Abstract
SR proteins and related factors play widespread roles in alternative pre-mRNA splicing and are known to promote splice site recognition through their Arg–Ser-rich effector domains. However, binding of SR regulators to some targets results in repression of splice sites through a distinct mechanism. Here, we investigate how activated and repressed targets of the Drosophila SR regulator Transformer2 elicit its differing effects on splicing. We find that, like activation, repression affects early steps in the recognition of splice sites and spliceosome assembly. Repositioning of regulatory elements reveals that Tra2 complexes that normally repress splicing from intronic positions activate splicing when located in an exon. Protein tethering experiments demonstrate that this position dependence is an intrinsic property of Tra2 and further show that repression and activation are mediated by separate effector domains of this protein. When other Drosophila SR factors (SF2 and Rbp1) that activate splicing from exonic positions were tethered intronically they failed to either activate or repress splicing. Interestingly, both activities of Tra2 favor the exonic identity of the RNA sequences that encompass its binding sites. This suggests a model in which these two opposite functions act in concert to define both the position and extent of alternatively spliced exons.
INTRODUCTION
Alternative pre-mRNA splicing is a widespread regulatory mechanism by which individual genes can express variant proteins with distinct functions. The selection of alternative splice sites depends on sequence-specific association of pre-mRNAs with splicing regulatory factors that promote or repress their recognition by the spliceosome. The sites bound by regulators can be located in either exons or introns and either adjacent to, or distant from, the affected splice site itself. Interestingly, number splicing regulators have the ability to either activate or repress splice sites depending on their target pre-mRNA (1–6). The position of binding in relation to the affected splice site is an important factor influencing these different effects of splicing regulators. For example recent transcriptome-wide mapping of RNA-binding sites of Nova, PTB and Fox2 proteins revealed that binding within the upstream or downstream intron tends to be associated with opposite effects on regulated exon skipping (5,7–9). At present the mechanisms responsible for such location-specific effects are in most cases poorly understood but are likely to involve location-specific interactions of these factors with the pre-spliceosomal complexes that are key to splice site recognition.
SR proteins and related splicing factors have important roles in exon definition and the regulation of alternative splicing (10,11).This is exemplified by the production of alternative mRNAs from Drosophila sex determination genes. In this system, the splicing of doublesex (dsx) and fruitless (fru) pre-mRNAs is under the control of the SR-related regulators Transformer (Tra) and Transformer-2 (Tra2) (12). These factors, along with other SR proteins, form complexes on exonic splicing enhancers (ESEs) in these pre-mRNAs that promote the use of sex-specific alternative splice sites (13–15). Tra2 is a required component of these enhancer complexes and is able to activate splicing independently of other SR factors when it is tethered downstream of a 3′-splice site (16,17). More generally, SR splicing factors activate splice sites when they are bound to ESEs. Notably, this occurs not only in cases of alternatively spliced exons but also in many constitutively spliced exons that depend on activation to prevent exon skipping. The ability to activate splicing results from the Arg–Ser-rich effector regions characteristic of SR factors. These domains interact directly with components of pre-spliceosomal complexes to facilitate their assembly at the affected splice sites (16,18,19).
In addition to these well-established functions, Tra2 and other SR factors also repress splicing of some targets (6,20–25). For example, Tra2 causes retention of the M1 intron in its own pre-mRNA as part of a negative feedback mechanism that limits its function in vivo (Supplementary Figure S1) (6,22,26). Repression is mediated by an intronic splicing silencer (ISS) region with multiple Tra2-binding sites (27). Both the repression of M1 and the activation of dsx splicing by Tra2 have been observed to occur together in the same cells (28) suggesting that cell type-specific factors are unlikely to explain the different effects on splicing in these targets. However, the ESE elements found in dsx and fru differ from the M1 ISS in both their exon/intron location and their component sequences (27) raising the possibility that the composition of regulatory complexes, or their positional relationship to affected splice sites is responsible for the different effects on splicing. Here, we investigate the way in which Tra2–ISS complexes affect spliceosome assembly and examine the requirements for their repressive function. Our results indicate that repression and activation are distinct and separable effector activities of the Tra2 protein itself and that its position of binding in the pre-mRNA determines how target splice sites are affected. We suggest that Tra2, and perhaps other SR regulators, utilize repression in concert with activation to define alternatively spliced exons.
MATERIALS AND METHODS
Transcription DNA templates and plasmids
The splicing substrate ftz-ISS contains wild-type (wt) sequences from the 78-nt ISS of the M1 intron flanked by ftz intron and exon sequences. Splicing substrate RNAs from this plasmid and ftz gene sequences were generated as described previously (27). To generate dsx-ISS hybrid splicing substrates lacking the dsx ESE in the female-specific exon, a 225-nt EcoRI–XmaI fragment containing dsx exon 3, intron 3 and 35 nt of the female-specific exon 4 was PCR-amplified from pdsx (29) and cloned into the pGEM2 plasmid vector (Promega). An 80-nt XmaI–XbaI sequence containing the ISS was inserted following the exon 4 segment to generate pdsx-ISS. An identical segment in which point mutations converted each of the five CAAGR repeats to CTGCT was used to generate pdsx-5mt.
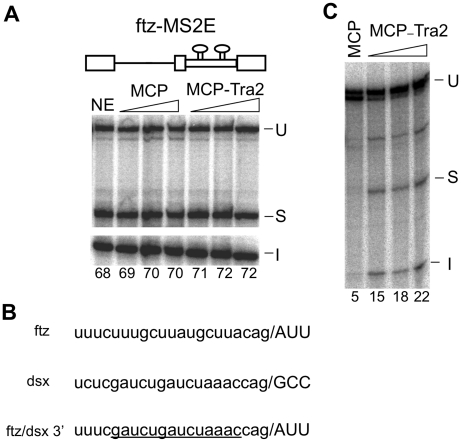
The dsxMS2 splicing substrate was generated from the plasmid pdsx70(MS2)2 (17) and contains two high affinity MS2 coat protein (MCP)-binding sequences separated by a 15-nt spacer. The same binding sites were used in the plasmid encoding the ftzM1-MS2 substrate. The MS2 sequences were inserted in place of the 78-nt ISS sequence of pftzM1-208 (27). To generate a series of ftz-MS2 hybrid RNA substrates, the plasmid pG6V21 (6) which carries sequences from the wt ftz gene was modified with two unique restriction sites (BsiWI and MluI) introduced at various locations in the intron or 3′ exon, A 60-nt BsiWI–MluI MS2-binding fragment with the same MS2 sequences and spacer as above was then inserted in the intron either 10 (ftzMS2-10) or 50 (ftzMS2-50) nt upstream of the branch point or 30-nt downstream (ftzMS2E) of the 3′-splice site. The bulged adenosines in these sites required for MS2 protein binding were deleted by PCR amplification from wt MS2-binding sequence with mutagenic primers. The mutant MS2-binding sequence were inserted 40-nt upstream of the 3′-splice site to create the plasmid ftzMS2-mt. The plasmid pftz3′dE was generated by replacing sequences upstream of the ftz 3′-splice site in pG6V21 with dsx sequences using the same approach.
Construction of the splicing reporter plasmids pActdsxISS and pActdsxISS-5mt was based on the plasmid pUC18-ActGFP (Drosophila Genomics Resource Center, DGRC). The same dsxISS and dsxISS-5mt DNA fragments described above for production of in vitro splicing substrates were amplified and inserted into GFP-coding region in pActGFP. The protein expression plasmid pAct-Flag-Tra2 was also derived from pActGFP by replacing GFP cDNA with a 792-bp BamH1–HindIII fragment of Tra2 cDNA encoding the full-length Tra2 protein and an N-terminal 3X Flag tag.
Protein expression and purification
Recombinant His-Tra2 protein corresponding to the Tra2-PA isoform was prepared using the same methods described previously (6). To prepare MCP fusion proteins, Tra2 coding sequences were inserted into the vector pHis-BIVT-MS2 (30) with BamH1 and HindIII sites. The MCP-coding sequences in this and all other constructs contain point mutations rendering MCP deficient in formation of capsid oligomers (31). Coding sequences for Rbp1, and ASF/SF2 and various subsegments of Tra2 were also cloned into pHis-BIVT-MS2 vector. Fusion protein expression constructs with mutations in the Tra2 RRM domain were recreated by amplification of a Tra2 cDNA with outwardly oriented mutagenic primers followed by insertion into pHis-BIVT-MS2 and complete resequencing. All MCP fusion proteins were expressed from baculovirus infected Sf9 cells as described previously (6). Cells were harvested 3 days after infection and the proteins were purified under denaturing conditions on Ni-NTA agarose and dialyzed against buffer BC850 (20 mM Tris–HCl, 850 mM KCl, 20% glycerol). The purified proteins were stained with coomassie blue after SDS–PAGE and binding activity was verified in gel shift experiments.
In vitro splicing assays
32P-labeled pre-mRNAs were synthesized by using Megascript T7 kit (Applied Biosystems) after the linearization. The substrates were incubated at 22°C with 50% Drosophila S2 nuclear extracts or at 30°C with 40% HeLa nuclear extracts under standard splicing conditions as described previously (17,27). Recombinant proteins were added to the reactions as indicated. The RNA splicing products and intermediates were separated on 3.5% denaturing polyacrylamide gels and visualized using Applied Biosystems Storm phosphorImager. The signals produced by individual bands were quantitated using Molecular Dynamics Imagequant software.
Spliceosome assembly assays
32P-labeled RNA was incubated in S2 nuclear extracts under in vitro splicing conditions. For control reactions in the absence of ATP, nuclear extracts were depleted of ATP by incubating at room temperature for 30 min. The 2′-O-methyl RNA oligonucleotide complementary to U2 small nuclear RNA (U2 snRNA) stem loop (5′-CGGUACUGCU-3′) was added into the reactions to block U2-branchpoint interaction, while another 2′-O-methyl RNA oligonucleotide complementary to U2 snRNA 5′-end (5′-CGAGAAGCGAU-3′) was added as a control. Reactions were terminated by adding heparin to the final concentration of 2 mg/ml at the specified time point and freezing in liquid nitrogen. Samples were loaded onto a 1.5% low-melting agarose gel immediately after thawing and the electrophoresis was conducted at 4°C at 10 V/cm for 2.5 h (32). Gels were fixed in 50% methanol/15% acidic acid for 30 min before dried at 55°C for 3 h.
RNA-binding assays
To verify the RNA-binding competence of various recombinant MCP fusion proteins they were incubated with 32P-labeled ftzMS2-10 or ftzMS2mt RNA as described (16) except that protein–RNA complexes were resolved by electrophoresis on a 3.5% non-denaturing polyacrylamide gel (49:1 bis).
Cell culture and transfections
S2 cells adapted to serum free medium (Dmel2) were grown at 28°C in Drosophila Schneider medium (Invitrogen) supplemented with 20 mM l-glutamine. A total of 2 × 106 cells were transfected in each well of six-well plates using the Cellfectin reagent (Invitrogen) and DNA mixes containing 0.5 µg of pActdsxISS-wt or pActdsxISS-5 mt and 1 µg of either pActGFP or pFlagTra2. All plasmids use the Act5C promoter. Cells were harvested 48 h after transfection for RNA extraction.
RNA and protein analysis
Total RNA was extracted from the transfected cells using the Trizol reagent (Invitrogen) based on the vendor's instructions. After DNase (Promega) treatment, samples were reverse transcribed using 0.5 µg RNA in 20-µl reaction containing 10 U MMLV Reverse Transcriptase (Roche), 5 U RNAase inhibitor (Roche), 2.5 mM dNTP and 10 pmol of a downstream primer complementary to the GFP coding region (5′-GAACGCTTCCATCTTCAATGTTG-3′). Products were then amplified in presence of the same downstream primer and 10 pmol/l 32P-labeled of a primer complementary to dsx Exon 3 (5′-CCGCTATCCTTGGGAGCTG-3′) and 5U Taq polymerase for 25 cycles. Radiolabeled RT–PCR products were fractionated on 5% non-denaturing polyacrylamide gel. Western blot were performed using the proteins extracted from the same transfected cells and the M2 anti-Flag antibody (Sigma). The results shown are representative of at least three independent transfections for each of the reporter genes.
RESULTS
Binding of Tra2 to the ISS inhibits formation of pre-spliceosomal complexes
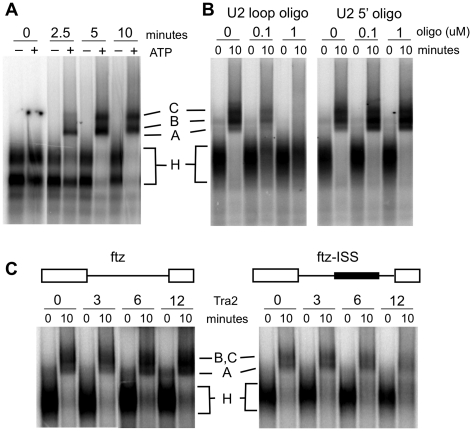
The interaction of Tra2 with ESE sequences facilitates committed pre-spliceosome complex formation and thus promotes splicing at steps prior to the first catalytic reaction (33). Likewise, Tra2 bound to the ISS is known to block splicing prior to the first catalytic reaction but the effects it has on spliceosome assembly are not known. To determine if binding of Tra2 to the ISS inhibits assembly of pre-spliceosomal complexes, we inserted the intact ISS into the intron of the fushi-tarazu (ftz) gene and tested how Tra2 affects its splicing. The ftz intron was chosen because it has high basal splicing efficiency in vitro allowing the visualization of splicing complexes. In addition, although the intron is not a normal target of Tra2, it has been shown previously to undergo Tra2-dependent repression when ISS sequences are inserted into it (27). Assembly reactions were first carried out on the native ftz intron in Drosophila Schneider-2 (S2) nuclear extracts and complexes were resolved on native agarose gels. As shown in Figure 1A, two ATP-dependent low mobility complexes are observed that are analogous to mammalian complexes A and C as previously identified in assembly reactions carried out in Drosophila extracts (34). Consistent with this assignment, we found that both of the complexes depend on the availability of the U2 snRNP loop region that is known to base pair with branch site sequences in the pre-spliceosome. Incubation of extracts with a 2′-O-methyl oligonucleotide directed at these pairing sequences inhibited assembly of the both complexes, while a control oligonucleotide directed at the 5′-end of the U2 snRNP did not (Figure 1B). When the ISS was inserted into the ftz intron, Tra2 was found to inhibit assembly of both complexes at protein concentrations that have previously been shown to block formation of splicing products (Figure 1C) (6). This effect is due to a specific interaction with the ISS as Tra2 did not inhibit complex assembly on the ftz intron alone in parallel controls. These results indicate that Tra2-dependent complexes bound to the ISS affect pre-spliceosome assembly in a manner opposite to that which has been observed for Tra2 dependent complexes formed on the dsx ESE.
Figure 1.
Inhibitory activity of Tra2 in pre-spliceosomal complex assembly. (A) Spliceosome assembly assays with a fushi tarazu (ftz) RNA splicing substrate were carried out in Drosophila Schneider 2 (S2) nuclear extract for the times indicated in the presence and absence of ATP. The positions of ATP-independent complexes (H), as well as ATP dependent A and C complexes are indicated. (B) Two 2′-O-methyl RNA oligos complementary to the U2 small nuclear RNA (snRNA) are tested for their effects on complex formation in the presence of ATP. The oligos were targeted at the loop region, essential for base pairing with the branchsite, and to the non-essential 20 nt at the 5′-end of the U2 snRNA. (C) Spliceosomal assembly assays on the ftz and ftz-ISS substrates were carried out with various amounts of recombinant Tra2 proteins in the presence of ATP. The dark bar in the schematic of ftz-ISS indicates the position of the 78-nt ISS element insertion in the ftz intron as previously described (27).
The ISS of the M1 intron activates splicing from an exonic position
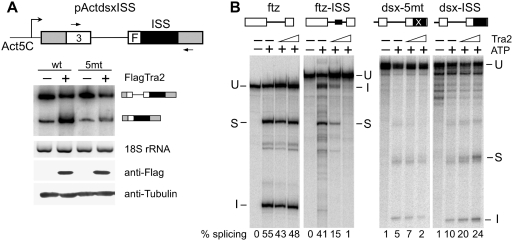
Tra2 dependent ISS and ESE complexes could affect splicing differently due to differences in their composition or position within the pre-mRNA. To determine if the ISS can carry out a repressive function from an exonic position, we first inserted this sequence into the downstream exon of a dsx minigene construct (ActdsxISS). This construct lacks the ESE normally present downstream of the female-specific 3′-splice site as illustrated in Figure 2A. The plasmid and controls were co-transfected into Drosophila S2 cells both with and without a plasmid driving expression of the Tra2 protein with an N-terminal Flag epitope tag (Flag-Tra2). Although the ISS has previously been shown to mediate Tra2-dependent repression of M1 splicing in S2 cells (28), when positioned exonically the wt ISS caused Tra2-dependent activation of splicing (Figure 2A). A similar minigene in which each of the critical CAAGR repeats of the ISS were altered (5 mt) was only slightly activated by Flag-Tra2. These results show that the ISS is capable of acting as an ESE and suggest that position is important in determining its effects on splicing in response to Tra2.
Figure 2.
ISS acts as an enhancer when positioned in dsx exon. (A) RT–PCR analysis showing enhancer activity of the ISS in transfected S2 cells. A schematic of splicing reporters dsxISS is shown. The white boxes represent dsx exon 3 and part of the female-specific exon (F) with the wt dsx intron sequences between them. The wt ISS or the same sequence with mutations at each of the CAAGR repeats (5 mt) was placed 40-nt downstream of female specific dsx 3′-splice site in the absence of the dsx splicing enhancer. Cells were co-transfected with each reporter construct and a FlagTra2 expression plasmid or pActGFP a control plasmid with the same promoter. RT–PCR was carried out with primers indicated in the diagram by arrows on total RNA from the co-transfections. Amplification products with or without intron retention are indicated. Control RT–PCR products from Drosophila 18S rRNA gene produced with the same RNA samples are shown. Expression of FlagTra2 was verified by western blot using anti-Flag antibody. (B) In vitro splicing assays carried out on constructs in which the ISS (black filled box) was inserted into either the intron of ftz or 40-nt downstream of the dsx 3′-splice site. Reactions with 0, 100 or 200 nM recombinant Tra2 are shown. The positions of unspliced RNA (U), spliced product (S) and the intron (I) are indicated. The percentage of splicing products and intermediates is indicated below each lane.
To further examine this issue, we next tested whether exonic position had a similar effect on ISS function in Drosophila S2 cell nuclear extracts supplemented with Tra2 protein. Again a dsx splicing target lacking its ESE sequence was replaced with either the ISS from the M1 intron or a version of this sequence with mutant repeat elements. In vitro splicing reactions were then carried out in the presence and absence of recombinant His tagged Tra2 protein. As shown in Figure 2B, Tra2 had no effect on the substrate with the mutant repeats (dsx-5mt) but when the ISS with wt repeats (dsx-ISS) was tested, splicing was activated. Control experiments with the ISS at an intronic position (ftz-ISS) resulted in repression as observed previously (27). These experiments show that the ISS can mediate either activation or repression in nuclear extracts and further supports the idea that ISS function depends on its position within the RNA.
Tethering of Tra2 to an intron is sufficient to elicit repression
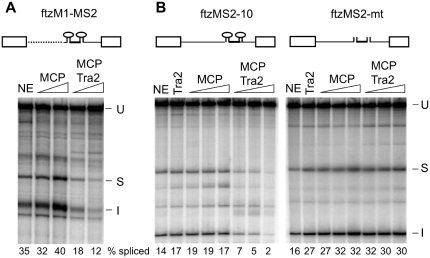
Repression of splicing from intronic positions might depend on multiple factors that bind to the ISS in combination with Tra2. Alternatively, it is possible that Tra2 binding alone within the M1 intron is sufficient to trigger repression. We therefore tested how splicing is affected when Tra2 is tethered within the M1 intron through fusion to the MS2 coat protein (MCP) in the absence of ISS sequences. We replaced ISS sequences in an M1-based splicing substrate ftzM1 (27) with those encoding two MCP-binding sites and carried out splicing reaction with this substrate (ftzM1-MS2) in S2 nuclear extracts in the presence of either the MCP alone or an MCP-Tra2 fusion protein. The fusion protein binds to specifically RNA substrates with intact MS2-binding sites (Supplementary Data S2). However, as shown in Figure 3A, MCP-Tra2 repressed splicing three fold while the MCP protein alone had no effect. This result suggested that the primary role of the ISS in repression is to recruit Tra2 and that the binding of other factors specified by the ISS is unnecessary.
Figure 3.
Tethering of Tra2 to an intron is sufficient to initiate splicing repression. (A) Splicing of ftzM1-MS2, in which the ISS of ftzM1 (27) was replaced by RNA sequences from the bacteriophage MS2 (shown as stem loops) that bind MCP and MCP fusion proteins, are shown. Other intron sequences upstream of the polypyrimidine tract (dotted line) derive from the M1 intron. The exons and 3′-end of the intron derive from ftz. Splicing was carried out in the presence of S2 nuclear extract (NE) or extracts supplemented with 100 and 300 nM MCP or MCP–Tra2 fusion protein. The mobilities of in vitro splicing products and the percentage of splicing are indicated as in Figure 2. (B) Shown are splicing reactions carried out on substrates in which two wt MCP-binding sites (ftzMS2-10), or mutant sites (ftzMS2-mt) were inserted into the intron of the ftz gene. In contrast to ftzM1, these splicing substrates lack any tra2 RNA sequences. Nuclear extracts (NE) were supplemented with 500 nM Tra2 or with 100, 200 or 400 nM MCP or MCP-Tra2 protein. The positions of unspliced RNA (U), spliced product (S) and spliced intron (I) are indicated. The percentage of total signal from splice products and intermediates is indicated below each lane.
Although the ISS is sufficient for Tra2-dependent repression and is deleted from the above substrate, it has previously been observed that redundant Tra2-binding sites located within tra2 transcripts but outside the ISS can mediate modest Tra2-dependent repression of splicing (6,27). Therefore, we next asked whether MCP-Tra2 could repress splicing in the context of a substrate based entirely on sequences from the ftz gene in which splicing is not normally regulated by Tra2. MCP-binding sites were inserted into the ftz intron at a position 10-nt upstream of the branch site which is similar to the position of the ISS in the M1 intron (ftzMS2-10). As shown in Figures 3B, the MCP–Tra2 protein significantly repressed splicing of this substrate while neither MCP nor the Tra2 protein alone did so. The same fusion protein did not affect splicing of a ftz substrate with mutant MCP-binding sites (ftzMS2-10mt). These results indicate that the observed repression occurs through binding of MCP and not Tra2 and further shows that the tethering of Tra2 to an unregulated intron is sufficient to trigger splicing repression.
Tethered Tra2 protein represses independently of position in the intron but activates splicing from an exon
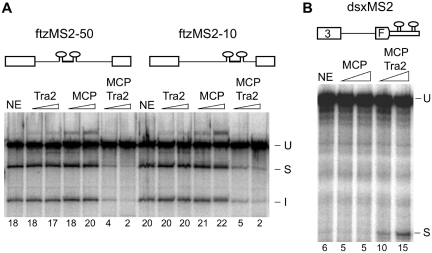
In the above experiment, the MCP-Tra2 fusion proteins were directed to a position just upstream of the branch site where pre-spliceosomal complexes assemble. However, previous studies on the ISS indicate that its function is not dependent on this proximity and that it effectively mediates repression when relocated to other positions in the M1 intron. To determine if a tethered Tra2 protein can cause repression at a distance, we performed parallel assays in which Tra2 is tethered 10 nt (ftzMS2-10) or 50 nt (ftzMS2-50) upstream of the branch site. As shown in Figure 4A MCP-Tra2 repressed splicing to a similar extent from these positions indicating that close proximity to the branch site is not required.
Figure 4.
Dependence of tethered Tra2 function on binding position. Splicing of RNAs with MCP-binding sequences at different positions are compared. S2 nuclear extracts were supplemented with 100 or 400 nM of Tra2, MCP or MCP-Tra2 as indicated. The mobility of precursors and products are indicated to the side of each gel as in other figures. The percentage of total signal from splice products and intermediates is indicated below each lane. The positions of unspliced RNA (U), spliced product (S) and spliced intron (I) are indicated. (A) Two MS2-binding site inserted either 50-nt (ftzMS2-50) or 10-nt (ftzMS2-10) upstream of the branch site are tested for the effect of MCP-Tra2. (B) Two MS2 sites were inserted 40-nt downstream of the dsx female-specific 3′-splice site in the absence of the dsx splicing enhancer.
Using a similar tethering approach, MCP-Tra2 fusion proteins have previously been shown to activate splicing from exonic positions (17), suggesting that exon/intron position determines whether activation or repression is observed. However, these previous studies have been carried out exclusively using nuclear extracts from human HeLa cells. Thus, it is possible that the difference in activity observed is due to a difference in experimental systems and not position of binding. In Figure 4B, we show that Drosophila S2 nuclear extracts support activation of splicing when MCP-Tra2 was tethered downstream of the dsx 3′-splice site as was previously reported in HeLa nuclear extracts.
To determine whether splicing of the ftz substrate was also activated by Tra2 bound at an exonic position, we directed MCP-Tra2 to a pair of binding MS2 sites in the exon immediately downstream of the ftz intron (ftzMS2E). MCP-Tra2 had little or no effect on splicing when tethered at this position (Figure 5A). However, unlike other substrates that have been tested for activation by tethered SR factors, a majority of the ftz RNA is spliced even in the absence of tethered protein and it is unclear if the residual unspliced molecules are available as splicing reactions do not usually go to completion in vitro. We therefore hypothesized that poor activation of ftz splicing could be due to the rather efficient basal splicing of the ftz intron in vitro. To address this issue, we reduced the substrate's basal splicing efficiency by substituting a weak polypyrimidine tract of dsx for that of the ftz 3′-splice site (Figure 5B). This resulted in little or no splicing of the intron in S2 nuclear extracts in the absence of recombinant protein and, as hypothesized, significant activation of splicing upon the addition of MCP-Tra2 (Figure 5C). Consistent with these observations, when splicing reactions were carried out in HeLa nuclear extracts, we observed that the fusion protein triggered activation of splicing by ftzMS2E and repression of ftzMS2-10 (Supplementary Data S3). These results support the above conclusions from fly extracts and further indicate that tethering of Tra2 to the RNA is sufficient to trigger activation or repression that is dependent on exon/intron position.
Figure 5.
Effects of Tra2 when tethered in the ftz exon. Splicing assays in S2 extracts with MCP and MCP-Tra2 performed on substrates containing two MS2-binding sites 30-nt downstream of the ftz 3′-splice site are shown. The positions of unspliced RNA (U), spliced product (S) and spliced intron (I) are indicated. (A) Splicing of ftzMS2E with the native ftz 3′-splice site. Amount of recombinant Tra2 proteins is as in previous figures. Note the high-level splicing observed in nuclear extract alone (NE). The percentage of total signal from splice products and intermediates is indicated below each lane. (B) Sequences near the ftz and dsx 3′-splice site illustrating the poor pyrimidine content of the latter. The dsx sequences substituted into the ftz intron in the modified substrate (ftz/dsx3′) are underlined. (C) Results of in vitro splicing in S2 nuclear extracts using ftz/dsx3′ with 400 nM MCP or 100, 200 and 400 nM Tra2.
MCP-Rbp1 and MCP-SF2 fusion proteins neither activate nor repress when tethered intronically
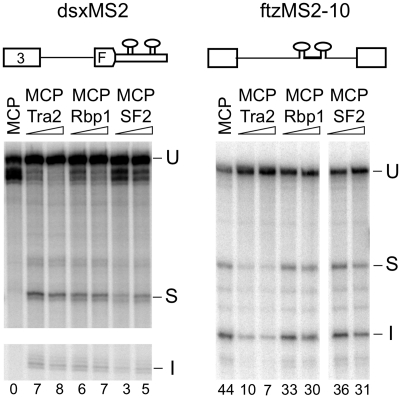
We next examined whether the ability to initiate splicing repression when tethered at an intronic position is shared by other SR splicing factors implicated in both activation and repression of splicing (15,21,27,35). MCP fusions were generated with two such Drosophila SR proteins (Rbp1 and SF2). As shown in Figure 6, these fusion proteins were able to activate splicing of dsxMS2 consistent with previous studies on tethered SR factors. Tethering of these factors to intronic positions in ftzMS2-10 did not increase splicing indicating that, like Tra2, they depend on exonic position for their activation function. However, in contrast to Tra2, both proteins only slightly repressed splicing from the intronic position. These results suggest that the repressive activity of intron-tethered Tra2 is not shared by all SR splicing factors.
Figure 6.
Exon and intron tethering of Drosophila SR proteins Rbp1 and ASF/SF2. In vitro splicing reactions with substrates dsxMS2 and ftzMS2-10 were carried out in S2 nuclear extracts supplemented with 400 nM MCP and 100 or 400 nM MCP-Tra2, MCP–Rbp1 and MCP–ASF/SF2 proteins. The positions of unspliced RNA (U), spliced product (S) and spliced intron (I) are indicated. The percentage of total signal from splice products and intermediates is indicated below each lane.
The activator and repressor functions of Tra2 require different effector sequences
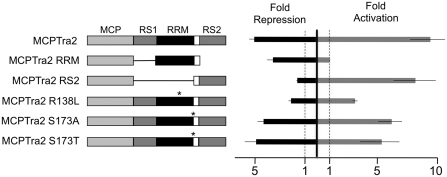
We next tested whether the repression function of Tra2 depends on an effector function of its Arg–Ser-rich regions are known to be effectors of splicing activation when Tra2 is bound at exonic locations. To determine if these sequences or other regions of the protein are effectors of splicing repression, we generated a fusion protein containing the carboxy-terminal Arg–Ser-rich region of Tra2 with MCP (MCP-RS2) and assayed its ability to activate splicing of dsxMS2 and to repress splicing of ftzMS2-10 in Drosophila extracts. Consistent with previous studies in HeLa extracts (17), the Arg–Ser sequences of Tra2 activated splicing almost as well as a fusion of the full-length Tra2 protein when tethered to dsxMS2 exonic positions (Figure 7). However, this fusion lacked the ability to repress splicing of ftzMS2-10. The effector activity for repression instead was mapped in the RRM-linker region of the Tra2 protein. A fusion of these sequences with MCP (MCP-RRM) repressed splicing of ftzMS2-50 but did not activate splicing of dsxMS2. To further test the role of these sequences in repression, we assayed fusions of the intact Tra2 protein that contain previously studied point mutations in this region (36). The ability to effect repression was reduced by a mutation (R138L) that is known to cause strong loss-of-function for sex determination and fertility in vivo. This mutation affects a conserved residue in the RNP1 consensus element of the RRM. In contrast, two mutations (S173A and S173T) that alter a highly conserved residue near the RRM-linker junction, and are known to cause partial loss of function phenotypes in vivo, had no effect on function of the tethered protein. Together these results indicate that the effector regions responsible for repression and activation of splicing can be separated and thus are likely to result from distinct molecular interactions of Tra2.
Figure 7.
Repression and activation of splicing by Tra2 depend on different effector domains. Diagram of various Tra2–MCP protein fusions tested for effector function are shown. MCP protein sequences, the Arg–Ser-rich regions (RS1 and RS2) as well as the RRM are denoted by various shades of gray as indicated. White indicates the linker region. Positions of point mutations are indicated with an asterisk. The amino acid changes of various point mutations are indicated based on their position in the 264 amino acid Tra2-PA isoform. Results are summarized to the right of the diagrams from quadruplicate splicing assays in S2 nuclear extracts using either ftzMS2-10 for repression (black bar) or dsxMS2 for activation (gray bar). Effects on splicing are expressed as the average fold activation or repression of splicing observed in each experiment relative to parallel control splicing reactions supplemented with the MCP protein. A value of 1 indicates no effect from the Tra2 sequences. All reactions were supplemented with 400 nM of the recombinant protein.
DISCUSSION
The results presented here indicate that Tra2 has two separable activities affecting splice site recognition. The activity observed depends on the position of Tra2-binding sites in relation to the affected splice site. Splice sites oriented to remove Tra2-binding sites (intronic orientation) are repressed, while splice sites oriented to include these binding sites as part of an exon are activated. The position dependence of these activities would account for differences previously observed in the effect of Tra2 on its known targets in Drosophila development.
The mechanism of activation by Tra2 and other SR factors bound to ESEs has been well studied and results from the ability of Arg–Ser-rich regions of these proteins to facilitate assembly of pre-spliceosomal complexes at splice sites that border the bound exon through interactions with general splicing factors and direct contact with canonical RNA signals near the splice site (16,19). Tra2 itself associates with several ESE elements within the female-specific exon of the dsx pre-mRNA in combination with Tra and other SR factors (13,14). While bound at the ESE these factors cooperate to promote recognition of the weak female-specific alternative 3′-splice site located upstream (18,37).
Interestingly, Tra2 and other SR factors can activate splicing even when they are bound at significant distances from the affected splice sites. For example, the ESE complexes in dsx RNA are located >290-nt downstream of the female specific 3′-splice site but are potent activators switching splicing almost exclusively to this site instead of a strong downstream 3′-splice site (38). Studies on how distance between ESEs and splice sites affects activation are consistent with the idea that pre-mRNA acts like a flexible tether connecting the regulatory complex with the splice site (30). The amount of splicing observed in these studies is directly proportional to the calculated effective concentration of the enhancer complex at the splice site. RNA looping is thus thought to allow SR factors bound internally in the exon to interact with pre-spliceosome complexes.
In its simplest form, this looping model would predict that Tra2 and other SR factors would also activate splice sites when bound within an intron over similar distances. However, as shown here, when Tra2 binds to the ISS within the M1 intron it prevents pre-spliceosome assembly and represses the use of the splice sites that flank the intron. Several Tra2-binding sites in the ISS are located immediately adjacent to the branch site which is a key signal in pre-spliceosome formation. In principle, the binding of Tra2 at this position might inhibit splicing through direct occlusion of factors that require access to the branch site. However, experiments in which the ISS was repositioned farther upstream in the intron yielded equal levels of repression indicating that the Tra2–ISS complexes can repress splicing at a distance (27). Consistent with this, we found that tethering of MCP-Tra2 fusion proteins to positions 10- or 50-nt upstream of the branchsite, resulted in similar repression. These results suggest that, like activation, splicing repression can occur at a distance from the splice site.
It is presently unclear whether the position sensitive repression activity of Tra2 is shared by other Drosophila SR splicing factors. MCP fusions of both Drosophila SF2 and Rbp1 activated splicing from exonic positions, but failed to either repress or activate when bound within the ftz intron. Thus while exon/intron position appears to be important for these factors, they do not show the same ability to repress splicing that Tra2 does. However, several studies in mammalian systems support the idea that intronic binding of SR factors leads to repression of splicing. In the adenovirus late pre-mRNA, it was found that binding of the human SR protein SRSF1 (formerly called ASF/SF2) to a series of sites located upstream of the IIIa 3′-splice site leads to repression of that site and selection of an alternative upstream site (21). As with the M1 ISS element studied here, the adenovirus elements functioned from different intronic positions and acted as splicing enhancer when relocated to an exon (21,39). Likewise, a repressive role for SR factors was inferred in studies on RNA substrates derived from the human β-globin gene (20). Based on a series of in vitro experiments in which the splice sites flanking the middle exon of a three exon substrate were mutated, it was found that ESE–SR complexes associated with the disabled exon acted to repress splicing events between the two outside exons. In this case the disabled exon is effectively part of the intron and the ESE acts like an ISS bound by SR factors. Thus it seems likely that these mammalian SR factors perform position sensitive repressive functions similar to those observed of Tra2.
Although the Arg–Ser-rich regions of Tra2 activate splicing, the effector region responsible splicing repression maps instead to its RRM domain. Repression is therefore likely to involve molecular interactions that are distinct from those involved in splicing activation. The primary activity ascribed to the single RRM of Tra2 is in binding of its pre-mRNA target sequences such as those in the ISS and dsx ESE regions (36,40). In the experiments presented here, effector function was detected in tethering experiments in which binding is mediated by MCP sequences and the splicing substrate lack these binding targets. Effector function therefore is likely to depend on a second RNA-binding activity of the RRM or its direct interaction with one or more protein factors. A number of RRM–protein interactions have now been described (41) and in some cases RRM domains are known to interact with RNA and another protein simultaneously (42,43). Given that Tra2 inhibits splicing complex assembly, it seems most likely that the Tra2 RRM participates in a ternary complex that directly interferes with early steps in the recognition of splice sites.
A number of SR factors are known to preferentially associate with exonic regions and it has recently been shown that a variety of hexamer sequences with demonstrated ESE activity are likewise highly enriched in exonic sequences across the transcriptome (44,45). In this regard, it is interesting that Tra2 promotes exonic identity of its binding sites in both retention of the M1 intron and inclusion of alternative exons in dsx and fru splicing. An attractive idea is that upon binding to multisite regulatory elements such as these, the activation and repression functions of Tra2, and perhaps other SR factors, act in concert to define both the position and the extent of exons in their target RNAs. The activation function would serve to ensure that splice sites at the ends of exons are efficiently recognized, while the repressive activity could simultaneously inhibit the use of oppositely oriented alternative or cryptic splice sites that would otherwise excise internal portions of the exons. The latter activity would thus ensure the continuity of the exon.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary figures 1–3.
FUNDING
National Institutes of Health (GM070892 to W.M., CA016672 to the M.D. Anderson Cancer Center DNA Analysis Facility). Funding for open access charge: M.D. Anderson Cancer Center.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Brent Graveley, Klemens Hertel and Andy McCullough for generously providing advice and materials used in this work.
REFERENCES
- 1.Han J, Cooper TA. Identification of CELF splicing activation and repression domains in vivo. Nucleic Acids Res. 2005;33:2769–2780. doi: 10.1093/nar/gki561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grammatikakis I, Goo YH, Echeverria GV, Cooper TA. Identification of MBNL1 and MBNL3 domains required for splicing activation and repression. Nucleic Acids Res. 2011;39:2769–2780. doi: 10.1093/nar/gkq1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lou H, Helfman DM, Gagel RF, Berget SM. Polypyrimidine tract-binding protein positively regulates inclusion of an alternative 3'-terminal exon. Mol. Cell. Biol. 1999;19:78–85. doi: 10.1128/mcb.19.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amir-Ahmady B, Boutz PL, Markovtsov V, Phillips ML, Black DL. Exon repression by polypyrimidine tract binding protein. RNA. 2005;11:699–716. doi: 10.1261/rna.2250405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandler DS, Qi J, Mattox W. Direct repression of splicing by transformer-2. Mol. Cell Biol. 2003;23:5174–5185. doi: 10.1128/MCB.23.15.5174-5185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Venables JP, Klinck R, Koh C, Gervais-Bird J, Bramard A, Inkel L, Durand M, Couture S, Froehlich U, Lapointe E, et al. Cancer-associated regulation of alternative splicing. Nat. Struct. Mol. Biol. 2009;16:670–676. doi: 10.1038/nsmb.1608. [DOI] [PubMed] [Google Scholar]
- 8.Llorian M, Schwartz S, Clark TA, Hollander D, Tan LY, Spellman R, Gordon A, Schweitzer AC, de la Grange P, Ast G, et al. Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nat. Struct. Mol. Biol. 2010;17:1114–1123. doi: 10.1038/nsmb.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 11.Shepard PJ, Hertel KJ. The SR protein family. Genome Biol. 2009;10:242. doi: 10.1186/gb-2009-10-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryner LC, Goodwin SF, Castrillon DH, Anand A, Villella A, Baker BS, Hall JC, Taylor BJ, Wasserman SA. Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell. 1996;87:1079–1089. doi: 10.1016/s0092-8674(00)81802-4. [DOI] [PubMed] [Google Scholar]
- 13.Lynch KW, Maniatis T. Synergistic interactions between two distinct elements of a regulated splicing enhancer. Genes Dev. 1995;9:284–293. doi: 10.1101/gad.9.3.284. [DOI] [PubMed] [Google Scholar]
- 14.Lynch KW, Maniatis T. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev. 1996;10:2089–2101. doi: 10.1101/gad.10.16.2089. [DOI] [PubMed] [Google Scholar]
- 15.Heinrichs V, Baker BS. The Drosophila SR protein RBP1 contributes to the regulation of doublesex alternative splicing by recognizing RBP1 RNA target sequences. EMBO J. 1995;14:3987–4000. doi: 10.1002/j.1460-2075.1995.tb00070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graveley BR, Maniatis T. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol. Cell. 1998;1:765–771. doi: 10.1016/s1097-2765(00)80076-3. [DOI] [PubMed] [Google Scholar]
- 17.Sciabica KS, Hertel KJ. The splicing regulators Tra and Tra2 are unusually potent activators of pre-mRNA splicing. Nucleic Acids Res. 2006;34:6612–6620. doi: 10.1093/nar/gkl984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graveley BR, Hertel KJ, Maniatis T. The role of U2AF35 and U2AF65 in enhancer-dependent splicing. RNA. 2001;7:806–818. doi: 10.1017/s1355838201010317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen H, Kan JL, Green MR. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol. Cell. 2004;13:367–376. doi: 10.1016/s1097-2765(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 20.Ibrahim EC, Schaal TD, Hertel KJ, Reed R, Maniatis T. Serine/arginine-rich protein-dependent suppression of exon skipping by exonic splicing enhancers. Proc. Natl Acad. Sci. USA. 2005;102:5002–5007. doi: 10.1073/pnas.0500543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanopka A, Muhlemann O, Akusjarvi G. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature. 1996;381:535–538. doi: 10.1038/381535a0. [DOI] [PubMed] [Google Scholar]
- 22.Mattox W, Baker BS. Autoregulation of the splicing of transcripts from the transformer-2 gene of Drosophila. Genes Dev. 1991;5:786–796. doi: 10.1101/gad.5.5.786. [DOI] [PubMed] [Google Scholar]
- 23.Feng Y, Chen M, Manley JL. Phosphorylation switches the general splicing repressor SRp38 to a sequence-specific activator. Nat. Struct. Mol. Biol. 2008;15:1040–1048. doi: 10.1038/nsmb.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin C, Manley JL. The SR protein SRp38 represses splicing in M phase cells. Cell. 2002;111:407–417. doi: 10.1016/s0092-8674(02)01038-3. [DOI] [PubMed] [Google Scholar]
- 25.Cook CR, McNally MT. SR protein and snRNP requirements for assembly of the Rous sarcoma virus negative regulator of splicing complex in vitro. Virology. 1998;242:211–220. doi: 10.1006/viro.1997.8983. [DOI] [PubMed] [Google Scholar]
- 26.Mattox W, McGuffin ME, Baker BS. A negative feedback mechanism revealed by functional analysis of the alternative isoforms of the Drosophila splicing regulator transformer-2. Genetics. 1996;143:303–314. doi: 10.1093/genetics/143.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi J, Su S, Mattox W. The doublesex splicing enhancer components Tra2 and Rbp1 also repress splicing through an intronic silencer. Mol. Cell. Biol. 2007;27:699–708. doi: 10.1128/MCB.01572-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi J, Su S, McGuffin ME, Mattox W. Concentration dependent selection of targets by an SR splicing regulator results in tissue-specific RNA processing. Nucleic Acids Res. 2006;34:6256–6263. doi: 10.1093/nar/gkl755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryner LC, Baker BS. Regulation of doublesex pre-mRNA processing occurs by 3'-splice site activation. Genes Dev. 1991;5:2071–2085. doi: 10.1101/gad.5.11.2071. [DOI] [PubMed] [Google Scholar]
- 30.Graveley BR, Hertel KJ, Maniatis T. A systematic analysis of the factors that determine the strength of pre-mRNA splicing enhancers. EMBO J. 1998;17:6747–6756. doi: 10.1093/emboj/17.22.6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LeCuyer KA, Behlen LS, Uhlenbeck OC. Mutants of the bacteriophage MS2 coat protein that alter its cooperative binding to RNA. Biochemistry. 1995;34:10600–10606. doi: 10.1021/bi00033a035. [DOI] [PubMed] [Google Scholar]
- 32.Lee CS, Das R, Reed R. Agarose gel separation/isolation of RNA-protein complexes. Curr Protoc. Mol. Biol. 2003 doi: 10.1002/0471142727.mb2701s63. Chapter 27, Unit 27 21. [DOI] [PubMed] [Google Scholar]
- 33.Tian M, Maniatis T. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell. 1993;74:105–114. doi: 10.1016/0092-8674(93)90298-5. [DOI] [PubMed] [Google Scholar]
- 34.Herold N, Will CL, Wolf E, Kastner B, Urlaub H, Luhrmann R. Conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosomal complexes. Mol. Cell Biol. 2009;29:281–301. doi: 10.1128/MCB.01415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar S, Lopez AJ. Negative feedback regulation among SR splicing factors encoded by Rbp1 and Rbp1-like in Drosophila. EMBO J. 2005;24:2646–2655. doi: 10.1038/sj.emboj.7600723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amrein H, Hedley ML, Maniatis T. The role of specific protein-RNA and protein-protein interactions in positive and negative control of pre-mRNA splicing by Transformer 2. Cell. 1994;76:735–746. doi: 10.1016/0092-8674(94)90512-6. [DOI] [PubMed] [Google Scholar]
- 37.Zuo P, Maniatis T. The splicing factor U2AF35 mediates critical protein-protein interactions in constitutive and enhancer-dependent splicing. Genes Dev. 1996;10:1356–1368. doi: 10.1101/gad.10.11.1356. [DOI] [PubMed] [Google Scholar]
- 38.Nagoshi RN, McKeown M, Burtis KC, Belote JM, Baker BS. The control of alternative splicing at genes regulating sexual differentiation in D. melanogaster. Cell. 1988;53:229–236. doi: 10.1016/0092-8674(88)90384-4. [DOI] [PubMed] [Google Scholar]
- 39.Dauksaite V, Akusjarvi G. Human splicing factor ASF/SF2 encodes for a repressor domain required for its inhibitory activity on pre-mRNA splicing. J. Biol. Chem. 2002;277:12579–12586. doi: 10.1074/jbc.M107867200. [DOI] [PubMed] [Google Scholar]
- 40.Clery A, Jayne S, Benderska N, Dominguez C, Stamm S, Allain FH. Molecular basis of purine-rich RNA recognition by the human SR-like protein Tra2-beta1. Nat. Struct. Mol. Biol. 2011;18:443–450. doi: 10.1038/nsmb.2001. [DOI] [PubMed] [Google Scholar]
- 41.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 42.Cho S, Hoang A, Sinha R, Zhong XY, Fu XD, Krainer AR, Ghosh G. Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proc. Natl Acad. Sci. USA. 2011;108:8233–8238. doi: 10.1073/pnas.1017700108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price SR, Evans PR, Nagai K. Crystal structure of the spliceosomal U2B”-U2A' protein complex bound to a fragment of U2 small nuclear RNA. Nature. 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- 44.Ke S, Shang S, Kalachikov SM, Morozova I, Yu L, Russo JJ, Ju J, Chasin LA. Quantitative evaluation of all hexamers as exonic splicing elements. Genome Res. 2011;8:1360–1374. doi: 10.1101/gr.119628.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J, Smith PJ, Krainer AR, Zhang MQ. Distribution of SR protein exonic splicing enhancer motifs in human protein-coding genes. Nucleic Acids Res. 2005;33:5053–5062. doi: 10.1093/nar/gki810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.