Abstract
Mgs1, the budding yeast homolog of mammalian Werner helicase-interacting protein 1 (WRNIP1/WHIP), contributes to genome stability during undisturbed replication and in response to DNA damage. A ubiquitin-binding zinc finger (UBZ) domain directs human WRNIP1 to nuclear foci, but the functional significance of its presence and the relevant ubiquitylation targets that this domain recognizes have remained unknown. Here, we provide a mechanistic basis for the ubiquitin-binding properties of the protein. We show that in yeast an analogous domain exclusively mediates the damage-related activities of Mgs1. By means of preferential physical interactions with the ubiquitylated forms of the replicative sliding clamp, proliferating cell nuclear antigen (PCNA), the UBZ domain facilitates recruitment of Mgs1 to sites of replication stress. Mgs1 appears to interfere with the function of polymerase δ, consistent with our observation that Mgs1 inhibits the interaction between the polymerase and PCNA. Our identification of Mgs1 as a UBZ-dependent downstream effector of ubiquitylated PCNA suggests an explanation for the ambivalent role of the protein in damage processing.
INTRODUCTION
Post-translational modification of a protein is an effective strategy to modulate its activity or its interactions with other cellular factors that selectively recognize the protein in its modified form. Ubiquitin is a particularly versatile modifier, due to its ability to form polymeric chains that can serve as a signal not only for proteasome-mediated degradation, but also for a variety of non-proteolytic events (1). Dedicated ubiquitin-binding domains, which differ in their selectivity for monoubiquitin or particular chain geometries, usually mediate the recognition of ubiquitylated target proteins by downstream effectors (2).
One of the areas of cellular metabolism that is heavily influenced by the ubiquitin system is the maintenance of genome stability (3,4). Several pathways for the recognition, repair and bypass of DNA damage are controlled or modulated by the ubiquitylation of key components, and ubiquitin-binding domains have been identified within relevant interaction partners (5). The RAD6 pathway, an ensemble of ubiquitin conjugation enzymes (E2) and ligases (E3), controls the replicative bypass of DNA lesions through modification of the eukaryotic sliding clamp, proliferating cell nuclear antigen (PCNA) (in yeast encoded by POL30). In response to DNA damage or replication stress, PCNA is mono- and polyubiquitylated at a highly conserved lysine (K) residue, K164 (6), at sites where replication problems have caused the accumulation of single-stranded (ss) DNA (7–9). Monoubiquitylation is achieved by the E2 Rad6 in cooperation with the E3 Rad18, and polyubiquitylation additionally involves the dimeric E2 complex Ubc13-Mms2 and the E3 Rad5 (6,10). While monoubiquitylation of PCNA activates a series of damage tolerant, highly mutagenic DNA polymerases for bypass via translesion synthesis (TLS) (11–13), polyubiquitylation is required for an error-free damage avoidance pathway that probably involves template switching to the undamaged sister chromatid (6,14). TLS polymerases of the Y family, polymerases η, κ, ι and Rev1, harbour ubiquitin-binding domains of the UBZ or UBM type, which mediate the recruitment to ubiquitylated PCNA at sites of replication problems (15,16). Ubiquitin-binding effector protein(s) that might recognize polyubiquitylated PCNA and thereby mediate error-free damage bypass are as yet unknown.
The RAD6 pathway operates in the context of other systems for genome maintenance (17). One of these is represented by Mgs1, a DNA-dependent ATPase of the AAA+ class (18). MGS1 from Saccharomyces cerevisiae was first identified as a gene contributing to the maintenance of genome stability both during undisturbed replication and in response to DNA damage (18), whereas its mammalian homolog, WRNIP1/WHIP, was independently discovered as an interactor of the Werner helicase, WRN (19). Gene dosage appears to be critical for MGS1 function, as both deletion and overexpression affect recombination and mutation rates, and overexpression—but not deletion—sensitizes cells to DNA-damaging agents or replication problems (18,20). Genetic and physical interactions have been observed with the polymerase δ–PCNA complex in yeast (20–22), and mammalian WRNIP1 was found to accumulate in PML bodies and RPA-associated nuclear foci (23). Although no direct effect of WRNIP1 on the WRN pathway was detected in mammalian cells (24), yeast mgs1 mutants exhibit a synthetic growth defect in combination with mutants of the WRN homolog SGS1 (25). An impact on the RAD6 pathway was postulated based on physical interactions between Mgs1 and PCNA and a synthetic lethality between mgs1 and rad18 mutants that could be suppressed by enhancing homologous recombination (21,26).
Recently, a ubiquitin-binding zinc finger (UBZ), related to those present in polymerases η and κ and in Rad18, was identified in mammalian WRNIP1 (23,27). We have now investigated the contributions of this domain to Mgs1 activity in yeast and find that UBZ-mediated binding of the protein to ubiquitylated PCNA specifically mediates the DNA damage-related functions of Mgs1. Physical interactions with ubiquitylated PCNA and a modulatory influence on the function of polymerase δ in ubiquitin-dependent DNA damage bypass implicate Mgs1 as a downstream effector of the RAD6 pathway and provide a functional link between an important ubiquitylation target and a downstream effector that recognizes the target in its modified form.
MATERIALS AND METHODS
Construction of yeast strains
Standard procedures were followed for growth and manipulation of Saccharomyces cerevisiae. All experiments were performed with sets of isogenic strains, which were derivatives of either DF5 (his1-1, leu2-3, 2-112, lys2-801, trp1-1, ura3-52) or W303 (ade2-1, ura3-1, his3-11, 15, trp1-1, leu2-3,112, can1-100), except for the isogenic series of strains used for analysis of the pol31 (hys2-1) mutant, a gift from D. Branzei (20) and originally isolated by K. Sugimoto (ade1, his2, his3-Δ200, trp1, ura3, leu2, hys2-1) and the strain used for two-hybrid assays, PJ69-4A (trp1-901, leu2-3,112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1-HIS3, GAL2-ADE2, met2::GAL7-lacZ). Mutants rad18, ubc13, pol30(K164R) and tlsΔ have been described (12,28–30). Deletion of MGS1, POL32 and SGS1 was accomplished by replacement with a PCR-generated cassette bearing the URA3, HisMX, KanMX or HphNT1 marker (31–33), and combinations of mutants were obtained by mating and tetrad dissection or by successive gene deletions. For analysis of spontaneous homologous recombination rates, diploid strains heterozygous for the HIS1 gene (his1-1/his1-7) were generated (34). The dna2Δ405N mutation (35) was obtained by replacement of the open reading frame with the N-terminally truncated allele in the genomic locus. Strains pol3ΔCt (L1094stop) and pol2-11 (S2193stop) (36) were constructed by introducing a PCR-generated cassette appending a STOP codon followed by the KanMX marker at the desired position. The pol30-79 mutant was obtained by transformation of WT cells with the plasmid pBL230-79, a gift from P. Burgers, and subsequent deletion of the endogenous POL30 as described (37). Mutant pol32ΔPIP (38) was generated by integration of the truncated allele, pol32(1-337), on a plasmid under control of its own promoter, into a pol32 deletion. A HisPOL30 derivative in pol32 was constructed by replacement of the POL30 open reading frame with the His-tagged allele, while in the pol31, pol2-11 and pol3ΔCt backgrounds the tagged allele was introduced on an integrative plasmid in addition to the endogenous POL30. Alleles of MGS1 tagged with the 9myc-epitope were generated by appending a PCR-generated cassette to the open reading frame. For genetic analysis of complementation, relevant MGS1 alleles were introduced into mgs1 strains on integrative vectors under control of the MGS1 promoter. For overexpression, integrative vectors with the GAL1 promoter or episomal vectors with the MGS1 promoter were used as indicated.
Construction of plasmids
Constructs encoding MGS1 and its mutants were generated in YIplac128, YIplac204, YEplac195 (39) or pRS424 (40) with the MGS1, GAL1 or ADH1 promoters, and the empty vectors were used as controls. Two-hybrid constructs were generated in pGAD424 or pGBT9 (Clontech) or derivatives thereof (41). Constructs for the UBZ domain only spanned amino acids 1–47. The series of plasmids encoding the linear ubiquitin–PCNA fusions and the Ub*-GFP* control have been described (30,42). Briefly, the Ub*n constructs are head-to-tail fusions of ubiquitin moieties carrying the mutations K29R, K48R, K63R and G76V, while the Ub*n(L) series incorporates a four-amino acid linker (VQIQ) between the ubiquitin moieties. The POL30 open reading frame in the respective PCNA* fusions bears the K127/164R mutations. Constructs Smt3*–PCNA* and PCNA*–Smt3* were created analogously. When used as C-terminal fusion partners, both Ub* and Smt3* moieties carried deletions of the two C-terminal glycine residues to prevent conjugation. For in vitro analysis, the Ub*n(L)–PCNA* fusions were subcloned into pET28c (Novagen), incorporating an N-terminal His6-tag, except for Ub*–PCNA* itself, which was in pQE-32 (Qiagen). Constructs for expression of GST-tagged proteins in Escherichia coli were created in pGEX-4T-1 (GE Healthcare).
Genetic assays in yeast
For analyses involving Gal-MGS1 overexpression, cells were pre-grown overnight at 30°C in glycerol medium, diluted into galactose or glucose medium and grown to exponential phase. Life span assays were performed microscopically by following the proliferation of 50 individual newly budded daughter cells per strain and repeatedly dissecting away their daughter (43). Spontaneous heteroallelic homologous recombination rates in the HIS1 locus (20) were measured by fluctuation analysis in diploid mgs1/mgs1 cells complemented with two copies of MGS1 or MGS1* as indicated. Briefly, 11 cultures of the respective diploids were grown in rich medium to a density of ~107 cells/ml, and aliquots were plated onto rich medium to determine total cell numbers and histidine-free medium to determine the number of recombinants. Rates and standard deviations were calculated according to the method of the median (44). Damage-induced mutation frequencies were measured in the CAN1 locus by incubating the relevant strains in 0.1% methyl methanesulfonate (MMS) in either glucose or galactose medium for 0–45 min, inactivation of the drug by dilution into 10% sodium thiosulfate and plating onto medium with and without 30 µg/ml canavanine to determine the ratio of canR cells to survivors. Each value was averaged from three independent cultures. Sensitivities to MMS were compared by spotting series of 10-fold dilutions of isogenic strains onto plates containing defined concentrations of the drug and recording growth after incubation for 3 days at 30°C (or 25°C for temperature-sensitive mutants) (12). Temperature sensitivities were assessed similarly by incubating the plates at differerent temperatures, or by following the optical density (600 nm) of exponentially growing liquid cultures.
Detection of PCNA modifications in vivo
HisPOL30 cultures were treated with 0.02% MMS for 60 min, and ubiquitylation of PCNA was detected by denaturing Ni–NTA affinity chromatography and western blot with PCNA- and ubiquitin-specific antibodies as described (7,28).
Preparation of recombinant proteins
Production of recombinant proteins was induced in E. coli BL21 by addition of 0.1 mM IPTG and incubation for 6–8 h at 37°C. GST- and His6-tagged proteins were purified by affinity chromatography on glutathione Sepharose or Ni–NTA resin, respectively, according to the manufacturer's instructions. Purification of untagged PCNA and the components for its in vitro ubiquitylation have been described (10). Human E1 was purchased from Biomol, and ubiquitin and its derivatives were from Boston Biochem.
Protein–protein interaction assays
Two-hybrid assays for detection of in vivo protein–protein interactions were performed in the reporter strain PJ69-4A as described (41), using fusions to the Gal4 activation (AD) and DNA-binding (BD) domains. For three-hybrid experiments, MGS1 and MGS1* were overexpressed under control of the ADH1 promoter from an episomal vector bearing the URA3 marker. In vitro interaction assays were performed in phosphate-buffered saline with 0.05% Triton X-100 by immobilizing the relevant GST-tagged protein on glutathione Sepharose for 1 h at 4°C, addition of the respective binding partner, further incubation for 2 h at 4°C, washing four times with the same buffer and eluting bound material by boiling in SDS sample buffer before analysing the samples by SDS gel electrophoresis and western blotting.
In vitro PCNA ubiquitylation
PCNA mono- and polyubiquitylation reactions were set up in the presence of primed, RPA-coated ΦX174 ssDNA as described previously (10). Control reactions contained PCNA, DNA, RPA and ATP and were incubated in the presence or absence of the clamp loader, RFC, but without ubiquitin conjugation factors. All reactions were treated with Benzonase (Novagen) to degrade the DNA before using them for GST pull-down assays.
Chromatin immunoprecipitations
Chromatin immunoprecipitation (ChIP) assays at an early-firing replication origin (ARS607) were performed in cells treated with 100 mM HU as described previously (28). The late origin ARS501 served as negative control. ChIP was performed from the same cultures with antibodies against PCNA and myc, and the signal relative to mock samples prepared without antibody was quantified by real-time PCR. Signals were averaged from two to three independent experiments.
RESULTS
DNA damage-related functions of Mgs1 require the UBZ domain
Deletion of MGS1, but also its overexpression, causes genome instability in budding yeast (18,20), and while some of the phenotypes associated with non-physiological protein levels are apparent in undamaged cells, others become evident only after treatment with DNA-damaging agents. In order to dissect the relevance of ubiquitin binding for Mgs1 function, we systematically analysed the contributions of its UBZ domain to a range of reported phenotypes. A UBZ mutant (designated MGS1*) was generated by changing an invariant aspartate residue, D31, to alanine. Analogous mutations were previously shown to abolish mono- and polyubiquitin binding in human WRNIP1 and human and yeast polymerase η (15,23,27,42). An mgs1 deletion strain was complemented with wild-type (WT) MGS1 or MGS1* at physiological levels in order to examine loss-of-function phenotypes, and MGS1 or MGS1* expressed under control of a galactose-inducible promoter or from a multicopy vector were used to assess the consequences of overexpression.
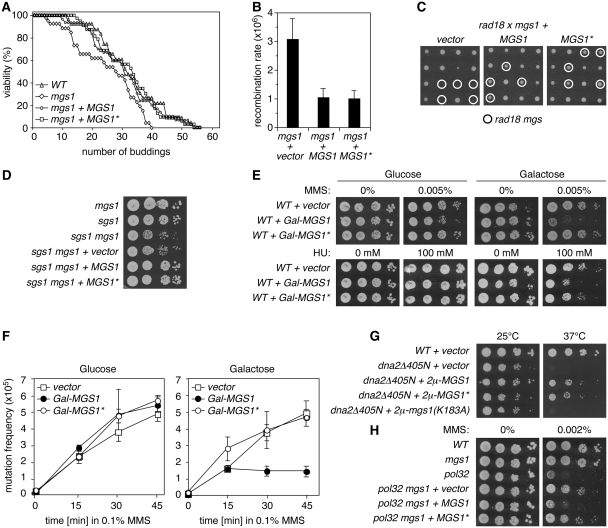
Inactivation of MGS1 causes a moderate reduction in life span (19). While we were able to demonstrate this ageing phenotype for the mgs1 deletion, mutation of the UBZ domain had no adverse effect (Figure 1A). Likewise, an elevated rate of spontaneous heteroallelic interchromosomal recombination, observable in diploid mgs1/mgs1 mutants (18,20), was not detectable in an MGS1*/MGS1* strain (Figure 1B). Finally, the MGS1* allele was as efficient as WT MGS1 in rescuing the synthetic lethality of a rad18 mgs1 double mutant (26) (Figure 1C) and the slight synthetic growth defect of an sgs1 mgs1 strain (25) (Figure 1D), indicating that the UBZ domain does not contribute to either of these aspects of MGS1 function.
Figure 1.
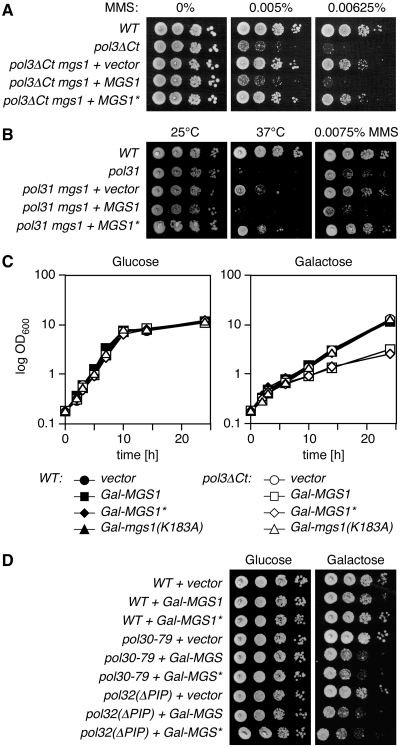
The UBZ domain is required specifically for the damage-related activities of Mgs1. (A) The UBZ domain is dispensable for longevity. Life span analysis of WT cells in comparison with mgs1 deletion mutants alone or complemented with either MGS1 or the UBZ mutant (D31A) allele, MGS1*, on an integrative plasmid under control of its own promoter. (B) The UBZ domain does not influence spontaneous recombination. Rates of heteroallelic recombination in the HIS1 locus (his1-1/his1-7) in diploid mgs1/mgs1 cells complemented with two copies of MGS1 or MGS1* as indicated. The empty vector served as control. Error bars indicate standard deviations. (C) The UBZ domain is irrelevant for the synthetic lethality of rad18 mgs1 mutants. Shown are progeny of genetic crosses between rad18 and mgs1 mutants, each carrying an empty vector, MGS1 or MGS1* as indicated. Tetrads are aligned in perpendicular orientation. (D) The UBZ domain is irrelevant for the synthetic growth defect of sgs1 mgs1 mutants. Growth of the indicated strains on rich medium was monitored by spot assays. (E) Overexpression of MGS1 sensitizes cells to DNA damage and replication problems in a UBZ-dependent manner. Spot assays for sensitivity to MMS and hydroxyurea (HU) at the indicated concentrations were performed with WT cells carrying vectors for overexpression of MGS1 alleles under control of a galactose-inducible promoter (Gal-MGS1 and Gal-MGS1*). The empty vector served as control. (F) Overexpression of MGS1 suppresses DNA damage-induced mutagenesis in a UBZ-dependent manner. Frequencies of MMS-induced mutations in the CAN1 locus were determined for WT strains harbouring Gal-MGS1 or Gal-MGS1*. The empty vector served as control. Error bars represent standard deviations from three independent measurements. (G) Suppression of the temperature sensitivity of dna2Δ405N by overexpression of MGS1 requires catalytic activity, but not the UBZ domain. Overexpression was achieved by means of episomal (2 µ) vectors with the MGS1 promoter. Growth at 25°C and 37°C was monitored by spot assays. (H) Inactivation of the Mgs1 UBZ domain is sufficient to suppress the damage sensitivity of pol32 mutants. Sensitivity of the indicated strains to MMS was assessed by spot assays.
Deletion of MGS1 does not result in a measurable DNA damage sensitivity, but overexpression sensitizes cells towards agents like ultraviolet (UV) radiation, MMS or hydroxyurea (HU) and suppresses damage-induced mutagenesis (18,20). We found that, in contrast to WT MGS1, overexpression of MGS1* resulted in neither increased sensitivity to MMS or HU nor suppression of induced mutagenesis after MMS treatment (Figure 1E and F). Protein levels of WT and mutant Mgs1 were indistinguishable (Supplementary Figure S1), indicating that a lack of protein stability was not likely responsible for the difference between the two alleles. Moreover, the notion that the ATPase activity of Mgs1 is required for mediating the effects caused by overexpression (18,20) suggested that the negative consequences for damage resistance and induced mutagenesis were due to a specific activity of Mgs1 and not simply to abnormally high protein levels.
These results indicated that ubiquitin binding is involved in some, but not all activities of Mgs1, but they did not differentiate whether the UBZ domain specifically affects damage-related functions or rather those that require overexpression. This issue was addressed by examining additional phenotypes that would uncouple the two effects. On one hand, MGS1 overexpression has previously been shown to suppress the temperature sensitivity of a truncation mutant of DNA2, a nuclease/helicase involved in Okazaki fragment processing (dna2Δ405N), in a damage-independent fashion (35). Figure 1G shows that MGS1*, in contrast to the catalytically inactive mutant mgs1(K183A), was functional in this assay. On the other hand, deletion of MGS1 is known to suppress the DNA damage sensitivity of strains lacking the non-essential subunit of polymerase δ, Pol32 (20). In this assay, inactivation of the UBZ domain had the same effect as an mgs1 deletion (Figure 1H). Hence, the conditions under which MGS1* exhibits defects correlate with the presence of exogenous DNA damage, but not with overexpression. Taken together, our data therefore support the idea that ubiquitin binding is important for those, and only those activities of Mgs1 related to the presence of exogenous DNA damage.
DNA damage-related functions of Mgs1 require PCNA ubiquitylation
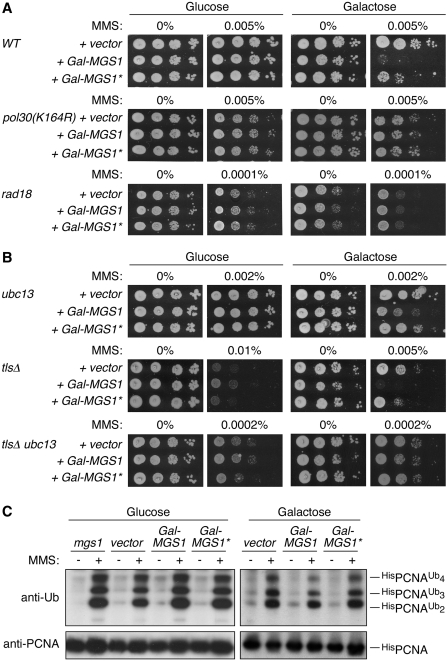
Given that physical interactions between Mgs1 and PCNA have been reported (21), and PCNA is ubiquitylated in response to DNA damage, we asked whether the remarkable damage dependence of the UBZ-associated Mgs1 activities was related to PCNA modification. To address this question, we examined the effect of MGS1 or MGS1* overexpression in mutants defective in various aspects of the RAD6 pathway. In pol30(K164R) and rad18 mutants, which are both completely devoid of damage-dependent PCNA ubiquitylation at K164 (6), the sensitization to MMS by overexpression of MGS1 was no longer observed, indicating that PCNA is indeed the ubiquitylation target relevant for the UBZ-dependent functions of Mgs1 (Figure 2A). Neither selective abolishment of polyubiquitylation (by deletion of UBC13) nor inactivation of all three TLS polymerases (rad30 rev1 rev3, designated as tlsΔ) was sufficient to abrogate the sensitizing effect of MGS1 overexpression, but the simultaneous inactivation of UBC13 and the TLS polymerases resulted in a situation comparable to the rad18 deletion (Figure 2B). Hence, the negative action of MGS1 relies on a combined effect on both branches of the RAD6 pathway.
Figure 2.
Ubiquitylation of PCNA is required for the UBZ-dependent activity of Mgs1. (A) UBZ-dependent sensitization to DNA-damaging agents by MGS1 overexpression requires PCNA ubiquitylation. MMS sensitivity assays were performed with the indicated strains harbouring Gal-MGS1 or Gal-MGS1*, as described in Figure 1E. The empty vector served as control. (B) The effect of MGS1 overexpression on DNA damage sensitivity is mediated by a combination of TLS and error-free damage bypass. MMS sensitivities upon MGS1 or MGS1* overexpression were assayed as in A in strains lacking the capacity for either error-free damage bypass mediated by UBC13-dependent PCNA polyubiquitylation (ubc13) or monoubiquitin-dependent TLS by damage-tolerant polymerases (rad30 rev1 rev3, designated as tlsΔ), or both pathways. (C) MGS1 deletion or overexpression does not affect PCNA ubiquitylation. Strains harbouring His6-tagged POL30 alleles and either an mgs1 deletion or the indicated vectors were incubated with or without 0.02% MMS for 60 min, and PCNA and its modified forms were isolated by Ni–NTA affinity chromatography under denaturing conditions and detected by western blotting.
These data are consistent with an action of Mgs1 either on events downstream of PCNA ubiquitylation or on the modification reaction itself. Our observation that neither deletion nor overexpression of MGS1 or MGS1* significantly affected damage-induced PCNA ubiquitylation (Figure 2C) ruled out the latter scenario. Hence, Mgs1 appears to interfere with an aspect of DNA damage bypass downstream of PCNA ubiquitylation.
Mgs1 preferentially interacts with polyubiquitylated PCNA via the UBZ domain
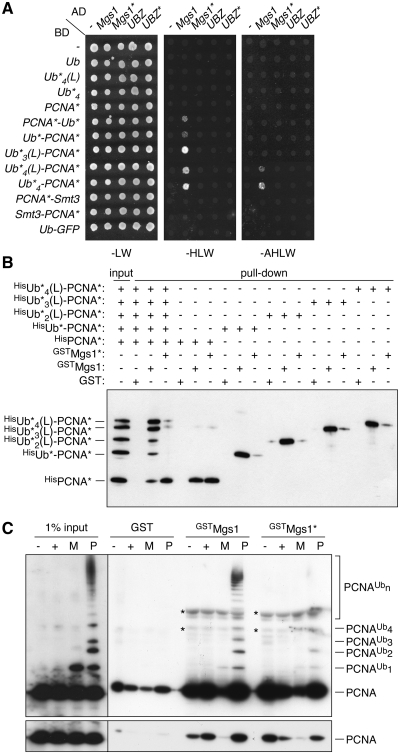
The notion that PCNA ubiquitylation is required for the damage- and UBZ-dependent activities of MGS1 strongly suggested a physical interaction of the UBZ domain with the ubiquitin moieties that are attached to PCNA in response to DNA damage. We examined this possibility in the two-hybrid system, using monoubiquitin and a series of linear tandem arrays as mimics of polyubiquitin chains, either free or fused to PCNA (30). Interactions between Mgs1 and either unmodified PCNA or free mono- or polyubiquitin were not observed in this assay, indicating that these associations are too weak or transient to be detected (Figure 3A). In contrast, monoubiquitin fused to the N- or C-terminus of PCNA resulted in a weak, but reproducible UBZ-dependent signal. The interaction was enhanced by increasing the number of ubiquitin moieties to three or four, suggesting that Mgs1 preferentially interacts with polyubiquitylated PCNA (Figure 3A). In contrast, PCNA fused to SUMO, or ubiquitin fused to an unrelated protein (GFP) did not interact measurably with Mgs1. Likewise, the UBZ domain in isolation did not bind any of the ubiquitin or PCNA constructs, indicating that a robust interaction requires the association of Mgs1 with PCNA in combination with UBZ-dependent binding of ubiquitin. Similar results were obtained with two-hybrid constructs arranged in the opposite orientation (Supplementary Figure S2A).
Figure 3.
Mgs1 preferentially interacts with ubiquitylated PCNA in a UBZ-dependent manner. (A) Protein–protein interactions with full-length Mgs1 and the isolated UBZ domain (amino acids 1–47) were analysed in the two-hybrid system, using fusions to the Gal4 activation (AD) and DNA-binding (BD) domains. Interactions were tested with monoubiquitin (Ub*) and linear ubiquitin arrays (Ub*n) either alone or fused to the N- or C-terminus of PCNA*. Asterisks indicate mutations in PCNA and ubiquitin that prevent further modification and processing by isopeptidases, and ‘L’ signifies a four-amino acid linker. Fusions of Ub* to GFP and of Smt3 (G98V, to prevent processing) to PCNA* were used as specificity controls. Empty vectors (–) served as negative controls. Presence of the constructs was confirmed by growth on medium lacking leucine and tryptophan (-LW), and positive signals were scored on medium additionally lacking histidine (-HLW) or—for strong interactions—histidine and adenine (-AHLW). (B) Interactions of Mgs1 with linear fusions of PCNA and ubiquitin were analysed in vitro. GST-tagged versions of Mgs1 and Mgs1* were immobilized on glutathione Sepharose and assayed for binding to the purified Ub*n(L)–PCNA* constructs described in A, produced as His6-tagged recombinant proteins, either in isolation or as an equimolar mixture. An anti-GST control blot is shown in Supplementary Figure S2B. (C) Interactions of Mgs1 were analysed in vitro as in B, but using PCNA subjected to in vitro mono- (M) or polyubiquitylation (P) at K164 prior to the assay. Unmodified PCNA that was either left unloaded (–) or loaded (+) onto DNA in parallel reactions served as controls. DNA was removed by nuclease treatment before performing the pull-down assays. Asterisks on the blot indicate cross-reactions of the anti-PCNA antibody with the GST fusion constructs. The bottom panel represents a weaker exposure of the blot.
In order to confirm that the observed interactions were direct, we performed in vitro pull-down assays with purified recombinant proteins (Figure 3B and Supplementary Figure S2B). When assayed in isolation, HisPCNA and the linear ubiquitin–PCNA fusions all interacted efficiently with immobilized GSTMgs1. Under these conditions, the isolated UBZ domain also bound polyubiquitin chains of K48- and—with some preference—K63 linkage (Supplementary Figure S2C), confirming previous observations with mammalian WRNIP1 (23,27). However, while HisPCNA bound with comparable affinity to GSTMgs1 and GSTMgs1*, interaction of the ubiquitin–PCNA fusions with GSTMgs1* was strongly reduced (Figure 3B). It is possible that ubiquitin fused to the N-terminus of PCNA interferes with binding to Mgs1 unless this effect is compensated by a productive ubiquitin–UBZ association. Within an equimolar mixture of unmodified HisPCNA and the ubiquitin–PCNA fusions, a clear preference of GSTMgs1 for the tri- and tetraubiquitin fusions was noted (Figure 3B, lane 3), confirming our two-hybrid data that implied a preferential interaction of Mgs1 with polyubiquitylated PCNA. In contrast, GSTMgs1* bound almost exclusively to unmodified HisPCNA.
These data indicated that linear arrays of ubiquitin effectively mediate the UBZ-dependent enhancement of Mgs1 binding to PCNA. However, we have previously shown that these constructs do not perfectly mimic the K63-linked polyubiquitin chains attached to K164 of PCNA in vivo (30). In order to confirm our results with physiologically K164-modified PCNA, in vitro mono- (M) and polyubiquitylated (P) PCNA (10) was also examined for binding to GSTMgs1 or GSTMgs1* (Figure 3C). As expected, both GSTMgs1 and GSTMgs1* effectively interacted with unmodified PCNA, while the polyubiquitylated forms were enriched only by GSTMgs1 and not by GSTMgs1*, thus largely confirming our results with the linear ubiquitin–PCNA fusions.
Ubiquitylated PCNA recruits Mgs1 to stalled replication intermediates
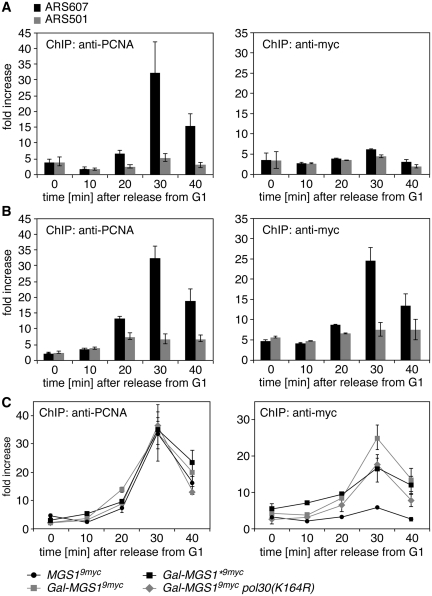
In order to determine whether the observed interaction of Mgs1 with ubiquitylated PCNA was relevant in vivo, we performed ChIP assays on synchronized cultures to follow the chromatin association of PCNA and Mgs1 at replication forks stalled by HU treatment. As shown previously (28), PCNA, which becomes ubiquitylated under these conditions, was readily detectable at an early replication origin with a timing that reflects the slow movement of the replication fork under these conditions (Figure 4A, left panel). While the signal of Mgs1 expressed under control of its endogenous promoter was insufficient for reliable detection (right panel), overexpression resulted in a pattern very similar to that of PCNA (Figure 4B), consistent with the recruitment of Mgs1 to those sites where PCNA accumulates under conditions of replication stress. A significant reduction in the signal upon mutation of either the UBZ domain of Mgs1 or the ubiquitin acceptor site on PCNA indicated that efficient Mgs1 association required UBZ-mediated binding to ubiquitylated PCNA (Figure 4C). The signal remaining in Mgs1* and pol30(K164R) likely reflects the basal affinity of Mgs1 for unmodified PCNA (21). These results suggest that ubiquitylation of PCNA contributes to the UBZ-mediated recruitment of Mgs1 to stalled replication intermediates in vivo.
Figure 4.
The UBZ domain contributes to the recruitment of Mgs1 to stalled replication intermediates. Association of Mgs19myc at an early replication origin was monitored by ChIP and quantitative PCR. G1-arrested cells were released into the cell cycle in the presence of 100 mM hydroxyurea to slow the progression of replication forks and induce PCNA ubiquitylation, and association of Mgs19myc with an early-firing replication origin (ARS607) was assessed at 10-min intervals (right panel). ChIP assays of PCNA from the same cultures served as a positive control for a fork-associated protein (left panel), and ChIP assays at a late origin (ARS501) that does not fire under these conditions served as a negative control. Signals were quantified relative to background samples prepared without antibody. Error bars indicate standard deviations from two to three experiments. (A) Mgs1 expressed at native levels is barely detectable by ChIP. Shown are signals of PCNA (left) and Mgs19myc (right) expressed at physiological levels. (B) When overexpressed, Mgs1 is detectable at stalled replication intermediates. Shown are ChIP signals of PCNA (left) and overexpressed Mgs19myc (right). (C) Mutating the UBZ domain of Mgs1 or the ubiquitylation site on PCNA reduces the association of Mgs1 with replication intermediates. Shown are the ChIP signals of PCNA and Mgs19myc in the indicated strains at ARS607. The signals at ARS501 are omitted for clarity.
Mgs1 modulates the activity of polymerase δ
The ubiquitin-mediated enhancement of the Mgs1–PCNA interaction in response to DNA damage or replication problems raised the question of what the function of Mgs1 at replication forks is. The effects of MGS1 overexpression, such as the DNA damage sensitivity, the suppression of damage-induced mutagenesis and the rescue of the temperature sensitivity of the dna2Δ405N mutant, strikingly resemble the phenotypes of a pol32 deletion (45–47), raising the possibility that Mgs1 interferes with the function of this particular polymerase δ subunit. As shown in Figure 1H, however, deletion of MGS1 suppresses the damage sensitivity of the pol32 deletion. This implies that Mgs1 has a negative activity even in the absence of Pol32. We therefore examined the effects of Mgs1 on other polymerase mutants with respect to its UBZ domain. As with pol32, the damage sensitivities of a truncation mutant of the catalytic polymerase δ subunit, pol3ΔCt (48), and a mutant allele of the essential second-largest subunit, pol31 (hys2-1), were suppressed by deletion of MGS1 or mutation of its UBZ domain (Figure 5A and B). Surprisingly, MGS1* also suppressed the temperature sensitivity of pol31 in the absence of exogenous DNA damage (Figure 5B). This was unexpected in light of our findings that linked the UBZ domain to damage-related functions. We reasoned that the effect might be due to a spontaneous ubiquitylation of PCNA that is observable in a number of polymerase mutants (49–51). Consistent with this notion, we found PCNA to be constitutively ubiquitylated in pol31 as well as other polymerase δ mutants (Supplementary Figure S3A). However, the polymerase ε mutant pol2-11, which also undergoes spontaneous PCNA ubiquitylation (Supplementary Figure S3A), and also the tlsΔ mutant remained unaffected by mgs1 or MGS1* (Supplementary Figure S3B and C). Hence, the inhibitory action of Mgs1 is directed specifically at polymerase δ, consistent with previous findings indicating a dosage effect on polymerase δ, but not ε or α (20). The negative action of Mgs1 on polymerase δ was further supported by a severe damage-independent growth inhibition conferred on pol3ΔCt by overexpression of MGS1 or MGS1*, but not the catalytically inactive mutant mgs1(K183A) (Figure 5C). A similar effect was also observed in a pol32(ΔPIP) mutant, encoding a protein that lacks the C-terminal PCNA interaction motif (38), and—reciprocally—in pol30-79, which expresses a mutant of PCNA defective in polymerase δ interaction (37) (Figure 5D). As before, overexpression of mgs1(K183R) had no influence on the growth rates of the host strains (data not shown), indicating that catalytic activity of Mgs1 was required for the effect.
Figure 5.
Mgs1 negatively acts on Polymerase δ and PCNA. (A) Deletion of MGS1 or inactivation of its UBZ domain suppresses the damage sensitivity of pol3ΔCt. MMS sensitivities were determined by spot assays in the indicated strains. (B) Deletion of MGS1 or inactivation of its UBZ domain suppresses the temperature and damage sensitivity of a pol31 (hys2-1) mutant. Spot assays were performed as above either on MMS-containing plates or by incubation at the indicated temperatures. (C) Overexpression of MGS1 inhibits growth of the pol3ΔCt mutant in a UBZ-independent manner. Growth of the indicated cultures in rich medium at 30°C was recorded by following optical densities at 600 nm. (D) Overexpression of MGS1 inhibits growth of mutants with defective PCNA–Pol32 interactions in a UBZ-independent manner. Growth of the indicated strains was monitored by spot assays on rich medium at 30°C.
Taken together, these data illustrate an overall negative impact of Mgs1 specifically on polymerase δ that becomes manifest in a dosage-dependent manner: on one hand, WT cells are affected only by elevated Mgs1 levels under conditions of DNA damage, when the protein is recruited to sites of replication problems via UBZ-dependent binding to ubiquitylated PCNA and appears to interfere with damage processing. On the other hand, cells with compromised polymerase δ activity are rendered damage- or temperature sensitive by physiological levels of Mgs1, while elevated levels now become inhibitory to growth even under permissive conditions. At these high concentrations only, Mgs1's toxic effect in the mutant strains no longer requires the UBZ domain.
Mgs1 competes with polymerase δ for interaction with PCNA
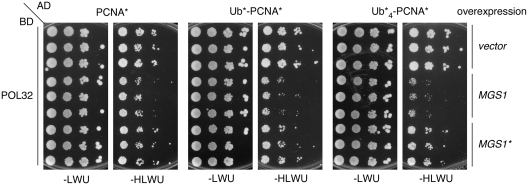
The notion that Mgs1 interacts with and is recruited to PCNA suggested that its impact on polymerase δ might be due to competitive physical interactions. We therefore examined the effect of Mgs1 on the association between PCNA and the polymerase δ subunits Pol32, which is known to make direct contacts with PCNA (38). As the Pol32–PCNA interaction is detectable by two-hybrid analysis, we monitored the consequences of overexpressing MGS1 in this system (Figure 6). Consistent with a competition between Pol32 and Mgs1 for PCNA binding, overexpression of MGS1 or MGS1* slightly reduced the signal corresponding to the Pol32–PCNA interaction. The inhibitory effect of MGS1 on the interaction of Pol32 with a monoubiquitin–PCNA fusion, Ub*–PCNA*, was considerably stronger, and in the case of the tetraubiquitin–PCNA fusion, Ub*4–PCNA*, the signal was virtually abolished. In contrast, overexpression of MGS1* had no additional effect on the interactions of the ubiquitin fusions with Pol32, consistent with the UBZ-dependent enhancement of Mgs1's affinity for PCNA by ubiquitylation of the clamp. Importantly, MGS1 overexpression did not disturb the interaction of PCNA or ubiquitin–PCNA fusions with polymerase η, which by itself gains additional affinity for the ubiquitylated forms of the clamp through a UBZ domain (Supplementary Figure S4) (15).
Figure 6.
Mgs1 competes with Pol32 for PCNA binding in the three-hybrid system. Fusions of the Gal4 activating domain to PCNA*, Ub*–PCNA* or Ub*4–PCNA* (as indicated above the panels) were combined with fusions of the DNA-binding domain to Pol32 as described in Figure 3A, and the effect of overexpressing MGS1 or MGS1* was assessed by spot assays, using 10-fold dilutions of three independent cultures per strain. An empty vector served as control. Uracil was omitted from the plates to maintain the additional plasmid (-LWU, -HLWU).
DISCUSSION
Mgs1 as an interactor of polyubiquitylated PCNA
The data presented above indicate clear functional and physical interactions of the Mgs1 UBZ domain with ubiquitin conjugated to PCNA upon exposure to replication stress or DNA damage. In combination with a basal affinity of Mgs1 for PCNA itself (21), the UBZ domain thus appears to act as a ubiquitylation-dependent ‘affinity enhancer’, similar to other ubiquitin-binding domains that mediate the recognition of relevant modified cellular targets (2). In this way, the protein is directed towards the replication fork in a damage-dependent manner, with PCNA ubiquitylation serving as the critical event controlling Mgs1 levels at the site of action.
Consistent with the requirements for target selectivity, we found the binding of the UBZ domain to free ubiquitin or ubiquitin chains to be weak and detectable only in vitro, thus minimizing in vivo interactions with ubiquitin species not conjugated to PCNA. Perhaps surprisingly, however, Mgs1 appears to exhibit some preference for polyubiquitylated over monoubiquitylated PCNA. This effect was apparent in the two-hybrid and pull-down experiments (Figure 3), but also in the genetic assays where deleting UBC13 had a stronger effect than inactivating TLS (Figure 2B). Given that UBZ domains tend to recognize single ubiquitin moieties (2), the preference of Mgs1 for polyubiquitylated PCNA may well be due to the proposed multimeric structure of the protein (52) that might result in a spatial arrangement of multiple UBZ domains conducive to oligoubiquitin binding. Indeed, accumulation of WRNIP1 in nuclear foci was found to depend on its oligomerization domain as well as the UBZ domain (23). In addition, Crosetto et al. found that the ubiquitin-binding domain derived from human Rad18, but not those from TLS polymerases η or ι were capable of substituting for the UBZ domain of WRNIP1, which is surprising in light of the ability of the polymerases, but not Rad18, to recognize ubiquitylated PCNA. While it was concluded from these data that a specific subtype of UBZ domain was required for correct localization of the protein, our results suggest that conformational or steric characteristics rather than the type of ubiquitin-binding domain may have been responsible for the lack of the WRNIP1 chimeras to localize correctly. Our identification of Mgs1 as the first protein reported to interact preferentially with polyubiquitylated PCNA raises the question of whether Mgs1 is indeed a mediator of error-free damage bypass.
Mgs1 as a downstream effector of the RAD6 pathway
Given that—in contrast to UBC13—deletion of MGS1 does not result in DNA damage sensitivity, Mgs1 is unlikely to act as the sole effector of polyubiquitylated PCNA in vivo. Instead, the genetic data described here and elsewhere (18,20,21) may even suggest a function as a negative regulator of DNA damage bypass. In fact, it was previously proposed that Mgs1 might inhibit the RAD6 pathway in the absence of damage by interfering with PCNA ubiquitylation (21). While our results (Figure 2C) clearly rule out this scenario, we cannot exclude that Mgs1 interferes with an event downstream of the modification reaction. However, for the following reasons we favour the idea that Mgs1 acts in a more subtle, regulatory fashion on damage processing: first, Mgs1 promotes rather than interferes with overall genome stability. Second, its inhibitory effects are apparent only at non-physiological expression levels or in genetic backgrounds with impaired polymerase activity, indicating that appropriate protein levels are critical for correct Mgs1 function. It is therefore conceivable that Mgs1 might affect the kinetics or timing of damage bypass in a way that we are unable to resolve with currently available methodology. Alternatively, Mgs1 may be redundant with another, yet to be identified factor downstream of ubiquitylated PCNA. Either scenario raises the question of what the molecular mechanism of Mgs1 action is.
Mgs1 as a UBZ-controlled modulator of polymerase δ
In a model that takes into account the physical interactions of Mgs1, the importance of protein levels as well as the negative effects on polymerase δ mutants, we envision Mgs1 as a ‘mobilizer’ for polymerase δ at the replication fork. By directly interfering with the PCNA–polymerase δ interaction, Mgs1 might facilitate release of the polymerase during a variety of DNA transactions, such as the polymerase switch associated with TLS, but also the events preceding strand exchange in preparation for template switching or other modes of replication restart that Mgs1 has been implicated in (20,26). In the context of damage bypass, where Mgs1 might facilitate or accelerate the reaction without being essential, the protein would be targeted to relevant sites by means of its UBZ-dependent interaction with ubiquitylated PCNA. In damage-unrelated situations, alternative, UBZ-independent targeting mechanisms might apply. This model offers an explanation for why excessive amounts of Mgs1 are detrimental for damage bypass, and why even physiological levels of the protein can interfere with replication of undamaged templates in mutants that are sensitized towards its actions through impaired polymerase δ function. Due to its modulatory function, loss of Mgs1 alone would not cause major defects, but its beneficial actions on genome stability become obvious and result in synergistic effects when other pathways of damage processing are compromised, for example in RAD6 pathway mutants (26) or in cells deficient in SGS1 (19,25) or mammalian BLM (53).
Unresolved questions
Our identification of the UBZ domain as a targeting module within Mgs1 provides an explanation for the dual action of the protein in damage-dependent versus -independent DNA transactions and places Mgs1 among the other reported PCNA effectors that react to modifications of the clamp, such as the TLS polymerases (15), the anti-recombinogenic helicase Srs2 (28,54) and the alternative clamp loader subunit Elg1 (55,56).
In contrast to its recruitment mechanism, Mgs1's exact mode of action still remains an open question. Our results suggest a competition between Mgs1 and polymerase δ for PCNA binding, but as Mgs1 has also been reported to associate with polymerase δ itself (22,25), the observed effect might be due to an interaction of Mgs1 with either PCNA or the polymerase or both. Based on the notion that fusion of ubiquitin to PCNA enhanced the competitive advantage of Mgs1 with respect to Pol32, we favour the first option, but further insight will have to come from biochemical experiments addressing the interactions in the context of DNA and the role of Mgs1's ATPase activity in the process.
Independently, the Mgs1 ATPase activity in combination with the UBZ domain had been postulated to play a role in the metabolism of ubiquitylated proteins related to that of the AAA+ ATPase Cdc48 (27). This model is based on experiments showing a reduced turnover of ubiquitin conjugates in mgs1 mutants, leading to a hyperresistance to the translation inhibitor cycloheximide. We have been unable to reproduce this effect in two different strain backgrounds (Supplementary Figure S5), and our data strongly suggest a function of the UBZ domain specifically in the recognition of ubiquitylated PCNA rather than a general action on bulk ubiquitin conjugates. Clearly, a biochemical characterization of the protein is needed to relate the nature of its ATPase activity to its biological function in vivo. In the context of DNA damage bypass, however, we are only beginning to understand the reactions following PCNA modification, and insight into the molecular mechanism of Mgs1 activity should therefore help us better understand the events involved in the replication of damaged DNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Cancer Research UK and a Research Training Network (Checkpoints and Cancer) from the European Commission (to H.D.U.). Funding for open access charge: Cancer Research UK.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Nicola Crosetto, Marzena Bienko and Ivan Dikic for valuable discussions and for communicating unpublished results, and Dana Branzei and Peter Burgers for reagents.
REFERENCES
- 1.Li W, Ye Y. Polyubiquitin chains: functions, structures, and mechanisms. Cell. Mol. Life. Sci. 2008;65:2397–2406. doi: 10.1007/s00018-008-8090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nat. Rev. Mol. Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–467. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- 4.Ulrich HD, Walden H. Ubiquitin signalling in DNA replication and repair. Nat. Rev. Mol. Cell Biol. 2010;11:479–489. doi: 10.1038/nrm2921. [DOI] [PubMed] [Google Scholar]
- 5.Hofmann K. Ubiquitin-binding domains and their role in the DNA damage response. DNA Repair. 2009;8:544–556. doi: 10.1016/j.dnarep.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 7.Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD. Activation of ubiquitin-dependent DNA damage bypass is mediated by Replication Protein A. Mol. Cell. 2008;29:625–636. doi: 10.1016/j.molcel.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang DJ, Lupardus PJ, Cimprich KA. Monoubiquitination of proliferating cell nuclear antigen induced by stalled replication requires uncoupling of DNA polymerase and mini-chromosome maintenance helicase activities. J. Biol. Chem. 2006;281:32081–32088. doi: 10.1074/jbc.M606799200. [DOI] [PubMed] [Google Scholar]
- 9.Niimi A, Brown S, Sabbioneda S, Kannouche PL, Scott A, Yasui A, Green CM, Lehmann AR. Regulation of proliferating cell nuclear antigen ubiquitination in mammalian cells. Proc. Natl Acad. Sci. USA. 2008;105:16125–16130. doi: 10.1073/pnas.0802727105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parker JL, Ulrich HD. Mechanistic analysis of PCNA poly-ubiquitylation by the ubiquitin protein ligases Rad18 and Rad5. EMBO J. 2009;28:3657–3666. doi: 10.1038/emboj.2009.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 12.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 13.Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Rad18 guides polη to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H, Lawrence CW. The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. Proc. Natl Acad. Sci. USA. 2005;102:15954–15959. doi: 10.1073/pnas.0504586102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 16.Plosky BS, Vidal AE, de Henestrosa AR, McLenigan MP, McDonald JP, Mead S, Woodgate R. Controlling the subcellular localization of DNA polymerases ι and η via interactions with ubiquitin. EMBO J. 2006;25:2847–2855. doi: 10.1038/sj.emboj.7601178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ulrich HD. Regulating post-translational modifications of the eukaryotic replication clamp PCNA. DNA Repair. 2009;8:461–469. doi: 10.1016/j.dnarep.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 18.Hishida T, Iwasaki H, Ohno T, Morishita T, Shinagawa H. A yeast gene, MGS1, encoding a DNA-dependent AAA(+) ATPase is required to maintain genome stability. Proc. Natl Acad. Sci. USA. 2001;98:8283–8289. doi: 10.1073/pnas.121009098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawabe Y, Branzei D, Hayashi T, Suzuki H, Masuko T, Onoda F, Heo SJ, Ikeda H, Shimamoto A, Furuichi Y, et al. A novel protein interacts with the Werner's syndrome gene product physically and functionally. J. Biol. Chem. 2001;276:20364–20369. doi: 10.1074/jbc.C100035200. [DOI] [PubMed] [Google Scholar]
- 20.Branzei D, Seki M, Onoda F, Enomoto T. The product of Saccharomyces cerevisiae WHIP/MGS1, a gene related to replication factor C genes, interacts functionally with DNA polymerase delta. Mol. Genet. Genomics. 2002;268:371–386. doi: 10.1007/s00438-002-0757-3. [DOI] [PubMed] [Google Scholar]
- 21.Hishida T, Ohya T, Kubota Y, Kamada Y, Shinagawa H. Functional and physical interaction of yeast Mgs1 with PCNA: impact on RAD6-dependent DNA damage tolerance. Mol. Cell. Biol. 2006;26:5509–5517. doi: 10.1128/MCB.00307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vijeh Motlagh ND, Seki M, Branzei D, Enomoto T. Mgs1 and Rad18/Rad5/Mms2 are required for survival of Saccharomyces cerevisiae mutants with novel temperature/cold sensitive alleles of the DNA polymerase delta subunit, Pol31. DNA Repair. 2006;5:1459–1474. doi: 10.1016/j.dnarep.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 23.Crosetto N, Bienko M, Hibbert RG, Perica T, Ambrogio C, Kensche T, Hofmann K, Sixma TK, Dikic I. Human Wrnip1 is localized in replication factories in a ubiquitin-binding zinc finger-dependent manner. J. Biol. Chem. 2008;283:35173–35185. doi: 10.1074/jbc.M803219200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawabe Y, Seki M, Yoshimura A, Nishino K, Hayashi T, Takeuchi T, Iguchi S, Kusa Y, Ohtsuki M, Tsuyama T, et al. Analyses of the interaction of WRNIP1 with Werner syndrome protein (WRN) in vitro and in the cell. DNA Repair. 2006;5:816–828. doi: 10.1016/j.dnarep.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 25.Branzei D, Seki M, Onoda F, Yagi H, Kawabe Y, Enomoto T. Characterization of the slow-growth phenotype of S. cerevisiae Whip/Mgs1 Sgs1 double deletion mutants. DNA Repair. 2002;1:671–682. doi: 10.1016/s1568-7864(02)00073-3. [DOI] [PubMed] [Google Scholar]
- 26.Hishida T, Ohno T, Iwasaki H, Shinagawa H. Saccharomyces cerevisiae MGS1 is essential in strains deficient in the RAD6-dependent DNA damage tolerance pathway. EMBO J. 2002;21:2019–2029. doi: 10.1093/emboj/21.8.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bish RA, Myers MP. Werner helicase-interacting protein 1 binds polyubiquitin via its zinc finger domain. J. Biol. Chem. 2007;282:23184–23193. doi: 10.1074/jbc.M701042200. [DOI] [PubMed] [Google Scholar]
- 28.Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 29.Ulrich HD, Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000;19:3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao S, Ulrich HD. Distinct consequences of posttranslational modification by linear versus K63-linked polyubiquitin chains. Proc. Natl Acad. Sci. USA. 2010;107:7704–7709. doi: 10.1073/pnas.0908764107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 32.Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 33.Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 34.von Borstel RC, Savage EA, Wang Q, Hennig UG, Ritzel RG, Lee GS, Hamilton MD, Chrenek MA, Tomaszewski RW, Higgins JA, et al. Topical reversion at the HIS1 locus of Saccharomyces cerevisiae. A tale of three mutants. Genetics. 1998;148:1647–1654. doi: 10.1093/genetics/148.4.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim JH, Kang YH, Kang HJ, Kim DH, Ryu GH, Kang MJ, Seo YS. In vivo and in vitro studies of Mgs1 suggest a link between genome instability and Okazaki fragment processing. Nucleic Acids Res. 2005;33:6137–6150. doi: 10.1093/nar/gki900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Navas TA, Zhou Z, Elledge SJ. DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell. 1995;80:29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- 37.Eissenberg JC, Ayyagari R, Gomes XV, Burgers PM. Mutations in yeast proliferating cell nuclear antigen define distinct sites for interaction with DNA polymerase delta and DNA polymerase epsilon. Mol. Cell. Biol. 1997;17:6367–6378. doi: 10.1128/mcb.17.11.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johansson E, Garg P, Burgers PM. The Pol32 subunit of DNA polymerase delta contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J. Biol. Chem. 2004;279:1907–1915. doi: 10.1074/jbc.M310362200. [DOI] [PubMed] [Google Scholar]
- 39.Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 40.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parker JL, Bielen AB, Dikic I, Ulrich HD. Contributions of ubiquitin- and PCNA-binding domains to the activity of Polymerase eta in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:881–889. doi: 10.1093/nar/gkl1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kennedy BK, Gotta M, Sinclair DA, Mills K, McNabb DS, Murthy M, Pak SM, Laroche T, Gasser SM, Guarente L. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell. 1997;89:381–391. doi: 10.1016/s0092-8674(00)80219-6. [DOI] [PubMed] [Google Scholar]
- 44.Lea DE, Coulson CA. The distribution of the numbers of mutants in bacterial populations. J. Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 45.Budd ME, Tong AH, Polaczek P, Peng X, Boone C, Campbell JL. A network of multi-tasking proteins at the DNA replication fork preserves genome stability. PLoS Genet. 2005;1:e61. doi: 10.1371/journal.pgen.0010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gerik KJ, Li X, Pautz A, Burgers PM. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. 1998;273:19747–19755. doi: 10.1074/jbc.273.31.19747. [DOI] [PubMed] [Google Scholar]
- 47.Huang ME, de Calignon A, Nicolas A, Galibert F. POL32, a subunit of the Saccharomyces cerevisiae DNA polymerase delta, defines a link between DNA replication and the mutagenic bypass repair pathway. Curr. Genet. 2000;38:178–187. doi: 10.1007/s002940000149. [DOI] [PubMed] [Google Scholar]
- 48.Brocas C, Charbonnier JB, Dherin C, Gangloff S, Maloisel L. Stable interactions between DNA polymerase delta catalytic and structural subunits are essential for efficient DNA repair. DNA Repair. 2010;9:1098–1111. doi: 10.1016/j.dnarep.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 49.Northam MR, Garg P, Baitin DM, Burgers PM, Shcherbakova PV. A novel function of DNA polymerase zeta regulated by PCNA. EMBO J. 2006;25:4316–4325. doi: 10.1038/sj.emboj.7601320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suzuki M, Niimi A, Limsirichaikul S, Tomida S, Miao Huang Q, Izuta S, Usukura J, Itoh Y, Hishida T, Akashi T, et al. PCNA mono-ubiquitination and activation of translesion DNA polymerases by DNA polymerase alpha. J. Biochem. 2009;146:13–21. doi: 10.1093/jb/mvp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karras GI, Jentsch S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 2010;141:255–267. doi: 10.1016/j.cell.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 52.Tsurimoto T, Shinozaki A, Yano M, Seki M, Enomoto T. Human Werner helicase interacting protein 1 (WRNIP1) functions as a novel modulator for DNA polymerase delta. Genes Cells. 2005;10:13–22. doi: 10.1111/j.1365-2443.2004.00812.x. [DOI] [PubMed] [Google Scholar]
- 53.Hayashi T, Seki M, Inoue E, Yoshimura A, Kusa Y, Tada S, Enomoto T. Vertebrate WRNIP1 and BLM are required for efficient maintenance of genome stability. Genes Genet. Syst. 2008;83:95–100. doi: 10.1266/ggs.83.95. [DOI] [PubMed] [Google Scholar]
- 54.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 55.Lee KY, Yang K, Cohn MA, Sikdar N, D'Andrea AD, Myung K. Human ELG1 regulates the level of ubiquitinated proliferating cell nuclear antigen (PCNA) through its interactions with PCNA and USP1. J. Biol. Chem. 2010;285:10362–10369. doi: 10.1074/jbc.M109.092544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parnas O, Zipin-Roitman A, Pfander B, Liefshitz B, Mazor Y, Ben-Aroya S, Jentsch S, Kupiec M. Elg1, an alternative subunit of the RFC clamp loader, preferentially interacts with SUMOylated PCNA. EMBO J. 2010;29:2611–2622. doi: 10.1038/emboj.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.