both exponential (P = 0.005) and stationary (P < 0.000001) phases of growth. Additionally, consistent with the reported higher levels of oxidative DNA damage in stationary phase, we observed that BER− cells in stationary phase yielded significantly shorter extended products (P = 0.008) than BER− cells in exponential phase (Figure 2A).
Figure 2.
Levels of endogenous DNA damage in WT and BER− cells. (A) Extension sizes (mean ± SD) observed for WT and BER− strains in exponential and stationary phases. Analysis of variance compared the natural log of extension sizes for the WT and BER− genotypes. *P = 0.005, **P = 0.008, ***P = 8.6 × 10−10. (B) Distribution of extension sizes observed for each sequenced extended product obtained from WT and BER− cells in stationary phase.(C) Observed extension sizes distributions for BER− cells in stationary phase before (unrepaired) and after (repaired) in vitro repair.
To confirm that we were indeed detecting DNA damage, we performed in vitro damage repair of the genomic DNA samples before subjecting them to f-PADDA analysis. The in vitro repair assay used has been reported to repair several types of DNA damage including abasic sites, deaminated cytosines, 8-oxoG and DNA nicks, however, it does not repair fragmented DNA [(46,47) and www.neb.com]. Remarkably, after in vitro damage repair, the length of primer extensions obtained from BER− cells in stationary phase increased significantly (P < 0.000001) attaining values similar to those observed for WT cells in exponential phase (Figure 2C and Supplementary Table S3). This demonstrates that PADDA detects most types of damage present in BER defective cells. Furthermore, in vitro repair also increased significantly the length of the primer extensions obtained from WT (P = 0.046) and BER− (P = 0.002) cells in exponential phase of growth (Supplementary Table S3). These results demonstrate that PADDA detects in vivo endogenous DNA damage and that the length of the primer extensions correlates inversely with the frequency of DNA damage.
Endogenous DNA damage of highly mutagenic potential increases significantly from exponential to stationary phase
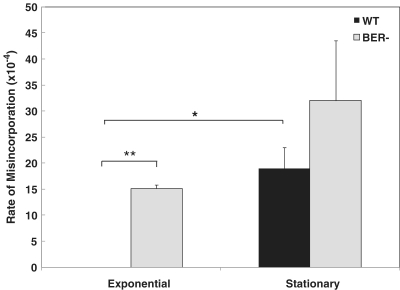
The application of f-PADDA to DNA extracted from cells in stationary phase led to a high rate of lesion-dependent misincorporations (LBMs and SMs), 19.4 × 10−4 in WT and 32 × 10−4 in BER−, respectively (Figure 3). In contrast, not a single misincorporation (LBM or SM) was observed when f-PADDA was applied to DNA extracted from WT cells in exponential phase (Figure 3). We also observed that, during exponential phase, DNA extracted from BER− cells yielded significantly more lesion-dependent misincorporations (P < 0.000001) than DNA extracted from WT cells in exponential phase (Figure 3). Additionally, we observed a significant (P < 0.000001) increase in lesion-dependent misincorporations in WT cells from exponential to stationary phase (Figure 3).
Figure 3.
Rate of nucleotide misincorporation (LBMs or SMs per base) during primer-extension in genomic DNA, modeled by logistic regression. In WT cells, the misincorporation rate was significantly higher in stationary than in exponential phase (*P = 2 × 10−151). In exponential phase, BER− extended products had a significantly higher rate of misincorporation than WT extended products (**P = 2 × 10−221).
The use of logistic regression to model the rate of misincorporation per base (Supplementary Table S4) showed that even conservative estimates of misincorporation rates for WT and BER− at stationary phase (defined by the lower bounds of 95% CI on the estimated rate) were significantly higher (P = 0.0002 and P < 0.000001, respectively) than the published misincorporation rate (27) for Vent exo– (1.9 × 10−4). These data, together with the high rate of LBMs and SMs observed for engineered oxidative DNA lesions, strongly support that LBMs and SMs represent lesion-dependent misincorporations opposite to a damaged base and therefore indicate directly the position of in vivo endogenous DNA damage. Together with the observed high sensitivity of the assay (Figure 1E), the capacity to detect by-passable DNA damage, such as the highly mutagenic 8-oxoG lesion (3), which are undetectable by other polymerase-based assays, suggest that f-PADDA will be an important tool to assess individual susceptibility to carcinogens and cancer risk.
Persistent endogenous DNA damage co-localizes with spontaneous mutations
To further determine the biological significance of our data, we compared the position of the identified nucleotide damage with the published CAN1 mutation distribution (Figure 4). Based on the results obtained with damaged synthetic oligonucleotides (Figure 1D and E), putatively damaged nucleotides on the genomic template were identified and mapped as follows: (i) nucleotides whose replication resulted in polymerase misincorporations, independently of whether the polymerase was stopped (SM) or able to bypass (LBM) the lesion; and (ii) nucleotides that blocked the polymerase progression without resulting in polymerase misincorporation (Stop). In the last case, the location of damage was assigned to the nucleotide on the template strand immediately adjacent and 5′ to the last replicated nucleotide. First, we performed a Chi-square goodness of fit test, which indicated that damage detection is not a purely random process (P < 0.000001). Then, we investigated the potential association between nucleotide damage detection and mutation using Fisher's Exact Test. We determined that f-PADDA is approximately five times more likely (Odds ratio = 5.1; exact 95% CI: 0.72–41.89; P = 0.105) to detect damage on nucleotides previously reported to be mutated than to detect damage on nucleotides not identified as mutated by the literature (Figure 4). The 5-fold magnitude of the Odds ratio is quite impressive because only lesions in the NTS of the CAN1 gene were mapped and only those mutations that disrupt the CAN1 function have been reported in the literature and considered in this analysis. These data strongly support the widely spread hypothesis that persistent endogenous DNA damage leads to mutation fixation.
Figure 4.
Co-localization of endogenous nucleotide damage detected in the NTS and spontaneous mutations in the CAN1 gene. Shown is a representative fragment of the CAN1 ORF (CAN1, X03784) nucleotides 1180–1215. Y-axis values represent number of instances of nucleotide damage mapped by f-PADDA (top panel) or mutations previously reported in literature (bottom panel).
Endogenous damage accumulates preferentially on the NTS
We showed that PADDA originates a pool of strand-specific biotin-tagged extended products, obtained from a non-cycled primer-extension, which reflect with high sensitivity the levels of strand-specific endogenous DNA damage. Therefore, the use of this pool of extended products as template for real-time PCR reactions (q-PADDA) should allow for the quantification of strand-specific endogenous DNA damage and clarify whether the preferential repair of oxidative damage occurs in vivo. To determine the levels of endogenous strand-specific DNA damage, we used q-PADDA to analyze the transcribed and NTS of an actively transcribed gene (CAN1) in WT cells in exponential and stationary growth phases.
Consistent with the data obtained by f-PADDA for the CAN1 gene and the reported higher levels of ROS in yeast cells in stationary phase than in exponential phase (11,48–50) independently of the genotype, we observed significantly more endogenous DNA damage in cells from stationary phase than from exponential phase (Supplementary Figure S5). These data demonstrate that q-PADDA allows for highly sensitive strand-specific quantification of endogenous DNA damage. To determine the relative levels of DNA damage between the transcribed and the NTS, we first repaired in vitro the DNA obtained from WT cells in exponential phase and then calculated the fold difference in damage per specific-strand relative to repaired WT. Our data shows that, independently of the growth phase, WT cells have significantly more endogenous DNA damage (P < 0.001) in the CAN1 NTS than the CAN1 transcribed strand (Figure 5). Furthermore, the data shows that in WT cells the rate of damage accumulation from exponential to stationary phase is significantly higher in the NTS (3.2-fold) than in the transcribed strand (1.9-fold) (Figure 5). Overall, our data indicate that in WT cells there is preferential repair of endogenous DNA damage on the transcribed strand of actively transcribed genes.
Figure 5.
Strand-specific DNA damage quantification by q-PADDA in WT and BER− cells. Fold difference reflects damage normalization against in vitro repaired DNA extracted from WT cells in exponential phase of growth. Data shown represents mean ± SD *P = 0.0001, **P = 0.001, ***P = 0.002, ****P = 2 × 10.−9, *****P = 0.007, ******P = 5 × 10−6.
BER repairs endogenous DNA damage preferentially in the transcribed strand
Consistent with the known role of BER in the repair of endogenous DNA damage, we observed that independent of growth phase BER− cells have significantly higher levels of endogenous DNA damage than WT cells (Supplementary Figure S5). Furthermore, by determining the relative levels of DNA damage between the transcribed and the NTS relative to exponential WT in vitro repaired DNA (Figure 5), we demonstrate that BER− cells in exponential phase have significantly more DNA damage in the NTS than in the transcribed strand (P < 0.01). However, and in contrast with WT cells, BER defective cells in stationary phase have more endogenous DNA damage in the CAN1 transcribed strand than in the CAN1 NTS (Figure 5). Our data demonstrate that in BER− cells, the rate of damage accumulation is significantly higher (P = 1 × 10−4) for the CAN1 actively transcribed strand (5-fold) than for the NTS (1.7-fold), establishing for the first time that BER plays a major role in the preferential repair of endogenous DNA damage localized in the transcribed strand of actively transcribed genes.
DISCUSSION
Despite the profound implications of endogenous DNA damage in human diseases (1–4) and mounting evidence that oxidative damage in the transcribed strand of actively transcribed genes block transcription (19,51), due to technical limitations it has been impossible to document transcription-coupled repair of endogenous DNA damage (19), or even establish the overall levels of endogenous DNA damage in different cell systems (7,10).
We have developed a novel DNA damage detection assay (PADDA) with sensitivity and specificity to map and quantify at the single nucleotide level in vivo endogenous base damage. To the best of our knowledge, PADDA is the first reported technique that has the sensitivity to quantify and differentiate strand-specific levels of endogenous DNA damage in <5 h after DNA extraction. PADDA belongs to a general class of approaches that take advantage of polymerase elongation properties as a sensor for damage on the template DNA (13–16,52,53). Particularly, PADDA can be included in a sub-class of assays that embrace TD-PCR (13,52), ss-QPCR, sslig-PCR (14) and QPCR (15). These polymerase based damage detection assays can map and quantify with some accuracy chemical and radiation induced DNA damage. However, due to multiple limitations they are neither widely used for the detection of induced DNA damage nor reliable for the detection of endogenous DNA damage. These limitations include: (i) low sensitivity (53), (ii) high background (14,53), (iii) capacity to identify only DNA damage blocking lesions or technically introduced DNA strand breaks (14), (iv) requirement of radioactive materials and multiple step optimization for each genomic area in study (14,15) and (v) end-point quantitative PCR analysis (14–16). In contrast, PADDA is highly sensitive, simple to perform, relatively inexpensive, and does not require hazardous reagents, sophisticated equipment or specialized skills.
The high sensitivity of PADDA results from the combination of several factors including: (i) a DNA extraction procedure that aims to reduce damage introduced during processing; (ii) a single non-cycled primer-extension step, which prevents the successive annealing of previously extended products to different template molecules, assuring that each extended product reflects nucleotide damage at a single strand and cell level, thus facilitating the detection of even very low levels of nucleotide lesions; (iii) the use of a 5′-biotin-tagged specific primer combined with several purification steps aimed at reducing spurious background; (iv) the use of a polymerase (Vent exo–) with high sensitivity to detect DNA damage and high primer-extension efficiency (22–24,54); (v) the use of an adapter–primer chemically modified to decrease artifacts; (vi) use of a highly efficient single-strand adapter ligase (25); (vii) the ability to retrieve information about damaged sites from sequenced individual primer extensions, allowing the mapping of lesions that lead to polymerase bypass or arrest with misincorporation opposite to a damaged nucleotide, extending the assay's damage detection capacity to a broader range of lesions than previously reported assays; (viii) the strategy to quantify damage using real-time PCR analysis, rather than end-point analysis, allowing the detection of subtle differences in damage levels, and opening the possibility for a practical and rapid high-throughput DNA damage detection assay relevant to population screenings; (ix) the optional use of in vitro repaired DNA, which theoretically contains minimal levels of DNA damage in both strands, permitting for the first time the ability to evaluate the relative levels of damage between the two strands of a given gene.
Here we used isogenic WT and BER− yeast cells in distinct phases of cell growth as a model to validate our novel assay. BER involves proteins with overlapping specificities. The BER− cells used in our study have deletions of ntg1 and ntg2, N-glycosylases required for the removal of oxidized pyrimidines and other lesions, and apn1, the major apurinic-apyrimidinic endonuclease in S. cerevisiae (11). BER− and WT cells have similar and very low rates of cell death ((11,55) and data not shown). Our data demonstrate that BER− cells have significantly higher levels of endogenous DNA damage than WT cells. We estimate that WT and BER− cells in exponential phase of growth have respectively ~3500 and 6300 lesions per genome, while WT and BER− stationary cells have ~12 900 and 18 700 lesions per genome, respectively. The fold difference in base damage for WT and BER− cells in stationary phase is similar to what is reported (11). However, our numbers are ~10- to 20-fold higher than those estimated for Ntg1p-recognized DNA lesions using a Southern blot technique (11). Two factors contribute for this difference: PADDA recognizes a much broader type of DNA lesions than Ntg1 and is much more sensitive than a Southern blot approach. Our data are consistent with the reported significantly higher levels of O2− (49,50) and Ntgp1 recognized DNA lesions (11) in BER− cells compared to the WT cells. Furthermore, consistent with the reported levels of Ntgp1 recognized DNA lesions (11) and ROS (11,48,56), we observed that for both genotypes, cells in stationary phase have significantly higher levels of damage in both DNA strands than cells in exponential growth phase. Overall, for the first time these results demonstrate that PADDA's high sensitivity provides a tool for strand-specific mapping and quantification of endogenous DNA damage.
Most of the identified putative damage on the CAN1 gene is represented as Stops. We observed a significant variation in the size of extended products that is consistent with the expected relative levels of damage for each strain and growth phase studied. This is consistent with previous reports that used Vent exo− polymerase to detect chemically distinct photoproducts (13,21,52) and the fact that multiple nucleotide lesions represent effective blocks to DNA replication (57). Consistent with the Stops representing authentic DNA lesions, we also observed a significant increase in the size of extended products after in vitro repair. The repair system has been used to repair a range of different lesions in vitro [(46,47) and www.neb.com] and contains several enzymes that have been shown to repair oxidatively damaged DNA (58–60), and share significant substrate overlap with Ntg1p and Ntg2p recognized lesions (59,60).
In addition to Stops, we observed high levels of polymerase misincorporation (SMs and LBMs) in BER− cells, which have been shown to accumulate high levels of ROS (48,49) and Ntg1p recognizable lesions (11,48–50). In contrast, SMs or LBMs were not observed in WT cells in exponential phase of growth, which are reported to have low levels of ROS and undetectable levels of Ntg1p recognizable lesions (48,49). This is consistent with LBMs and SMs representing lesion-dependent misincorporations opposite to a damaged base, and therefore being direct indicators of the position of in vivo DNA damage. The observation of misincorporation at the stop-site (SM), or polymerase bypass with a misincorporation opposite the damaged base (LBM) in non-genomic DNA is well established (42). However, to the best of our knowledge, f-PADDA is the first DNA damage detection methodology that documents the in vivo position of genomic damage leading to lesion-dependent misincorporations (SMs and LBMs). This capacity to expose the potential mutagenicity of the damaged DNA, usually only revealed during its replication by a DNA polymerase (61,62), is a unique and important advantage of our approach.
The observation that persistent endogenous nucleotide damage in the CAN1 gene co-localizes with CAN1 spontaneous gene mutations strongly reveals the importance of PADDA in the detection of damage with highly mutagenic potential. In this regard, the detected LBMs suggest that PADDA detects lesions that may lead to in vivo error-prone translesion DNA synthesis, and ultimately to mutation fixation in vivo. Error-prone replicative by-pass of chemically modified DNA bases (LBM in our data) is a biologically significant process in mammalian cells leading to hyper-mutability and subsequent cancer predisposition, such as in the disease xeroderma pigmentosum variant form (42). Therefore, f-PADDA's ability to precisely map by-passable DNA lesions of high mutagenic potential opens new opportunities for multiple studies, including the early detection of cancer precursor DNA lesions in high risk populations.
Of major importance, we documented that WT cells have significantly lower levels of endogenous DNA damage in the transcribed strand than in the NTS of the actively transcribed CAN1 gene. This data strongly supports that WT cells repair endogenous damage preferentially on the transcribed strand, and strengthens a link between repair of endogenous damage and transcription. Given the functional overlap between BER and NER in the repair of oxidative damage, the preferential repair of endogenous damage on the transcribed strand could potentially occur through NER and/or BER. For example, transcription-dependent repair of 8-oxoG has been reported in E. coli (63) and mouse embryonic fibroblasts (64,65), and was shown to require CSA and CSB, proteins with a known role in TC-NER, as well as Ogg1, a BER DNA N-glycosylase (63–65). Here we show that the rate of endogenous damage accumulation is significantly higher in the transcribed than in the NTS of BER− cells. Additionally, and in contrast with WT cells, BER− cells in stationary phase have more DNA damage in the transcribed strand than in the NTS. These data are consistent with recent biochemical and in vitro data (19) and strongly support a major role for BER in the preferential repair of endogenous DNA damage in the transcribed strand of actively transcribed genes. Recently, using a reporter system, it was shown that NER can preferentially remove abasic sites in the transcribed strand of yeast DNA (66). This suggests that in our system, the damage in the transcribed strand resulting directly from the lack of BER is underestimated due to a probable NER compensatory role. However, we cannot exclude that other repair pathways compensate for the loss of BER more on the NTS than on the transcribed strand.
In theory, q-PADDA should be applicable for the detection of DNA damage in virtually any nucleic acid sequence from virtually any specie. For example, we have demonstrated that q-PADDA is able to quantify endogenous and induced DNA damage in the TRP53 gene of WT and repair defective mice (unpublished data). PADDA's unprecedented sensitivity to identify a broad spectrum of DNA lesions and its ability to quantify on a high-throughput setting in vivo endogenous and induced DNA damage makes it quite attractive for large population studies. Therefore, it may become a critical tool for assessing differential susceptibility to specific exposures as well as cancer risk, prevention and treatment outcome.
To the best of our knowledge, this is the first study to document differences in levels of endogenous DNA damage between strands in WT cells. Importantly, and consistent with the fact that we are detecting a variety of lesions with high sensitivity, we established significant differences between WT and BER− cells on a per strand basis even during exponential phase, when the levels of ROS and DNA damage are expected to be the lowest. Furthermore, our data for the first time provide direct evidence for the preferential repair of endogenous DNA damage in the transcribed strand of actively transcribed genes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Presbyterian Health Foundation (C5019101 to A.R.); National Institutes of Health (R03 CA117316-01 to A.R.). L.Q. holds a Presbyterian Health Foundation Endowed Chair in Otorhinolaryngology Position. Funding for open access charge: Department of Otorhinolaryngology of the University of Oklahoma Health Sciences Center.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Paul Doetsch (Emory University, Atlanta, GA) for kindly providing the yeast strains used in this study. Contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of NIH. This article is dedicated to the memory of António Manuel Corga Reis who developed the PADDA assay and died tragically of cancer at the age of 51.
REFERENCES
- 1.Doll R, Peto R. The causes of cancer: quantitative estimates of avoidable risks of cancer in the United States today. J. Natl Cancer. Inst. 1981;66:1191–1308. [PubMed] [Google Scholar]
- 2.Hasty P, Campisi J, Hoeijmakers J, van Steeg H, Vijg J. Aging and genome maintenance: lessons from the mouse? Science. 2003;299:1355–1359. doi: 10.1126/science.1079161. [DOI] [PubMed] [Google Scholar]
- 3.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein C, Bernstein H, Payne CM, Garewal H. DNA repair/pro-apoptotic dual-role proteins in five major DNA repair pathways: fail-safe protection against carcinogenesis. Mutat. Res. 2002;511:145–178. doi: 10.1016/s1383-5742(02)00009-1. [DOI] [PubMed] [Google Scholar]
- 5.Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 6.Swanson RL, Morey NJ, Doetsch PW, Jinks-Robertson S. Overlapping specificities of base excision repair, nucleotide excision repair, recombination, and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol. Cell Biol. 1999;19:2929–2935. doi: 10.1128/mcb.19.4.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cadet J, Douki T, Gasparutto D, Ravanat JL. Oxidative damage to DNA: formation, measurement and biochemical features. Mutat. Res. 2003;531:5–23. doi: 10.1016/j.mrfmmm.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Beckman KB, Ames BN. Oxidative decay of DNA. J. Biol. Chem. 1997;272:19633–19636. doi: 10.1074/jbc.272.32.19633. [DOI] [PubMed] [Google Scholar]
- 9.Anson RM, Mason PA, Bohr VA. Gene-specific and mitochondrial repair of oxidative DNA damage. Methods Mol. Biol. 2006;314:155–181. doi: 10.1385/1-59259-973-7:155. [DOI] [PubMed] [Google Scholar]
- 10.(ESCODD), T.E.S.C.o.O.D.D. Measurement of DNA oxidation in human cells by chromatographic and enzymic methods. Free Radic. Biol. Med. 2003;34:1089–1099. doi: 10.1016/s0891-5849(03)00041-8. [DOI] [PubMed] [Google Scholar]
- 11.Evert BA, Salmon TB, Song B, Jingjing L, Siede W, Doetsch PW. Spontaneous DNA damage in Saccharomyces cerevisiae elicits phenotypic properties similar to cancer cells. J. Biol. Chem. 2004;279:22585–22594. doi: 10.1074/jbc.M400468200. [DOI] [PubMed] [Google Scholar]
- 12.Johansson C, Moller P, Forchhammer L, Loft S, Godschalk RWL, Langie SAS, Lumeij S, Jones GDD, Kwok RWL, Azqueta A, et al. An ECVAG trial on assessment of oxidative damage to DNA measured by the comet assay. Mutagenesis. 2010;25:125–132. doi: 10.1093/mutage/gep055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfeifer GP, Chen HH, Komura J, Riggs AD. Chromatin structure analysis by ligation-mediated and terminal transferase-mediated polymerase chain reaction. Methods Enzymol. 1999;304:548–571. doi: 10.1016/s0076-6879(99)04032-x. [DOI] [PubMed] [Google Scholar]
- 14.Grimaldi KA, McGurk CJ, McHugh PJ, Hartley JA. PCR-based methods for detecting DNA damage and its repair at the sub-gene and single nucleotide levels in cells. Mol. Biotechnol. 2002;20:181–196. doi: 10.1385/MB:20:2:181. [DOI] [PubMed] [Google Scholar]
- 15.Ayala-Torres S, Chen Y, Svoboda T, Rosenblatt J, Van Houten B. Analysis of gene-specific DNA damage and repair using quantitative polymerase chain reaction. Methods. 2000;22:135–147. doi: 10.1006/meth.2000.1054. [DOI] [PubMed] [Google Scholar]
- 16.Kovalenko OA, Santos JH. Analysis of oxidative damage by gene-specific quantitative PCR. Curr. Protoc. Hum. Genet. 2009 doi: 10.1002/0471142905.hg1901s62. Chapter 19, Unit 19 11. [DOI] [PubMed] [Google Scholar]
- 17.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 18.Tornaletti S. DNA repair in mammalian cells. Cell. Mol. Life Sci. 2009;66:1010–1020. doi: 10.1007/s00018-009-8738-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banerjee D, Mandal SM, Das A, Hegde ML, Das S, Bhakat KK, Boldogh I, Sarkar PS, Mitra S, Hazra TK. Preferential repair of oxidized base damage in the transcribed genes of mammalian cells. J. Biol. Chem. 2011;286:6006–6016. doi: 10.1074/jbc.M110.198796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frosina G. The current evidence for defective repair of oxidatively damaged DNA in Cockayne syndrome. Free Radic. Biol. Med. 2007;43:165–177. doi: 10.1016/j.freeradbiomed.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 21.Smith CA, Baeten J, Taylor JS. The ability of a variety of polymerases to synthesize past site-specific cis-syn, trans-syn-II, (6-4), and Dewar photoproducts of thymidylyl-(3′–>5′)-thymidine. J. Biol. Chem. 1998;273:21933–21940. doi: 10.1074/jbc.273.34.21933. [DOI] [PubMed] [Google Scholar]
- 22.Vigneault F, Drouin R. Optimal conditions and specific characteristics of Vent exo- DNA polymerase in ligation-mediated polymerase chain reaction protocols. Biochem. Cell Biol. 2005;83:147–165. doi: 10.1139/o04-134. [DOI] [PubMed] [Google Scholar]
- 23.Garrity PA, Wold BJ. Effects of different DNA polymerases in ligation-mediated PCR: enhanced genomic sequencing and in vivo footprinting. Proc. Natl Acad. Sci. USA. 1992;89:1021–1025. doi: 10.1073/pnas.89.3.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Angers M, Cloutier JF, Castonguay A, Drouin R. Optimal conditions to use Pfu exo(-) DNA polymerase for highly efficient ligation-mediated polymerase chain reaction protocols. Nucleic Acids Res. 2001;29:E83. doi: 10.1093/nar/29.16.e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blondal T, Hjorleifsdottir SH, Fridjonsson OF, Aevarsson A, Skirnisdottir S, Hermannsdottir AG, Hreggvidsson GO, Smith AV, Kristjansson JK. Discovery and characterization of a thermostable bacteriophage RNA ligase homologous to T4 RNA ligase 1. Nucleic Acids Res. 2003;31:7247–7254. doi: 10.1093/nar/gkg914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Mattila P, Korpela J, Tenkanen T, Pitkanen K. Fidelity of DNA synthesis by the Thermococcus litoralis DNA polymerase–an extremely heat stable enzyme with proofreading activity. Nucleic Acids Res. 1991;19:4967–4973. doi: 10.1093/nar/19.18.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lang GI, Murray AW. Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics. 2008;178:67–82. doi: 10.1534/genetics.107.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohnishi G, Endo K, Doi A, Fujita A, Daigaku Y. Spontaneous mutagenesis in haploid and diploid Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2004;325:928–933. doi: 10.1016/j.bbrc.2004.10.120. [DOI] [PubMed] [Google Scholar]
- 30.Winnier AR, Meir JY, Ross JM, Tavernarakis N, Driscoll M, Ishihara T, Katsura I, Miller DM., 3rd UNC-4/UNC-37-dependent repression of motor neuron-specific genes controls synaptic choice in Caenorhabditis elegans. Genes Dev. 1999;13:2774–2786. doi: 10.1101/gad.13.21.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sikorsky JA, Primerano DA, Fenger TW, Denvir J. Effect of DNA damage on PCR amplification efficiency with the relative threshold cycle method. Biochem. Biophys. Res. Commun. 2004;323:823–830. doi: 10.1016/j.bbrc.2004.08.168. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez H, Akman SA. Mapping oxidative DNA damage at nucleotide level. Free Radic. Res. 1998;29:499–510. doi: 10.1080/10715769800300551. [DOI] [PubMed] [Google Scholar]
- 33.Akman SA, O'Connor TR, Rodriguez H. Mapping oxidative DNA damage and mechanisms of repair. Ann. NY Acad. Sci. 2000;899:88–102. doi: 10.1111/j.1749-6632.2000.tb06178.x. [DOI] [PubMed] [Google Scholar]
- 34.Heaton PR, Ransley R, Charlton CJ, Mann SJ, Stevenson J, Smith BH, Rawlings JM, Harper EJ. Application of single-cell gel electrophoresis (comet) assay for assessing levels of DNA damage in canine and feline leukocytes. J. Nutr. 2002;132:1598S–1603S. doi: 10.1093/jn/132.6.1598S. [DOI] [PubMed] [Google Scholar]
- 35.Rothfuss O, Gasser T, Patenge N. Analysis of differential DNA damage in the mitochondrial genome employing a semi-long run real-time PCR approach. Nucleic Acids Res. 2010;38:e24. doi: 10.1093/nar/gkp1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beranek DT. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat. Res. 1990;231:11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- 37.Encell L, Shuker DE, Foiles PG, Gold B. The in vitro methylation of DNA by a minor groove binding methyl sulfonate ester. Chem. Res. Toxicol. 1996;9:563–567. doi: 10.1021/tx9501849. [DOI] [PubMed] [Google Scholar]
- 38.Guillet M, Boiteux S. Endogenous DNA abasic sites cause cell death in the absence of Apn1, Apn2 and Rad1/Rad10 in Saccharomyces cerevisiae. Eur. Mol. Biol. Organization. 2002;21:2833–2841. doi: 10.1093/emboj/21.11.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greagg MA, Fogg MJ, Panayotou G, Evans SJ, Connolly BA, Pearl LH. A read-ahead function in archaeal DNA polymerases detects promutagenic template-strand uracil. Proc. Natl Acad. Sci. USA. 1999;96:9045–9050. doi: 10.1073/pnas.96.16.9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Regenberg B, Düring-Olsen L, Kielland-Brandt MC, Holmberg S. Substrate specificity and gene expression of the amino-acid permeases in Saccharomyces cerevisiae. Curr. Genet. 1999;36:317–328. doi: 10.1007/s002940050506. [DOI] [PubMed] [Google Scholar]
- 41.Ahmad M, Bussey H. Yeast arginine permease: nucleotide sequence of the CAN1 gene. Curr. Genet. 1986;10:587–592. doi: 10.1007/BF00418125. [DOI] [PubMed] [Google Scholar]
- 42.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd edn. Washington, DC: American Society Microbiology; 2005. [Google Scholar]
- 43.You HJ, Swanson RL, Harrington C, Corbett AH, Jinks-Robertson S, Senturker S, Wallace SS, Boiteux S, Dizdaroglu M, Doetsch PW. Saccharomyces cerevisiae Ntg1p and Ntg2p: broad specificity N-glycosylases for the repair of oxidative DNA damage in the nucleus and mitochondria. Biochemistry. 1999;38:11298–11306. doi: 10.1021/bi991121i. [DOI] [PubMed] [Google Scholar]
- 44.Meadows KL, Song B, Doetsch PW. Characterization of AP lyase activities of Saccharomyces cerevisiae Ntg1p and Ntg2p: implications for biological function. Nucleic Acids Res. 2003;31:5560–5567. doi: 10.1093/nar/gkg749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bruner SD, Nash HM, Lane WS, Verdine GL. Repair of oxidatively damaged guanine in Saccharomyces cerevisiae by an alternative pathway. Curr. Biol. 1998;8:393–403. doi: 10.1016/s0960-9822(98)70158-7. [DOI] [PubMed] [Google Scholar]
- 46.Stein V, Hollfelder F. An efficient method to assemble linear DNA templates for in vitro screening and selection systems. Nucleic Acids Res. 2009;37:e122. doi: 10.1093/nar/gkp589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teng Y, Bennett M, Evans KE, Zhuang-Jackson H, Higgs A, Reed SH, Waters R. A novel method for the genome-wide high resolution analysis of DNA damage. Nucleic Acids Res. 2011;39:e10. doi: 10.1093/nar/gkq1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laun P, Pichova A, Madeo F, Fuchs J, Ellinger A, Kohlwein S, Dawes I, Frohlich KU, Breitenbach M. Aged mother cells of Saccharomyces cerevisiae show markers of oxidative stress and apoptosis. Mol. Microbiol. 2001;39:1166–1173. [PubMed] [Google Scholar]
- 49.Rowe LA, Degtyareva N, Doetsch PW. DNA damage-induced reactive oxygen species (ROS) stress response in Saccharomyces cerevisiae. Free Radic. Biol. Med. 2008;45:1167–1177. doi: 10.1016/j.freeradbiomed.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salmon TB, Evert BA, Song B, Doetsch PW. Biological consequences of oxidative stress-induced DNA damage in Saccharomyces cerevisiae. Nucleic Acids Res. 2004;32:3712–3723. doi: 10.1093/nar/gkh696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kitsera N, Stathis D, Luhnsdorf B, Muller H, Carell T, Epe B, Khobta A. 8-Oxo-7,8-dihydroguanine in DNA does not constitute a barrier to transcription, but is converted into transcription-blocking damage by OGG1. Nucleic Acids Res. 2011;39:5926–5934. doi: 10.1093/nar/gkr163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Komura J, Riggs AD. Terminal transferase-dependent PCR: a versatile and sensitive method for in vivo footprinting and detection of DNA adducts. Nucleic Acids Res. 1998;26:1807–1811. doi: 10.1093/nar/26.7.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McGurk CJ, McHugh PJ, Tilby MJ, Grimaldi KA, Hartley JA. Measurement of covalent drug-DNA interactions at the nucleotide level in cells at pharmacologically relevant doses. Methods Enzymol. 2001;340:358–376. doi: 10.1016/s0076-6879(01)40431-9. [DOI] [PubMed] [Google Scholar]
- 54.Luu H, Zagaja G, Dubauskas Z, Chen S, Smith R, Watabe K, Ichikawa Y, Ichikawa T, Davis E, Le Beau M, et al. Identification of a novel metastasis-suppressor region on human chromosome 12. Cancer Res. 1998;58:3561–3565. [PubMed] [Google Scholar]
- 55.Pawar V, Jingjing L, Zhang H, Patel N, Kaur N, Doetsch P, Shadel G, Siede W. Checkpoint kinase phosphorylation in response to endogenous oxidative DNA damage in repair-deficient stationary-phase Saccharomyces cerevisiae. Mech. Ageing Dev. 2009;130:501–508. doi: 10.1016/j.mad.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Godschalk R, Nair J, van Schooten FJ, Risch A, Drings P, Kayser K, Dienemann H, Bartsch H. Comparison of multiple DNA adduct types in tumor adjacent human lung tissue: effect of cigarette smoking. Carcinogenesis. 2002;23:2081–2086. doi: 10.1093/carcin/23.12.2081. [DOI] [PubMed] [Google Scholar]
- 57.Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 58.Senturker S, Auffret van der Kemp P, You H, Doetsch P, Dizdaroglu M, Boiteux S. Substrate specificities of the ntg1 and ntg2 proteins of Saccharomyces cerevisiae for oxidized DNA bases are not identical. Nucleic Acids Res. 1998;26:5270–5276. doi: 10.1093/nar/26.23.5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boiteux S, Gajewski E, Laval J, Dizdaroglu M. Substrate specificity of the Escherichia coli Fpg protein formamidopyrimidine-DNA glycosylase: excision of purine lesions in DNA produced by ionizing radiation or photosensitization. Biochemistry. 2002;31:106–110. doi: 10.1021/bi00116a016. [DOI] [PubMed] [Google Scholar]
- 60.Wiederholt CJ, Patro JN, Jiang YL, Haraguchi K, Greenberg MM. Excision of formamidopyrimidine lesions by endonucleases III and VIII is not a major DNA repair pathway in Escherichia coli. Nucleic Acids Res. 2005;33:3331–3338. doi: 10.1093/nar/gki655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feig DI, Loeb LA. Oxygen radical induced mutagenesis is DNA polymerase specific. J. Mol. Biol. 1994;235:33–41. doi: 10.1016/s0022-2836(05)80009-9. [DOI] [PubMed] [Google Scholar]
- 62.Wang D, Kreutzer DA, Essigmann JM. Mutagenicity and repair of oxidative DNA damage: insights from studies using defined lesions. Mutat. Res. 1998;400:99–115. doi: 10.1016/s0027-5107(98)00066-9. [DOI] [PubMed] [Google Scholar]
- 63.Bregeon D, Doddridge ZA, You HJ, Weiss B, Doetsch PW. Transcriptional mutagenesis induced by uracil and 8-oxoguanine in Escherichia coli. Mol. Cell. 2003;12:959–970. doi: 10.1016/s1097-2765(03)00360-5. [DOI] [PubMed] [Google Scholar]
- 64.Spivak G, Hanawalt PC. Host cell reactivation of plasmids containing oxidative DNA lesions is defective in Cockayne syndrome but normal in UV-sensitive syndrome fibroblasts. DNA Repair. 2006;5:13–22. doi: 10.1016/j.dnarep.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 65.Pastoriza-Gallego M, Armier J, Sarasin A. Transcription through 8-oxoguanine in DNA repair-proficient and Csb /Ogg1 DNA repair-deficient mouse embryonic fibroblasts is dependent upon promoter strength and sequence context. Mutagenesis. 2007;22:343–351. doi: 10.1093/mutage/gem024. [DOI] [PubMed] [Google Scholar]
- 66.Kim N, Jinks-Robertson S. Abasic sites in the transcribed strand of yeast DNA are removed by transcription-coupled nucleotide excision repair. Mol. Cell. Biol. 2010;30:3206–3215. doi: 10.1128/MCB.00308-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.