Abstract
The Sox10 transcription factor is a central regulator of vertebrate neural crest and nervous system development. Its expression is likely controlled by multiple enhancer elements, among them U3 (alternatively known as MCS4). Here we analyze U3 activity to obtain deeper insights into Sox10 function and expression in the neural crest and its derivatives. U3 activity strongly depends on the presence of Sox10 that regulates its own expression as commonly observed for important developmental regulators. Sox10 bound directly as monomer to at least three sites in U3, whereas a fourth site preferred dimers. Deletion of these sites efficiently reduced U3 activity in transfected cells and transgenic mice. In stimulating the U3 enhancer, Sox10 synergized with many other transcription factors present in neural crest and developing peripheral nervous system including Pax3, FoxD3, AP2α, Krox20 and Sox2. In case of FoxD3, synergism involved Sox10-dependent recruitment to the U3 enhancer, while Sox10 and AP2α each had to bind to the regulatory region. Our study points to the importance of autoregulatory activity and synergistic interactions for maintenance of Sox10 expression and functional activity of Sox10 in the neural crest regulatory network.
INTRODUCTION
Sox10 is one of the central transcriptional regulators of neural crest development (1–4). Following its loss or mutation many aspects of neural crest development are disturbed, leading to defects in many of the neural crest-derived cell types and structures of the vertebrate body including melanocytes, peripheral glia, the sympathetic and the enteric nervous system. Its essential role is highly conserved among vertebrates as evident from loss- and gain-of-function studies and the analyses of spontaneous mutations in a wide range of species such as zebrafish, Xenopus, chicken, mouse and man (5–14). Considering the importance of Sox10 for neural crest development and the numerous contributions of the neural crest to the vertebrate body plan, it is essential to understand how Sox10 functions and how it is regulated in the neural crest.
Hallmarks of such central regulators are their ability to functionally interact with many other co-expressed transcription factors in a regulatory network and to reinforce their own expression in a positive autoregulatory loop. Additionally, central regulators of development are often governed in their expression by the combined action of multiple, evolutionarily conserved enhancers in the vicinity of their genes (15,16). Analysis of these enhancers is therefore not only crucial to understand the expression pattern of these regulatory factors. It also provides an opportunity to see these factors at work in the context of their regulatory network. Enhancer studies may also reveal insights into human disease as many disease-causing or predisposing genetic alterations in the human genome affect non-coding regions.
Sox10 enhancers have been identified in recent years by two complementary approaches. BAC transgenesis has first served to delineate the part of the genome in which the enhancers are present, before bioinformatics was employed to identify potential enhancers by way of their evolutionary conservation (17–22). Consecutive functional analyses of reporter transgenes have led to the identification of more than 10 different regions with enhancer activity that are spread over a distance of at least 120 kb and localized both upstream as well as downstream of the Sox10 gene. Alterations that affect these enhancers such as spontaneously occurring deletions in the distal upstream region or immediately in front of the promoter have been shown to disturb neural crest development in mouse and chicken (17,23). Considering that heterozygous mutations in the SOX10 coding region also lead to neural crest defects (so-called neurocristopathies) in humans including Waardenburg syndrome (in which melanocyte development is disturbed) and Hirschsprung disease (in which enteric nervous system development remains incomplete) (10,24,25), it seems plausible that cases with unclear molecular origin may result from such SOX10 enhancer mutations that may cause or predispose to disease.
Two nomenclatures exist for the enhancers: one in which the regulatory regions are designated as MCS (Multiple-species Conserved Sequences) (18) and an alternative one in which the regions are referred to as U/D (upstream/downstream) enhancers (20). Two of these enhancers are particularly active in the early neural crest along most parts of the rostrocaudal axis including the vagal and trunk neural crest. These are the U1/MCS7 and the U3/MCS4 regions that for the remainder will be referred to as U1 and U3 regions (18,20). Both enhancers continue to function during later times of embryonic development in several derivatives of the neural crest including the developing enteric nervous system and Schwann cells as the glial cells along peripheral nerves arguing that their impact on Sox10 expression is not restricted to early phases of neural crest development. U3 is 0.4 kb long and situated ~28 kb in front of the Sox10 gene; U1 is even further upstream.
Both U1 and U3 contain many potential binding sites for transcription factors with occurrence in the early neural crest and in neural crest derivatives. Without mapping the exact binding sites we have shown in an earlier study that the neural crest transcription factors Pax3, AP2α, Lef1, Sox9 and Sox10 bind to U1 and U3 in vitro (20). To get further insight into the regulatory network that is built from these factors, the underlying functional interactions and the ability of Sox10 to regulate its own expression, we here study the mechanisms by which the U3 element regulates Sox10 expression.
MATERIALS AND METHODS
Transgenic animals
Sox10+/lacZ and Sox10+/rtTA mice in which Sox10 coding sequences were replaced on one allele by lacZ marker or reverse tetracycline-controlled transactivator (rtTA) sequences, and mice carrying the lacZ transgene under control of the hsp68 minimal promoter and the mouse U3 enhancer (U3-lacZ) have been described before (7,20,26).
U3m2-lacZ resembled the previously described U3-lacZ transgene but carried a mutation in site 2 that prevented Sox10 binding (Figure 3A). In U3null-lacZ, inactivating mutations were simultaneously introduced into sites 2, 5, 7 and 9. After separation from the vector backbone and purification, the U3-lacZ, U3m2-lacZ and U3null-lacZ transgenes were each separately microinjected as NotI/KpnI fragments into the male pronucleus of fertilized FVB oocytes. Transgenic mice were generated from injected oocytes according to standard techniques. Founder mice and transgenic offspring were identified and genotyped by PCR analysis of DNA prepared from tail biopsies using the U3-specific forward primer 5′-GAGCCCTGCTCATAAACAAG-3′ and the lacZ-specific reverse primer 5′-AGTAGCTGTCAGCGTCTGGT-3′ (20). Determination of copy numbers in transgenic mice was performed by quantitative PCR with lacZ-specific primers followed by normalization to Sox10+/lacZ mice.
Figure 3.
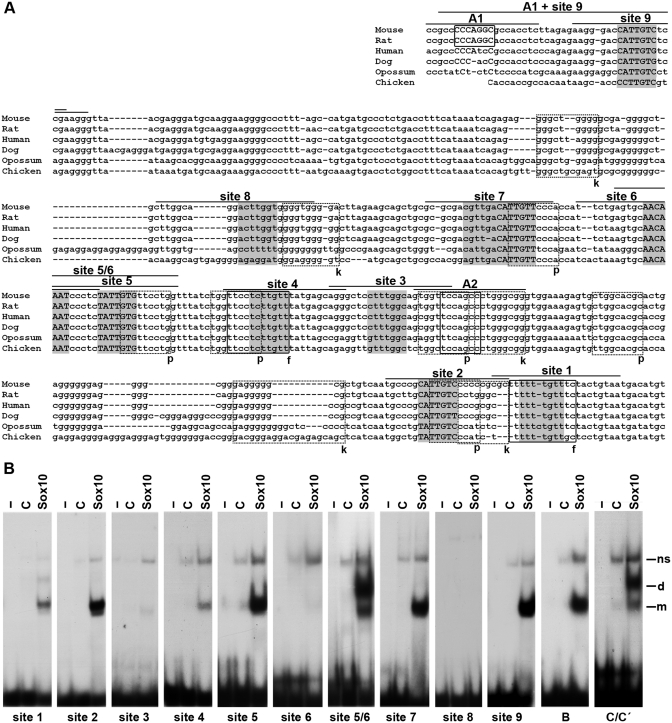
The U3 enhancer contains multiple binding sites for Sox10. (A) Comparative alignment of U3 sequences from mouse, rat, human, dog, opossum and chicken. Putative Sox binding sites 1–9 are marked in gray. Putative other binding sites are boxed with solid lines for AP2α and FoxD3 (f) and with stippled lines for Pax3 (p) and Krox20 (k). Confirmed binding sites are additionally highlighted by capital letters. The lines above the sites indicate the regions used as oligonucleotides for EMSA. (B) EMSA with radiolabeled, double-stranded oligonucleotides encompassing sites 1–9 as indicated below the gels. Oligonucleotides were incubated in the absence (−), or presence (C, Sox10) of protein extracts before gel electrophoresis as indicated above the lanes. Extracts were from mock-transfected 293 cells (C) or 293 cells expressing Sox10 MIC (Sox10). Oligonucleotides with sites B and C/C′ from the Mpz promoter (41) served as control for high-affinity monomeric and dimeric Sox10 binding. All oligonucleotides were size-matched. m, bound monomer; d, bound dimer; ns, non-specific complex.
Tissue preparation, histological staining, immunohistochemistry and documentation
Embryos from 9.5 days post-coitum (dpc) to 11.5 dpc underwent fixation in 1% paraformaldehyde and were stained for β-galactosidase activity by incubation in 1% X-gal for several hours at 37°C either immediately as whole mounts or after being frozen at −80°C and processed to 20-µm transverse cryosections.
Alternatively, fixation was in 4% paraformaldehyde followed by cryoprotection in sucrose and storage at −80°C in tissue freezing medium (Leica, Bensheim, Germany). For immunohistochemistry, 10 -µm cryotome sections from the forelimb level of genotyped, age-matched mouse embryos were incubated with anti-β-galactosidase goat antiserum (1 : 500 dilution, Biotrend), anti-Brn3.0 rabbit antiserum (1 : 300 dilution, gift of Dr E. Turner, UCSD, San Diego, CA, USA) and anti-Sox10 guinea pig antiserum (1 : 2,000 dilution) (27) as primary antibodies. Secondary antibodies conjugated to Cy2 and Cy3 immunofluorescent dyes (Dianova) were used for detection.
Hematoxylin–eosin staining was performed on transverse sections of paraffin-embedded material. Samples were analyzed and documented either with a Leica DMR microscope equipped with a Leica DFC 420C CCD camera (Wetzlar, Germany) or with a Leica MZFLIII stereomicroscope equipped with an Axiocam (Zeiss, Oberkochem, Germany).
Plasmids
Expression plasmids were derived from pCMV5 or pCAGGS-IRES-nGFP, and thus carried coding sequences under control of the cytomegalovirus immediate early promoter or the chicken β-actin promoter. The eukaryotic pCMV5 expression plasmids for AP2α, Pax3, FoxD3, Lef1, β-catenin, Slug, Krox20, Sox2, Sox9 and Sox10, and pCAGGS-Sox10-IRES-nGFP have been described previously (12,20,28). As reporter gene construct the U3-luc plasmid was used in wildtype or mutagenized form. It contained the luciferase reporter gene under control of the 0.4-kb U3 region (20) upstream of the β-globin minimal promoter. SoxE and AP2α binding sites within U3 were mutated using the QuickChange XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA).
Chicken in ovo electroporation, RNA preparation, RT–PCR
Fertilized chicken eggs were obtained from Lohmann (Cuxhaven, Germany) and incubated in a humidified incubator at 37.8°C. Embryos were staged according to Hamburger and Hamilton (29) and at stage 10–11 injected into the lumen of the neural tube with pCAGGS-IRES-nGFP-based expression plasmids at a concentration of 2 µg/µl. Electrodes were placed at either side of the neural tube, and electroporation was carried out using a BTX ECM830 electroporator delivering five 50-ms pulses of 30 V. Then electrodes were reversed and electroporation repeated to transfect both sides of the neural tube. Transfected embryos were allowed to develop for 24 h before electroporated regions of the neural tube were identified by GFP expression, dissected and processed to RNA. RNA was reverse transcribed and the resulting cDNA used for quantitative PCR (30). For detection of chicken Sox10 the species-specific primers 5′-CTCGCCGGGACCATCCATGGCA-3′ and 5′-AGGCAGCCCCGCCAGCCTCCCC-3′ were used at an annealing temperature of 57°C.
Cell culture, transient transfection, extract preparation and electrophoretic mobility shift assays
HEK 293 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum (FCS) and transfected by the polyethylenimine (PEI) technique using 10 µg of empty pCMV5 plasmid, pCMV5-Sox10 MIC (coding for amino acids 1–189 of Sox10) or pCMV5-Sox10 per 100-mm plate. Forty-eight hours post-transfection, cells were harvested for extract preparation (31) and electrophoretic mobility shift assays (EMSA) were performed with these extracts as described (32) using 32P-labeled double-stranded oligonucleotides containing putative SoxE binding sites (for sequences, see Figure 3A). In some cases, antibodies against Sox10 and AP2α (12) were added during incubation.
Luciferase assay
N2a cells were maintained in DMEM containing 10% FCS and transfected with SuperFect® Transfection Reagent (Qiagen, Hilden, Germany) on 24-well tissue culture plates using varying amounts (2–500 ng, usually 100 ng) of pCMV5-based expression vectors and 500 ng of luciferase reporter plasmid. Cells were harvested 48 h post-transfection. Luciferase activity was determined in the presence of luciferin substrate by detection of chemiluminescence.
Chromatin immunoprecipitation
Chromatin immunoprecipitation assays were performed as described (33) on dissected, trypsinized 11.5-dpc-old mouse embryos. Proteins were crosslinked to DNA in 1% formaldehyde. After addition of glycine, chromatin was prepared and sheared to an avarage fragment length of 300–600 bp using a Sonoplus HD2070 homogenizer (Bandelin, Berlin, Germany). Immunoprecipitations were performed overnight at 4°C using guinea pig polyclonal α-Sox10 antibodies as well as control pre-immune serum and protein A sepharose beads. DNA was purified from input and precipitated chromatin after crosslink reversal and subjected to PCR. The following primer pairs were used for detection in 33 cycles of standard PCR using an annealing temperature of 57°C: 5′-GGAGCCCTGCTCATAAACAA-3′ and 5′-CCATTGTCTCCGAAGGGTTA-3′ for mouse U3 and 5′-GACGAAGGAGAAGCCTTGTC-3′ and 5′-ACTGCGACTCTGCTGTCTCT-3′ for mouse U4 (20).
GST pulldowns and western blotting
For pulldown experiments, glutathione-S-transferase (GST) or GST fusion proteins with parts of Sox10 were produced in Escherichia coli strain BL21 DE3 pLysS and bound in the presence of DNaseI to glutathione sepharose 4B beads (28). An aliquot of the washed and equilibrated beads, now carrying GST or GST fusion protein, was incubated with extracts from 293 cells transfected with AP2α or FoxD3 expression plasmids in interaction buffer (20 mM HEPES pH 7.9, 10 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 5% glycerol, 1 mM dithiothreitol, 0.05% Triton-X 100 and 100 mM NaCl). After centrifugation and washing, bead-bound proteins from the 293 extracts were separated by SDS–PAGE, transferred to a nitrocellulose membrane and detected by antibodies directed against the specific tag attached to AP2α (T7-tag) and FoxD3 (HA-tag) (12) as described (28).
RESULTS
The U3 enhancer is activated by many transcription factors with occurrence in neural crest and peripheral glia
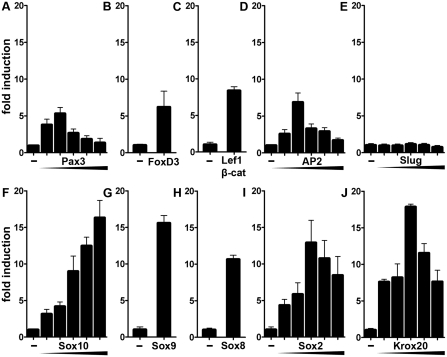
Previous EMSA had shown that the U3 enhancer binds in vitro several transcription factors that influence neural crest development (20). To address the functional relevance of this in vitro binding activity, we analyzed whether these transcription factors also influence the ability of U3 to activate expression of a luciferase reporter gene under control of a minimal promoter in transiently transfected cultured cells. We used N2a neuroblastoma cells for our analyses as these tumor cells are derived from and still exhibit some similarities to neural crest cells. Among the neural crest transcription factors tested, Pax3, FoxD3, the combination of Lef1 and β-catenin, and AP2α each activated U3-dependent luciferase expression (Figure 1A–D). Activation rates were sensitive to the exact amount of transcription factor present in the cells and maximal rates were between 6- and 8-fold. Co-transfection with the neural crest transcription factor Slug did not lead to reporter gene activation at any concentration tested (Figure 1E). High reporter gene induction rates were also obtained in the presence Sox10 or its close relatives Sox9 and Sox8 with maximal induction rates above 15-fold for Sox10 and Sox9, and above 10-fold for Sox8 (Figure 1F–H). These results argue that many, but not all transcription factors with influence on neural crest development have the ability to activate U3 in tissue culture.
Figure 1.
U3 activity in transiently transfected neuroblastoma cells is modulated by many transcription factors with occurrence in the neural crest and its derivatives. Transient transfections were performed in N2a cells with a luciferase reporter under control of the Sox10 U3 enhancer and the β-globin minimal promoter (U3-luc, 500 ng per well). Empty pCMV expression plasmids (−) or expression plasmids for various transcription factors were co-transfected as indicated below the bars. Expression plasmids coded for Pax3 (A, increasing amounts of 10, 25, 100, 200 or 500 ng per well as indicated by the triangle), FoxD3 (B, 100 ng per well), Lef1 and β-catenin (C, 200 and 25 ng per well, respectively), AP2α (D, 10, 25, 100, 200 or 500 ng per well), Slug (E, 10, 25, 100, 200 or 500 ng per well), Sox10 (F, 10, 25, 100, 200 or 500 ng per well), Sox9 (G, 25 ng per well), Sox8 (H, 500 ng per well), Sox2 (I, 10, 25, 100, 200 or 500 ng per well) and Krox20 (J, 10, 25, 100, 200 or 500 ng per well). Luciferase activities in extracts from transfected cells were determined in three experiments each performed in duplicates. The luciferase activity obtained for U3-luc in the absence of ectopic transcription factor was arbitrarily set to 1. Fold inductions in the presence of transcription factors were calculated and are presented as mean ± SEM. Determined inductions were statistically significant according to Student's t-test (P ≤ 0.001).
Because of the continued activity of U3 in neural crest derivatives during later phases of development, we also included Sox2 and Krox20 in our study as two transcription factors with phase-specific occurrence and relevance in developing Schwann cells (34–37). Again, we found both transcription factors to strongly activate U3 and correspondingly increase luciferase expression in transiently transfected N2a cells with maximal induction rates of 13- and 18-fold, respectively (Figure 1I and J). We conclude that U3 can mediate the effects of many transcription factors that are expressed during the right time and in the right cells to be bona fide regulators of Sox10 expression.
The U3 enhancer may participate in Sox10 autoregulation
Intriguingly, U3 is strongly activated by four different Sox proteins. Sox9-dependent activation may be important for Sox10 induction in the early emigrating neural crest (11), while Sox2 activity may be important for Sox10 expression in immature Schwann cells (35). Sox10 as a key regulator of neural crest development may use the U3 enhancer to maintain its expression in an autoregulatory loop.
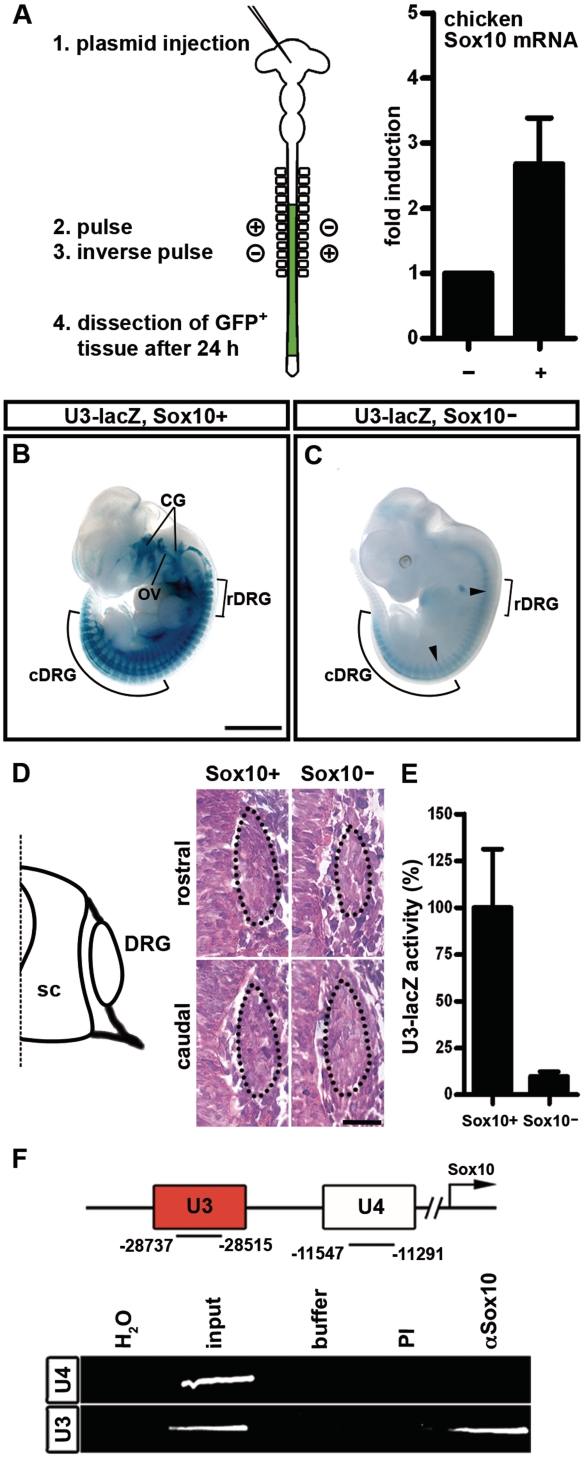
To see whether Sox10 has the capability to regulate the expression of its own gene, we electroporated neural tubes of chicken embryos with a rat Sox10 cDNA construct, and analyzed the impact of ectopic expression of rat Sox10 on the endogenous expression of chicken Sox10 by RT–PCR using chicken-specific primers. These studies revealed a 2.7-fold average induction of endogenous Sox10 expression 24 h after electroporation (Figure 2A). This strongly argues for an autoregulatory ability of Sox10.
Figure 2.
Sox10 autoregulates its own expression at least partly through the U3 enhancer. (A) pCAGGS-IRES-nGFP-based expression plasmids were injected into HH10-11 chicken neural tubes before double-sided electroporation. Electroporated, GFP-expressing regions of the neural tube were dissected 24 h later and processed to RNA as outlined on the left. Expression levels of endogenous Sox10 were determined by RT–PCR in chicken neural tubes electroporated with empty expression plasmid (−; arbitrarily set to 1) and compared to those in neural tubes electroporated with rat Sox10 expression plasmids (+). Shown is the mean ± SEM from five values. (B, C) LacZ activity was detected colorimetrically using X-gal substrate in whole embryos at 11.5 dpc. Embryos carried the lacZ reporter as a U3-lacZ transgene on wildtype (Sox10+, B) or Sox10-deficient Sox10rtTA/rtTA (Sox10−, C) background. Whole-mount stainings were documented from the side. DRG in rostral (rDRG) and caudal regions (cDRG) are marked. Size bar corresponds to 2 mm and is valid for (B, C). (D) Hematoxylin–eosin staining of transverse sections of wildtype (Sox10+) and Sox10-deficient (Sox10−) 10.5 dpc-old embryos at rostral and caudal levels corresponding to the positions marked by arrowheads in (C). The DRG are next to the spinal cord (sc) as indicated in the diagram and encircled by dotted lines. Size bar corresponds to 25 µm. (E) Chemiluminescent determination of U3-lacZ transgene activity in extracts from caudal regions of wildtype (Sox10+) and Sox10-deficient Sox10rtTA/rtTA (Sox10−) 10.5-dpc-old embryos. (F) Immunoprecipitation was performed on formaldehyde crosslinked chromatin obtained from 11.5-dpc-old wildtype embryos in the absence (buffer) and presence of antibodies (PI, pre-immune serum; α-Sox10, anti-Sox10 antibodies). PCR was applied on the immunoprecipitate to detect U3 and the evolutionarily conserved U4 region located between U3 and the transcriptional start of Sox10. These regions of the Sox10 locus were also amplified from 1/10 of the material used for immunoprecipitation (input). H2O, water control.
To find evidence for a role of the U3 enhancer in this autoregulation, we next compared expression of a U3-lacZ transgene (20) in wildtype and Sox10-deficient (i.e. Sox10rtTA/rtTA) embryos at 11.5 dpc (26). This comparison proved that transgene expression in the neural crest-derived dorsal root ganglia (DRG) is strongly reduced in vivo in the absence of Sox10 (Figure 2B and C). Hematoxylin–eosin staining showed that DRG at rostral trunk levels are already hypomorphic at mid-embryogenesis in a Sox10-deficient background (Figure 2D). Reduced transgene expression at rostral levels may therefore be secondary to neural crest cell loss. At more caudal levels, however, DRG have a normal size and still contain the normal number of neural crest cells (Figure 2D) as previously reported in the literature (7,38). The strong reduction of lacZ expression in caudal DRG of U3-lacZ, Sox10rtTA/rtTA embryos (Figure 2C) can thus not be attributed to neural crest cell loss and indicates that Sox10 is indeed required to maintain U3 activity at least in these structures. Direct determination of U3-lacZ transgene activity in extracts from caudal regions of wildtype and Sox10-deficient embryos confirmed this conclusion (Figure 2E).
In all likelihood, autoregulation requires Sox10 to directly bind to the U3 enhancer. To address this question, we prepared crosslinked chromatin from 11.5-dpc-old embryos and performed chromatin immunoprecipitations. Indeed, we were able to enrich the U3 enhancer in chromatin precipitated by antibodies directed against Sox10 (Figure 2F). Such enrichment was not observed for U3 with control IgG antibodies, nor was it seen for another evolutionarily conserved control fragment from the Sox10 genomic locus (U4) after precipitation with Sox10-specific antibodies. We therefore conclude that Sox10 influences U3 activity by directly binding to the enhancer.
The Sox10 U3 enhancer contains multiple conserved binding sites for Sox proteins
Standard chromatin immunoprecipitations do not have the resolution to exactly map the binding sites for Sox10 within this 0.4-kb long enhancer. Bioinformatic analysis and visual inspection revealed the presence of nine sites within U3 that conformed in sequence to the Sox consensus 5′-(A/T)(A/T)CAA(A/T)G-3′ and were conserved among various mammalian species as well as between mouse and chicken (Figure 3A). As binding site predictions are not very reliable for Sox proteins (39), we tested in EMSA whether these sites really bind Sox10 with high affinity. Like all SoxE proteins, Sox10 exhibits both a dimeric as well as a monomeric mode of binding (31,40). This is evident from the formation of complexes with distinct mobilities on the prototypic monomeric binding site B and the dimeric binding site C/C′ (Figure 3B) (41). Of the nine potential sites present within U3, sites 2, 5, 7 and 9 exhibited the strongest binding, whereas sites 1, 3, 4, 6 and 8 exhibited only weak or no Sox10 binding at all (Figure 3B). As the CAA core of site 6 was approximately 1 helix turn away from the CAA core of site 5, it seemed possible that sites 5 and 6 function as a dimeric site. To test this possibility, we analyzed Sox10 binding to an oligonucleotide that contained both sites. When such a site 5/6 oligonucleotide was used, we indeed obtained a complex with lower mobility (Figure 3B). The fact that this complex was dominant over the fast migrating monomer-specific complex in the presence of sufficient remaining free probe is indicative of cooperative binding of two Sox10 molecules, i.e. dimer binding. The U3 enhancer thus contains the three preferred monomeric sites 2, 7 and 9 and the dimeric site 5/6.
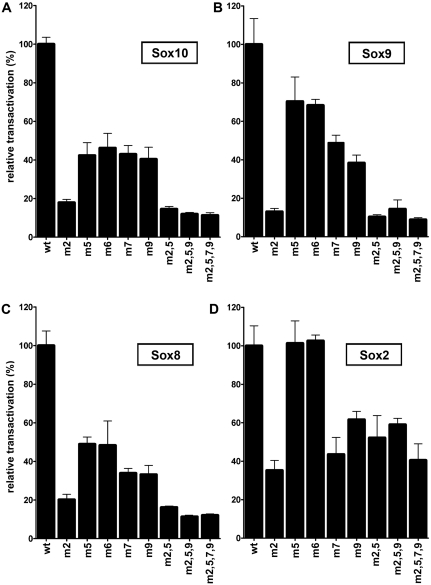
To test the functional significance of these binding sites, we introduced mutations into each of these sites that completely destroyed the CAA core. EMSA verified that the mutated sites were no longer able to bind Sox10 (data not shown). Mutant U3 sequences were then tested in reporter gene assays for their ability to respond to Sox proteins. All U3 enhancer sequences with a single mutant site were less inducible by Sox10 than the wildtype in transiently transfected N2a cells (Figure 4A). Mutation of site 2 furthermore exhibited the most severe effect with Sox10-dependent transactivation being reduced to <20% of the wildtype U3 enhancer (Figure 4A). In contrast, mutations of site 5, site 7 or site 9 reduced Sox10-dependent activation rates only to 45% of the wildtype. Despite the poor ability of site 6 to bind Sox10 on its own, a site 6 mutation caused a similar decrease in Sox10-dependent activation rate as the site 5 mutation (Figure 4A). This supports the notion that sites 5 and 6 function as a dimer site. Even after mutation of all three monomeric and the dimeric site, Sox10 retained ~10% of its transactivation capacity. This may be due to the presence of additional low affinity sites in U3. Analogous transfections with Sox9 or Sox8 yielded similar results with site 2 mutations again having the strongest effect (Figure 4B and C). In case of Sox2, the effect of a site 2 mutation was less pronounced, and mutation of site 5 or site 6 had no effect on Sox2-dependent U3 activity (Figure 4D). Even after mutation of all monomeric and the dimeric site, Sox2-dependent transactivation of U3 was only reduced by half. This may be taken as evidence that Sox2 has different binding site preferences than Sox9 and Sox10 and may thus function through some of the sites that bound Sox10 only in limited amounts.
Figure 4.
U3 responsiveness to Sox proteins in transiently transfected neuroblastoma cells diminishes after mutation of Sox binding sites. Transient transfections were performed in N2a cells with the U3-luc reporter in wildtype or various mutant versions (500 ng per well). Mutant U3 either carried mutations in one of the three major monomeric binding sites 2, 7 and 9, in site 5 or site 6 as halves of the dimeric site or in successively increasing numbers of binding sites as indicated below the bars. All U3-luc variants were co-transfected with empty expression plasmids or expression plasmids for Sox10 (500 ng per well, A), Sox9 (25 ng per well, B), Sox8 (500 ng per well, C) and Sox2 (100 ng per well, D). Luciferase activities in extracts from transfected cells were determined in three experiments each performed in duplicates. The fold induction obtained with a particular Sox protein for wildtype U3-luc was arbitrarily set to 100% and transactivation rates obtained for the U3-luc mutants are presented relative to this value as mean ± SEM. A one-way analysis of variance (ANOVA) with Tukey's multiple comparison post-test confirmed the statistical significance of the detected differences in Sox10-dependent activation rates between wildtype and mutant U3-luc reporters (P ≤ 0.001).
The Sox protein binding sites within the U3 enhancer are functional in vivo
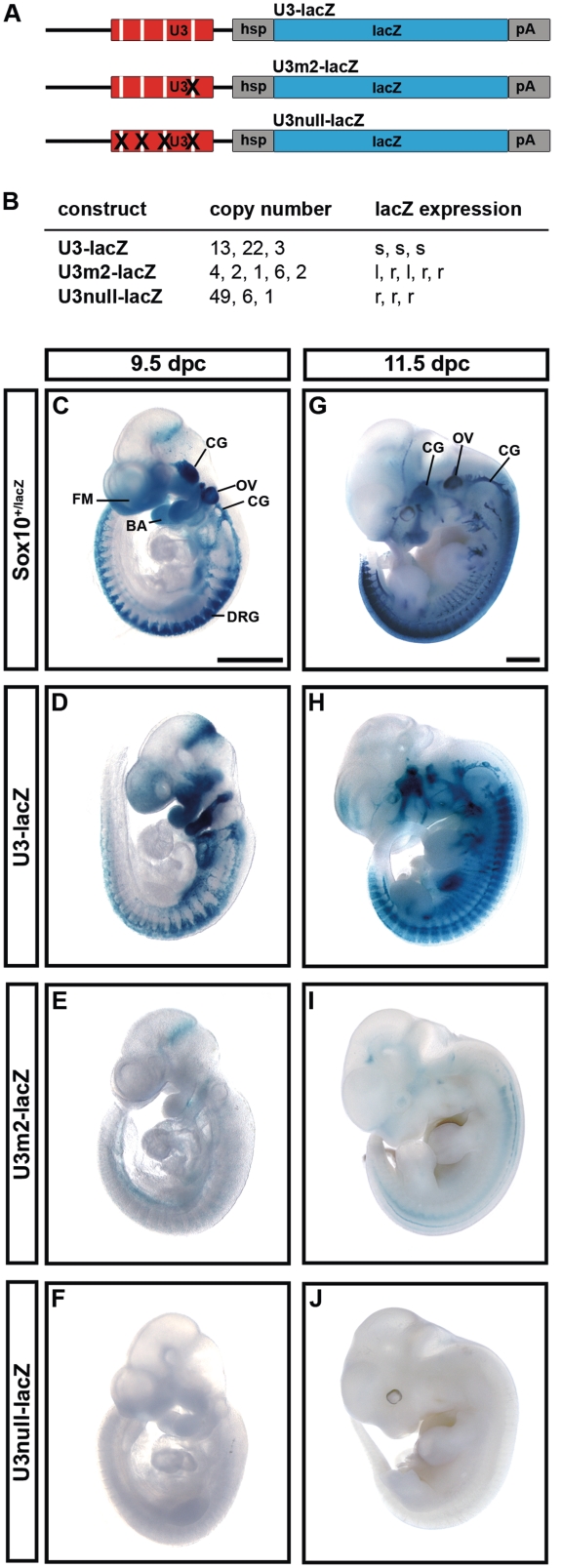
To confirm the functional relevance of the mapped Sox binding sites for activity of the U3 enhancer during embryonic development, we generated transgenic mice with mutant versions of the U3-lacZ reporter (Figure 5A). As Sox binding site 2 had the strongest effect on Sox10-dependent enhancer activity in N2a cells, we generated the U3m2-lacZ transgene in which solely site 2 was mutated. U3null-lacZ, in contrast, contained inactivating mutations in all three monomeric and the single dimeric Sox binding site. For each of the transgenes, at least three different founders were obtained that transmitted the transgene to their offspring (Figure 5B). Transgene numbers varied from 3 to 22 copies for wildtype U3-lacZ, from 1 to 6 copies for U3m2-lacZ and from 1 to 49 copies for U3null-lacZ. Copy numbers had no influence on expression patterns and only little influence on expression levels that were well comparable among progenies from different founders for each transgenic construct.
Figure 5.
Early embryonic activity of the U3 enhancer depends on intact Sox binding sites. (A) The wildtype U3-lacZ transgene and its two mutant versions U3m2-lacZ and U3null-lacZ are shown. Each construct contains U3 sequences, the hsp68 minimal promoter (hsp), the lacZ marker gene (lacZ) and a SV40 polyA signal (pA). (B) Summary of copy numbers and expression levels in the different lines obtained for each transgenic construct. s, strong expression; l, low expression; r, residual expression. (C–J) LacZ activity was detected colorimetrically using X-gal substrate on whole embryos at 9.5 dpc (C–F) and at 11.5 dpc (G–J). Embryos carried a lacZ reporter integrated into the Sox10 genomic region (Sox10+/lacZ) or one of the transgenes depicted in A. Whole-mount stainings were documented from the side and are representative of strong (D, H), low (E, I) and residual (F, J) staining intensities observed for each transgenic construct. BA, branchial arches; CG, cranial ganglia; FM, facial mesenchyme; OV, otic vesicle. Size bars in (C) and (G) correspond to 1 mm and are valid for (C–F) and (G–J), respectively.
Whereas the wildtype U3-lacZ reporter exhibited strong expression in the branchial arches, facial mesenchyme and throughout the forming peripheral nervous system including cranial ganglia and DRG at 9.5 dpc (Figure 5D) and thus resembled the Sox10 expression pattern (Figure 5C), there was only very low expression of the U3m2-lacZ reporter in branchial arches and peripheral nervous system when analyzed at the level of the whole embryo (Figure 5E). The U3null-lacZ reporter showed even less residual expression at 9.5 dpc (Figure 5F).
When whole-mount analysis was performed 2 days later, very similar results were obtained. At this level of resolution the expression pattern of the wildtype U3-lacZ reporter closely resembled Sox10 expression at 11.5 dpc. The main difference was the lack of expression of the U3-lacZ transgene in the otic vesicle despite it being a major expression site of Sox10 (compare Figure 5G with Figure 5H). The U3m2-lacZ reporter, in contrast, exhibited only low level expression throughout most of the typical Sox10 expression sites. Some residual staining was detectable in the DRG (Figure 5I). At 11.5 dpc, the U3null-lacZ reporter showed virtually no expression (Figure 5J). These findings confirm that the Sox binding sites are absolutely essential for U3 activity during early and mid-embryonic development. The monomeric site 2 obviously has a significant contribution to that function.
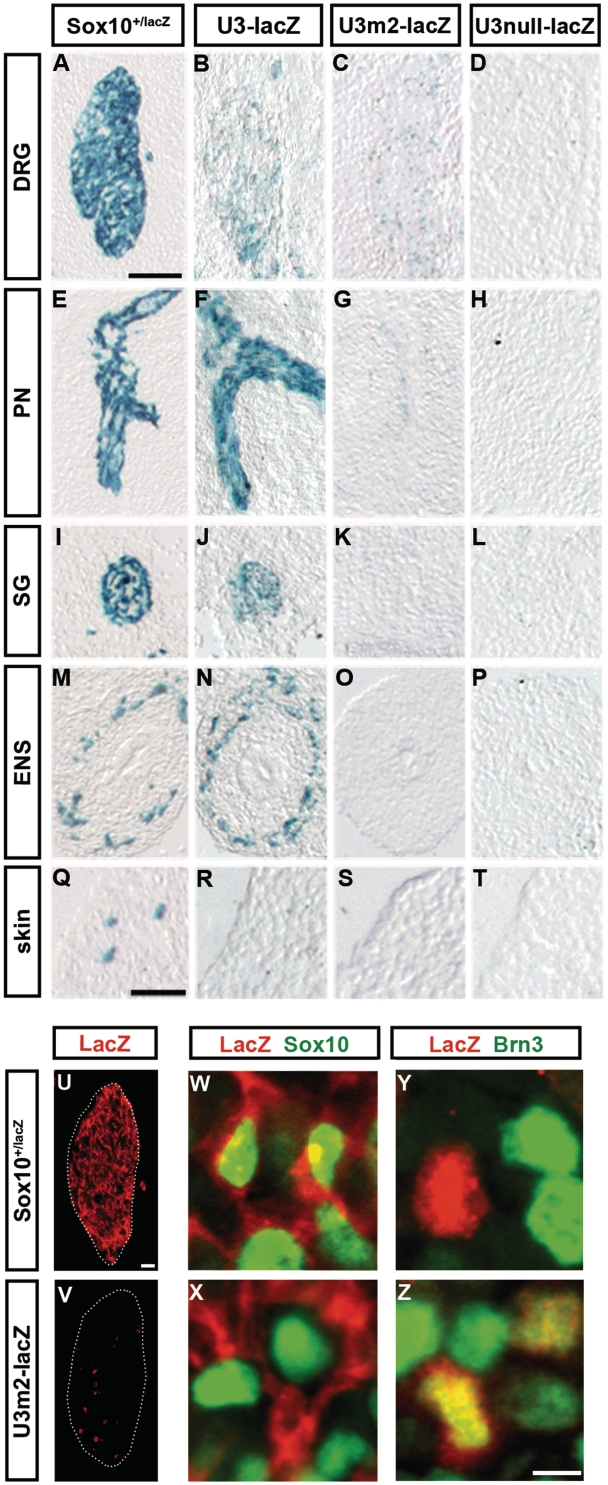
The results from whole-mount stainings were complemented by stainings of embryo sections at 11.5 dpc to increase resolution. These stainings confirmed previous findings on the expression of the U3 enhancer (20) with strong lacZ reporter expression along peripheral nerves, in sympathetic ganglia and the enteric nervous system (compare Figure 6E, I and M with Figure 6F, J and N). Although DRG strongly express Sox10, U3-lacZ expression was relatively low in this structure of the peripheral nervous system, consistent with loss of U3 activity around this time (compare Figure 6A with Figure 6B). Similarly, melanoblasts of the skin expressed Sox10, but not the U3-lacZ transgene (compare Figure 6Q with Figure 6R).
Figure 6.
Loss of Sox binding sites decreases proper U3 activity and causes weak ectopic activity. (A–T) LacZ reporter activity was detected colorimetrically using X-gal substrate in transverse sections of the trunk of 11.5-dpc-old Sox10+/lacZ embryos (A, E, I, M, Q) and age-matched embryos carrying the wildtype U3-lacZ transgene (B, F, J, N, R), the U3m2-lacZ transgene (C, G, K, O, S) or the U3null-lacZ transgene (D, H, L, P, T). Specific parts of the peripheral nervous system including DRG (A–D), peripheral nerves (PN) (E–H), sympathetic ganglia (SG) (I–L) and enteric nervous system (ENS) (M–P) are shown as well as a skin region containing melanoblasts (Q–T). (U–Z) Co-immunohistochemistry was performed on transverse sections of the trunk of 11.5-dpc-old Sox10+/lacZ embryos (U, W, Y) and age-matched embryos carrying the U3m2-lacZ transgene (V, X, Z) using antibodies directed against β-galactosidase (LacZ, in red) in combination with antibodies directed against Sox10 as a marker for glia (W, X) and Brn-3.0 as a marker for sensory neurons (Y, Z) (both in green). Only DRG are shown. Size bars: 200 µm in (A) (valid for A–P), 50 µm in (Q) (valid for Q–T), 25 µm in (U) (valid for U, V) and 5 µm in (Z) (valid for W–Z).
Neither U3m2-lacZ nor U3null-lacZ showed significant expression in any of the sites where wildtype U3-lacZ occurs at 11.5 dpc (Figure 6G, H, K, L, O and P). Melanocytes in the skin were as much negative for U3m2-lacZ and U3null-lacZ expression as they were negative for U3-lacZ expression (Figure 6R–T). The already weak expression of the U3-lacZ transgene in DRG was even further reduced for the U3m2-lacZ transgene and again completely missing for the U3null-lacZ transgene (Figure 6B–D).
Immunohistochemistry with antibodies directed against Sox10 as a marker for glial cells within the DRG and against Brn-3.0 as a marker for sensory neurons additionally indicated that the few lacZ-expressing cells that remained in DRG of U3m2-lacZ transgenic mice (compare Figure 6U with Figure 6V) were almost exclusively Brn-3.0-positive sensory neurons and not Sox10-positive glia (Figure 6X and Z). LacZ-expressing cells in DRG of Sox10+/lacZ mice, in contrast, were instead glia (Figure 6W and Y). We thus have to conclude that the low residual activity in the U3m2 mutant enhancer is in a cell type in which Sox10 is normally not expressed at this time. Loss of site 2 thus not only strongly decreases the proper activity of the U3 enhancer but also causes weak ectopic activity.
Sox10 cooperates with other neural crest transcription factors in regulating U3 activity
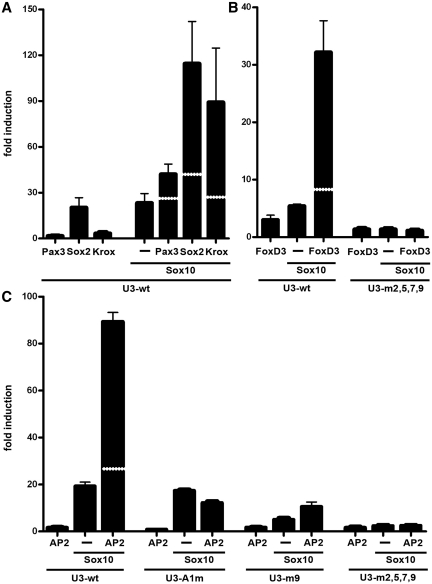
Considering that U3 responds not only to Sox10 and closely related Sox proteins, but additionally to many other transcription factors, we wanted to find out whether U3 responds to combinations of these factors and thereby integrates their effects. When we transfected the U3-luc reporter with combinations of Sox10 and a second activating transcription factor, we achieved a more than additive effect on U3 activity in many of these combinations. Pax3 and Sox10 led to a 40-fold increase in U3-luc activity on average (Figure 7A), while Sox10 and FoxD3 activated U3 activity 30-fold (Figure 7B). Even more impressive was the joint effect of Sox10 and Krox20, Sox10 and Sox2 or Sox10 and AP2α with reporter gene expression increasing 85-, 115- and 90-fold, respectively (Figure 7A and C). The U3 enhancer can thus be synergistically activated by many transcription factors in combination with Sox10. At the same time, we failed to detect synergistic activations of the U3 enhancer when Pax3, FoxD3, AP2α and Krox20 were paired in different combinations in the absence of Sox10 (data not shown). While this may be due to technical reasons, it is tempting to speculate that the presence of a Sox protein is essential for synergistic activation.
Figure 7.
U3 is synergistically activated by transcription factor combinations in transiently transfected neuroblastoma cells. Transient transfections were performed in N2a cells with wildtype (wt) or mutant U3-luc reporter plasmids (500 ng per well) as indicated below the lanes. Mutant versions include U3 enhancers with mutated AP2α site (A1m), mutated Sox10 binding site 9 (m9) and mutated Sox10 binding sites 2, 5, 7 and 9 (m2,5,7,9). Sox10 expression plasmid (500 ng in A–C) was co-transfected with the reporter, alone (−) or in combination with plasmids for Pax3, Sox2, Krox20 (all 500 ng, A), FoxD3 (100 ng, B) and AP2α (10 ng, C) as indicated below the bars. Luciferase activities in extracts from transfected cells were determined in three experiments each performed in duplicates. The luciferase activity obtained for luciferase reporter in the absence of ectopic transcription factor was arbitrarily set to 1. Fold inductions in the presence of transcription factors were calculated and are presented as mean ± SEM. Determined inductions were statistically significant according to Student's t-test (P ≤ 0.001). The white stippled line in the bars from transfections with transcription factor combinations represent the sum of activation rates obtained with both transcription factors alone.
To determine the mechanistic basis for synergistic activation, we concentrated on the interplay of FoxD3 and AP2α with Sox10. Binding site prediction and visual inspection revealed only two potential binding sites each for FoxD3 and AP2α (Figure 3A). However, when we performed EMSA with FoxD3 and either one of its potential binding sites, we failed to obtain a complex (data not shown). This is in agreement with our previous inability to detect FoxD3 binding to larger fragments of the U3 enhancer (20). At the same time, we made an interesting observation in luciferase assays with a reporter under the control of a U3 enhancer with mutations in all of the four prominent Sox10 binding sites. Not only were the Sox10-dependent activation and the synergistic effect with FoxD3 abolished, but additionally, the FoxD3-dependent activation was also strongly reduced (Figure 7B). We conclude from these findings that FoxD3 may be recruited to the U3 enhancer by Sox10 without having to bind to the enhancer on its own. This is somewhat at odds with the observation that FoxD3 activates the U3 enhancer in N2a cells up to 7-fold in the absence of co-transfected Sox10 (Figure 1B). However, it has to be taken into account that N2a cells express low levels of endogenous Sox9 that may be capable of recruiting FoxD3 to U3 in the absence of Sox10 (data not shown).
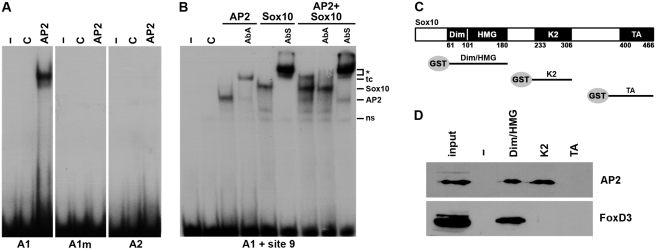
Analogous EMSA with AP2α revealed that the putative site A1 shows binding, but not A2 (Figure 8A). Interestingly, A1 is located at one of the U3 ends in close vicinity of Sox10 binding site 9 and exhibits only limited evolutionary conservation (Figure 3A). Mutation of the core of A1 abolished binding of AP2α (Figure 8A). When analyzed for its effects on U3 enhancer activity in luciferase assays, this mutation turned U3 unresponsive to AP2α while it did not interfere with Sox10-dependent activation (Figure 7C and data not shown). Additionally, the mutant U3 enhancer could no longer be synergistically activated by the combination of Sox10 and AP2α (Figure 7C).
Figure 8.
Mechanistic aspects of the synergistic activation of the U3 enhancer by Sox10, AP2α and FoxD3. (A) EMSA with radiolabeled double-stranded oligonucleotides encompassing the AP2α binding sites A1 and A2 (see Figure 3A). For A1, a wildtype and mutant version (A1m) were used, in which the CCAGGC core was altered to a CCAATT. Oligonucleotides were incubated in the absence (−) or presence (C, AP2) of protein extracts before gel electrophoresis as indicated above the lanes. Extracts were from mock-transfected 293 cells (C) or 293 cells expressing AP2α (AP2). (B) EMSA with a radiolabeled probe encompassing the AP2α binding site A1 and the Sox10 binding site 9 (A1 + site9). The probe was incubated before gel electrophoresis without extract (-) or with extracts from mock-transfected 293 cells (C) or 293 cells expressing AP2α (AP2), full-length Sox10 (Sox10) or a combination of both (AP2 + Sox10). Antibodies against AP2α (AbA) or Sox10 (AbS) were added to some reactions as indicated above the lanes. ns, non-specific complex; AP2, AP2α-containing complex; Sox10, Sox10-specific complex; tc, ternary complex; asterisk, supershifted complex. (C) Schematic diagram of the conserved regions of Sox10 which were fused to GST and used for pulldown experiments. (D) GST pulldown experiments were performed on extracts from 293 cells expressing AP2α (AP2) or FoxD3. In addition to GST, proteins were used for pulldown that contained the dimerization (Dim), HMG, K2 or transactivation (TA) domain of Sox10. Detection of the precipitated AP2α and FoxD3 proteins was in western blots by antibodies directed against the T7-tag present in AP2α and the HA-tag present in FoxD3.
As already mentioned, the identified AP2α site is near Sox10 binding site 9. Therefore, we also tested how Sox10 and AP2α behave on a U3 enhancer with mutant site 9. As already reported, Sox10-dependent enhancer activity was reduced, but not abolished, while there was no effect on AP2α-dependent activation. Intriguingly, the rate of synergistic activation was also dramatically decreased (Figure 7C). When the site 9 mutant of the U3 enhancer was replaced by a version with all prominent Sox10 binding sites mutated, the effect was even more pronounced with Sox10-dependent and synergistic activation completely gone (Figure 7C). These results indicate that both AP2α and Sox10 have to bind to the enhancer to achieve synergistic activation.
Considering the importance of the identified AP2α site and the adjacent Sox10 binding site 9 for the synergism, we next performed EMSA with a probe that spanned both sites (Figure 8B). This larger probe already interacted with proteins from control extracts (ns in Figure 8B). Extracts containing either full-length AP2α or full-length Sox10 yielded additional complexes of specific mobility whose identity was confirmed by the application of specific antibodies. Both the AP2α-specific antibody and the Sox10-specific antibody supershifted the corresponding binary complexes. When both AP2α and Sox10 were present in the extract, a novel band appeared with a mobility lower than each of the two binary complexes. Incubation with either AP2α- or Sox10-specific antibodies identified this novel band as a ternary complex between both proteins and the probe.
As there may be protein–protein contacts between Sox10 and AP2α in the ternary complex, we performed GST pulldown assays in which full-length AP2α was tested for its ability to bind to various regions of Sox10 that were produced as GST fusions and immobilized on sepharose beads (Figure 8C). These pulldown experiments revealed that AP2α interacts both with a fragment containing the HMG domain and the adjacent dimerization region of Sox10 and with a fragment containing the conserved K2 domain (Figure 8D). No interaction was observed between AP2α and the transactivation domain of Sox10 (Figure 8D). We conclude that AP2α interacts with multiple regions of Sox10 that may in turn facilitate its binding to the U3 enhancer so that cooperative binding may constitute at least part of the mechanism by which synergism is achieved in this case. Finally, it remains to be mentioned that FoxD3 also interacted in GST pulldown experiments with the fragment containing HMG domain and dimerization region (Figure 8D) further confirming that Sox10 likely has the capability to recruit FoxD3 to the U3 enhancer.
DISCUSSION
We have shown in this study that the U3 enhancer strongly responds to Sox10 both in cultured cells and in vivo. As we have also provided evidence for autoregulation of Sox10 in vivo, it is reasonable to assume that the U3 enhancer is involved in mediating this Sox10 autoregulation during development of the neural crest and its derivatives. Such autoregulation constitutes a convenient regulatory mechanism for a central transcriptional regulator that is expressed in a particular cell type for extended times or continuously after specification, and whose expression needs to be maintained. Sox10 is such a regulator as its expression begins in the emerging neural crest at the time when these cells start to emigrate and continues in several neural crest-derived cell types for quite some time after specification (7–9, 42–44). In peripheral glia such as Schwann cells along nerves and enteric glia in the gastrointestinal tract, Sox10 remains expressed even into the fully matured and terminally differentiated state (30,42).
In addition to Sox10, several other neural crest and glial transcription factors activate the U3 enhancer. Our studies indicate that these transcription factors function synergistically with Sox10. While U3 enhancer activity in transfected N2a cells can be induced by single factors, it is very likely that U3 activity in vivo requires their concerted action. This would explain why mutation of Sox binding sites is sufficient to inactivate U3 almost completely in transgenic mice. The central role of Sox10 for U3 activity is also supported by our observation that synergistic effects were readily detected in transcription factor combinations with Sox10, but not in combinations without Sox10 or a related Sox protein.
Synergistic action is an important functional principle for Sox proteins and several Sox proteins have been reported to switch cooperating transcription factors during the course of development (39,45,46). While Pax3, AP2α and FoxD3 may help Sox10 to maintain U3 activity in neural crest cells, these factors are no longer present in peripheral glia. Instead, immature peripheral glia express Sox2 and glia maturing to myelinating Schwann cells induce Krox20. Now these transcription factors may help Sox10 to maintain activity of the U3 enhancer.
In the course of this study, we have analyzed two synergistic interactions in closer detail. Intriguingly, the underlying mechanisms seem to vary, although physical interactions between Sox10 and other transcription factors seem to be generally important. That many such interactions exist has been shown before (28). Krox20 is, for instance, a known interaction partner of Sox10 (28).
Considering that we detected a physical interaction between Sox10 and FoxD3, but failed to see direct binding of FoxD3 to specific sites in U3 enhancer, the synergism between Sox10 and FoxD3 appears to be based on the Sox10-dependent recruitment of FoxD3 to the enhancer. In contrast, synergism between AP2α and Sox10 required both proteins to bind to their own sites in U3. Existing physical interactions between AP2α and Sox10 may facilitate this binding. Interestingly, AP2α contacts two separate Sox10 regions. It is actually the first protein shown to interact with the K2 domain, a region that contributes to the function of Sox10, but whose mode of action is still poorly understood (12,47).
In the absence of transcription factors that are able to synergize with Sox10, U3 activity likely subsides. This will for instance be the case in sensory neurons of the peripheral nervous system in which Sox10 and U3 activity are not maintained despite the origin of these cells from Sox10-positive neural crest cells (7). Our current study and the resulting model thus gives important insights into the mechanisms by which maintenance of Sox10 expression may be achieved, but also provides important clues how the positive autoregulatory loop may be broken and Sox10 expression be extinguished.
It needs to be emphasized that autoregulation cannot solely be responsible for Sox10 expression. This is for instance evident from the fact that some residual expression of the Sox10lacZ reporter remains detectable in Sox10-deficient mice, as long as the normally Sox10-expressing cells survive in its absence (7). There are two likely explanations. Some of the other enhancers (i.e. U1 and U5) whose activities overlap with U3 may not depend as strictly on autoregulation as U3.
Additionally, it has to be taken into consideration that the Sox10 binding sites in U3 and the other enhancers will also be recognized by other Sox proteins, in particular by the closely related Sox8 and Sox9 that have very similar, if not identical binding characteristics and form the SoxE subgroup with Sox10 (31,32,40,48). This assumption is confirmed by our reporter studies in transiently transfected cells. Particularly Sox9 may be important in vivo, as it precedes Sox10 in the neural crest and has been shown to induce Sox10 expression (11,49). Sox9 may thus induce Sox10 in the emerging neural crest via U3. At later times Sox10 appears to be the dominant SoxE protein in such neural crest derivatives as peripheral glia (1) so that Sox8 and Sox9 are unlikely to contribute as much to maintenance of Sox10 expression as they do to its induction.
Our studies in transgenic mice did not only show the importance of the identified Sox protein binding sites in general, but also pointed to an essential role of binding site 2 in agreement with tissue culture studies. Site 2 binds a single molecule of Sox10. This is noteworthy because previous studies have placed greater emphasis on the role of dimeric sites. Dimeric sites can only be properly used by SoxE and a few other Sox proteins and are therefore more specific than the standard monomeric sites. It has even been postulated that the occurrence of dimeric sites can be used to predict regulatory regions with neural crest activity bioinformatically (18). While our study does not question the importance of dimeric sites, of which one is indeed present in U3, it clearly shows that monomeric sites are likewise essential to guarantee Sox10-dependent activity in neural crest and glial cells. Future studies may also address the question whether all four of the identified Sox10 binding sites are needed in vivo for activity or whether some sites (such as site 2) dominate functionally. The presence of multiple binding sites and the ability of Sox proteins to change the topology of bound DNA as architectural factors (39,50) at least permits the attractive hypothesis that multiple binding is required to alter the three-dimensional conformation of U3 from an inactive into an active state.
Finally, it would be interesting to screen patients with neurocristopathies of unexplained cause for sequence polymorphisms in the Sox10 binding sites of U3 as identified in this study. Such sequence polymorphisms may turn out to be inactivating mutations that may lead to developmental defects in neural crest or peripheral glia.
FUNDING
Deutsche Forschungsgemeinschaft grant (We1326/9; to Mi.W.) and FAU graduate school fellowship (Förderung der Chancengleichheit für Frauen in Forschung und Lehre; to Ma.W.); Funding for open access charge: Deutsche Forschungsgemeinschaft grant – Universität Erlangen.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank J. Behrens, P. Labosky, H. Schorle and E. Turner for providing cDNAs and antibodies.
REFERENCES
- 1.Stolt CC, Wegner M. SoxE function in vertebrate nervous system development. Int. J. Biochem. Cell Biol. 2010;42:437–440. doi: 10.1016/j.biocel.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 2.Kelsh RN. Sorting out Sox10 functions in neural crest development. Bioessays. 2006;28:788–798. doi: 10.1002/bies.20445. [DOI] [PubMed] [Google Scholar]
- 3.Haldin CE, LaBonne C. SoxE factors as multifunctional neural crest regulatory factors. Int. J. Biochem. Cell Biol. 2010;42:441–444. doi: 10.1016/j.biocel.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- 5.Dutton KA, Pauliny A, Lopes SS, Elworthy S, Carney TJ, Rauch J, Geisler R, Haffter P, Kelsh RN. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development. 2001;128:4113–4125. doi: 10.1242/dev.128.21.4113. [DOI] [PubMed] [Google Scholar]
- 6.Aoki Y, Saint-Germain N, Gyda M, Magner-Fink E, Lee YH, Credidio C, Saint-Jeannet JP. Sox10 regulates the development of neural crest-derived melanocytes in Xenopus. Dev. Biol. 2003;259:19–33. doi: 10.1016/s0012-1606(03)00161-1. [DOI] [PubMed] [Google Scholar]
- 7.Britsch S, Goerich DE, Riethmacher D, Peirano RI, Rossner M, Nave KA, Birchmeier C, Wegner M. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 2001;15:66–78. doi: 10.1101/gad.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herbarth B, Pingault V, Bondurand N, Kuhlbrodt K, Hermans-Borgmeyer I, Puliti A, Lemort N, Goossens M, Wegner M. Mutation of the Sry-related Sox10 gene in Dominant megacolon, a mouse model for human Hirschsprung disease. Proc. Natl. Acad. Sci. USA. 1998;95:5161–5165. doi: 10.1073/pnas.95.9.5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Southard-Smith EM, Kos L, Pavan WJ. Sox10 mutation disrupts neural crest development in Dom Hirschsprung mouse model. Nat. Genet. 1998;18:60–64. doi: 10.1038/ng0198-60. [DOI] [PubMed] [Google Scholar]
- 10.Pingault V, Bondurand N, Kuhlbrodt K, Goerich DE, Prehu M-O, Puliti A, Herbarth B, Hermans-Borgmeyer I, Legius E, Matthijs G, et al. Sox10 mutations in patients with Waardenburg-Hirschsprung disease. Nat. Genet. 1998;18:171–173. doi: 10.1038/ng0298-171. [DOI] [PubMed] [Google Scholar]
- 11.Cheung M, Chaboissier M-C, Mynett A, Hirst E, Schedl A, Briscoe J. The transcriptional control of trunk neural crest induction, survival, and delamination. Dev. Cell. 2005;8:179–192. doi: 10.1016/j.devcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 12.Cossais F, Wahlbuhl M, Kriesch J, Wegner M. SOX10 structure-function analysis in the chicken neural tube reveals important insights into its role in human neurocristopathies. Hum. Mol. Genet. 2010;19:2409–2420. doi: 10.1093/hmg/ddq124. [DOI] [PubMed] [Google Scholar]
- 13.McKeown SJ, Lee VM, Bronner-Fraser M, Newgreen DF, Farlier PG. Sox10 overexpression induces neural crest-like cells from all dorsoventral levels of the neural tube but inhibtis differentiation. Dev. Dyn. 2005;233:430–444. doi: 10.1002/dvdy.20341. [DOI] [PubMed] [Google Scholar]
- 14.Stanchina L, Baral V, Robert F, Pingault V, Lemort N, Pachnis V, Goossens M, Bondurand N. Interactions between Sox10, Edn3 and Ednrb during enteric nervous system and melanocyte development. Dev. Biol. 2006;295:232–249. doi: 10.1016/j.ydbio.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 15.Sandelin A, Bailey P, Bruce S, Engstrom PG, Klos JM, Wasserman WW, Ericson J, Lenhard B. Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genomics. 2004;5:99. doi: 10.1186/1471-2164-5-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antonellis A, Bennett WR, Menheniott TR, Prasad AB, Lee-Lin SQ, Green ED, Paisley D, Kelsh RN, Pavan WJ, Ward A. Deletion of long-range sequences at Sox10 compromises developmental expression in a mouse model of Waardenburg-Shah (WS4) syndrome. Hum. Mol. Genet. 2006;15:259–271. doi: 10.1093/hmg/ddi442. [DOI] [PubMed] [Google Scholar]
- 18.Antonellis A, Huynh JL, Lee-Lin S-Q, Vinton RM, Renaud G, Loftus SK, Elliot G, Wolfsberg TG, Green ED, McCallion AS, et al. Identification of neural crest and glial enhancers at the mouse Sox10 locus through transgenesis in zebrafish. PLoS Genet. 2008;4:e1000174. doi: 10.1371/journal.pgen.1000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deal KK, Cantrell VA, Chandler RL, Saunders TL, Mortlock DP, Southard-Smith EM. Distant regulatory elements in a Sox10-betaGEO BAC transgene are required for expression of Sox10 in the enteric nervous system and other neural crest-derived tissues. Dev. Dyn. 2006;235:1413–1432. doi: 10.1002/dvdy.20769. [DOI] [PubMed] [Google Scholar]
- 20.Werner T, Hammer A, Wahlbuhl M, Bösl MR, Wegner M. Multiple conserved regulatory elements with overlapping functions determine Sox10 expression in mouse embryogenesis. Nucleic Acids Res. 2007;35:6526–6538. doi: 10.1093/nar/gkm727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Betancur P, Bronner-Fraser M, Sauka-Spengler T. Genomic code for Sox10 activation reveals a key regulatory enhancer for cranial neural crest. Proc. Natl Acad. Sci. USA. 2010;107:3570–3575. doi: 10.1073/pnas.0906596107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Küspert M, Hammer A, Bosl MR, Wegner M. Olig2 regulates Sox10 expression in oligodendrocyte precursors through an evolutionary conserved distal enhancer. Nucleic Acids Res. 2011;39:1280–1293. doi: 10.1093/nar/gkq951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gunnarsson U, Kerje S, Bed'hom B, Sahlqvist AS, Ekwall O, Tixier-Boichard M, Kampe O, Andersson L. The Dark brown plumage color in chickens is caused by an 8.3-kb deletion upstream of SOX10. Pigment Cell. Melanoma Res. 2011;24:268–274. doi: 10.1111/j.1755-148X.2011.00825.x. [DOI] [PubMed] [Google Scholar]
- 24.Southard-Smith EM, Angrist M, Ellison JS, Agarwala R, Baxevanis AD, Chakravarti A, Pavan WJ. The Sox10(Dom) mouse: modeling the genetic variation of Waardenburg- Shah (WS4) syndrome. Genome Res. 1999;9:215–225. [PubMed] [Google Scholar]
- 25.Pingault V, Ente D, Dastot-Le Moal F, Goossens M, Marlin S, Bondurand N. Review and update of mutations causing Waardenburg syndrome. Hum. Mutat. 2010;31:391–406. doi: 10.1002/humu.21211. [DOI] [PubMed] [Google Scholar]
- 26.Ludwig A, Schlierf B, Schardt A, Nave KA, Wegner M. A Sox10 rtTA mouse line for tetracycline-inducible expression of transgenes in neural crest cells and oligodendrocytes. Genesis. 2004;40:171–175. doi: 10.1002/gene.20083. [DOI] [PubMed] [Google Scholar]
- 27.Maka M, Stolt CC, Wegner M. Identification of Sox8 as a modifier gene in a mouse model of Hirschsprung disease reveals underlying molecular defect. Dev. Biol. 2005;277:155–169. doi: 10.1016/j.ydbio.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 28.Wissmüller S, Kosian T, Wolf M, Finzsch M, Wegner M. The High-mobility-group domain of Sox proteins interacts with the DNA-binding domains of many transcription factors. Nucleic Acids Res. 2006;34:1735–1744. doi: 10.1093/nar/gkl105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamburger H, Hamilton HL. A series of normal stages in the development of the chick embryo. J. Morphol. 1953;88:49–92. [PubMed] [Google Scholar]
- 30.Finzsch M, Schreiner S, Kichko T, Reeh P, Tamm ER, Bosl MR, Meijer D, Wegner M. Sox10 is required for Schwann cell identity and progression beyond the immature Schwann cell stage. J. Cell. Biol. 2010;189:701–712. doi: 10.1083/jcb.200912142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlierf B, Ludwig A, Klenovsek K, Wegner M. Cooperative binding of Sox10 to DNA: requirements and consequences. Nucleic Acids Res. 2002;30:5509–5516. doi: 10.1093/nar/gkf690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stolt CC, Schlierf A, Lommes P, Hillgärtner S, Werner T, Kosian T, Sock E, Kessaris N, Richardson WD, Lefebvre V, et al. SoxD proteins influence multiple stages of oligodendrocyte development and modulate SoxE protein function. Dev. Cell. 2006;11:697–710. doi: 10.1016/j.devcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 33.Schlierf B, Werner T, Glaser G, Wegner M. Expression of Connexin47 in oligodendrocytes is regulated by the Sox10 transcription factor. J. Mol. Biol. 2006;361:11–21. doi: 10.1016/j.jmb.2006.05.072. [DOI] [PubMed] [Google Scholar]
- 34.Topilko P, Schneider MS, Levi G, Baron VEA, Chennoufi AB, Seitanidou T, Babinet C, Charnay P. Krox-20 controls myelination in the peripheral nervous system. Nature. 1994;371:796–799. doi: 10.1038/371796a0. [DOI] [PubMed] [Google Scholar]
- 35.Le N, Nagarajan R, Wang JY, Araki T, Schmidt RE, Milbrandt J. Analysis of congenital hypomyelinating Egr2Lo/Lo nerves identifies Sox2 as an inhibitor of Schwann cell differentiation and myelination. Proc. Natl. Acad. Sci. USA. 2005;102:2596–2601. doi: 10.1073/pnas.0407836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jessen KR, Mirsky R. The origin and development of glial cells in peripheral nerves. Nat. Rev. Neurosci. 2005;6:671–682. doi: 10.1038/nrn1746. [DOI] [PubMed] [Google Scholar]
- 37.Decker L, Desmarquet-Trin-Dinh C, Taillebourg E, Ghislain J, Vallat JM, Charnay P. Peripheral myelin maintenance is a dynamic process requiring constant Krox20 expression. J. Neurosci. 2006;26:9771–9779. doi: 10.1523/JNEUROSCI.0716-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sonnenberg-Riethmacher E, Miehe M, Stolt CC, Goerich DE, Wegner M, Riethmacher D. Development and degeneration of dorsal root ganglia in the absence of the HMG-domain transcription factor Sox10. Mech. Dev. 2001;109:253–265. doi: 10.1016/s0925-4773(01)00547-0. [DOI] [PubMed] [Google Scholar]
- 39.Wegner M. All purpose Sox: the many roles of Sox proteins in gene expression. Int. J. Biochem. Cell Biol. 2010;42:381–390. doi: 10.1016/j.biocel.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 40.Peirano RI, Wegner M. The glial transcription factor Sox10 binds to DNA both as monomer and dimer with different functional consequences. Nucleic Acids Res. 2000;28:3047–3055. doi: 10.1093/nar/28.16.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peirano RI, Goerich DE, Riethmacher D, Wegner M. Protein zero expression is regulated by the glial transcription factor Sox10. Mol. Cell. Biol. 2000;20:3198–3209. doi: 10.1128/mcb.20.9.3198-3209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M. Sox10, a novel transcriptional modulator in glial cells. J. Neurosci. 1998;18:237–250. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim J, Lo L, Dormand E, Anderson DJ. SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron. 2003;38:17–31. doi: 10.1016/s0896-6273(03)00163-6. [DOI] [PubMed] [Google Scholar]
- 44.Paratore C, Goerich DE, Suter U, Wegner M, Sommer L. Survival and glial fate acquisition of neural crest cells are regulated by an interplay between the transcription factor Sox10 and extrinsic combinatorial signaling. Development. 2001;128:3949–3961. doi: 10.1242/dev.128.20.3949. [DOI] [PubMed] [Google Scholar]
- 45.Kamachi Y, Uchikawa M, Kondoh H. Pairing SOX off: with partners in the regulation of embryonic development. Trends Genet. 2000;16:182–187. doi: 10.1016/s0168-9525(99)01955-1. [DOI] [PubMed] [Google Scholar]
- 46.Wilson M, Koopman P. Matching SOX: partner proteins and co-factors of the SOX family of transcriptional regulators. Curr. Opin. Genet. Dev. 2002;12:441–446. doi: 10.1016/s0959-437x(02)00323-4. [DOI] [PubMed] [Google Scholar]
- 47.Schreiner S, Cossais F, Fischer K, Scholz S, Bösl MR, Holtmann B, Sendtner M, Wegner M. Hypomorphic Sox10 alleles reveal novel protein functions and unravel developmental differences in glial lineages. Development. 2007;134:3271–3281. doi: 10.1242/dev.003350. [DOI] [PubMed] [Google Scholar]
- 48.Sock E, Pagon RA, Keymolen K, Lissens W, Wegner M, Scherer G. Loss of DNA-dependent dimerization of the transcription factor SOX9 as a cause for campomelic dysplasia. Hum. Mol. Genet. 2003;12:1439–1447. doi: 10.1093/hmg/ddg158. [DOI] [PubMed] [Google Scholar]
- 49.Cheung M, Briscoe J. Neural crest development is regulated by the transcription factor Sox9. Development. 2003;130:5681–5693. doi: 10.1242/dev.00808. [DOI] [PubMed] [Google Scholar]
- 50.Wegner M. From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res. 1999;27:1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]