Abstract
Routine methods for assaying steady-state mRNA levels such as RNA-seq and micro-arrays are commonly used as readouts to study the role of transcription factors (TFs) in gene expression regulation. However, cellular RNA levels do not solely depend on activity of TFs and subsequent transcription by RNA polymerase II (Pol II), but are also affected by RNA turnover rate. Here, we demonstrate that integrated analysis of genome-wide TF occupancy, Pol II binding and steady-state RNA levels provide important insights in gene regulatory mechanisms. Pol II occupancy, as detected by Pol II ChIP-seq, was found to correlate better with TF occupancy compared to steady-state RNA levels and is thus a more precise readout for the primary transcriptional mechanisms that are triggered by signal transduction. Furthermore, analysis of differential Pol II occupancy and RNA-seq levels identified genes with high Pol II occupancy and relatively low RNA levels and vice versa. These categories are strongly enriched for genes from different functional classes. Our results demonstrate a complementary value in Pol II chip-seq and RNA-seq approaches for better understanding of gene expression regulation.
INTRODUCTION
Extrapolation of transcriptional changes in response to signal transduction to molecular mechanisms and regulatory networks remains a major challenge. Over the past years, the increase in microarray densities and quality and the development of massively parallel sequencing of transcriptomes (RNA-seq) allowed affordable genome-wide readout of gene expression over multiple samples with high accuracy and reproducibility. These techniques have proven to be very useful for studying and understanding regulatory networks controlled by different transcriptional programs. However, the above-mentioned techniques measure accumulated levels of RNA that do not necessarily fully reflect transcriptional status of a gene under the given conditions, because steady-state RNA levels are the result of a tightly regulated balance between RNA synthesis and degradation rate (1) with certain classes of genes having different rates of mRNA degradation (2–4).
Other more direct alternatives for measuring transcription are based on ‘nuclear run-on’ (5), dynamic transcriptome analysis (6) or sequencing of nascent transcripts from immunoprecipitated RNA polymerase II (7). However, these techniques require a relatively laborious experimental setup (e.g. metabolic RNA labeling with 4-thiouridine in living cells) or rely on expression of tagged versions of proteins, making it difficult to use in organism-based studies. Other methods such as GRO-seq (8) require the isolation of viable nuclei, which may affect the transcriptional programs in response to stimuli that would normally not occur in intact cells. In addition, these techniques are not compatible with frozen or formalin fixed paraffin embedded (FFPE) archived material.
To address these issues and to obtain a more direct readout of gene expression in a simple and unbiased way, we applied RNA polymerase II (Pol II) chromatin immunoprecipitation (ChIP-seq) (9) as a versatile complementary approach to RNA-seq and microarrays. We demonstrate that this approach, which is based on commonly used ChIP-seq and RNA-seq protocols, provides detailed insight in transcriptional processes. While we demonstrated utility in cultured cells, ChIP-seq and RNA-seq have been shown to work on frozen or FFPE archived material as well as on very small numbers of cells (10–14), providing unique opportunities for studying transcriptional processes where other methods that more directly measure transcriptional rates have limitations or are even impossible.
Applied to the colon cancer model system used here, we were able to identify subclasses of genes that appear regulated by different mechanisms upon WNT-induced signal transduction. These findings illustrate the complementarity of techniques in further dissecting gene regulatory networks.
MATERIALS AND METHODS
Cells
We used Ls174T human colon cancer cells carrying an activating point mutation in β-catenin and Ls174T-pTER-β-catenin cell line carrying a doxycyclin-inducible short hairpin RNA (shRNA) against β-catenin (15). Cells were grown in the presence or absence of doxycyclin (1 µg/ml) for 72 h.
Microarray analyses
We used publicly available data of doxycyclin-treated and -untreated Ls174T-pTER-β-catenin performed on HG-U133 Plus 2.0 microarrays (Affymetrix) (9). CEL files (GEO accession number: GSE18560) were processed by RMA method (16) using rma() function from Bioconductor Affy library with standard settings. Gene expression is defined as direct value from RMA analysis. Expressed gene is gene with expression higher than 16. Differentially transcribed genes were set as genes with at least 2-fold intensity change in all three biological replicates with normalized intensity higher than 16 in all six samples.
RNA-seq
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. To deplete for non-informative ribosomal RNA, 5 µg of total RNA were purified using Ribominus kit (Invitrogen) according to manufacturer's instructions. Ribosome-depleted RNA was resuspended in 50 µl of diethylpyrocarbonate (DEPC)-treated water and fragmented for 60 s using the Covaris sonicator (6 ×16 mm AFA fiber Tube, duty cycle: 10%, intensity: 5, cycles/burst: 200, frequency sweeping). Sheared RNA fragments were phosphorylated using 30 U of Polynucleotide Kinase (Promega) with 0.5 mM adenosine triphosphate (ATP) for 30 min at 37°C. Phosphorylated RNA was purified using TRIzol according to the manufacturer's instruction and resuspended in 1.5 µl of DEPC-treated water with 1 µl of Adaptor mix A and 1.5 µl of Hybridization solution from SOLiD Small RNA Expression Kit (SREK) (Ambion). The mixture was incubated at 65°C in a thermocycler for 5 min and quickly cooled on ice. RNA with hybridized adaptors was mixed with 5 µl of SREK ligation buffer and 1 µl of SREK ligation enzyme and incubated at room temperature for 4 h. Ligated sample was mixed with 10 µl of denaturing buffer (90% formamide, bromphenol blue, crysol red) and size selected on 10% denaturing urea gel for the appropriate size fraction (150–300 bp). The piece of gel containing selected fragments was shredded and RNA was eluted in 300 µl 300 mM NaCl with gentle agitation for 4 h at room temperature. The eluate was separated from gel debris using SPIN-X centrifuge tube filters (Costar); the RNA was precipitated by isopropanol and resuspended in 5 µl of DEPC-treated water. Size-selected RNA was mixed with 2 µl of reverse transcription (RT) buffer, 1.5 µl of dNTPs (10 mMeach), 0.5 µl of barcode RT primer (10 µM), incubated in a thermocycler at 72°C for 4 min, 62°C for 2 min and then put on ice. The sample with hybridized barcode RT primer was mixed with MMLV-RT enzyme (Promega) and incubated at 37°C for 30 min. The library was amplified by ligation-mediated polymerase chain reaction (LM-PCR) by adding 2 µl of RT mix from the previous reaction into 100 µl of PCR mix from the SREK kit (Ambion) with P1 and P2 primer compatible with SOLiD/AB sequencing and cycled in a thermocycler with the following program: 95°C for 2 min; 15 cycles of 95°C for 30 s, 62°C for 30 s, 72°C for 30 s; 72°C for 7 min. The library was size selected on a 2% agarose gel for 150–400-bp long fragments and sequenced on SOLiD/AB sequencer in a multiplexed way to produce 50-bp long reads.
Sequencing reads were mapped against the reference genome (hg18 assembly, NCBI build 36) with Maq package (17), with following settings: -c -n 3, -e 170. Reads with mapping quality zero were discarded. To set gene expression from RNA-seq data, we counted the number of the sequencing tags aligned to exons and untranslated regions (UTRs) with the same strand orientation as the annotated transcripts. To avoid transcripts with zero mapped tags to interfere with logarithmic transformation of read counts, one read per every 10 million sequencing tags was added to each transcript. Raw read counts were normalized to the length of mature transcript RNA and sequencing depth. All six samples (three biological replicates of two experimental conditions) were quantile normalized using normalizeQuantiles() function (17) from limma (18) and are presented as normalized read counts per transcript per 10 kb of transcript per million sequencing tags (NRP10KM). Expressed gene is gene with expression higher than 4 NRP10KM. Differentially transcribed genes were set as genes with at least 2-fold NRP10KM change in all three biological replicates with absolute NRP10KM higher than four in all six samples.
Pol II ChIP-seq
Approximately 30 × 106 Ls174T-pTER-β-catenin cells grown 72 h in the presence or absence of doxycyclin (1 µg/ml) were used for ChIP-seq procedure. Chromatin immunoprecipitation (9,19). In brief, cells were crosslinked with 1% formaldehyde for 20 min at room temperature. The reaction was quenched with glycine and the cells were successively washed with phosphate-buffered saline, buffer B [0.25% Triton-X 100, 10 mM ethylenediaminetetraacetic acid (EDTA), 0.5 mM ethylene glycol tetraacetic acid (EGTA), 20 mM HEPES (pH 7.6)] and buffer C [0.15 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES (pH 7.6)]. The cells were then resuspended in ChIP incubation buffer [0.3% sodium dodecyl sulfate (SDS), 1% Triton-X 100, 0.15 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES (pH 7.6)] and sheared using Covaris S2 (Covaris) for 8 min with the following settings: duty cycle: max, intensity: max, cycles/burst: max, mode: Power Tracking. The sonicated chromatin was diluted to 0.15 SDS, incubated for 12 h at 4°C with 10 µl of the anti RBP1 (PB-7G5) antibody (Euromedex) per IP with 100 µl of protein G beads (Upstate). The beads were successively washed two times with buffer 1 [0.1% SDS, 0.1% deoxycholate, 1% Triton-X 100, 0.15 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES (pH 7.6)], one time with buffer 2 [0.1% SDS, 0.1% sodium deoxycholate, 1% Triton-X 100, 0.5 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES (pH 7.6)], one time with buffer 3 (0.25 M LiCl, 0.5% sodium deoxycholate, 0.5% NP-40, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES (pH 7.6)], and two times with buffer 4 (1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES (pH 7.6)] for 5 min each at 4°C. Chromatin was eluted by incubation of the beads with elution buffer (1% SDS, 0.1 M NaHCO3), the eluted fraction was reconstituted to 0.15% SDS with ChIP incubation buffer and the immunoprecipitation repeated for a second time with half the amount of antibody. After washing and elution, the immunoprecipitated chromatin was de-cross-linked by incubation at 65°C for 5 h in the presence of 200 mM NaCl, extracted with phenol–chloroform, and ethanol precipitated. Immunoprecipitated chromatin was additionally sheared, end-repaired, sequencing adaptors were ligated and the library was amplified by LMPCR. After LMPCR, the library was purified and checked for the proper size range and for the absence of adaptor dimers on a 2% agarose gel and sequenced on SOLiD/AB sequencer to produce 50-bp long reads. Sequencing reads were mapped against the reference genome (hg18 assembly, NCBI build 36) using the Maq package(17), with following settings: -c -n 3, -e 170. Reads with mapping quality zero were discarded. To set gene expression from Pol II ChIP-seq data, we counted the number of the sequencing tags aligned to annotated transcript coordinates. To avoid transcripts with zero mapped tags to interfere with logarithmic transformation of read counts, one read per every 10 million sequencing tags was added to each transcript. Raw read counts were normalized to the transcript length (from TSS to TES) and sequencing depth. All six samples (three biological replicates of two experimental conditions) were quantile normalized using normalizeQuantiles() function (17) from limma (18) and are presented as normalized read counts per transcript per 100 kb of transcript per million sequencing tags (NRP100KM). Expressed gene is gene with an expression higher than 16 NRP100KM. Differentially transcribed genes were set as genes with at least 2-fold NRP100KM change in all three biological replicates with absolute NRP100KM higher than 16 in all six samples.
ChIP-seq of transcription factors
We used publically available datasets for TCF4 and TBP (GEO database accession number: GSE18481) produced in our lab (9) in Ls174T cells. ChIP, library preparation and sequencing of other transcription factors (TFs; Supplementary Data SI) in Ls174T cells were performed as for the Pol II ChIP-seq with modifications: Approximately 50.106 cells were used per IP. For β-catenin ChIP-seq, cells were crosslinked for 40 min using ethylene glycol-bis(succinimidylsuccinate) (Thermo scientific) at 12.5 mM final concentration, with the addition of formaldehyde (1% final concentration) after 20 min of incubation. Cisgenome software package (20) was used for the identification of binding peaks from the ChIP-seq data.
Data files from ENCODE project
ChIP-seq data of 21 TFs and Pol II (Supplementary Data SII) were produced in by the Myers Lab at the HudsonAlpha Institute for Biotechnology and downloaded from http://genome.ucsc.edu/ENCODE/downloads.html website (21). RNA-seq data (Supplementary Data SII) were produced by the Wold Group at the California Institute of Technology and downloaded from http://genome.ucsc.edu/ENCODE/downloads.html website (21).
Calculation of TF–transcript association score
TF–transcript association score (TTAS) represents the relative likelihood of transcript j to be regulated by TF i. To calculate TTASij we first calculated a raw score (tkj) that reflects the likelihood of transcript j being regulated by TF i via binding site k. We adapted the published method that was used previously to calculate TF-Gene association scores (22) to calculate this raw score for each transcript-binding site tkj separately. We first calculated the distribution of binding sites k of TF i with the closest transcriptional start site (TSS) g and created histograms Hist of distances l(k,g) consisting of 18 location bins separated by {−200 k bp, −100 k bp, −50 k bp, −20 k bp, −10 k bp, −5 k bp, −2 k bp, −1 k bp, 0 bp, 1 k bp, 2 k bp, 5 k bp, 10 k bp, 20 k bp, 50 k bp, 100 k bp, 200 k bp}. Next, we randomized positions of TF binding sites k and calculated the distribution of random sites with respect to TSSs in the same way as distribution of real sites. Let m be the index of bin corresponding to l(k,g) the raw score t (Supplementary Figure S1) for binding site k and transcript j is calculated by
Next, since peak intensity was shown to be a factor that contributes to the prediction of expression (23) we included this principle in our final TTAS. Final TTAS of transcript–TF pair is then calculated as log2 value of the sum of all above zero raw scores multiplied by number of tags mapped to the peak coordinates:
where tk is the raw score of the kth binding site of TF i in the vicinity of transcript j and nk is the number of sequencing tags mapped to the kth peak coordinates. This approach takes into account TF–DNA interaction strength (reflected by the number of reads in a peak), distance from TSS, the number of individual peaks in the vicinity of TSS and the distribution of binding sites. To extract principal components from TTAS we used R language princomp (24) command.
Classification trees
We used individual principal components as inputs to train the classification tree to distinguish between expressed and non-expressed genes. Training was performed by the CART algorithm (25) using the rpart() command from R package rpart (26) (http://CRAN.R-project.org/package=rpart). To avoid overfitting of the data, we prune back the tree to select the tree size with complexity parameter that associates with the smallest 10-cross validation error.
Calculation of TAS
TAS of represents normalized enrichment of Pol II tags mapped within 300 bp from TSS to tags mapped to the transcript body (excluding 3′UTR) of transcript i:
where ti represents number of tags mapped within 300 bp from TSS of transcript i, bi represents number of tags mapped to transcript i excluding first 300 bp and 3′UTR and li represent length of transcript i excluding first 300 bp and 3′UTR. Transcripts with change in TAS after doxycyclin induction are transcripts with at least 0.6 log 2-fold change of TAS in all three biological replicates. We assayed only transcripts with at least 50 aligned reads combined from all three replicates and with gene expression defined by POL II higher than 16 in all three replicates.
RESULTS AND DISCUSSION
WNT/β-catenin model system
For studying molecular mechanisms downstream of a defined signal transduction pathway, we used the human colon cancer cell line Ls174T-pTER-β-catenin (15). These cells carry a doxycyclin-inducible shRNA targeting β-catenin, which allows for complete and specific blocking of the—in these cells constitutively active—Wnt pathway, causing major changes in the expression of many genes; including direct Wnt target genes (15). We performed microarray expression analysis, RNA-seq and Pol II ChIP-seq (Supplementary Table 1) in triplicate on untreated cells and cells 72 h after doxycyclin induction to determine which measurement correlates best with the activity of TFs that are associated with the Wnt signal transduction pathway or the core transcriptional machinery.
Pol II ChIP-seq
We used the well-characterized monoclonal antibody 1PB-7G5, raised against the heptad repeat CTD-containing peptide of the RPB1 subunit, which recognizes both hyper- and hypo-phosphorylated forms of Pol II with high affinity (27). Although the use of antibodies specific for hyper-phosphorylated forms of actively transcribing polymerase could potentially further improve on the specificity of the method, the antibody used here has the advantage of a very high affinity, which results in high yields of chromatin and a very robust and reproducible procedure for routine operation what is crucial especially when source of material is limited. To calculate the Pol II DNA occupancy based on Pol II ChIP-seq data, we counted all sequencing tags aligned between the annotated TSSs and the transcription termination sites. Read counts were normalized for transcript gene length and sequencing depth and quantile normalization was performed across all six samples (three biological replicates of two conditions).
Correlation of Pol II ChIP-seq with RNA levels
To determine the RNA levels from the RNA-seq data, we used a comparable approach as for Pol II ChIP-seq. However, we only counted sequencing tags aligned to exons and UTRs with the same strand orientation as the annotated transcript and normalized the read count to the length of mature transcript mRNA. Pol II occupancy and RNA levels assayed by RNA-seq and microarrays of 21 854 RefSeq genes showed good correlation among biological replicates within the same method (Figure 1 and Supplementary Figure S2), although a significant and reproducible lower correlation was observed between different methods as compared to triplicates. These results show that the reproducibility of Pol II ChIP-seq is at least comparable to other methods for measuring gene expression, but does not reveal which method best reflects the transcriptional processes. Interestingly, the Pol II Chip-seq results cluster separately from the microarrays and RNA-seq results (Figure 1A). This indicates that Pol II occupancy and RNA-seq measurements have a different information content that could be exploited to obtain mechanistic and biological insights.
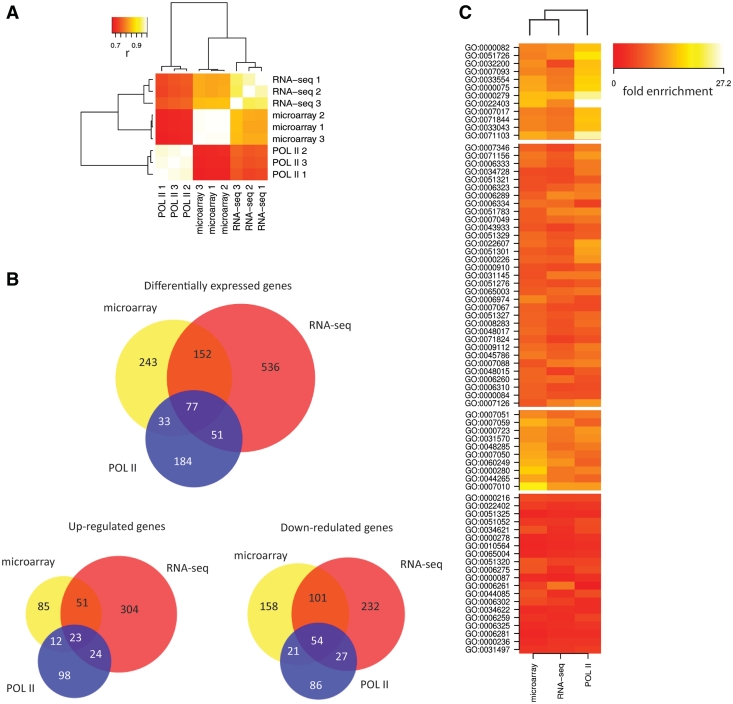
Figure 1.
Comparison of Pol II ChIP-seq, RNA-seq and microarrays. (A) Clustering of three different methods (RNA-seq, gene-expression microarray, Pol-II ChIP-seq) and biological replicates based on correlations of absolute gene-expression levels as measured by Pol II occupancy, RNA-seq and normalized probe intensities from microarrays. (B) Overlap between differentially expressed genes as determined by POL II occupancy, RNA-seq and microarrays. (C) Gene ontology analysis of down-regulated genes (Wnt target genes) identified by the same three methods. Each displayed term was found significantly enriched by at least one method. Functional classes in the first cluster are less enriched in genes identified by methods based on measuring RNA-levels, compared to genes identified based on POL II occupancy.
Identification of differentially expressed genes
Next, to explore the ability of Pol II ChIP-seq to identify differentially expressed genes we compared changes in Pol II occupancy in Wnt-active and non-active cells with differences of RNA levels assayed by RNA-seq and microarrays. Based on Pol II Chip-seq, we identified 345 differentially expressed genes (157 up and 188 down-regulated) with at least 2-fold change in Pol II occupancy in all three replicates. These genes show overlap with genes that are differentially expressed (>2-fold) as detected by microarrays (110 out of 505) and RNA-seq (128 out of 816). Overlap for down-regulated genes, which represent possible Wnt target genes, is better than for up-regulated genes (Figure 1B). A total of 77 genes were identified by all three methods, but a set of 152 genes commonly identified by RNA-seq and microarrays was not detected as differentially expressed by Pol II Chip-seq. These discrepancies could reflect different mechanisms of RNA level regulation. For example, induced changes in RNA stability will result in changed RNA levels as measured by RNA-seq or microarrays, but with smaller or no differences in Pol II occupancy (Supplementary Figure S3). However, these features do allow us to distinguish the regulatory mechanisms that act through production of RNA from those regulating RNA stability. Interestingly, gene ontology analysis revealed different gene classes in down-regulated genes (Wnt-target genes) identified by POL II as compared to methods measuring changes in RNA levels, indicating different types of regulation for different functional gene categories (Figure 1C, Supplementary Data SIII).
Correlation of Pol II ChIP-seq with the presence of transcriptional regulators
To determine which method correlates best with TF-mediated regulatory processes, we compared the Pol II DNA occupancy and RNA levels with TF presence. We used ChIP-seq profiles of TCF4 and TBP (9), and generated additional ChIP-seq profiles of three other TFs (β-catenin, E2A and c-Myc) in LS174 cells (Supplementary Data SI). These TFs are part of, or are related to, the Wnt-pathway (28–30), which is constitutively active in LS174 cells, or are part of the basic transcriptional machinery. To link binding sites to individual genes, we determined the likelihood of a gene being regulated by a particular TF based on genome-wide TF-gene distribution patterns. Without the information about long-range chromatin interactions, this score only has a probabilistic character, since many TFs can regulate genes that are up to several hundreds of kilobases away from a binding site, while on the other hand neighboring genes do not necessarily have to be regulated (31,32). We therefore defined a TTAS in which we combined previously described scores based on TF–DNA interaction strength, distance to TSS and genome-wide binding site distribution with respect to TSSs (22,23) (Supplementary Figure S1). Linear regression analysis of individual TTAS together with Pol II occupancy or RNA levels and principal component regression analysis (PCRA) of individual principal components extracted from TTAS (Figure 2A and B) shows a better correlation of TF occupancy with Pol II occupancy as compared to RNA-seq and microarray-based RNA-level measurements.
Figure 2.
Linear regression analysis of Pol II occupancy, RNA-seq and microarray probe intensities with (A) transcription factor occupancy represented by individual TTAS scores separately for each transcription factor and with (B) individual principal components extracted from TTAS of five transcription factors. TTAS scores reflect the likelihood of a gene being regulated by a given transcription factor. (C) Loadings of TTAS scores in the four major principal components that explain most of the variability in Pol II occupancy. Individual loadings were multiplied by −1 when the correlation of principal component with Pol II occupancy was negative.
More than half of the variation in Pol II occupancy can be explained by the first principal component. In this principal component, all five TFs have positive loadings (Figure 2C), indicating a major role in the activation of gene expression (23). More importantly, the high degree of co-linearity also explains why the first principal component does not reflect more of the Pol II occupancy variation than cMyc alone. The other principal components explain only a minority of variation in Pol II occupancy and RNA expression levels. However they may reveal existence of genes with different mechanisms of regulation, e.g. in a small subgroup of genes with high third principal component values, the presence of TCF4, β-catenin and even TBP may lead to transcriptional repression. The second principal component suggests the existence of mechanisms where TF regulates RNA levels without changing Pol II occupancy. Positive loadings of cMyc in all five principal components support its role as pure transcriptional activator in cancer cells, supporting its role in regulation of transcriptional pause release (33).
ENCODE datasets
To strengthen these findings, we repeated the analysis using publically available ChIP-seq profiles of 21 different TFs performed in duplicates, Pol II ChIP-seq and RNA-seq datasets from a lymphoblastoid cell line (GM12878) that was generated as a part of the ENCODE project (21,34) (Supplementary Data SII). Both linear regression analysis of individual TTAS and PCRA showed very similar results as for our datasets, with Pol II ChIP-seq correlating better with TF occupancy than cellular RNA levels (Supplementary Figure S4).
Poised polymerase
Since our model for readout of gene expression expects all DNA bound Pol II to be processive, we next evaluated the potential effects of poised polymerase (35) and polymerase that is accumulated at 3′-UTRs on the readout and its correlation with TFs. We repeated the analysis while excluding the information from sequencing tags mapped close to 3′-UTRs and close to TSS where the majority of the poised polymerase is located. However, similar correlations with transcription factor occupancy as for total Pol II occupancy were obtained (Supplementary Figure S5), suggesting that non-processive poised polymerase and polymerase accumulated at 3′-UTR reflect only a minor fraction of DNA-bound Pol II compared to processive polymerase and thus does not have a major influence on the overall readout of gene expression based on Pol II ChIP-seq.
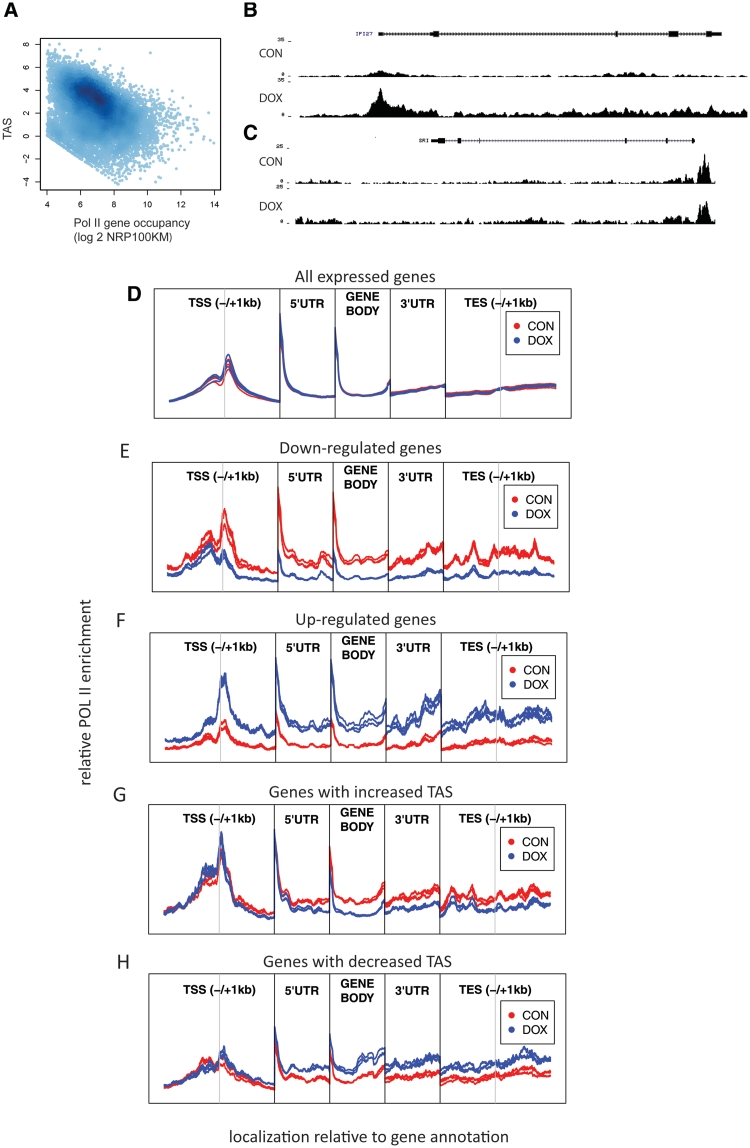
Nevertheless, quantification of changes in poised polymerase levels could be useful for dissection of regulatory mechanisms of individual genes as regulation of gene transcription after recruitment of RNA polymerase II to TSSs is a widely accepted mechanism of regulation of mRNA levels (35). In a simplified model, lowly expressed genes have a relatively high accumulation of poised Pol II around their TSSs compared to actively transcribed genes, where Pol II is spread over the complete genomic region. Thus, a change in Pol II accumulation close to the TSS compared to accumulation in gene body can reflect changes in transcription. We therefore explored the possibility of using changes in TSS accumulation scores (TAS) to determine differentially expressed genes. The TAS score represents the relative enrichment of Pol II at the promoter compared to the gene body. Indeed, the reduction of Pol II in TSS reflected by decreased TAS correlates (r = −0.366) with an increase in Pol II occupancy of the gene (Figure 3A). Next, we identified 481 genes that reproducibly changed TAS after WNT signaling inhibition by doxycyclin treatment in all three replicates. Seventy eight (23%) of the genes that were identified by Pol II ChIP-seq as differentially expressed based on a change in the number of tags aligned to the whole transcript overlap with the set of genes with change in TAS score. Even though regulation of gene expression is typically mediated by an increase or decrease in POL II recruitment to TSS and a subsequent change in Pol II occupancy in downstream parts of genes (Figure 3D–F), our data indicate that in particular genes changed Pol II occupancy in the gene body is not accompanied by a change in Pol II at TSS. Regulation of these genes may thus be mediated via a change in frequency of releasing paused polymerase without the need for regulation of POL II recruitment by TFs (Figure 3G and H). This mechanism has previously been shown as a major regulator mechanisms of cMyc (33), which is also active in the model system we used here. In conclusion, additionally to the quantitative aspect of Pol II ChIP-seq, the spatial distribution information of Pol II can be used as additional information to develop more reliable models for the identification of specific subsets of differentially expressed genes that are predominantly regulated by releasing paused polymerase (33).
Figure 3.
Poised polymerase and metagene analysis of Pol II occupancy. (A) Correlation of total Pol II occupancy with TAS score. The TAS score represents the relative enrichment of POL II at the promoter compared to the gene body and reflects the fraction of poised polymerase. Genes with lower expression have more Pol II deposited on their transcription start site and less processing polymerase in the gene body compared to genes with higher expression (r = −366). (B, C) Pol II coverage for up-regulated genes with (B) increased density over TSS and gene body and (C) increased density in gene body without change in TSS. (D) Relative enrichment of POL II sequencing tags in Wnt plus (CON) and Wnt minus (DOX) samples with respect to gene annotation in (D) all annotated genes (E) down-regulated and (F) up-regulated genes. POL II enrichment changes simultaneously in TSS and gene body, suggesting that a substantial proportion of transcription regulation is mediated by changes in POL II recruitment. In subclass of genes, with increase (G) or decrease (H) in relative enrichment of POL II at the promoter compared to gene body; difference of POL II accumulation in downstream part of genes is not accompanied by change in enrichment on TSS. Every individual region is normalized separately against input and average enrichment of CON samples.
Bimodal distribution of Pol II occupancy
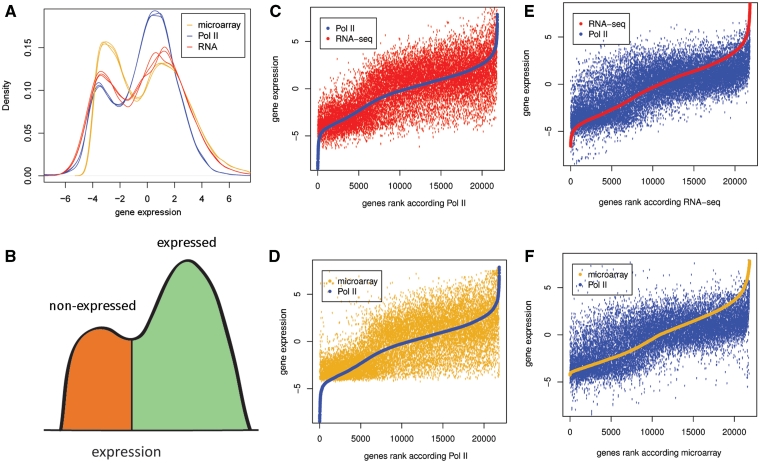
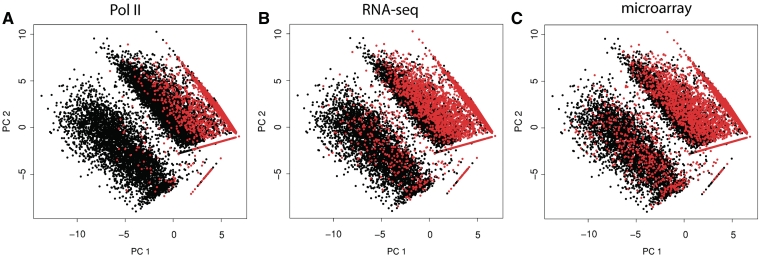
Pol II DNA occupancy and RNA levels measured by RNA-seq and microarrays show a bimodal distribution pattern that is specific and reproducible for every method (Figure 4A). A similar picture is observed at the genome-wide level of Pol II occupancy where more than half of the genome is occupied by Pol II with a domain-like distribution (Supplementary Figure S6), most likely reflecting open and closed chromatin regions. This pattern can be used to split genes into expressed and non-expressed groups (Figure 4B). Pol II ChIP-seq was found to classify more genes as expressed compared to the other methods. Many genes that are defined as expressed by Pol II ChIP-seq appear as non-expressed by the other two methods (Figure 4C and D). This is, however, not true in the opposite direction: genes defined as expressed by RNA-seq or microarray are virtually always categorized as transcribed by Pol II ChIP-seq (Figure 4E and F). In line with this, expressed and non-expressed genes form more distinct clusters in a principal component analysis (Figure 5) when these classes are defined from Pol II ChIP-seq than from RNA levels. Furthermore, expression classes defined from Pol II occupancy are more accurately predicted by classification and regression tree (CART) algorithm (25), when using principal components extracted from TTAS scores as input to predict expression status of all genes. In our dataset, only 11.8% of genes were wrongly predicted when Pol II was used to define expression status of a gene, with 14.8% and 17.6% of wrongly predicted genes when RNA-seq and microarrays were used to determine expression status.
Figure 4.
Bimodal distribution of Pol II occupancy, RNA-seq and microarray probe intensities (A) All methods reveal a bimodal pattern of gene expression indicative of (B) expressed (green) and non-expressed (red) genes. (C–F) Rank analysis of Pol II occupancy, RNA-seq and microarray probe intensities. Genes are ranked according to Pol II ChIP-seq results (C, D), according to RNA-seq (E) or microarrays (F). Many transcripts classified as expressed according the Pol II ChIP-seq are called as not expressed according to the RNA-seq and microarray data. In contrast, only a very limited number of transcripts that are called transcribed by RNA-seq and microarrays are called as non-transcribed by Pol II ChIP-seq. All expression values represent median centered and log2 transformed NR100KM (Pol II), NR10KM (RNA-seq) and normalized microarray probe intensity.
Figure 5.
Clustering of expressed (black) and non-expressed (red) genes defined by POL II chip-seq, RNA-seq and microarrays. Genes are plotted according to principal components extracted from TTAS scores, which reflect the likelihood of a gene being regulated by a given transcription factor. Genes categorized into expressed and non-expressed according to bimodal distribution of Pol II results (A) form more defined clusters compared to genes categorized according to RNA levels (B and C).
Gene ontology analysis
To determine if the observed differences in RNA levels and POL II occupancy reflect functional gene classes, we performed a gene ontology (GO) term enrichment analysis (36). We empirically defined three classes containing genes that are expressed as defined by Pol II ChIP-seq but as non-expressed (class I), lowly expressed (class II) and very highly expressed (class III) by RNA-seq (Figure 6, Supplementary Figure S7). Interestingly, a significant enrichment of particular GO terms in different gene classes is observed (Supplementary Data SIV), which overlaps with previously described gene classes that are characterized by particular low or high mRNA turnover rates (2–4). Genes in class I are enriched in secreted proteins and plasma membrane receptors and reflect regulated genes for which mRNA is synthesized but rapidly degraded. Class II genes are enriched in transcriptional regulators, while the third class includes genes that are involved in basal translational homeostasis and energy metabolisms, characterized by high mRNA levels. These analyses indicate that gene expression of particular gene classes with short-living mRNA can be underestimated by methods measuring solely RNA levels, while the expression of gene classes with more stable transcripts can be systematically overestimated.
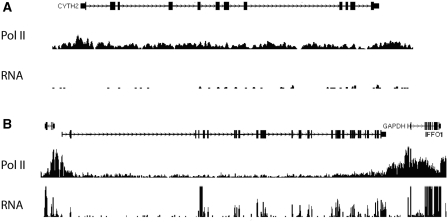
Figure 6.
Examples of Pol II and RNA-seq coverage over of genes with low (A) and high (B) RNA stability. The vertical axis represents relative sequencing tag coverage per position (A) A gene (CYTH2) with high density of sequencing tags over the gene body with very low levels of RNA. Pol II ChIP-seq classifies the depicted gene as expressed; however, both RNA-seq and microarrays classify the gene as non-expressed (Class I gene). (B) Genes (IFFO1 and GAPDH) with high density of Pol II ChIP-seq tags mapping to the gene body and with high numbers of sequencing RNA-seq tags mapping to annotated exons.
Concluding remarks
Taken together, we have shown that routine Pol II ChIP-seq approaches provide valuable information that is complementary to total mRNA-level measurements. We also show that the combination of data modalities allows for the dissection of mechanisms involved in gene expression regulation. Data analysis within the WNT/β-catenin model system showed a highly variable balance between RNA stability and gene expression dynamics of specific gene classes. This indicates that regulatory processes are systematically over- or underestimated when steady-state RNA levels are used as the only determinant for gene expression.
We showed that Pol II ChIP-seq correlates very well with the binding of transcriptional regulators to promoter elements, but also found that in a limited number of genes, TFs may regulate RNA levels without affecting Pol II occupancy. We were able to recapitulate these findings using publicly available datasets with different combinations of TFs, supporting the universal nature of our findings. Finally, we showed that spatial distribution and dynamics of Pol II over promoter and gene body can be used to identify specific subsets of differentially expressed genes that are predominantly regulated by releasing paused polymerase instead of increasing the rate of polymerase recruitment.
In sum, our results demonstrate that changes in RNA production and mechanisms responsible for RNA stability can be discerned by routine techniques in any sample of interest, allowing for the dissection of regulatory mechanisms in a wide variety of model systems.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary table, Supplementary figures 1–7, Supplementary material I-IV and Supplementary reference(37)
FUNDING
The Netherlands Bioinformatics Center (NBIC); the Netherlands Center for Systems Biology (NCSB); and the Cancer Genomics Center (CGC) program of the Netherlands Genomics Initiative (NGI). Funding for open access charge: Publication charges will be paid from institutional resources (Hubrecht Institute).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
M.M., J.S., P.H., H.C. and E.C. designed the study, J.S. and P.H. performed chromatin immunoprecipitations, M.M. performed library preparation and analyzed the data, N.L. performed deep sequencing, F.P.R. performed mapping of the sequencing data and contributed to figure generation, M.M. and E.C. wrote the manuscript.
REFERENCES
- 1.Shyu AB, Wilkinson MF, van Hoof A. Messenger RNA regulation: to translate or to degrade. EMBO J. 2008;27:471–481. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raghavan A, Ogilvie RL, Reilly C, Abelson ML, Raghavan S, Vasdewani J, Krathwohl M, Bohjanen PR. Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acids Res. 2002;30:5529–5538. doi: 10.1093/nar/gkf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharova LV, Sharov AA, Nedorezov T, Piao Y, Shaik N, Ko MS. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 2009;16:45–58. doi: 10.1093/dnares/dsn030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO. Precision and functional specificity in mRNA decay. Proc. Natl Acad. Sci. USA. 2002;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia-Martinez J, Aranda A, Perez-Ortin JE. Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol. Cell. 2004;15:303–313. doi: 10.1016/j.molcel.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Miller C, Schwalb B, Maier K, Schulz D, Dumcke S, Zacher B, Mayer A, Sydow J, Marcinowski L, Dolken L, et al. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol. Syst. Biol. 2011;7:458. doi: 10.1038/msb.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mokry M, Hatzis P, de Bruijn E, Koster J, Versteeg R, Schuijers J, van de Wetering M, Guryev V, Clevers H, Cuppen E. Efficient double fragmentation ChIP-seq provides nucleotide resolution protein–DNA binding profiles. PLoS ONE. 2010;5:e15092. doi: 10.1371/journal.pone.0015092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adli M, Zhu J, Bernstein BE. Genome-wide chromatin maps derived from limited numbers of hematopoietic progenitors. Nat. Methods. 2010;7:615–618. doi: 10.1038/nmeth.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahl JA, Collas P. A rapid micro chromatin immunoprecipitation assay (microChIP) Nat. Protoc. 2008;3:1032–1045. doi: 10.1038/nprot.2008.68. [DOI] [PubMed] [Google Scholar]
- 12.Dahl JA, Collas P. MicroChIP–a rapid micro chromatin immunoprecipitation assay for small cell samples and biopsies. Nucleic Acids Res. 2008;36:e15. doi: 10.1093/nar/gkm1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahl JA, Reiner AH, Collas P. Fast genomic muChIP-chip from 1,000 cells. Genome Biol. 2009;10:R13. doi: 10.1186/gb-2009-10-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fanelli M, Amatori S, Barozzi I, Soncini M, Dal Zuffo R, Bucci G, Capra M, Quarto M, Dellino GI, Mercurio C, et al. Pathology tissue-chromatin immunoprecipitation, coupled with high-throughput sequencing, allows the epigenetic profiling of patient samples. Proc. Natl Acad. Sci. USA. 2010;107:21535–21540. doi: 10.1073/pnas.1007647107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van de Wetering M, Oving I, Muncan V, Pon Fong MT, Brantjes H, van Leenen D, Holstege FC, Brummelkamp TR, Agami R, Clevers H. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 2003;4:609–615. doi: 10.1038/sj.embor.embor865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smyth G. In: Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S, editors. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 19.Hatzis P, van der Flier LG, van Driel MA, Guryev V, Nielsen F, Denissov S, Nijman IJ, Koster J, Santo EE, Welboren W, et al. Genome-wide pattern of TCF7L2/TCF4 chromatin occupancy in colorectal cancer cells. Mol. Cell. Biol. 2008;28:2732–2744. doi: 10.1128/MCB.02175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP–chip and ChIP-seq data. Nat. Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myers RM, Stamatoyannopoulos J, Snyder M, Dunham I, Hardison RC, Bernstein BE, Gingeras TR, Kent WJ, Birney E, Wold B, et al. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 23.Ouyang Z, Zhou Q, Wong WH. ChIP-Seq of transcription factors predicts absolute and differential gene expression in embryonic stem cells. Proc. Natl Acad. Sci. USA. 2009;106:21521–21526. doi: 10.1073/pnas.0904863106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Development Core Team. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2009. [Google Scholar]
- 25.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. New York: Springer; 2003. [Google Scholar]
- 26.Therneau TM, Atkinson B. rpart: Recursive Partitioning. 2009. http://stat.ethz.ch/CRAN/ [Google Scholar]
- 27.Besse S, Vigneron M, Pichard E, Puvion-Dutilleul F. Synthesis and maturation of viral transcripts in herpes simplex virus type 1 infected HeLa cells: the role of interchromatin granules. Gene Expr. 1995;4:143–161. [PMC free article] [PubMed] [Google Scholar]
- 28.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 29.Graham TA, Weaver C, Mao F, Kimelman D, Xu W. Crystal structure of a beta-catenin/Tcf complex. Cell. 2000;103:885–896. doi: 10.1016/s0092-8674(00)00192-6. [DOI] [PubMed] [Google Scholar]
- 30.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 31.Kooren J, Palstra RJ, Klous P, Splinter E, von Lindern M, Grosveld F, de Laat W. Beta-globin active chromatin Hub formation in differentiating erythroid cells and in p45 NF-E2 knock-out mice. J. Biol. Chem. 2007;282:16544–16552. doi: 10.1074/jbc.M701159200. [DOI] [PubMed] [Google Scholar]
- 32.Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, et al. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Margaritis T, Holstege FC. Poised RNA polymerase II gives pause for thought. Cell. 2008;133:581–584. doi: 10.1016/j.cell.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 36.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anders S. Visualization of genomic data with the Hilbert curve. Bioinformatics. 2009;25:1231–1235. doi: 10.1093/bioinformatics/btp152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.