Abstract
Promoter recognition and local melting of DNA are key steps of transcription initiation catalyzed by RNA polymerase and initiation factors. From single molecule fluorescence resonance energy transfer studies of the yeast (Saccharomyces cerevisiae) mitochondrial RNA polymerase Rpo41 and its transcription factor Mtf1, we show that the pre-initiation complex is highly dynamic and undergoes repetitive opening–closing transitions that are modulated by Mtf1 and ATP. We found that Rpo41 alone has the intrinsic ability to bend the promoter but only very briefly. Mtf1 enhances bending/opening transition and suppresses closing transition, indicating its dual roles of nucleating promoter opening and stabilizing the open state. The cognate initiating ATP prolongs the lifetime of the open state, plausibly explaining the ‘ATP sensing mechanism’ suggested for the system. We discovered short-lived opening trials upon initial binding of Rpo41-Mtf1 before the establishment of the opening/closing equilibrium, which may aid in promoter selection before the formation of stable pre-initiation complex. The dynamics of open complex formation provides unique insights into the interplay between RNA polymerase and transcription factors in regulating initiation.
INTRODUCTION
Transcription of the DNA to synthesize RNA starts with the sequence-specific formation of an initiation complex at promoter sites. Transcription initiation is a multistep process catalyzed by the RNA polymerase (RNAP) that begins with the recognition of the promoter sequence, the formation of competent initiation complex, and the synthesis of initial short transcript, followed by transition to the elongation phase (1). Disrupting several base-pairs around the start site is critical for the formation of pre-initiation open complex, which enables the search for correct nucleotides for RNA synthesis by base-pairing. Promoter melting by the well-studied single-subunit T7 RNA polymerase is synchronized on base-pairs adjacent to the transcription start site and does not require other factors (2). In comparison, promoter melting by bacterial, archaeal, or eukaryotic multi-subunit RNAPs is a multi-step isomerization process, requiring coordinated actions between the core catalytic unit and accessory protein factors that are not necessarily synchronized (3,4). Although the kinetics of initial promoter binding and opening steps has been studied for various transcription systems (2,5–7), little is known about the dynamics of the open complex. Hence, our understanding of how this process is regulated by the core RNA polymerase and transcription factors is limited.
Studies of the relatively simple transcription system of the mitochondria can be used to understand the interplay of the core RNAP and transcription factors. Mitochondrial RNA polymerases from yeast to human are homologous to those encoded by bacteriophages T3 and T7 (8–10) with the major difference that they require accessory proteins for transcription initiation (11–13). Yeast (Saccharomyces cerevisiae) mitochondrial RNAP, Rpo41, can initiate transcription by itself on supercoiled or pre-melted templates (14), but needs the transcription factor, Mtf1, for sequence-specific transcription initiation on duplex promoter (15,16). Similarly, specific transcription initiation on human mitochondrial genome promoters LSP and HSP1 requires both the human mitochondrial RNAP, POLRMT, and the specific factor, TFB2M, an analog of Mtf1 (17). 2-aminopurine fluorescence and protein–DNA cross-linking studies indicate that Mtf1 interacts with the entire nonanucleotide consensus sequence including the melted region of the promoter and facilitates promoter opening (18–20). From these studies, it was suggested that Mtf1 contributed to promoter opening by trapping the melted non-template strand. Similar role of stabilizing the open complex has been suggested for the initiation factors TFE in archaea (21–23) and TFIIE in eukaryote (24,25). However, the mechanisms by which these transcription factors facilitate promoter opening is unclear. For example, it is not known whether the transcription factors nucleate promoter opening or simply stabilize the open complex. Also, the conformational dynamics of the pre-initiation complex and how it is affected by the initiation factors remain uncharacterized.
The pre-initiation open complex of the single-subunit T7 RNAP is unstable and it collapses back to the closed form competing against successful initiation (2,26). This instability was proposed to be the kinetic mechanism that allows the polymerase to achieve promoter specificity while still allowing efficient promoter release (26). Similar instability of the open complex has been reported for the Escherichia coli RNAP (27). In the above systems and mitochondrial RNAP as well, the initiating NTPs play an important role and regulate transcription by stabilizing the open complex and promoting forward reaction (28–30). Based on the observation that the Km for ATP, the initiating nucleotide for the yeast mitochondrial transcription, correlates with in vivo transcript level for a variety of promoters, an ATP sensing mechanism was proposed for the yeast mitochondria (31), by which ATP concentration regulates transcription. However, the mechanism for ATP sensing has not been established.
In this study, we have used single molecule Fluorescence Resonance Energy Transfer (smFRET) (32) to investigate the roles of the core subunit Rpo41 and the transcription factor Mtf1 in transcription initiation. smFRET has been used previously to study transcription initiation by E. coli RNAP (33) and T7 RNAP (34,35). Here we show that the pre-initiation complex oscillates between the bent/open and unbent/closed conformations whether Mtf1 is present or not, showing that Rpo41 has the intrinsic ability to bend the promoter. Mtf1 facilitates promoter opening by both increasing the opening rate and decreasing the closing rate, which indicates that it both nucleates promoter opening and stabilizes the open complex. The initiating nucleotide has a marked effect on the dynamics and further prolongs the lifetime of the open conformation, which provides a plausible explanation for the ATP sensing mechanism. We discovered a short-lived open intermediate that is formed in the presence of Mtf1 before the establishment of the opening/closing equilibrium, which may aid in promoter selection.
MATERIALS AND METHODS
Preparation of Rpo41, Mtf1, and DNA templates
Proteins were expressed and purified as described earlier (19). DNA oligos were custom-synthesized and purified by HPLC (Integrated DNA Technologies, Coralville, IA, USA).
Vesicle encapsulation
Encapsulation in lipid vesicles was performed as described earlier (36). Lipid films were prepared by mixing 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(Cap Biotinyl) with dimyristoyl phosphatidylcholine (DMPC) (Avanti Polar Lipids, Inc.) dissolved in chloroform (1:100 molar ratio) and gently drying with nitrogen gas. The lipids were hydrated with buffer containing 10 mM Tris–acetate, pH 7.5 and 50 mM sodium chloride. Lipid solution was repeatedly frozen in liquid nitrogen and thawed seven times. DNA template was incubated with proteins as necessary at room temperature for 1 min in the protein buffer, which contains 50 mM Tris–acetate, pH 7.5, 100 mM potassium glutamate and 10 mM magnesium acetate. These solutions were mixed 1:1 in volume so that the mixture has 12.5 mg/ml lipids and 400 nM DNA. It was extruded through track-etched membrane with 200 nm holes (Whatman, catalog number 800281) seven times to induce encapsulation.
Single molecule detection and analysis
We used total internal reflection fluorescence microscopy for imaging as described previously (36). We immobilized lipid vesicles or DNA template directly on polyethylene glycol coated quartz slide using biotin-neutravidin interaction. Imaging buffer contained 1 mg/ml glucose oxidase, 0.04 mg/ml catalase, 0.8% dextrose, and saturated trolox (~3 mM), in addition to the protein buffer. From the fluorescence intensity measured by EMCCD camera, we calculated FRET efficiency defined with leakage correction as EFRET = (IA − 0.08 × ID)/(ID + IA) where ID and IA are the measured intensity of the donor and acceptor, respectively.
RESULTS
Initiation complex exhibits opening–closing dynamics
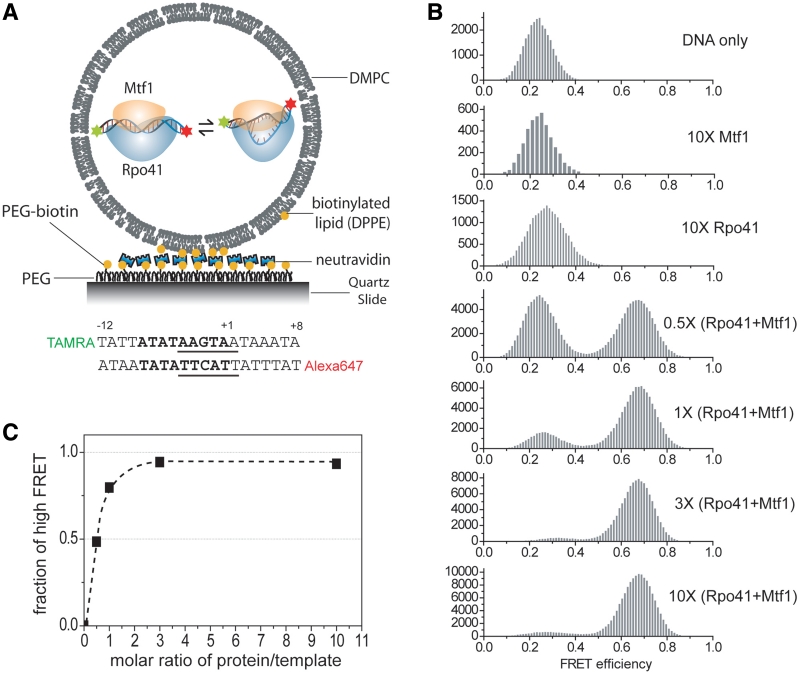
We designed a 20 bp DNA (Figure 1A), with the mitochondrial 14S rRNA sequence from −12 to +8, labeled with tetramethylrhodamine as the donor (D) and Alexa Fluor 647 as the acceptor (A) fluorescent probes at the 5′-ends of the non-template and template strand, respectively. Rpo41-Mtf1 can bind to such a short template tightly at sub-nanomolar dissociation constant with 1:1 stoichiometry, melt the duplex, and efficiently initiate specific transcription initiation in the presence of nucleotides (Supplementary Figure S1). Extending the template such as to −25 and +20 did not enhance the binding affinity of Rpo41 and Mtf1. These results suggest that the −12/+8 template retains specific interactions with the Rpo41-Mtf1 heterodimer important for transcription initiation.
Figure 1.
smFRET titration of the promoter with Rpo41-Mtf1 inside lipid vesicles. (A) Schematic of surface-immobilized vesicles containing the initiation complex. Below is the design of promoter template. Promoter sequence (−8 to +1) is shown in bold. Transcription start site is defined as +1. Melting region (−4 to +2) is underlined. (B) Change of smFRET histogram upon adding proteins as indicated. DNA concentration was kept at 400 nM during vesicle preparation which corresponds to roughly one molecule in the volume of 200 nm diameter sphere. Protein concentration is represented as multiples of DNA concentration. (C) Change of the fraction of high FRET population as a function of the molar ratio of (Rpo41 + Mtf1)/DNA.
To minimize hindrance from surface immobilization, we used lipid vesicles with 200 nm diameter to encapsulate and immobilize the molecules. The promoter DNA was encapsulated with or without proteins and smFRET values from individual surface-immobilized vesicles were measured. The FRET efficiency histogram of DNA alone shows a peak at 0.22 (Figure 1B). Each histogram was built by analyzing vesicles showing single photobleaching steps for both fluorophores and each count represents signal acquired for 100 ms. Upon co-encapsulation with excess Rpo41, we observed an emerging peak at a higher FRET efficiency of 0.28, consistent with a decrease in the D-A distance and a slight bend of the DNA. Mtf1 alone did not induce any noticeable FRET change. When we added a mixture of Rpo41 and Mtf1, we observed a clearly separated FRET peak with a much higher efficiency of 0.68, indicating a much greater decrease in D–A distance.
The high FRET state population as a function of the molar ratio of the proteins to the DNA follows a simple titration curve with inflection close to 1:1 (Figure 1C), consistent with the 1:1 complex formation between Rpo41-Mtf1 and DNA in the transcription complex. A control DNA template with a random sequence did not show any FRET change (Supplementary Figure S2), confirming that sequence-specific interaction with Rpo41-Mtf1 is responsible for the FRET change observed with the promoter template. Note that even at saturating protein concentration within vesicles, there remains a small low FRET population (Figure 1B and C). This means that some DNAs are not affected by the proteins or each complex populates the low FRET state for a certain fraction of time.
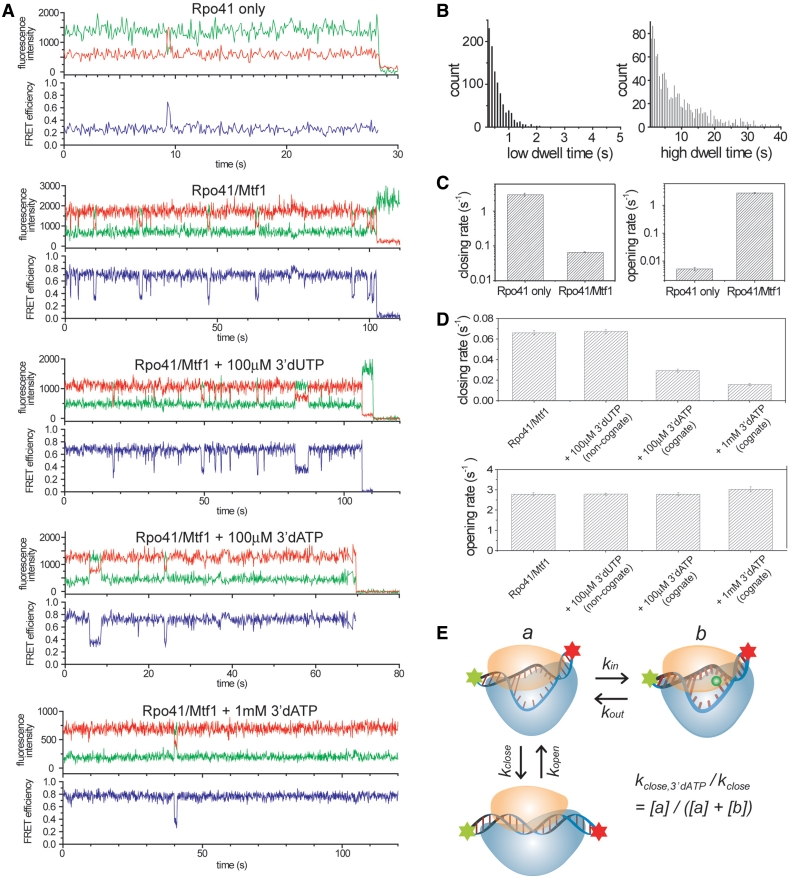
Single molecule time traces provide a direct measure of the dynamic behavior of each complex (Figure 2A). In the presence of Rpo41 alone, the complex remains mainly in the low FRET state but makes rare and brief excursions to the high FRET state (more examples in Supplementary Figure S3). The transient high FRET state has the same FRET level as the high FRET state found in Rpo41-Mtf1-DNA complex, suggesting that Rpo41 alone can bend the DNA to a similar geometry, but with a low frequency. It is not evident yet whether the DNA is also open in this short-lived bent conformation. Adding both Rpo41 and Mtf1 stabilizes the high FRET state, but we still see frequent fluctuations to the low FRET state (Figure 2A), which is consistent with the residual low FRET population seen at saturating protein concentration (Figure 1B). Based on the previous 2-aminopurine fluorescence studies that showed DNA melting with Rpo41-Mtf1 (19), we propose that the sharply bent DNA with both proteins is also melted, hence we refer to the high FRET state as bent/open and the low FRET state as unbent/closed. If so, the FRET dynamics observed would represent the promoter opening–closing transition of the pre-initiation complex which we further confirmed as follows.
Figure 2.
Opening–closing dynamics revealed in single molecule traces and regulation of the dynamics by Mtf1 and NTPs. (A) Representative single molecule traces for each condition. Rpo41 only: 10× higher concentration of Rpo41 (4 µM) than DNA (400 nM). Rpo41/Mtf1: same concentration as DNA. (B) Dwell time histograms of complexes with both Rpo41 and Mtf1 constructed from 484 molecules. (C) Comparison of the unbending/closing rate (high to low FRET) and the bending/opening rate (low to high FRET) between Rpo41 only and Rpo41/Mtf1. 255 molecules for Rpo41 only and 484 molecules for Rpo41/Mtf1 were used. Closing rate of Rpo41 only and opening rate of Rpo41/Mtf1 were obtained by fitting the dwell time distribution with single exponential excluding first two bins because there will be missed events. Opening rate of Rpo41 only and closing rate of Rpo41/Mtf1 were obtained by dividing total dwell time with total number of transitions because the dwell time is comparable to the photobleaching lifetime of the dyes, which will bias exponential fitting toward smaller time constant. (D) Change of closing and opening rates upon adding nucleotides. Number of traces analyzed to obtain each value was 484, 477, 500, and 554, from left to right. Same method of analysis was used as described in (C) for Rpo41/Mtf1. The measurements were repeated with two independent sample preparations and the rate constants showed < 10% of variation. (E) Proposed model to explain the suppression of closing upon nucleotide addition.
Two other possible origins for the observed FRET fluctuations were considered: (i) the instability of short DNA duplex and (ii) protein binding and dissociation. To rule out the effects of DNA instability, we designed a longer template (Supplementary Figure S4), −20 to +10 with the fluorophores at each end. The overall FRET efficiency was lower due to the longer D–A distance, but the complexes exhibited similar fluctuating behavior as seen in the example traces (Supplementary Figure S4). Transition rates in both directions are comparable to those of the 20 bp DNA. With Rpo41 alone, the 30 bp DNA also shows similar bending trials to those of the 20 bp DNA (Supplementary Figure S5). The unbending rate is comparable to that of the minimal template. The bending rate is higher for this longer template possibly due to multiple Rpo41 interacting with one DNA.
In order to test if the fluctuations are due to protein binding and dissociation, we measured the opening rate (low to high FRET) by exponential fitting of the low FRET dwell time at different protein concentrations (Supplementary Figure S6). If the transition to the low FRET state were due to protein dissociation, we would expect the opening rate to increase with increasing protein concentration, which was not observed. On the contrary, we observed a slight decrease of the opening rate at 10 times higher protein concentration, possibly due to multiple polymerases interacting with one template. More direct evidence was provided by washing away free proteins in the solution (Supplementary Figure S7). Using a DNA template that could be tethered to the surface, we were able to remove free proteins in solution. The remaining DNA/protein complexes still showed the same fluctuating behavior and their opening and closing rates were very close to those of vesicle-encapsulated complexes.
These results indicate that the dynamics is an inherent property of the promoter-protein complex. We assign the observed promoter dynamics to the opening and closing transitions of the transcription bubble based on the following observations: (i) The DNA fluctuates to the same low FRET level as bare duplex DNA. (ii) Addition of the initiating nucleotide decreases the transition rate to the low FRET state without affecting the transition rate to the high FRET state (see below). (iii) Higher temperature (from 22°C to 37°C) or lower salt concentrations (from 100 to 10 mM potassium phosphate or from 10 to 1 mM magnesium chloride), all of which would stabilize the melted DNA structure, resulted in 2–3 times decrease in the transition rate to the low FRET state (Supplementary Figure S8).
The dynamic behavior demonstrates the reversibility of the pre-initiation open complex (2,26,27) and the single molecule method allows us to determine the transition rates. We analyzed several hundred molecules to create dwell time distributions of the low and high FRET states. These distributions show close to exponential decay implying that there is a single rate-limiting step for each transition (Figure 2B). The DNA bending/opening rate, kopening, by Rpo41 alone was 0.0052 s−1 and this increased to 2.8 s−1 in the presence of Mtf1. The DNA unbending/closing rate, kclosing, was 3.0 s−1 with Rpo41 alone and it decreased to 0.066 s−1 in the presence of Mtf1. Therefore, compared to Rpo41 alone, the presence of both Rpo41 and Mtf1 increases kopening by ~500-fold and decreases the kclosing by ~45-fold. From the ratios of kopening and kclosing, we can determine that Mtf1 stabilizes the bent/open complex by ~23,000-fold, which is equivalent to ~10kT binding free energy contribution from Mtf1.
Initiating nucleotide slows down promoter closing
Next, we investigated how ATP, the initiating nucleotide for Rpo41 in yeast mitochondria, affects the dynamics of pre-initiation complex. In order to limit the number of nucleotides inserted, we used 3′-deoxy nucleotide analogs, which can bind at +1 and +2 templating positions, but cannot react to make the 2-mer RNA. The lipid vesicle made with DMPC is porous at room temperature and permeable to small molecules such as nucleotides (36). Addition of the incorrect nucleotide, 3′-dUTP, to the Rpo41-Mtf1-DNA did not change the transition rates. In contrast, addition of the correct nucleotide, 3′-dATP, at 100 µM reduced kclosing from 0.066 to 0.029 s−1, without affecting kopening (Figure 2A and D). 1 mM 3′-dATP resulted in a further decrease of kclosing to 0.016. Similar effects of 3′-dATP were also observed with the 30 bp template (Supplementary Figure S4).
The above results reveal the mechanism of how the correct initiating nucleotide can aid transcription initiation by extending the lifetime of the open complex so that the complex can proceed to the subsequent steps. The fact that the initial transcribing complex is still dynamic and the frequency of fluctuation in individual molecules depends on the nucleotide concentration implies that the initiating nucleotide leaves and re-enters the complex frequently. A simple model shows how the nucleotide insertion could be coupled with the promoter opening/closing dynamics (Figure 2E). Relative closing rate at a given nucleotide concentration is determined by the ratio of kin and kout, which are the rate constants of nucleotide’s entering and leaving the open complex.
Rpo41 alone forms a stable open complex on pre-melted promoter
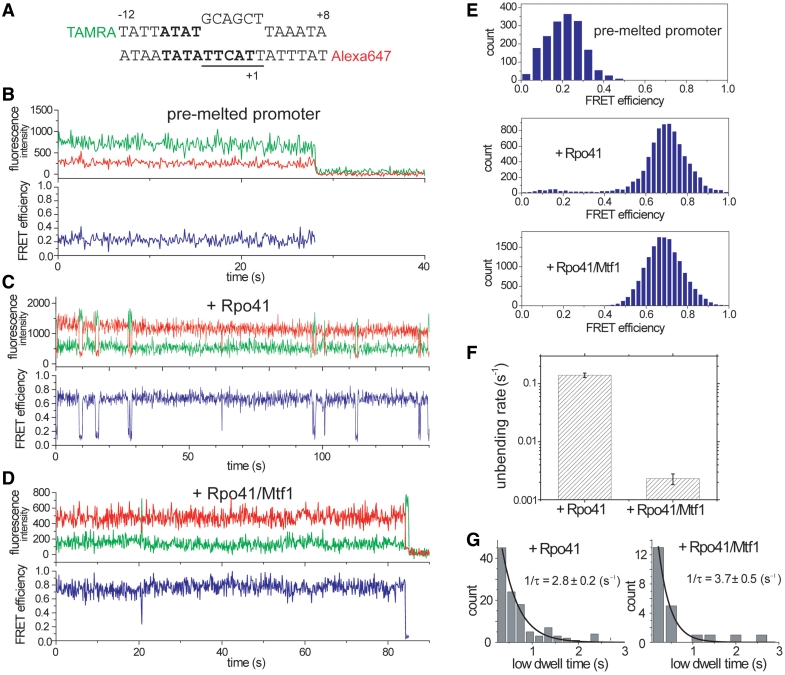
Rpo41 alone can initiate transcription efficiently on a pre-melted promoter prepared by introducing base changes in the non-template sequence around the initiation region (14). We designed a 20 bp pre-melted DNA (Figure 3A) with a mismatched sequence (−4 to +2) to examine the dynamics of this particular initiation complex. In contrast to the duplex promoter for which the bent/open state was rarely populated with Rpo41 alone, the pre-melted promoter showed an almost complete shift to the high FRET state with Rpo41 alone (Figure 3C and E). The pre-melted promoter bound to Rpo41 still made transient excursions to a low FRET state, which we interpret as unbending without Rpo41 dissociation. This low FRET state exhibits a FRET value (0.15) lower than that of the free DNA (0.23), which indicates that the fluctuating low FRET state has a DNA geometry different from that of the free pre-melted form. The unbending rate of the Rpo41-pre-melted DNA complex is 0.14 s−1 (Figure 3F), which is ~20 times lower than the kclosing of the Rpo41-duplex promoter complex, whereas its bending rate of 2.8 s−1 (Figure 3G) is ~500-fold higher than the kopening of the Rpo41-duplex promoter complex. In fact, the bending and unbending rates of the pre-melted promoter Rpo41 alone were close to kopening and kclosing of the duplex promoter with Rpo41 and Mtf1 (compare Figures 2C, 3F and G), suggesting that Mtf1 lowers the energy barrier against promoter bending/melting and thus disrupts the melting region (−4 to +2) of the duplex promoter.
Figure 3.
Bending-unbending dynamics of pre-melted promoter. (A) Design of the pre-melted template with mismatches at −4 to +2 positions. (B–D) Example traces of DNA only, DNA + Rpo41, and DNA + Rpo41/Mtf1, respectively. All samples were prepared as equimolar mixture and incubated at room temperature for 1 min before vesicle encapsulation. (E) smFRET histograms of (B–D). With Rpo41, small population exists at low FRET, which is much suppressed by the addition of Mtf1. (F) Comparing the unbending rate between Rpo41 and Rpo41/Mtf1 reveals ~60-fold difference. It is 0.14 ± 0.01 s−1 with Rpo41 alone and 0.0023 ± 0.0005 s−1 with both Rpo41 and Mtf1. (G) Bending rate can be determined from the dwell time distribution of the low FRET state. It does not show significant change upon Mtf1 addition.
When we added both Rpo41 and Mtf1 to the pre-melted DNA, the unbending rate decreased to 0.0023 s−1, which is ~60-fold lower compared to Rpo41 alone (Figure 3D and F). Therefore, Mtf1 plays an additional role of raising the barrier against promoter unbending. The bending rate, on the other hand, was not significantly influenced by Mtf1 (Figure 3G). Our data overall provide direct evidence that Rpo41 has the intrinsic ability to induce the bent/open conformation, but it is ineffective in maintaining the open state. Mtf1 facilitates promoter opening by both lowering the energy barrier against promoter opening and raising the barrier against promoter closing.
Instability of the initially open promoter
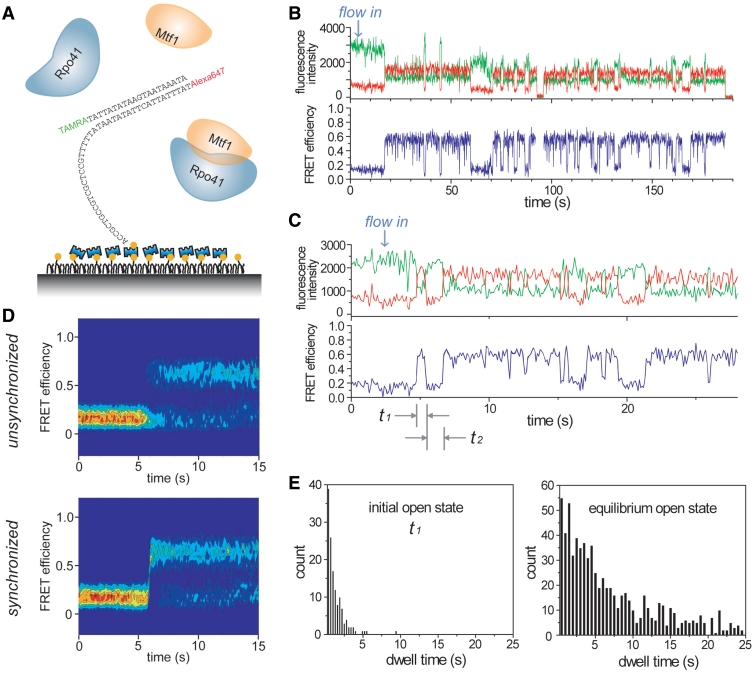
The dwell times of both the open and closed states follow an exponential distribution once an equilibrium is established (Figure 2B), indicating one major rate-liming step in each direction. However, multiple steps have been observed during the initial promoter opening process in other RNAPs (2,5–7). Thus, the initial promoter opening dynamics immediately after proteins bind the DNA may be different from that observed at equilibrium. To monitor the initial opening process, we designed another DNA construct with the same 20-mer sequence but with a 5′ single-stranded overhang in the template strand that can be specifically tethered to the surface (Figure 4A). By flowing in a mixture of Rpo41 and Mtf1 into the imaging chamber while recording the fluorescence signals, we could observe the initial promoter opening events in real time. Time traces showed a single-step increase of FRET followed by repeated opening-closing transitions (Figure 4B and C). Interestingly, the first high FRET state has a much shorter average lifetime (t1 in Figure 4C) than the average lifetime determined after the equilibrium is established (Figure 4E). More examples showing such transient melting trials are listed in Supplementary Figure S9.
Figure 4.
Instability of the initially open promoter. (A) Schematic of surface-tethered template shown with protein mixture being flown in. (B) Typical time trace showing successful promoter opening. (C) Typical time trace demonstrating transient opening trial in the beginning. (D) Time evolution of FRET distribution obtained from 177 molecules when 100 nM Rpo41/Mtf1 was flowed in. Lower plot was synchronized at the moment of FRET exceeding 0.45. (E) Lifetime distributions of the first open state, t1 in (C), and the equilibrium open state.
The above results indicate that Rpo41-Mtf1 forms an initial high FRET state that is different from the high FRET state visited at equilibrium. The initial high FRET state is less stable and collapses faster to the closed conformation. In order to identify the FRET level of this state, we constructed the time evolution of FRET density by overlapping the traces (Figure 4D). From the moment of protein addition, the high FRET population grows rapidly in the unsynchronized plot. As we synchronize the traces at the moment of FRET exceeding a threshold of 0.45, we observe a single-step increase of FRET but no additional FRET level for the initial open state distinct from that of the equilibrium open state. The distribution of the time gap between the first two opening events (t2 in Figure 4C) does not depend on the protein concentrations (Supplementary Figure S9), indicating that in most cases the proteins remain bound between opening events. These results indicate that the DNA can be readily bent upon protein binding, but the initial bent intermediate is less stable than the open state at equilibrium. Perhaps, Rpo41-Mtf1 is positioned incorrectly in the initial bent intermediate or the DNA is opened at wrong positions, which then corrects itself by rapid closing and reopening without dissociation. Thus, the initial transient opening trials may represent attempts to form the proper pre-initiation complex at the promoter site. Another possibility is that the short-lived high FRET state represents one of the intermediate states in the initiation pathway and it is reversible as suggested by multiple short-lived states in several examples shown in Supplementary Figure S9.
DISCUSSION
We have characterized the dynamics of pre-initiation and initially transcribing complexes of the yeast mitochondrial transcription machinery. This study enabled us to investigate the roles of the core subunit Rpo41 and the transcription factor Mtf1 in transcription initiation. Rpo41 alone does not catalyze promoter-specific transcription from duplex promoter and it is believed that this is because Rpo41 by itself cannot open the promoter. However, our smFRET time traces show that Rpo41 alone has the ability to at least bend and possibly open the promoter with detectable frequency. But, even if the promoter is open in this state, its lifetime is too short and transcription initiation by Rpo41 alone may not be kinetically feasible.
Upon addition of Mtf1, the bending transition becomes more frequent and the Rpo41-Mtf1-DNA complex stays in the high FRET state with a longer lifetime. The mechanism by which Mtf1 promotes open complex formation was not known. Our results clearly show that Mtf1 increases the DNA bending rate and decreases the unbending rate, which indicates that Mtf1 both facilitates promoter bending and stabilizes the bend complex. Recent studies have shown that Mtf1 contacts the promoter DNA bound to Rpo41 (18,20). Hence, Mtf1 may nucleate DNA bending/opening by direct interaction with the DNA, although it remains to be determined if Mtf1 makes sequence-specific contacts with the promoter DNA.
When the promoter is pre-melted, Rpo41 initiates transcription without Mtf1 (14). We have shown that Rpo41 forms the high FRET state on its own on the pre-melted promoter. This explains the promoter-specific transcription activity of Rpo41 on pre-melted promoter. Moreover, our results suggest that the structure of the open complex, especially the bending angle, is likely to be determined largely by the interaction of Rpo41 with DNA. When Mtf1 was added to the pre-initiation open complex of the pre-melted DNA, the unbending rate was lowered by ~60-fold compared to the normal duplex DNA. Thus, Mtf1 further stabilizes the bent conformation of the pre-melted promoter, which is consistent with the suggested role of Mtf1 as interacting with the melted DNA strand and trapping the complex in the bent/open state (18). We expect that the increased stability of the pre-melted promoter in the presence of Mtf1 may inhibit the release of Mtf1 that is required to make the transition from initiation to elongation (37). This is consistent with the observation that the synthesis of longer RNA products is inhibited on pre-melted promoter in the presence of Mtf1 (14).
The frequent FRET fluctuations due to DNA opening and closing imply the reversibility of the pre-initiation complex. Reversibility of the pre-initiation open complex has been postulated as a mechanism for correctly locating the promoter sequence in several different transcription machineries (2,26,27). When we measured promoter dynamics immediately after adding the proteins, we detected an initial unstable intermediate that underwent rapid closing transition. We propose that this initial bent intermediate could represent incorrect positioning of the promoter, which then corrects itself by rapid closing and reopening. Thus, the reversibility could be beneficial in searching for the correct promoter sequence and not transcribing at wrong positions. Only the DNA sequence that can form a stable open complex would be long-lived to recruit the initiating nucleotides before the DNA closes. This may work as a mechanism utilized by the transcription system in locating the promoter sites with minimal errors. Once the correct NTPs are bound, the fluctuation decreases and the complex will stay longer in the open transcribing state.
RNAP regulation by the initiating nucleotide had been shown for T7, bacterial, and mitochondrial RNAP (26,28–31). The +1 site of mitochondrial promoter is highly conserved for ATP addition (38) and the Km for this initial nucleotide is higher than the subsequent nucleotides by an order of magnitude, leading to the proposal of an ATP sensing mechanism inherent in the transcription machinery of yeast mitochondria (31). Our observation that the correct initiating nucleotide decreases the unbending/closing transition presents a possible structural mechanism for ATP sensing: the inserted nucleotide interferes with the DNA duplex re-zipping or locks the complex in the open conformation. Then, this significantly raises the chance of transcription initiation by suppressing the promoter closing and the dissociation of Rpo41-Mtf1 from the promoter. In our measurements, the initiating ATP did not affect the bending/opening rate, but only decreased the unbending/closing rate in a concentration-dependent manner, suggesting that ATP can enter the complex only in the open state and the complex is able to close only when the ATP leaves it. Based on these observed kinetics of the structural transitions, our results provide an explanation for the ATP sensing mechanism.
It would be interesting to investigate whether the dynamics of pre-initiation open complex and its regulation by initiating nucleotides also exist in the transcription machinery of the mammalian mitochondria. In the early model, specific transcription initiation in human and other mammalian mitochondria was believed to require the coordinated actions of POLRMT, TFB2M, and a High-Mobility-Group (HMG) box protein, TFAM, which is different from the two-component transcription system of the yeast mitochondria (39–42). With its ability of binding and bending a DNA region of 14–35 bp upstream of promoters LSP and HSP1, TFAM was proposed to assist in the recruitment of POLRMT-TFB2M to the promoter and the initial melting of the promoter (43). Its yeast analog, Abf2, is not required for yeast mitochondrial transcription and instead functions in genome packaging and maintenance (44,45). A most recent study, however, strongly argued against TFAM as a requisite of the human transcription machinery at least for in vitro transcription initiation on either LSP or HSP1 promoters (17). In such a new two-component human mitochondrial transcription complex, POLRMT and TFB2M could act as a heterodimer like the Rpo41-Mtf1 complex by working coordinately to recognize the promoter sequence and to form a dynamic pre-initiation open complex that is responsive to initiating nucleotides. Further investigations are required to address these questions.
We have demonstrated smFRET as a powerful tool to directly observe the dynamics of transcription initiation in the two-component yeast mitochondrial transcription system. The method can be applied to further understand how the core RNAP and transcription factor select the specific promoter site by studying the DNA dynamics on promoter variants. Our study may provide a framework to investigate more complicated protein–DNA dynamics in multi-subunit transcription systems. For example, it can be extended to explore the archaeal and eukaryotic transcription systems to observe how TFE and TFIIE modulate the conformational dynamics of the pre-initiation complex.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (GM51966 to S.S.P., GM065367 to T.H.); National Science Foundation (0822613 to T.H.); Korean Research Foundation (KRF-2006-352-C00019 to H.K.). Funding for open access charge: Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Swaroopa Paratkar for providing purified Rpo41 and Mtf1 proteins.
REFERENCES
- 1.DeHaseth PL, Zupancic ML, Record MT., Jr RNA polymerase-promoter interactions: the comings and goings of RNA polymerase. J. Bacteriol. 1998;180:3019–3025. doi: 10.1128/jb.180.12.3019-3025.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bandwar RP, Patel SS. Peculiar 2-aminopurine fluorescence monitors the dynamics of open complex formation by bacteriophage T7 RNA polymerase. J. Biol. Chem. 2001;276:14075–14082. doi: 10.1074/jbc.M011289200. [DOI] [PubMed] [Google Scholar]
- 3.DeHaseth PL, Helmann JD. Open complex formation by Escherichia coli RNA polymerase: the mechanism of polymerase-induced strand separation of double helical DNA. Mol. Microbiol. 1995;16:817–824. doi: 10.1111/j.1365-2958.1995.tb02309.x. [DOI] [PubMed] [Google Scholar]
- 4.Roy S, Lim HM, Liu M, Adhya S. Asynchronous basepair openings in transcription initiation: CRP enhances the rate-limiting step. EMBO J. 2004;23:869–875. doi: 10.1038/sj.emboj.7600098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wellington SR, Spiegelman GB. The kinetics of formation of complexes between Escherichia coli RNA polymerase and the rrnB P1 and P2 promoters of Bacillus subtilis. Effects of guanosine tetraphosphate on select steps of transcription initiation. J. Biol. Chem. 1993;268:7205–7214. [PubMed] [Google Scholar]
- 6.Holstege FC, van der Vliet PC, Timmers HT. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMBO J. 1996;15:1666–1677. [PMC free article] [PubMed] [Google Scholar]
- 7.Record MT, Reznikoff WS, Craig ML, McQuade KL, Schlax PJ. In: Escherichia coli and Salmonella Cellular and Molecular Biology. Neidhardt FC, Curtiss R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Washington, DC: ASM Press; 1996. pp. 792–821. [Google Scholar]
- 8.Masters BS, Stohl LL, Clayton DA. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987;51:89–99. doi: 10.1016/0092-8674(87)90013-4. [DOI] [PubMed] [Google Scholar]
- 9.Gaspari M, Larsson NG, Gustafsson CM. The transcription machinery in mammalian mitochondria. Biochim. Biophys. Acta. 2004;1659:148–152. doi: 10.1016/j.bbabio.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Cermakian N, Ikeda TM, Cedergren R, Gray MW. Sequences homologous to yeast mitochondrial and bacteriophage T3 and T7 RNA polymerases are widespread throughout the eukaryotic lineage. Nucleic Acids Res. 1996;24:648–654. doi: 10.1093/nar/24.4.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schinkel AH, Koerkamp MJ, Touw EP, Tabak HF. Specificity factor of yeast mitochondrial RNA polymerase. Purification and interaction with core RNA polymerase. J. Biol. Chem. 1987;262:12785–12791. [PubMed] [Google Scholar]
- 12.Fisher RP, Clayton DA. Purification and characterization of human mitochondrial transcription factor 1. Mol. Cell. Biol. 1988;8:3496–3509. doi: 10.1128/mcb.8.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tracy RL, Stern DB. Mitochondrial transcription initiation: promoter structures and RNA polymerases. Curr. Genet. 1995;28:205–216. doi: 10.1007/BF00309779. [DOI] [PubMed] [Google Scholar]
- 14.Matsunaga M, Jaehning JA. Intrinsic promoter recognition by a “core” RNA polymerase. J. Biol. Chem. 2004;279:44239–44242. doi: 10.1074/jbc.C400384200. [DOI] [PubMed] [Google Scholar]
- 15.Jang SH, Jaehning JA. The yeast mitochondrial RNA polymerase specificity factor, MTF1, is similar to bacterial sigma factors. J. Biol. Chem. 1991;266:22671–22677. [PubMed] [Google Scholar]
- 16.Winkley CS, Keller MJ, Jaehning JA. A multicomponent mitochondrial RNA polymerase from Saccharomyces cerevisiae. J. Biol. Chem. 1985;260:14214–14223. [PubMed] [Google Scholar]
- 17.Shutt TE, Lodeiro MF, Cotney J, Cameron CE, Shadel GS. Core human mitochondrial transcription apparatus is a regulated two-component system in vitro. Proc. Natl Acad. Sci. USA. 107:12133–12138. doi: 10.1073/pnas.0910581107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paratkar S, Patel SS. Mitochondrial transcription factor Mtf1 traps the unwound non-template strand to facilitate open complex formation. J. Biol. Chem. 2010;285:3949–3956. doi: 10.1074/jbc.M109.050732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang G-Q, Paratkar S, Patel SS. Fluorescence mapping of the open complex of yeast mitochondrial RNA polymerase. J. Biol. Chem. 2009;284:5514–5522. doi: 10.1074/jbc.M807880200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savkina M, Temiakov D, McAllister WT, Anikin M. Multiple functions of yeast mitochondrial transcription factor Mtf1p during initiation. J. Biol. Chem. 2010;285:3957–3964. doi: 10.1074/jbc.M109.051003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grünberg S, Bartlett MS, Naji S, Thomm M. Transcription factor E is a part of transcription elongation complexes. J. Biol. Chem. 2007;282:35482–35490. doi: 10.1074/jbc.M707371200. [DOI] [PubMed] [Google Scholar]
- 22.Grohmann D. Molecular mechanisms of archaeal RNA polymerase. Biochem. Soc. Trans. 2009;37:12. doi: 10.1042/BST0370012. [DOI] [PubMed] [Google Scholar]
- 23.Werner F, Weinzierl ROJ. Direct modulation of RNA polymerase core functions by basal transcription factors. Mol. Cell. Biol. 2005;25:8344–8355. doi: 10.1128/MCB.25.18.8344-8355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen H-T, Warfield L, Hahn S. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat. Struct. Mol. Biol. 2007;14:696–703. doi: 10.1038/nsmb1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forget D, Langelier M-F, Therien C, Trinh V, Coulombe B. Photo-cross-linking of a purified preinitiation complex reveals central roles for the RNA polymerase II mobile clamp and TFIIE in initiation mechanisms. Mol. Cell. Biol. 2004;24:1122–1131. doi: 10.1128/MCB.24.3.1122-1131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Villemain J, Guajardo R, Sousa R. Role of open complex instability in kinetic promoter selection by bacteriophage T7 RNA polymerase. J. Mol. Biol. 1997;273:958–977. doi: 10.1006/jmbi.1997.1358. [DOI] [PubMed] [Google Scholar]
- 27.Gourse RL. Visualization and quantitative analysis of complex formation between E.coli RNA polymerase and an rRNA promoter in vitro. Nucleic Acids Res. 1988;16:9789–9809. doi: 10.1093/nar/16.20.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stano NM, Levin MK, Patel SS. The +2 NTP binding drives open complex formation in T7 RNA polymerase. J. Biol. Chem. 2002;277:37292–37300. doi: 10.1074/jbc.M201600200. [DOI] [PubMed] [Google Scholar]
- 29.Gaal T, Bartlett MS, Ross W, Turnbough CL, Gourse RL. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 30.Amiott EA, Jaehning JA. Sensitivity of the yeast mitochondrial RNA polymerase to +1 and +2 initiating nucleotides. J. Biol. Chem. 2006;281:34982–34988. doi: 10.1074/jbc.M608638200. [DOI] [PubMed] [Google Scholar]
- 31.Amiott EA, Jaehning JA. Mitochondrial transcription is regulated via an ATP “sensing” mechanism that couples RNA abundance to respiration. Mol. Cell. 2006;22:329–338. doi: 10.1016/j.molcel.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 32.Ha T, Enderle T, Ogletree DF, Chemla DS, Selvin PR, Weiss S. Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl Acad. Sci. USA. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kapanidis AN, Margeat E, Ho SO, Kortkhonjia E, Weiss S, Ebright RH. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science. 2006;314:1144–1147. doi: 10.1126/science.1131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang G-Q, Roy R, Bandwar RP, Ha T, Patel SS. Real-time observation of the transition from transcription initiation to elongation of the RNA polymerase. Proc. Natl Acad. Sci. USA. 2009;106:22175–22180. doi: 10.1073/pnas.0906979106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang G-Q, Roy R, Ha T, Patel SS. Transcription initiation in a single-subunit RNA polymerase proceeds through DNA scrunching and rotation of the N-terminal subdomains. Mol. Cell. 2008;30:567–577. doi: 10.1016/j.molcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cisse I, Okumus B, Joo C, Ha T. Fueling protein? DNA interactions inside porous nanocontainers. Proc. Natl Acad. Sci. USA. 2007;104:12646–12650. doi: 10.1073/pnas.0610673104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mangus DA, Jang SH, Jaehning JA. Release of the yeast mitochondrial RNA polymerase specificity factor from transcription complexes. J. Biol. Chem. 1994;269:26568–26574. [PubMed] [Google Scholar]
- 38.Biswas TK, Edwards JC, Rabinowitz M, Getz GS. Characterization of a yeast mitochondrial promoter by deletion mutagenesis. Proc. Natl Acad. Sci. USA. 1985;82:1954–1958. doi: 10.1073/pnas.82.7.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaspari M, Falkenberg M, Larsson N-G, Gustafsson CM. The mitochondrial RNA polymerase contributes critically to promoter specificity in mammalian cells. EMBO J. 2004;23:4606–4614. doi: 10.1038/sj.emboj.7600465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol. Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 41.Sologub M, Litonin D, Anikin M, Mustaev A, Temiakov D. TFB2 Is a transient component of the catalytic site of the human mitochondrial RNA polymerase. Cell. 2009;139:934–944. doi: 10.1016/j.cell.2009.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Litonin D, Sologub M, Shi Y, Savkina M, Anikin M, Falkenberg M, Gustafsson CM, Temiakov D. Human mitochondrial transcription revisited. J. Biol. Chem. 285:18129–18133. doi: 10.1074/jbc.C110.128918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dairaghi DJ, Shadel GS, Clayton DA. Human mitochondrial transcription factor A and promoter spacing integrity are required for transcription initiation. Biochim. Biophys. Acta. 1995;1271:127–134. doi: 10.1016/0925-4439(95)00019-z. [DOI] [PubMed] [Google Scholar]
- 44.Diffley JF, Stillman B. A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc. Natl Acad. Sci. USA. 1991;88:7864–7868. doi: 10.1073/pnas.88.17.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dairaghi DJ, Shadel GS, Clayton DA. Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J. Mol. Biol. 1995;249:11–28. doi: 10.1006/jmbi.1995.9889. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.