Abstract
Bacterial chromosome replication is initiated by binding of DnaA to a DnaA-box cluster (DBC) within the replication origin (oriC). In Bacillus subtilis, six additional DBCs are found outside of oriC and some are known to be involved in transcriptional regulation of neighboring genes. A deletion mutant lacking the six DBCs (Δ6) initiated replication early. Further, inactivation of spo0J in Δ6 cells yielded a pleiotropic phenotype, accompanied by severe growth inhibition. However, a spontaneous suppressor in soj or a deletion of soj, which stimulates DnaA activity in the absence of Spo0J, counteracted these effects. Such abnormal phenotypic features were not observed in a mutant background in which replication initiation was driven by a plasmid-derived replication origin. Moreover, introduction of a single DBC at various ectopic positions within the Δ6 chromosome partly suppressed the early-initiation phenotype, but this was dependent on insertion location. We propose that DBCs negatively regulate replication initiation by interacting with DnaA molecules and play a major role, together with Spo0J/Soj, in regulating the activity of DnaA.
INTRODUCTION
Bacterial chromosome replication initiates at a single origin (oriC) and proceeds bidirectionally to a terminus (terC) located on the opposite side of the circular chromosome. DnaA is a protein that initiates replication by binding to multiple DnaA-binding sequences, termed DnaA-boxes, in the oriC region, to open an AT-rich segment, termed the DNA-unwinding element (DUE), forming single-stranded DNA that recruits replication machinery (1).
The chromosome must be replicated only once per cell cycle to ensure that each chromosome within a cell is faithfully transmitted to daughter cells. To this end, initiation of replication is tightly regulated by redundant systems, principally by negative feedback controls that inhibit DnaA activity (1). In Escherichia coli, four such systems are known: autoregulation of dnaA transcription via direct binding of DnaA to DnaA-boxes in the promoter region; sequestration of newly replicated origins by the SeqA protein; the action of the ‘regulatory inactivation of DnaA’ (RIDA) system which promotes hydrolysis of ATP bound to DnaA by a complex composed of DnaA homolog protein (Hda) and the DnaN clamp; and titration of DnaA to a specific locus termed datA harboring five high-affinity DnaA-boxes, which trap DnaA molecules and prevent their functioning at oriC.
The E. coli oriC is located about 42 kb from the dnaA gene (2), whereas oriC of Bacillus subtilis lies between the dnaA and dnaN genes (encoding the β-clamp subunit of DNA polymerase III, respectively) (3). B. subtilis oriC also contains multiple DnaA-boxes and AT-rich stretches. In vitro experiments have shown that DnaA binding induces melting at one AT-rich region between dnaA and dnaN (oriC2), with a requirement for ATP (3,4). Regulation of replication initiation in B. subtilis features autoregulation of dnaA transcription, as in E. coli (5), but no homologs of the E. coli Hda and SeqA proteins have yet been identified. Rather, B. subtilis uses different proteins in this autoregulation, namely the YabA protein and the genome-encoded parABS system.
YabA is conserved in Gram-positive bacteria of low (G+C) content, and has been shown to interact with both DnaA and DnaN, as does E. coli Hda (6). Notably, chromosomal deletion of YabA induces overinitiation and replication asynchrony, as does removal of Hda (6,7), although the regulatory mechanism appears to be distinct from that involving Hda. We have proposed that YabA inhibits replication initiation via competitive inhibition of the binding of the helicase loader component DnaD to DnaA (8). In addition, Graumann et al. have suggested that YabA sequesters DnaA molecules from oriC units that migrate to cell poles after replication initiation, by tethering DnaA to a replisome that is retained in the central cellular region, via a tertiary interaction between DnaN, YabA and DnaA (9).
The parABS system was originally identified in low copy-number plasmids of E. coli and was found to be essential for accurate plasmid partitioning (10). The system has three components: a Walker Box ATPase (ParA), a DNA-binding protein (ParB) and a ParB-binding sequence that acts as a centromere (parS). Interestingly, not only ParA and ParB, but also putative parS sites, have been identified on the chromosomes of 69% of studied strains from all bacterial phyla (11). Further, the majority of these loci are present in origin-proximal regions, suggesting that the parABS system is primarily involved in the regulation of processes that involve the origins of bacterial chromosomes (11).
The B. subtilis chromosome harbors eight parS sites in the oriC-proximal region, and involvement of Soj (ParA) and Spo0J (ParB) in control of both replication initiation and chromosome segregation has been described, although the molecular mechanisms remain unclear (12,13). Recently, it has been shown that Spo0J binds to parS sites and promotes chromosome segregation via recruitment of the SMC (structural maintenance of chromosome) protein to the oriC region (14,15). Gruber and Errington have proposed that SMC recruited to the oriC-proximal region acts an ‘organization center’ to promote efficient chromosome segregation via compaction of chromosomal DNA emerging from the replisome (14). Importantly, the association between Spo0J and SMC is not affected by the absence of Soj, indicating that Spo0J plays an SMC-dependent, but Soj-independent, role in chromosome segregation.
On the other hand, Murray and Errington have recently shown that Soj functions as a spatially regulated molecular switch, capable of either inhibiting or activating DnaA depending on Soj subcellular localization (16). Both localization and activity of Soj were controlled by Spo0J. The cited authors clearly showed that a monomer of Soj directly interacted with DnaA and inhibited DnaA activity. It was also shown in a spo0J deletion mutant and in an ATP hydrolysis-deficient Soj mutant (SojD40A), that co-operative and non-specific DNA binding by ATP-Soj occurred and positively regulated DnaA activity. However, it remains unclear how Soj activates DnaA, because a direct interaction between DnaA and SojD40A was not detected by two-hybrid analysis or using an in vivo pull-down assay. Recently, it has been demonstrated that Spo0J inhibits Soj dimerization by stimulating the intrinsic ATPase activity of Soj and thus controls the DnaA activation function of this protein (17).
Mutation of DNA replication initiation genes in both prokaryotes and eukaryotes leads to pleiotropic phenotypes, featuring defects in chromosomal segregation, cell division, cell cycle progression and transcriptional regulation (18). Indeed, several genes involved in such processes have been shown to be regulated by DnaA in B. subtilis (19,20). In addition, we have recently demonstrated that in exponentially growing B. subtilis cells DnaA stably binds not only to the oriC region (upstream of dnaA [oriC1] and the intergenic region between dnaA and dnaN [oriC2]), but also to six DnaA-box clusters (DBCs) located in the intergenic regions of gcp-ydiF, yqeG-sda, ywlC-ywlB, ywcI-vpr, yydA-yycS and thdF-jag (21). Very recently, binding of DnaA to these DBCs was also demonstrated by Grossman and co-workers (19,22,23). Notably, these regions are generally located around oriC, except for yqeG-sda (Figure 1A). In addition, DnaA has been shown to directly modulate the expression of genes located downstream of DnaA-boxes in some of these regions; DnaA positively regulates sda expression and negatively regulates that of dnaA, ywlC and yydA (21,24). Interestingly, searches for DBCs in Bacillus genomes that could possibly bind DnaA stably have indicated that while potential DnaA-binding sequences are located upstream of different genes in each species examined, almost all are in close proximity to oriC. This suggests that DBCs may play additional role(s), such as controlling the amount of the functional chromosome replication initiator, ATP-DnaA, by titration to these extra binding sites and thus regulating initiation of replication. It has been proposed that E. coli datA functions in this way (2). Recently, two DBCs on the E. coli genome, DARS1 and DARS2, were identified to reactivate DnaA for replication initiation by exchanging bound ADP with ATP in vitro, and deletion of DARS1, DARS2 or both repressed initiation frequency in vivo (25). Thus, it is also possible that B. subtilis DBCs have similar activity. Another possibility is that DnaA may organize the domain structure of the oriC region to facilitate appropriate initiation of chromosome replication by Spo0J in B. subtilis.
Figure 1.
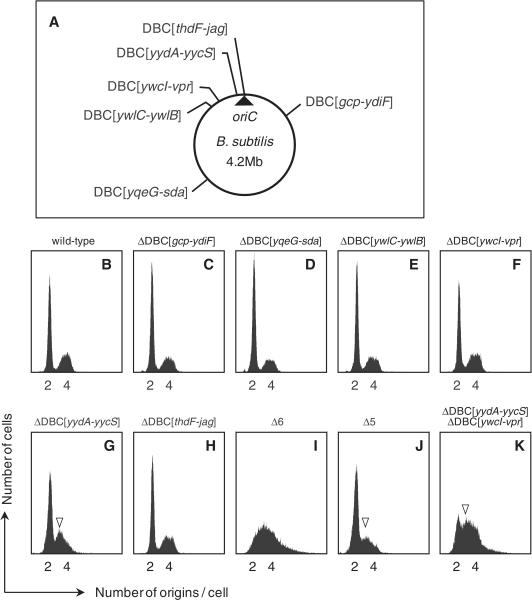
Flow cytometry profiles of DBC-deleted strains. (A) The positions from which DBCs were deleted in the MYA143 (Δ6) strain are indicated on the B. subtilis circular chromosome. (B–K) Flow cytometric profiles of wild-type and mutant strains; (B) 168 (wild-type), (C) MYA155 (ΔDBC[gcp-ydiF]), (D) MYA156 (ΔDBC[yqeG-sda]), (E) MYA157 (ΔDBC[ywlC-ywlB]), (F) MYA018 (ΔDBC [ywcI-vpr]), (G) MYA019 (ΔDBC[yydA-yycS]), (H) MYA158 (ΔDBC[thdF-jag]), (I) MYA143 (Δ6), (J) HO1009 (Δ5), and (K) HO1233 (ΔDBC[yydA-yycS] ΔDBC[ywcI-vpr]). All strains were grown in SMM at 30°C, and cells were subjected to flow cytometry as described in ‘Materials and Methods’ section. Location of 3N peaks are indicated by arrowheads.
In the present study, we investigated the roles of DBCs located outside of oriC. We found that simultaneous deletion of the six DBCs (in the Δ6 strain) resulted in early initiation of replication, and that reintroduction of a single DBC partly complemented the defect, even when the DBC was inserted into the replication-terminus region. These data indicate that DBCs function not only in transcriptional control of neighboring genes but also in negative regulation of replication initiation. Further, inactivation of Spo0J in Δ6 cells yielded a pleiotropic phenotype, accompanied by severe growth inhibition. However, a spontaneous suppressor in soj or a deletion of soj counteracted these effects. We propose that DBCs negatively regulate replication initiation by interacting with DnaA molecules, to sequestrate active ATP-DnaA and/or inactivate DnaA through promotion of the ATP-hydrolysis, and play a major role, together with Spo0J/Soj, in regulating the activity of DnaA to ensure correct and tight regulation of the initiation of chromosome replication.
MATERIALS AND METHODS
Bacterial strains, plasmids and primers
The bacterial strains, plasmids and primers used in the present study are listed in Supplementary Table S1 (strains), Supplementary Table S2 (plasmids) and Supplementary Table S3 (primers). All B. subtilis strains of the present work were derived from the wild-type strain 168 (a laboratory stock), as described in Supplementary Materials. The E. coli strains DH5α and C600 were used for plasmid construction. Because a Δ6–Δspo0J mutant (HO1073) could not be maintained stably, we used a freshly prepared transformant of the Δ6 mutant (MYA143) with the genomic DNA of Δspo0J mutant (HO1072).
Bacterial growth conditions
When bacteria were to be subjected to microscopic observation or flow cytometry, Spizizen's minimal medium (SMM) (26) supplemented with 0.5% (w/v) glucose, trace elements, and a required amino acid (50 mg l−1 l-tryptophan) was employed. In addition, l-glutamic acid (1 g l−1) was added when bacteria were grown for microscopic observation, and l-glutamic acid (3 g l−1), l-asparagine (3 g l−1), and Casamino acids (500 mg l−1) were added for growth prior to flow cytometry. In all other experiments, Luria-Bertani (LB) medium was used for cell growth, supplemented with appropriate antibiotic(s) if necessary (50 µg ml−1 ampicillin, 0.5 µg ml−1 erythromycin, 5 µg ml−1 chloramphenicol, 10 µg ml−1 tetracycline, 100 µg ml−1 spectinomycin and 5 µg ml−1 kanamycin).
Transcriptosome analysis
Transcriptosome profiles of wild-type 168 and mutant cells were examined using a customized Affymetrix tiling array, as described in Supplementary Data.
Flow cytometry
Chloramphenicol at a final concentration of 200 µg ml−1 was added to exponentially growing cells (OD600 = 0.3) in SMM at 30°C, and the cells were further incubated for 5 h to allow completion of ongoing rounds of chromosome replication, but without any further replication initiation or cell division, followed by fixation in 70% (v/v) ethanol overnight at 4°C. After removal of ethanol by centrifugation, fixed cells were suspended in buffer (10 mM Tris–HCl, pH 7.5; 15 mM NaCl; and 1 mM EDTA) with 250 µg ml−1 RNaseA, and incubated at 37°C for 1 h with shaking. To dissociate cell chains into single cells, samples were sonicated using a Bioruptor UCD-250 (Cosmo Bio Co., Ltd.) for 2 min (2 s ‘on’ and 8 s ‘off’, at the low output level). The OD600 values of cell suspensions were adjusted to 0.008 in the same buffer. Next, SYTO16 (Molecular Probes) was added to each cell suspension to a final concentration of 1 µM, and each mixture was incubated at room temperature for 1 h in the dark. The number of replication origins per cell was measured using a FACScan (Becton Dickinson), and data were analyzed with CellQuest software (Becton Dickinson).
Measurement of the oriC/terC ratio by qPCR
To measure the levels of oriC and terC DNA, real-time quantitative PCR (qPCR) was conducted using a LightCycler 480 System (Roche), according to the supplier's protocol. The primers used to amplify the oriC and terC regions were designed using Primer3 software (http://frodo.wi.mit.edu/primer3/input.htm) (Supplementary Table S3). Chromosomal DNA purified from exponentially growing cells, under conditions identical to those used for flow cytometric analysis, was evaluated, and chromosomal DNA purified from chloramphenicol-treated wild-type cells, in which the levels of oriC and terC were equalized by completion of any ongoing rounds of chromosome replication, was used as a standard.
Fluorescence microscopy
Cell morphology, nucleoid structure and protein foci were examined by fluorescence microscopy after staining for DNA with DAPI (Dojin; 1–5 ng ml−1) and the cell membranes with FM4-64 (Invitrogen; 1 µg ml−1) or MitoTracker Green FM (Invitrogen; 100 nM). Fluorescence was visualized using a DMRE-HC microscope (Leica Microsystems) and a cooled digital CCD camera (model 1300Y; Roper Scientific), equipped with appropriate filters (DAPI, Filter Cube A4 [Leica Microsystems]; MitoTracker Green FM, Filter Cube L5 [Leica Microsystems]; and mCherry and FM4-64, Filter Cube N3 [Leica Microsystems]). The intracellular location of fluorescent foci was analyzed using MetaMorph software (Universal Imaging).
Examination of the SOS response
The SOS response was examined by monitoring expression of the bgaB gene encoding B. stearothermophilus β-galactosidase under the control of a damage-inducible promoter (PdinC), at the amyE locus on the chromosome, as previously described (5).
RESULTS
Simultaneous deletion of six DBCs lying outside of oriC induces overinitiation of chromosome replication
We have recently shown that some DBCs lying outside of oriC are involved in transcriptional regulation of neighboring genes (21). In addition, it raises the possibility as to whether these DBCs play roles in the initiation of chromosome replication and/or chromosome organization around oriC. Thus, to understand the effects of stable binding of DnaA protein to DBCs lying outside of oriC, we created the MYA143 (Δ6) strain, in which six DBCs outside of oriC (Figure 1A) were deleted, in the absence of residual drug-resistant marker cassettes, as described in ‘Methods’ of Supplementary Data. The detailed structure of each DBC deletion is shown in Supplementary Figure S1.
The growth rate and the average cell length of Δ6 cells were comparable to those of wild-type cells, as described later. We examined the replication initiation frequency of Δ6 cells during exponential growth, and compared it to that of wild-type and mutant cells harboring single deletions of each DBC. We used two methods to determine the initiation frequency. Flow cytometry was employed to compare the number of oriC units per cell (Figure 1), whereas qPCR was used to determine the copy-number ratio of oriC to terC (Table 1). Flow cytometry showed that most wild-type cells growing in SMM, supplemented with l-glutamic acid, l-asparagine and Casamino acids, at 30°C (generation time of ~45 min) contained two oriC units per cell (as indicated by the major 2N peak and the minor 4N peak) (Figure 1B), and similar flow cytometry patterns were obtained for the single DBC deletion mutants (Figure 1C–H), although a mild but significant decrease in the 2N peak and emergence of 3N peak were evident in the single deletion mutant of DBC in the yydA-yycS region (ΔDBC[yydA-yycS]), suggesting that this strain exhibited slight overinitiation of chromosome replication (Figure 1G). However, this was barely evident by qPCR analysis (Table 1).
Table 1.
Ratio of oriC to terC
| Strain | Relevant genotype | oriC/terCa | Relative ratiob |
|---|---|---|---|
| 168 | Wild-type | 2.32 ± 0.43 | 1.00 |
| MYA155 | ΔDBC[gcp-ydiF] | 2.30 ± 0.13 | 0.99 |
| MYA156 | ΔDBC[yqeG-sda] | 2.23 ± 0.20 | 0.96 |
| MYA157 | ΔDBC[ywlC-ywlB] | 2.34 ± 0.25 | 1.01 |
| MYA018 | ΔDBC[ywcI-vpr] | 2.16 ± 0.21 | 0.93 |
| MYA019 | ΔDBC[yydA-yycS] | 2.22 ± 0.18 | 0.96 |
| MYA158 | ΔDBC[thdF-jag] | 2.11 ± 0.15 | 0.91 |
| HO1009 | Δ5DBC | 2.38 ± 0.32 | 1.03 |
| MYA143 | Δ6DBC | 3.17 ± 0.28 | 1.37 |
| HO1019 | Δ6DBC, 7°::DBC[yydA-yycS] | 2.45 ± 0.42 | 1.06 |
| HO1020 | Δ6DBC, 90°::DBC[yydA-yycS] | 2.25 ± 0.46 | 0.97 |
| HO1021 | Δ6DBC, 200°::DBC[yydA-yycS] | 2.49 ± 0.20 | 1.07 |
| HO1022 | Δ6DBC, 270°::DBC[yydA-yycS] | 2.67 ± 0.17 | 1.15 |
| HO1023 | Δ6DBC, 353°::DBC[yydA-yycS] | 2.14 ± 0.30 | 0.92 |
| HO1242 | Δ6DBC, 200°::DBC[yydA-yycS],180°::DBC[yydA-yycS] | 2.48 ± 0.59 | 1.07 |
| HO1241 | Δ6DBC, 200°::DBC[yydA-yycS],180°::DBC[yydA-yycS], 190°::DBC[yydA-yycS] | 2.13 ± 0.22 | 0.92 |
| HO1015 | Δ6DBC, pO2HC | 3.15 ± 0.50 | 1.36 |
| HO1016 | Δ6DBC, pBOXyydA-yycS | 3.13 ± 0.59 | 1.35 |
| HO1230 | ΔDBC[yydA-yycS], ΔDBC[gcp-ydiF] | 2.24 ± 0.18 | 0.97 |
| HO1231 | ΔDBC[yydA-yycS], ΔDBC[yqeG-sda] | 2.37 ± 0.25 | 1.02 |
| HO1232 | ΔDBC[yydA-yycS], ΔDBC[ywlC-ywlB] | 2.48 ± 0.21 | 1.07 |
| HO1233 | ΔDBC[yydA-yycS], ΔDBC[ywcI-vpr] | 2.71 ± 0.28 | 1.17 |
| HO1234 | ΔDBC[yydA-yycS], ΔDBC[thdF-jag] | 2.76 ± 0.25 | 1.19 |
| HO1247 | Δ6DBC, 353°::DBC[gcp-ydiF] | 3.13 ± 0.14 | 1.35 |
| HO1243 | Δ6DBC, 353°::DBC[yqeG-sda] | 3.22 ± 0.24 | 1.39 |
| HO1244 | Δ6DBC, 353°::DBC[ywlC-ywlB] | 2.98 ± 0.05 | 1.28 |
| HO1245 | Δ6DBC, 353°::DBC[ywcI-vpr] | 2.68 ± 0.15 | 1.16 |
| HO1246 | Δ6DBC, 353°::DBC[thdF-jag] | 3.02 ± 0.28 | 1.30 |
| HO1034 | Δspo0J | 2.46 ± 0.18 | 1.06 |
| HO1030 | Δ(soj–spo0J) | 2.43 ± 0.15 | 1.05 |
| HO1035 | Δ6DBC, Δspo0J | 5.02 ± 0.33 | 2.17 |
| HO1225 | Δ6DBC, Δspo0J, sojT88M | 3.54 ± 0.31 | 1.53 |
| HO1031 | Δ6DBC, Δ(soj–spo0J) | 3.88 ± 0.56 | 1.67 |
aRatio of oriC to terC was determined by qPCR assay from at least five independent experiments and shown with the standard deviation.
bRelative value to the wild-type value.
In contrast, no distinguishable peak was detected in the flow cytometry profile of Δ6 cells. Rather, broad peaks expanding to regions above 4N were evident (Figure 1I). This strongly suggested that completion of ongoing replication did not occur in Δ6 cells during chloramphenicol treatment. We repeated the flow cytometry analysis using a 10-fold greater concentration (thus, 2 mg ml−1) of the drug, but the peak profile did not change. Thus, the initiation frequency of chromosome replication in Δ6 cells could not be precisely estimated by flow cytometry. However, the qPCR results indicated that the copy number of oriC relative to that of terC in Δ6 cells was 1.37-fold higher than in wild-type cells, strongly suggesting that overinitiation of chromosome replication occurred in the former cells (Table 1). Thus, the abnormal flow cytometry profile of Δ6 cells may be attributed to the combined effects of replication overinitiation and incomplete termination of replication during chloramphenicol treatment. In addition, the profile may also reflect a wide variation of initiation frequency in cells within the population.
The phenotypic features of Δ6 cells are due to by multiple deletions of DBC
The flow cytometry data suggested that the abnormal flow cytometry peak profile and overinitiation of replication in Δ6 cells resulted from a cumulative effect of multiple deletions of DBC, as no single deletion of DBC induced a drastic change in profile. Interestingly, restoration of the deleted DBC[yydA-yycS] sequence in Δ6 cells (to form the HO1009 [Δ5] strain, Supplementary Figure S2) partly rescued the abnormal flow cytometry profile and decreased the oriC/terC ratio to resemble those of ΔDBC[yydA-yycS] cells (Figure 1G and 1J, and Table 1), indicating that the deletion of the DBC[yydA-yycS] sequence is partly responsible for the phenotype of the Δ6 strain.
To examine the contribution of the DBCs other than DBC[yydA-yycS] on the overinitiation phenotype of Δ6 strain, double DBC deletion mutants were constructed by introduction of another DBC deletion into the ΔDBC[yydA-yycS] strain and examined by flow cytometry. Interestingly, only when ΔDBC[ywcI-vpr] was introduced did the initiation frequency increase to a similar level of the Δ6 strain (Figure 1K). Further, a significant restoration of the overinitiation phenotype of Δ6 strain was observed when DBC[ywcI-vpr] was introduced into the oriC-proximal sites (353°) where DBC[yydA-yycS] was removed, although to a lesser extent than that of DBC[yydA-yycS] (Figure 2H and Supplementary Figure S3).
Figure 2.
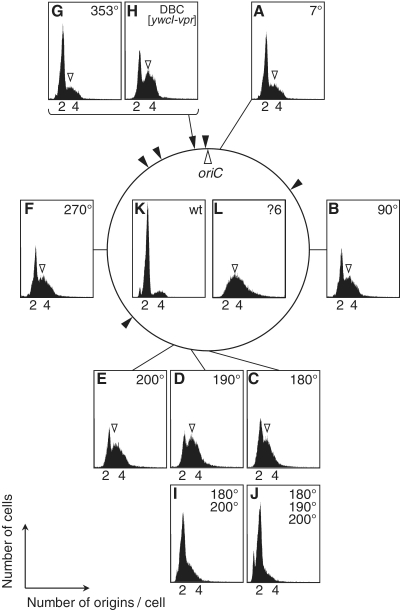
Flow cytometry profiles of DBC-deleted and –reintroduced strains. Flow cytometry profiles of strains with an insertion of DBC in various ectopic positions on the B. subtilis chromosome. (A) HO1019 (Δ6 7°::DBC[yydA-yycS]), (B) HO1020 (Δ6 90°::DBC[yydA-yycS]), (C) HO1238 (Δ6 180°::DBC[yydA-yycS]), (D) HO1239 (Δ6 190°::DBC[yydA-yycS]), (E) HO1021 (Δ6 200°::DBC[yydA-yycS]), (F) HO1022 (Δ6 270°::DBC[yydA-yycS]), (G) HO1023 (Δ6 353°::DBC[yydA-yycS]), (H) HO1245 (Δ6 ΔDBC[yydA-yycS]::DBC[ywcI-vpr]), (I) HO1242 (Δ6 180°::DBC[yydA-yycS] 200°::DBC[yydA-yycS]) and (J) HO1241 (Δ6 180°::DBC[yydA-yycS] 190°::DBC[yydA-yycS] 200°::DBC[yydA-yycS]). Profiles of the (K) 168 (wild-type) and (L) MYA143 (Δ6) strains of Figure 1 are also shown for comparison. Location of 3N peaks are indicated by arrowheads.
These results indicate that the overinitiation phenotype of the Δ6 strain is mainly attributable to the absence of DBC[yydA-yycS] and DBC[ywcI-vpr]. However, the flow cytometry profile of the double mutant was still different from that of the Δ6 strain; the 2N peak is still detectable in the double mutant. Thus, it is plausible that some of the remaining DBCs also contribute to the overinitiation profile of the Δ6 strain, even though the change in flow cytometry profile was barely detectable in other double DBC-deletion mutants (Supplementary Figure S3).
Rescue of the overinitiation of replication in Δ6 cells DBC sequences at ectopic positions on the chromosome
Deletion of the datA sequence located close to oriC, and harboring five high-affinity DnaA-boxes, on the E. coli chromosome results in overinitiation of chromosome replication, although the growth rate was not affected, and the phenotype was suppressed by ectopic introduction of datA in either the chromosome or plasmids. Thus, the datA sequence was proposed to titrate free DnaA molecules after replication initiation, to prevent earlier-than-normal reinitiation (2). DBCs outside of the oriC were also predicted to titrate DnaA in B. subtilis, although no experimental examination of this possibility has been explored (22).
We further explored whether the abnormal phenotypic features of Δ6 cells were rescued by a DBC inserted at ectopic positions on the chromosome. To this end, we inserted the DBC[yydA-yycS] fragment into various positions on the chromosome of the Δ6 strain (Figure 2). As expected from the DnaA titration model, flow cytometry profiles demonstrated that insertion of a single DBC suppressed overinitiation of chromosome replication in Δ6 cells. Interestingly the effect was dependent on the locus of insertion (Figure 2A–G). When the DBC[yydA-yycS] fragment was inserted into oriC-proximal sites (at 7° or 353°), overinitiation was suppressed to a level similar to that observed in Δ5 cells. However, insertion of a DBC into a terC-proximal site (at 180°, 190° and 200°), or between oriC and terC (90° and 270°), also suppressed overinitiation, but to a lesser extent. The oriC/terC ratios estimated by qPCR in such cells also support the idea that suppression of overinitiation of chromosome replication was achieved by introduction of a single DBC copy into an ectopic position on the Δ6 chromosome (Table 1). In exponentially growing B. subtilis cells, relative gene dosage continuously decreases from oriC to terC, and the ratio of oriC/terC is more than two under our experimental conditions (Table 1). Importantly, we found that the increase in copy number of DBC[yydA-yycS] fragments to two or three at the terC region suppressed the overinitiation of Δ6 cells to a level similar to that observed in Δ5 cells (Figure 2I and 2J, and Table 1). Thus the locus-dependence of the effect of DBC[yydA-yycS] reintroduction would be mainly due to the difference in gene dosage of the inserted region.
However, introduction of a multicopy plasmid harboring the DBC of the intergenic region between yydA-yycS into Δ6 cells did not complement the overinitiation phenotype (Table 1 and Supplementary Figure S4) even though DnaA binding to the plasmid was detected by qPCR (Supplementary Figure S4). In contrast, overinitiation of chromosome replication induced by deletion of datA in E. coli cells was suppressed by the introduction of plasmids harboring datA (2), suggesting that the characteristics of interaction between DnaA and DnaA-box cluster sequences differ between B. subtilis and E. coli, at least in terms of regulation of initiation of chromosome replication.
Initiation of chromosome replication occurs earlier in Δ6 cells compared to wild-type cells
To further characterize the phenotype of the Δ6 strain, we examined cell and nucleoid morphology of slowly growing Δ6 cells (generation time of ~120 min) upon cultivation in SMM at 25°C, and compared it with that of wild-type cells. In addition, we analyzed spo0J-inactivated cells (the HO1034 strain), in which earlier timing of initiation of replication and a defect in chromosomal segregation have been reported (12–15,27).
Comparison of membrane-stained images of wild-type and Δ6 cells suggested that cell length was not affected in Δ6 cells, and indeed, analysis of cell length distribution (Supplementary Figure S5) showed that the average length of Δ6 cells (2.82 µm) was similar to that of wild-type cells (2.96 µm). However, Δspo0J mutant cells were slightly elongated (average cell length of 3.23 µm), as reported previously (13,28).
We measured the relative positions of oriC regions on nucleoids using the tetO-TetR fluorescent repressor operator system (FROS), by inserting a tetO array near oriC (at 345°C, thus about 167 kb from oriC), and labeling of the insertion with the TetR-mCherry fusion protein (29) (Supplementary Table S4, see also Figure 4F–H). Most wild-type cells (95.2%) harbored single replicating nucleoids; a minor proportion of long cells (4.8%) contained two replicating nucleoids (Supplementary Table S4). A similar distribution of nucleoids was observed in Δspo0J mutant and Δ6 mutant cells, indicating that chromosome segregation and cell division are coupled normally in these strains, and cell division occurs immediately after segregation of duplicated nucleoids in our cultivation condition.
Figure 4.
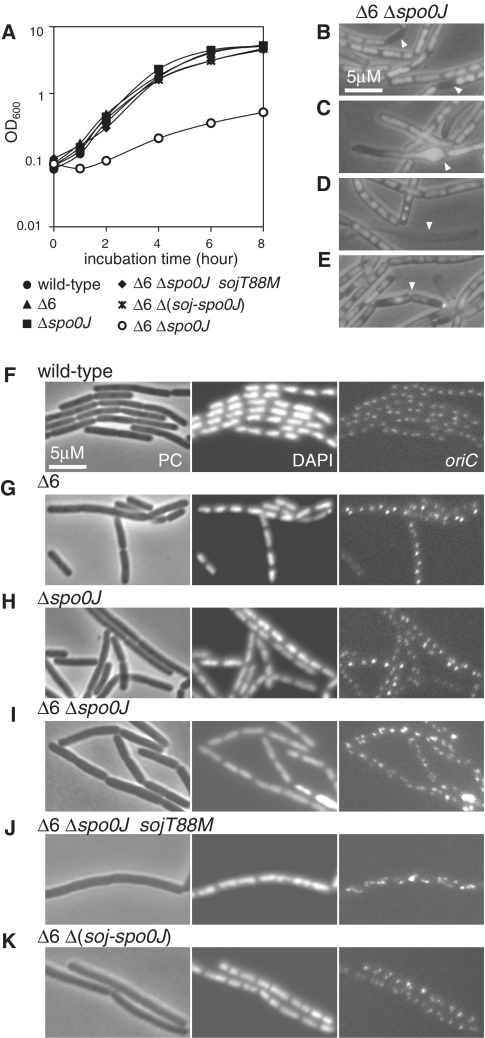
Growth and morphological defects in Δ6 and Δspo0J double-mutant cells, and suppression thereof by soj inactivation. (A) Growth curves of strain 168 (wild-type, closed circles), MYA143 (Δ6, closed triangles), HO1034 (Δspo0J, closed squares), HO1035 (Δ6 Δspo0J, open circles), HO1225 (Δ6 Δspo0J sojT88M, closed diamonds) and HO1031 (Δ6 Δ(soj-spo0J), asterisks) in SMM at 37°C. (B–E) Overlays of phase contrast and DAPI-stained micrographs of HO1035 cells grown in SMM at 25°C, showing abnormal morphology; (B) anucleate cell, (C) bulging cell, (D) ghost cell and, (E) cell with a guillotined chromosome. (F–K) Phase contrast (PC), DAPI (DAPI) and oriC focal (oriC) images of derivatives of 168, MYA143, HO1034, HO1035, HO1225 and HO1301 harboring the tetO array and the tetR-mCherry gene (HO1065 [F], HO1067 [G], HO1069 [H], HO1226 [I], HO1071 [J] and HO1200 [K], respectively), grown in SMM at 25°C.
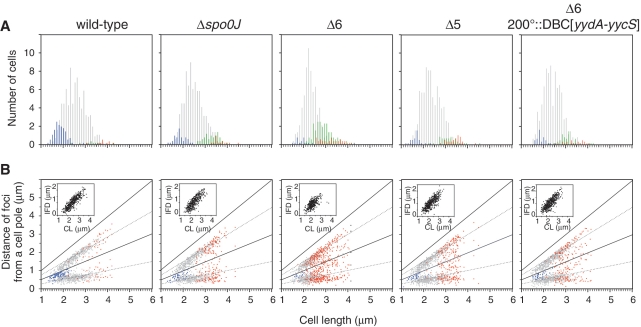
First, we analyzed the relationship between the number of oriC foci and cell length by visualizing the data in histogram format (Figure 3A, see also Supplementary Table S4). It has been shown that duplicated oriC sequences separate immediately after initiation of replication in B. subtilis cells (30), and we observed that most wild-type cells (81.5%) contained two oriC foci (2-oriC cells, gray bars), whereas short cells tended to harbor a single oriC focus (14.8%) (1-oriC cells, blue bars). Although a few long cells harbored three (green bars) or four (red bars) oriC foci (3.8%), the data clearly indicate that, under the slow growth conditions employed in this experiment, wild-type cells essentially completed chromosome replication before cell division and, following completion of cell division, initiated a new round of replication from a single oriC locus in each cell. In contrast, cells containing one copy of oriC were barely detectable (3.1%) in Δ6 cells. Instead, the proportion of 3- or 4-oriC-containing cells was increased (27.4%) (Supplementary Table S4), indicating that Δ6 cells mostly initiate replication in cells containing two origins prior to cell division. Thus the mutant cells initiate chromosome replication earlier than that of the wild-type cells relative to the timing of cell division. These results strongly suggest that the increase of the oriC/terC ratio in Δ6 cells is attributable to earlier initiation of chromosome replication in the cell cycle, compared to wild-type cells. In Δspo0J cells, we observed that proportions of 1-oriC cells (7.2%) and 3- or 4-oriC cells (12.3%) are similar, indicating that replication initiation in Δspo0J cells was also shifted to an earlier time, as suggested in previous reports (12–15,27), but to a lesser extent than that seen in Δ6 cells. Thus, initiation of replication overlaps with cell division in Δspo0J cells, to produce nearly equal amounts of 1-oriC cells (a population that has not yet initiated replication after cell division) and 3- or 4-oriC cells (a population that initiated replication before cell division). It should be noted that the abundance of cells with 3 foci in Δspo0J mutant and Δ6 mutant cells suggests that only one origin often initiates chromosome replication in cells harboring two origins competent for initiation. It is also possible that separation of replicated origin regions were impaired in these mutants. We prefer the former possibility because flow cytometric profiles of ΔDBC[yydA-yycS] (Figure 1G) and some Δ6 mutants with ectopic insertion of DBC (Figure 2) suggest the existence of the 3N peak between 2N and 4N regions, although that is not clear in Δ6 mutant and Δspo0J mutant cells (Figure 1I and Supplementary Figure S6).
Figure 3.
Subcellular locations of origin foci in cells harboring single nucleoids. The origin region was visualized using TetR-mCherry bound to a tetO array at the oriC-proximal region of the chromosomes of wild-type (HO1065), Δ6 (HO1067), Δspo0J (HO1069), HO1176 (Δ5), and HO1175 (Δ6 200°::DBC[yydA-yycS]) strains. The bacteria were grown in SMM at 25°C, and MitoTracker Green FM (membrane) and mCherry (oriC) images were acquired as described in ‘Materials and Methods’ section. (A) Histograms of cell length at 0.1 µm intervals for cells harboring one, two, three, four or more than four foci (in a single nucleoid) are shown using blue, gray, green and red bars, respectively. The x-axes are scaled identically in panels A and B and numbers are indicated in panel B. (B) oriC positioning was statistically analyzed, with plotting of the distance from each focus to the same pole in each cell (on the x-axis) against cell length (on the y-axis). The positions of foci in cells harboring one, two or more than two foci (in a single nucleoid) are indicated by blue, gray and red points, respectively. The distance between the two oriC foci in each cell (interfocal distance, IFD) is indicated in the inset as a function of cell length (CL).
Next, we analyzed the relative positions of oriC foci in various strains (Figure 3B). In wild-type cells, oriC foci were located close to the mid-cell region in 1-oriC cells (blue points), and, when a cell length that triggers replication initiation was obtained, duplicated oriC foci began to appear in cell quarters (in 2-oriC cells, gray points). Further, in Δspo0J mutant and Δ6 mutant cells, 3- or 4-oriC cells, in which oriC foci were located in individual one-eighth regions of cells (red points), emerged when replication was initiated from two oriC loci within a single cell. Thus, although initiation of replication is induced earlier in Δspo0J and Δ6 cells, oriC positioning is apparently not disturbed under slow growth conditions. In support of this conclusion, the distance between the two oriC foci of each cell (the interfocal distance) was similar in the three strains examined (inset of Figure 3B), although longer cells harboring two oriC foci were absent in the Δ6 strain, further suggesting that no segregation defect was notably induced under our experimental conditions. It has been demonstrated that oriC positioning is disturbed in Δspo0J cells grown in S7 minimal medium at 30°C because of the observed segregation defect (12). However, it was also reported that the segregation defect of the Δspo0J mutant was suppressed under slow growth conditions (28). Thus, the segregation defect due to the spo0J incativation would be negligible in our experimental condition.
As judged by tetO-TetR FROS analysis, Δ6 cells with restoration of DBC[yydA-yycS] to the authentic position (Δ5) and Δ6 cells with a 200°::DBC[yydA-yycS] insertion increased 1-oriC cells and decreased 3- or 4-oriC cells (Figure 3A). Thus, recovery from the overinitiation phenotype of Δ6 mutant by introduction of the DBC[yydA-yycS] sequence detected by flow cytometry and qPCR analysis correlates with the partial suppression of the earlier timing of replication initiation seen in Δ6 cells.
Δ6 and Δspo0J mutations synergistically enhance overinitiation of chromosome replication, resulting in inhibition of nucleoid segregation and development of a severe growth defect
The phenotype of the Δ6 mutant, showing earlier initiation of replication, was also observed in an spo0J deletion mutant, although initiation timing differed in the two mutants, as described above. Further, both DBCs and Spo0J-binding sites (parS sites) are distributed in the origin-proximal region of the chromosome (21). It seemed possible that the phenotypic features of the Δ6 mutant were attributable to impairment of Spo0J function, and/or that DnaA bound to DBCs outside of oriC could play role(s) overlapping with that of Spo0J. To examine these possibilities, we introduced a spo0J deletion into the Δ6 mutant.
We found that the Δ6 and Δspo0J defects acted synergistically to severely reduce cell growth rate, not only under relatively fast growth conditions (SMM at 37°C, Figure 4A), but also under conditions of slow growth (SMM at 25°C, Figure 4B–E), during which growth defect of neither Δ6 mutant nor Δspo0J mutant cells was observed. In double-mutant cells grown in SMM at 25°C, dispersed and unsegregated nucleoids were prominent, compared to the parental stains (Figure 4F–I). In addition, at least 10% of cells failed to inherit chromosomes, yielding anucleate cells (Figure 4B). The frequency of formation of such aberrant cells was much higher than previously reported for a Δspo0J mutant (27,28). Thus, nucleoid segregation was markedly impaired in the double-mutant strain. The average length of double-mutant cells (8.67 µm) was more than twice that of wild-type cells (Supplementary Figure S5), indicating that cell division was also severely impaired. Significant numbers of abnormally bulging and ghost cells were also observed (Figure 4C and D). In addition, we detected cell division over the nucleoid (the guillotine effect) (Figure 4E). As the cell and nucleoid morphology of Δ6–Δspo0J cells was highly disorganized, and many cells were dead, we could not analyze the distribution of oriC foci. However, the number of foci appeared to be significantly greater in Δ6–Δspo0J cells compared to Δ6 or Δspo0J cells (Figure 4I versus Figure 4G and H). qPCR analysis showed that the oriC/terC ratio of the Δ6–Δspo0J cells was twice that of wild-type cells (Table 1). Thus, we conclude that overinitiation is also enhanced in the double mutant.
Inactivation of Soj suppressed the severe growth defect of Δ6–Δspo0J double mutant cells
Interestingly, we could easily isolate a spontaneous suppressor mutant (in strain HO1225) from Δ6–Δspo0J cells. In this mutant the growth rate was comparable to that of wild-type cells (Figure 4A). The chromosome of the suppressor mutant had a point mutation in the soj gene that changed threonine 88 to methionine in the Soj protein. Although we did not characterize the SojT88M protein in detail, the sojT88M mutation should be a loss-of-function mutation in nature, because a complete deletion of soj from the Δ6–Δspo0J double mutant also suppressed the growth defect to an extent similar to that afforded by the sojT88M mutation (Figure 4A). Soj has been shown to activate DnaA, and this activity is suppressed by Spo0J (16,17). Flow cytometry profiles confirmed that the overinitiation phenotype of the spo0J mutant was suppressed by simultaneous inactivation of soj in the genetic background of the cells used in the present study (Supplementary Figure S6). Thus our observations indicate that further overinitiation of chromosome replication in the Δ6–Δspo0J double mutant was caused by unregulated activation of DnaA by Soj, and that this contributed to the severe growth defects of Δ6–Δspo0J cells.
In parallel with the recovery in cell growth rate, cell elongation was essentially suppressed in Δ6–Δspo0J–sojT88M and Δ6–Δ(soj–spo0J) cells (cell lengths were 3.48 and 3.10 µm, respectively) when grown in SMM at 25°C (Supplementary Figure S5). However, the oriC/terC ratios of Δ6–Δspo0J–sojT88M and Δ6–Δ(soj–spo0J) cells (3.83 and 3.79, respectively) remained higher than that of Δ6 cells (3.09) (Table 1). Comparison of flow cytometric profiles (Supplementary Figure S6) and distribution of oriC foci (Supplementary Figure S7) in wild-type and Δ(soj–spo0J) cells indicated that a slight increase in 3- or 4-oriC cell numbers was evident among Δ(soj–spo0J) cells compared to wild-type cells, although the oriC/terC ratio of Δ(soj–spo0J) cells as determined by qPCR was comparable to that of wild-type cells (Table 1). Overinitiation in the Δ(soj–spo0J) background has also been reported by Lee and Grossman (12), and this would enhance earlier initiation of replication in Δ6–Δ(soj–spo0J) cells, compared to Δ6 cells. It has recently been reported that the balance between DnaA inhibition and activation activity of Soj is regulated by Spo0J (17). Thus it is possible that Soj is in a form that inhibits DnaA activity during most of the cell cycle, as proposed by Murray and Errington (16), and that inactivation of Soj would result in constitutive activation of DnaA. In sum, cell elongation of Δ6–Δspo0J cells was rescued by inactivation of soj, but overinitiation remained as a cumulative effect of Δ6 and loss of the Spo0J–Soj system.
Introduction of DBCs suppress the severe growth defect of Δ6–Δspo0J double mutant cells depending on the DBC copy number
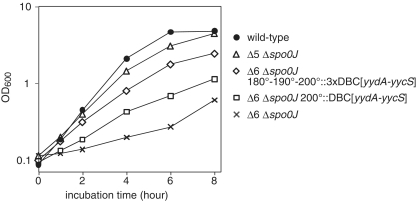
In the present study, we have shown that the introduction of DBCs suppresses the overinitiation of chromosome replication observed in the Δ6 mutant, depending on chromosome loci and copy number of DBC. Thus, we speculated that DBCs would also complement the growth defect of the Δ6–Δspo0J mutant. Consistent with this idea, reintroduction of DBC[yydA-yycS] in the authentic position (353°, Δ5–Δspo0J) restored the severe growth defect of the Δ6–Δspo0J double-mutant cells to a level similar to that of wild-type cells, while insertion at terC-proximal region (200°) rescued the growth defect only partially (Figure 5). In addition, when the copy number of the DBC[yydA-yycS] at the terC-proximal was increased to three (Δ6–Δspo0J-terC::3DBC[yydA-yycS]), the growth rate was increased close to that of wild-type. It should also be noted that even in the Δ5–Δspo0J mutant strain, cell elongation without division was still evident (Supplementary Figure S5) and oriC foci were diffusely distributed (Supplementary Figure S7).
Figure 5.
Suppression of growth defects of Δ6–Δspo0J double mutant cells by ectopic integration of DBC[yydA-yycS]. Growth curves of wild-type (168, closed circles), HO1035 (Δ6 Δspo0J, crosses), HO1041 (Δ5 Δspo0J, open triangles), HO1178 (Δ6 Δspo0J 200°::DBC[yydA-yycS], open squares) and HO1254 (Δ6 Δspo0J 180°::DBC[yydA-yycS] 190°::DBC[yydA-yycS] 200°::DBC[yydA-yycS], open diamonds) in SMM at 37°C.
The growth defect in the Δ6–Δspo0J mutant requires DnaA-dependent initiation of chromosome replication from oriC
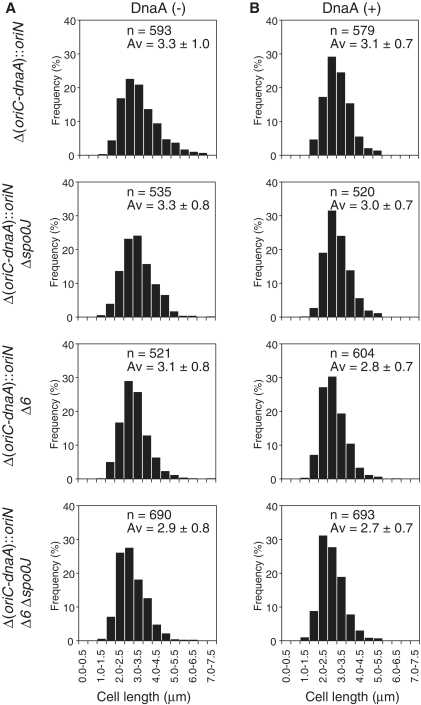
Suppression of the severe growth defect of Δ6–Δspo0J double-mutant cells by introduction of the soj deletion strongly suggested that hyperactivation of DnaA was responsible, at least in part, for several phenotypic features of Δ6–Δspo0J cells. To directly demonstrate the effects of such hyperactivation, we constructed a strain in which the entire oriC sequence and dnaA gene were substituted with a plasmid-derived replication origin (MU01 [Δ(oriC-dnaA)::oriN]; Supplementary Figure S8), and transferred the plasmid oriN sequence into Δspo0J, Δ6 and Δ6–Δspo0J strains, replacing the native oriC and dnaA loci on the chromosome. In addition, we introduced the dnaA gene under the control of the Pxyl promoter at the amyE locus of these strains (Supplementary Figures S9 and S10). As a result, the growth defect of Δ6–Δspo0J double mutant cells was rescued independent of DnaA induction with xylose (Supplementary Figure S11). Examination of cell lengths and division septa by membrane staining showed that the morphology of cells harboring the Δ6 mutation became indistinguishable from that of cells without the mutation (Figure 6A). Further, overproduction of DnaA from an ectopic gene position on the chromosome of such cells was without notable effect (Figure 6B and Supplementary Figure S11). These results imply that the phenotypes induced by multiple DBC deletions in Δ6 cells require DnaA-dependent initiation of chromosome replication, suggesting that hyperactivation of DnaA is the principal cause of the observed phenotypic features.
Figure 6.
Cell length distribution of wild-type, Δ6, Δspo0J and Δ6–Δspo0J derivatives in which chromosome replication is driven by the oriN replication system. MU03 (Δ[oriC-dnaA]::oriN amyE::Pxyl-dnaA), HO1201 (Δ[oriC-dnaA]::oriN Δspo0J amyE::Pxyl-dnaA), HO1203 (Δ[oriC-dnaA]::oriN Δ6 amyE::Pxyl-dnaA) and HO1206 (Δ[oriC-dnaA]::oriN Δ6 Δspo0J amyE::Pxyl-dnaA) cells were grown in SMM without (A) and with (B) xylose at 25°C to mid-exponential phase, and membrane-stained (FM4-64) images are taken. Glucose in SMM was substituted by glycerol (0.5%, v/v) to fully induce DnaA from the xylose promoter. Histograms of cell length of indicated number of cells at 0.5 µm intervals are shown.
DISCUSSION
Here, we report new and important findings on the control of initiation of chromosome replication in B. subtilis. We demonstrate that DBCs lying outside of oriC play an important role in the regulation of the initiation of chromosome replication in this organism, together with the Soj–Spo0J system.
Simultaneous deletion of all six stable DnaA-binding sequences in DBC regions lying remote from oriC on the B. subtilis chromosome induced apparent overinitiation of chromosome replication. DBC[yydA-yycS] was the sole region whose single deletion showed a weak but significant overinitiation phenotype, and its reintroduction into the chromosome of Δ6 cells clearly remedied the defect in replication initiation control. In addition, although overinitiation was not clearly detected when DBC[ywcI-vpr] was singly deleted, it becomes prominent when deleted with DBC[yydA-yycS], to a level similar to that of Δ6 mutant. Since reintroduction of DBC[ywcI-vpr] into Δ6 mutant also rescued the overinitiation phenotype, but to a lesser extent than that of DBC[yydA-yycS], we conclude that DBC[ywcI-vpr] is a secondary contributor to the phenotype of Δ6 strain. However, the flow cytometry profile of the double mutant was still different from that of the Δ6 strain, indicating that some of the remaining DBCs would also contribute to the overinitiation phenotype of the Δ6 strain.
In Δ6 cells, genes repressed by the transition state regulator AbrB were strongly derepressed as in the deletion mutant of abrB (31), and the σD regulon appeared not to be fully activated as in the deletion mutant of sigD (the gene encoding σD) (32). However, neither inactivation of abrB nor sigD in wild-type cells affected the flow cytometry profile (Supplementary Figure S12), suggesting that any of these regulons would not the cause of the overinitiation phenotype of Δ6 cells. In addition, the DBC[yydA-yycS] sequence harbors the promoter sequence of the yydA gene and its expression is suppressed in Δ6 cells, but the yydA inactivation in wild-type cells displayed no apparent phenotype (Supplementary Figure S13). These results, together with fact that ectopic reintroduction of either DBC[yydA-yycS] or DBC[ywcI-vpr] into Δ6 mutant rescued the overinitiation phenotype, although partially, indicate that the overinitiation of replication in Δ6 cells would not be the consequence of alteration of expression of specific gene(s), but would be induced directly through alteration of interaction of DnaA proteins with DnaA-boxes in DBCs. However, we have not yet succeeded in identifying structural feature(s) of DBCs that discriminate DBCs those having the ability to regulate initiation of replication and those without this ability.
Further, the introduction of a spo0J deletion into the Δ6 mutant resulted in severe disorganization of nucleoid structure and abnormal cell division, and these phenotypic features caused a major reduction in the growth rate of Δ6–Δspo0J double-mutant cells. Importantly, we showed that inactivation of soj, which activates DnaA in the absence of Spo0J, remedied the growth defect of Δ6–Δspo0J double-mutant cells, indicating that hyperactivation of DnaA contributed to the observed growth aberrations in the double mutant. Indeed, replacement of the DnaA-dependent initiation system for chromosome replication from oriC with a DnaA-independent plasmid-derived system (oriN) rescued the growth defects, and overproduction of DnaA did not affect this observation (Figure 6 and Supplementary Figure S11). Thus, our results strongly suggest that DBCs located outside of oriC would also contribute to control of chromosome replication initiation by regulating DnaA activity functional for initiation of replication at oriC, via interaction between DBCs and DnaA molecules.
Analysis of the correlation between the number of oriC foci and cell length in wild-type, Δspo0J and Δ6 cells indicated that chromosome replication was initiated earlier in Δspo0J mutant and Δ6 mutant cells, compared to wild-type cells, assuming that duplicated oriC sequences separate immediately after initiation of replication, as reported previously (30). In wild-type cells, initiation of replication occurred after cell division, while initiation of replication arose before cell division in Δ6 cells and overlapped with cell division in Δspo0J cells under the growth conditions used here. It has been proposed that in wild-type cells of B. subtilis, the initiation potential for chromosome replication is determined by the number of active DnaA molecules available to bind to oriC, and such numbers are controlled so that a threshold amount of DnaA is reached at an appropriate time in the cell cycle (33). The level of DnaA available for binding to oriC would be expected to increase in the absence of the six DBCs or Spo0J, leading to early accumulation of initiation potential.
What was particularly surprising was that this earlier-than-normal initiation of replication observed with the Δspo0J or Δ6 mutations was not accompanied by changes to the doubling time or cell length distribution. One exception is the slight elongation of Δspo0J cells. Further, segregation of nucleoids seemed to be coupled to cell division in all three strains (Δspo0J, Δ6 or double Δspo0J–Δ6 mutant). Although no information on the timing of replication termination during the B. subtilis cell cycle is currently available, our data raise the possibility that the timing of all three processes, termination of replication, segregation of the replicated chromosome and cell division are essentially unaffected by the Δspo0J and Δ6 mutation. If this is correct, the implication is that early initiation of replication slows down the replication elongation rate, resulting in an increase in the oriC/terC ratio in Δspo0J mutant and Δ6 mutant cells. How could this occur? One possibility is that the nucleotide substrates for chromosome replication are limited in number under the culture conditions employed, leading to stalling of replication forks. How might this be? Recently, a direct link between DNA replication and central carbon metabolism has been genetically demonstrated in B. subtilis and E. coli (34,35). Although the exact mechanism is still unclear, replication rate may be regulated by a feedback system to ensure synchrony with cellular metabolic activities. It should be noted that, if elongation of DNA replication in mutant cells proceeds at the same rate as in wild-type cells, it is difficult to explain the increase in the oriC/terC ratio and the constant growth rate seen when replication is initiated earlier.
We found that double-mutant Δ6–Δspo0J cells suffered from a severe growth defect that was almost lethal. This defect was DnaA-dependent and rescued by deletion of soj, confirming an earlier finding that Soj activates DnaA (16,17). These results indicate that unrepressed DnaA activity is fatal, and DnaA activity should thus be multiply regulated. In B. subtilis, it has been reported that overexpression of DnaA from an ectopic locus induces the SOS response, which is elicited by depletion of DnaN because of transcriptional repression of the dnaA-dnaN operon by autoregulation mechanism by DnaA; and the SOS response results in cell elongation due to the inhibition of cell division (5). Actually, we found that the SOS response was not induced in Δ6 cells, but induced in Δ6–Δspo0J cells at very high levels (Supplementary Figure S14). However, division was frequently observed in nucleoid-free regions and even over unsegregated nucleoids in Δ6–Δspo0J cells, leading to the production of anucleate cells and a nucleoid guillotine effect. Thus, SOS response in the Δ6–Δspo0J strain may not be directly induced as a consequence of hyperinitiation of replication from oriC, but indirectly induced by a sever segregation defect and subsequent guillotining of the nucleoids inducing further DNA damage. In E. coli, overexpression of DnaA leads to overinitiation and stalling of replication forks, accompanied by filamentation attributable to a cell division defect, nucleoid elongation, and a decrease in viability, in an SOS response-independent manner (36). As lack of a double-strand break (DSB) repair system further decreased the viability of such cells, DSBs, probably generated by collision between new forks and stalled or collapsed forks, have been suggested to be the cause of the viability decrease in overinitiating E. coli cells. Interestingly, incomplete chromosome replication upon overproduction of DnaA has been detected as a broad flow cytometric pattern in E. coli cells (36), as observed also in the B. subtilis cells of the present study. In addition, the lethal effect of DnaA overproduction required initiation from oriC in E. coli (37). These similarities between earlier reports and our current data suggest that stalling and collapse of replication forks, resulting in accumulation of DSBs, occurs in a lethal level in the Δ6–Δspo0J strain, although further work is required to prove this hypothesis. It is interesting to note that, upon hyperinitiaiton of replication, E. coli induces inhibition of cell division and stalling of replication fork in an SOS-independent manner (36), providing a chance to escape from fatal nucleoid disorganization. However, such a system has not been known in B. subtilis cells. In addition, it has been shown that DSBs provoke global SOS induction in E. coli but not in B. subtilis (38). Thus, it seems reasonable to assume that accumulation of DSBs is a direct reason for the lethality in Δ6–Δspo0J cells. However, SOS response in them would be induced by DNA damage other than DSBs, such as fragmentation of the chromosome caused by guillotining of the nucleoids.
Our results showed that DBCs lying outside of oriC, together with the Soj protein, play major roles in regulation of the activity of DnaA acting on oriC in B. subtilis cells. Differences in the observed phenotypes of single and double mutants suggest that the DBCs and Soj are complementary in function; one takes over when the other is inactivated. Under our culture conditions, a yabA deletion mutant, constructed by transformation of a wild-type 168 strain with genomic DNA of NIS6050 (CRK6000: yabA::spec), did not show a growth defect, as previously reported (7,8). Interestingly, a preliminary experiment revealed that double mutation of Δ6–ΔyabA induced further overinitiation compared to that of the parental strain, but also did not cause a growth defect (our unpublished data). This result suggests that YabA negatively controls DnaA activity via a mechanism distinct from that employed by DBCs lying outside of oriC, although hyperactivation of replication initiation is milder in Δ6–ΔyabA cells compared to Δ6–Δspo0J cells. Comprehensive analysis of the contributions of DBCs lying outside of oriC, Soj and YabA to regulation of initiation of replication is now in progress.
We have shown herein that DBC sequences located at the intergenic regions between yydA-yycS and ywcI-vpr regulate replication initiation negatively, and that the extent of such negative regulation was dependent on the location of DBCs in the chromosome. Introduction of three copies of DBCs into the terC-proximal region restored the overinitiation phenotype of the Δ6 mutant and rescued the growth defect of the Δ6–Δspo0J mutant to a level similar to those of strains with a single DBC insertion at the oriC-proximal site. We therefore conclude that the locus dependence correlates with relative dosage of DBC to that of oriC in exponentially growing cells. However, it is also conceivable that local higher-order structures may affect the extent and stability of DnaA molecules bound to a DBC. The inability of the same DBC sequence present in plasmids to suppress overinitiation of replication in Δ6 cells strongly suggests that regulating the activity of DnaA by DBC depends on a higher-order structure of DNA.
Deletion of datA of E. coli resulted in a similar level of replication overinitiation [~1.3-fold increase in the oriC/terC ratio compared to that of wild-type cells (2)] as seen in Δ6 cells (1.37-fold). Overinitiation of chromosome replication induced by the datA deletion in E. coli cells was suppressed by the introduction of plasmids harboring datA (2). In contrast, introduction of a multicopy plasmid harboring DBC[yydA-yycS] into Δ6 cells did not complement the overinitiation phenotype. These differences suggest that features of interaction between DnaA molecules and DnaA-box cluster sequences, as well as mechanisms of negative regulation of replication initiation via these interactions, differ between B. subtilis and E. coli. It is possible that the DBCs trap a large number of DnaA molecules sequestering them from the replication origin as proposed for E. coli datA (39). However, the result that introduction of a multicopy plasmid harboring DBC[yydA-yycS] into Δ6 cells did not complement the overinitiation phenotype seems to argue against this model. Recently, DBCs named DARS that reactivate DnaA for replication initiation by exchanging bound ADP with ATP have been identified in E. coli (25). Thus, it would be also possible that B. subtilis DBC[yydA-yycS] and DBC[ywcI-vpr] play opposite roles on the DnaA activity to that of E. coli DARS, inactivate ATP-DnaA by inducing the ATP hydrolysis. Although similar levels of stable DnaA binding were detected at all DBCs by in vivo ChIP-chip experiments (19,21–23), the present study demonstrated that only two of them have the ability to effectively regulate replication initiation negatively. These results support the idea that specific DnaA–DBC interactions that modify the DnaA activity are involved in the regulation of initiation of replication. Further studies to elucidate the molecular mechanism of the DBC action in B. subtilis cells are warranted.
We found that transcription of the dnaA-dnaN operon in Δ6 cells was reduced 0.7-fold relative to that in wild-type cells (Supplementary Table S5), suggesting that dnaA transcription is repressed by autoregulation mechanism (5). However, western blotting analyses to determine DnaA amounts per constant cell mass in various strains did not permit us to conclude that DnaA protein level is reduced in parallel with the reduced level the dnaA transcription, due to limitation of the accuracy of the assay (Supplementary Figure S15).
The fundamental question that remains is, how are the three effects, differential binding of DnaA molecules to three types of DBCs, DBC at the dnaA promoter (to keep the DnaA concentration constant), regulatory DBC(s) (to control DnaA activity functioning to oriC and dnaA promoter), and DBCs in the oriC sequence (to initiate replication), regulated? The mechanism(s) underlying these processes requires further study.
DBCs are usually located near oriC not only in Bacillus species (21), but also in many bacteria of the Firmicutes and Actinobacteria. In fact, it has been reported that oriC-proximal DBCs in Streptomyces coelicolor are involved in the regulation of chromosome replication (40). Our results suggest that regulatory interaction between DBCs and DnaA would be effective in this regard when DnaA-binding sequences are located in the oriC-proximal regions. Thus, in bacteria lacking SeqA and Hda systems, DBCs in oriC-proximal regions, that regulate DnaA activity negatively by titration, inactivation or other unknown mechanism, would be expected to be common, and to be important in regulation of replication initiation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant-in-aid for scientific research in the Priority Area ‘Systems Genomics’ awarded by the Ministry of Education, Culture, Sports, Science and Technology of Japan. Funding for open access charge: Research fund from Nara Institute of Science and Technology.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Elizabeth Harry for critical reading of the manuscript.
REFERENCES
- 1.Katayama T, Ozaki S, Keyamura K, Fujimitsu K. Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat. Rev. Microbiol. 2010;8:163–170. doi: 10.1038/nrmicro2314. [DOI] [PubMed] [Google Scholar]
- 2.Kitagawa R, Ozaki T, Moriya S, Ogawa T. Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 1998;12:3032–3043. doi: 10.1101/gad.12.19.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moriya S, Firshein W, Yoshikawa H, Ogasawara N. Replication of a Bacillus subtilis oriC plasmid in vitro. Mol. Microbiol. 1994;12:469–478. doi: 10.1111/j.1365-2958.1994.tb01035.x. [DOI] [PubMed] [Google Scholar]
- 4.Fukuoka T, Moriya S, Yoshikawa H, Ogasawara N. Purification and characterization of an initiation protein for chromosomal replication, DnaA, in Bacillus subtilis. J. Biochem. 1990;107:732–739. doi: 10.1093/oxfordjournals.jbchem.a123117. [DOI] [PubMed] [Google Scholar]
- 5.Ogura Y, Imai Y, Ogasawara N, Moriya S. Autoregulation of the dnaA-dnaN operon and effects of DnaA protein levels on replication initiation in Bacillus subtilis. J. Bacteriol. 2001;183:3833–3841. doi: 10.1128/JB.183.13.3833-3841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noirot-Gros MF, Dervyn E, Wu LJ, Mervelet P, Errington J, Ehrlich SD, Noirot P. An expanded view of bacterial DNA replication. Proc. Natl Acad. Sci. USA. 2002;99:8342–8347. doi: 10.1073/pnas.122040799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayashi M, Ogura Y, Harry EJ, Ogasawara N, Moriya S. Bacillus subtilis YabA is involved in determining the timing and synchrony of replication initiation. FEMS Microbiol. Lett. 2005;247:73–79. doi: 10.1016/j.femsle.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 8.Cho E, Ogasawara N, Ishikawa S. The functional analysis of YabA, which interacts with DnaA and regulates initiation of chromosome replication in Bacillus subtilis. Genes Genet. Syst. 2008;83:111–125. doi: 10.1266/ggs.83.111. [DOI] [PubMed] [Google Scholar]
- 9.Soufo CD, Soufo HJ, Noirot-Gros MF, Steindorf A, Noirot P, Graumann PL. Cell-cycle-dependent spatial sequestration of the DnaA replication initiator protein in Bacillus subtilis. Dev. Cell. 2008;15:935–941. doi: 10.1016/j.devcel.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 10.Hiraga S. Dynamic localization of bacterial and plasmid chromosomes. Annu. Rev. Genet. 2000;34:21–59. doi: 10.1146/annurev.genet.34.1.21. [DOI] [PubMed] [Google Scholar]
- 11.Livny J, Yamaichi Y, Waldor MK. Distribution of centromere-like parS sites in bacteria: insights from comparative genomics. J. Bacteriol. 2007;189:8693–8703. doi: 10.1128/JB.01239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee PS, Grossman AD. The chromosome partitioning proteins Soj (ParA) and Spo0J (ParB) contribute to accurate chromosome partitioning, separation of replicated sister origins, and regulation of replication initiation in Bacillus subtilis. Mol. Microbiol. 2006;60:853–869. doi: 10.1111/j.1365-2958.2006.05140.x. [DOI] [PubMed] [Google Scholar]
- 13.Ogura Y, Ogasawara N, Harry EJ, Moriya S. Increasing the ratio of Soj to Spo0J promotes replication initiation in Bacillus subtilis. J. Bacteriol. 2003;185:6316–6324. doi: 10.1128/JB.185.21.6316-6324.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruber S, Errington J. Recruitment of condensin to replication origin regions by ParB/Spo0J promotes chromosome segregation in B. subtilis. Cell. 2009;137:685–696. doi: 10.1016/j.cell.2009.02.035. [DOI] [PubMed] [Google Scholar]
- 15.Sullivan NL, Marquis KA, Rudner DZ. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell. 2009;137:697–707. doi: 10.1016/j.cell.2009.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murray H, Errington J. Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell. 2008;135:74–84. doi: 10.1016/j.cell.2008.07.044. [DOI] [PubMed] [Google Scholar]
- 17.Scholefield G, Whiting R, Errington J, Murray H. Spo0J regulates the oligomeric state of Soj to trigger its switch from an activator to an inhibitor of DNA replication initiation. Mol. Microbiol. 2011;79:1089–1100. doi: 10.1111/j.1365-2958.2010.07507.x. [DOI] [PubMed] [Google Scholar]
- 18.Scholefield G, Veening JW, Murray H. DnaA and ORC: more than DNA replication initiators. Trends Cell Biol. 2011;21:188–194. doi: 10.1016/j.tcb.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 19.Breier AM, Grossman AD. Dynamic association of the replication initiator and transcription factor DnaA with the Bacillus subtilis chromosome during replication stress. J. Bacteriol. 2009;191:486–493. doi: 10.1128/JB.01294-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goranov AI, Katz L, Breier AM, Burge CB, Grossman AD. A transcriptional response to replication status mediated by the conserved bacterial replication protein DnaA. Proc. Natl Acad. Sci. USA. 2005;102:12932–12937. doi: 10.1073/pnas.0506174102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishikawa S, Ogura Y, Yoshimura M, Okumura H, Cho E, Kawai Y, Kurokawa K, Oshima T, Ogasawara N. Distribution of stable DnaA-binding sites on the Bacillus subtilis genome detected using a modified ChIP-chip method. DNA Res. 2007;14:155–168. doi: 10.1093/dnares/dsm017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smits WK, Merrikh H, Bonilla CY, Grossman AD. Primosomal proteins DnaD and DnaB are recruited to chromosomal regions bound by DnaA in Bacillus subtilis. J. Bacteriol. 2011;193:640–648. doi: 10.1128/JB.01253-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smits WK, Goranov AI, Grossman AD. Ordered association of helicase loader proteins with the Bacillus subtilis origin of replication in vivo. Mol. Microbiol. 2010;75:452–461. doi: 10.1111/j.1365-2958.2009.06999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burkholder WF, Kurtser I, Grossman AD. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell. 2001;104:269–279. doi: 10.1016/s0092-8674(01)00211-2. [DOI] [PubMed] [Google Scholar]
- 25.Fujimitsu K, Senriuchi T, Katayama T. Specific genomic sequences of E. coli promote replicational initiation by directly reactivating ADP-DnaA. Genes Dev. 2009;23:1221–1233. doi: 10.1101/gad.1775809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spizizen J. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl Acad. Sci. USA. 1958;44:1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ireton K, Gunther NWt, Grossman AD. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J. Bacteriol. 1994;176:5320–5329. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Autret S, Nair R, Errington J. Genetic analysis of the chromosome segregation protein Spo0J of Bacillus subtilis: evidence for separate domains involved in DNA binding and interactions with Soj protein. Mol. Microbiol. 2001;41:743–755. doi: 10.1046/j.1365-2958.2001.02551.x. [DOI] [PubMed] [Google Scholar]
- 29.Veening JW, Murray H, Errington J. A mechanism for cell cycle regulation of sporulation initiation in Bacillus subtilis. Genes Dev. 2009;23:1959–1970. doi: 10.1101/gad.528209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Webb CD, Graumann PL, Kahana JA, Teleman AA, Silver PA, Losick R. Use of time-lapse microscopy to visualize rapid movement of the replication origin region of the chromosome during the cell cycle in Bacillus subtilis. Mol. Microbiol. 1998;28:883–892. doi: 10.1046/j.1365-2958.1998.00808.x. [DOI] [PubMed] [Google Scholar]
- 31.Chumsakul O, Takahashi H, Oshima T, Hishimoto T, Kanaya S, Ogasawara N, Ishikawa S. Genome-wide binding profiles of the Bacillus subtilis transition state regulator AbrB and its homolog Abh reveals their interactive role in transcriptional regulation. Nucleic Acids Res. 2011;39:414–428. doi: 10.1093/nar/gkq780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serizawa M, Yamamoto H, Yamaguchi H, Fujita Y, Kobayashi K, Ogasawara N, Sekiguchi J. Systematic analysis of SigD-regulated genes in Bacillus subtilis by DNA microarray and Northern blotting analyses. Gene. 2004;329:125–136. doi: 10.1016/j.gene.2003.12.024. [DOI] [PubMed] [Google Scholar]
- 33.Moriya S, Kato K, Yoshikawa H, Ogasawara N. Isolation of a dnaA mutant of Bacillus subtilis defective in initiation of replication: amount of DnaA protein determines cells’ initiation potential. EMBO J. 1990;9:2905–2910. doi: 10.1002/j.1460-2075.1990.tb07481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jannière L, Canceill D, Suski C, Kanga S, Dalmais B, Lestini R, Monnier AF, Chapuis J, Bolotin A, Titok M. Genetic evidence for a link between glycolysis and DNA replication. PLoS ONE. 2007;2:e447. doi: 10.1371/journal.pone.0000447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maciag M, Nowicki D, Janniere L, Szalewska-Palasz A, Wegrzyn G. Genetic response to metabolic fluctuations: correlation between central carbon metabolism and DNA replication in Escherichia coli. Microb. Cell Fact. 2011;10:19. doi: 10.1186/1475-2859-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grigorian AV, Lustig RB, Guzmán EC, Mahaffy JM, Zyskind JW. Escherichia coli cells with increased levels of DnaA and deficient in recombinational repair have decreased viability. J. Bacteriol. 2003;185:630. doi: 10.1128/JB.185.2.630-644.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Felczak MM, Kaguni JM. DnaAcos hyperinitiates by circumventing regulatory pathways that control the frequency of initiation in Escherichia coli. Mol. Microbiol. 2009;72:1348–1363. doi: 10.1111/j.1365-2958.2009.06724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simmons LA, Goranov AI, Kobayashi H, Davies BW, Yuan DS, Grossman AD, Walker GC. Comparison of responses to double-strand breaks between Escherichia coli and Bacillus subtilis reveals different requirements for SOS induction. J. Bacteriol. 2009;191:1152–1161. doi: 10.1128/JB.01292-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nozaki S, Yamada Y, Ogawa T. Initiator titration complex formed at datA with the aid of IHF regulates replication timing in Escherichia coli. Genes Cells. 2009;14:329–341. doi: 10.1111/j.1365-2443.2008.01269.x. [DOI] [PubMed] [Google Scholar]
- 40.Smulczyk-Krawczyszyn A, Jakimowicz D, Ruban-Osmialowska B, Zawilak-Pawlik A, Majka J, Chater K, Zakrzewska-Czerwinska J. Cluster of DnaA boxes involved in regulation of Streptomyces chromosome replication: from in silico to in vivo studies. J. Bacteriol. 2006;188:6184–6194. doi: 10.1128/JB.00528-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.