Abstract
β-D-3′-Azido-2′,3′-dideoxyguanosine (3′-azido-ddG) is a potent inhibitor of HIV-1 replication with a superior resistance profile to zidovudine. Recently, we identified five novel 6-modified-3′-azido-ddG analogs that exhibit similar or superior anti-HIV-1 activity compared to 3′-azido-ddG in primary cells. To gain insight into their structure–activity–resistance relationships, we synthesized their triphosphate (TP) forms and assessed their ability to inhibit HIV-1 reverse transcriptase (RT). Steady-state and pre-steady-state kinetic experiments show that the 6-modified-3′-azido-ddGTP analogs act as adenosine rather than guanosine mimetics in DNA synthesis reactions. The order of potency of the TP analogs against wild-type RT was: 3′-azido-2,6-diaminopurine >3′-azido-6-chloropurine; 3′-azido-6-N-allylaminopurine > 2-amino-6-N,N-dimethylaminopurine; 2-amino-6-methoxypurine. Molecular modeling studies reveal unique hydrogen-bonding interactions between the nucleotide analogs and the template thymine base in the active site of RT. Surprisingly, the structure–activity relationship of the analogs differed in HIV-1 RT ATP-mediated excision assays of their monophosphate forms, suggesting that it may be possible to rationally design a modified base analog that is efficiently incorporated by RT but serves as a poor substrate for ATP-mediated excision reactions. Overall, these studies identify a promising strategy to design novel nucleoside analogs that exert profound antiviral activity against both WT and drug-resistant HIV-1.
INTRODUCTION
Most combination therapies used to treat antiretroviral-naïve and -experienced HIV-1 infected patients include at least two nucleoside reverse transcriptase (RT) inhibitors (NRTIs). NRTIs are analogs of 2′-deoxyribonucleosides that lack a 3′-OH group on the ribose sugar/pseudosugar. Once metabolized by host cell kinases to their active triphosphate (TP) forms, they inhibit viral reverse transcription by acting as chain terminators of HIV-1 RT DNA synthesis (1). Although combination therapies that contain two or more NRTIs have substantially reduced mortality from HIV-1 associated disease, the approved NRTIs have significant limitations that include acute and chronic toxicity, and the selection of drug-resistant variants of HIV-1 that exhibit cross-resistance to other NRTIs. Accordingly, there is a need to develop potent and safe NRTIs that demonstrate activity against a broad-range of drug-resistant HIV-1.
To date, most NRTIs were discovered by an empirical approach in which novel sugar modified nucleoside analogs were synthesized without a priori knowledge of their activity against wild-type (WT) or drug-resistant HIV-1. To specifically identify NRTIs that retain activity against a broad spectrum of drug-resistant HIV-1, our group adopted a rational discovery approach that utilized structure–activity–resistance relationships to identify NRTI base and sugar moieties that retain potent activity against drug-resistant HIV-1 (2,3). Importantly, this approach identified 3′-azido-2′,3′-dideoxyguanosine (3′-azido-ddG) as a lead compound (4) that exhibits potent activity against HIV-1 variants that contain the discrimination mutations K65R, L74V or M184V and against HIV-1 that contains multiple thymidine analog mutations (TAMs) that enhance nucleotide excision. Furthermore, 3′-azido-ddG does not exhibit cytotoxicity in primary lymphocytes or epithelial and T-cell lines, and does not decrease the mitochondrial DNA content of HepG2 cells (4).
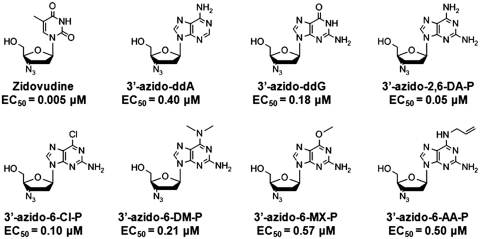
To further improve the antiviral activity of 3′-azido-ddG, we recently synthesized and characterized a series of novel base modified 3′-azido-2′,3′-dideoxypurine analogs (5). Of the 26 analogs studied, five [namely, 3′-azido-2′,3′-dideoxy-2,6-diaminopurine (3′-azido-2,6-DA-P), 3′-azido-2′,3′-dideoxy-2-amino-6-chloropurine (3′-azido-6-Cl-P), 3′-azido-2′,3′-dideoxy -2-amino-6- N,N-dimethylaminopurine (3′-azido-6-DM-P), 3′-azido-2′,3′-dideoxy-2-amino-6-methoxypurine (3′-azido-6-MX-P) and 3′-azido-2′,3′-dideoxy-2-amino-6-N-allylaminopurine (3′-azido-6-AA-P)] exhibited superior or equivalent activity to 3′-azido-ddG in primary lymphocytes (Figure 1). To gain additional insight into the structure–activity–resistance relationships of these compounds, we have now synthesized their TP forms and have assessed their ability to inhibit DNA synthesis by recombinant purified HIV-1 RT. Importantly, we show for the first time that HIV-1 RT recognizes and incorporates these 6-modified-3′-azido-ddGTP nucleotides as adenosine analogs. These findings reveal new avenues for developing novel ambiguous nucleoside analogs that could be used to treat HIV-1 infection.
Figure 1.
Structures of zidovudine (ZDV), 3′-azido-2′,3′-dideoxyadenosine (3′-azido-ddA), 3′-azido-2′,3′-dideoxyguanosine (3′-azido-ddG), 3′-azido-2′,3′-dideoxy-2,6-diaminopurine (3′-azido-2,6-DA-P), 3′-azido-2′,3′-dideoxy-2-amino-6-chloropurine (3′-azido-6-Cl-P), 3′-azido-2′,3′-dideoxy-2-amino-6-N,N-dimethylaminopurine (3′-azido-6-DM-P), 3′-azido-2′,3′-dideoxy-2-amino-6-methoxypurine (3′-azido-6-MX-P) and 3′-azido-2′,3′-dideoxy-2-amino-6-N-allylaminopurine (3′-azido-6-AA-P). The EC50 values, adapted from Zhang et al. (5), are for WT HIV-1 and were determined in peripheral blood mononuclear (PBM) cells.
MATERIALS AND METHODS
Reagents
The WT, K65R, L74V, M184V, A62V/V75I/F77L/F116Y/Q151M, M41L/L210W/T215Y (TAM41) and D67N/K70R/T215F/K219Q (TAM67) HIV-1 RTs were purified as described previously (6,7). The protein concentration of the purified enzymes was determined spectrophotometrically at 280 nm using an extinction coefficient (ε280) of 260 450 M−1 cm−1, and by Bradford protein assays (Sigma-Aldrich, St. Louis, MO, USA). The RNA- and DNA-dependent DNA polymerase activities of the purified WT and mutant enzymes were similar (data not shown). The triphosphate (TP) forms of each of the 6-modified-3′-azido-ddG analogs were synthesized and purified using the methods of Ludwig and Eckstein (8). 3′-Azido-ddGTP, 3′-azido-ddATP, 3′-azido-ddCTP and zidovudine-TP (ZDV-TP) were purchased from TriLink Biotechnologies, Inc. (San Diego, CA, USA). [γ-32P]ATP was obtained from PerkinElmer Life Sciences (Boston, MA, USA). All DNA oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA, USA).
Inhibition of HIV-1 RT DNA synthesis under steady-state conditions by the 6-modified-3′-azido-ddGTP analogs
The ability of the 6-modified-3′-azido-ddGTP analogs to inhibit HIV-1 RT DNA- or RNA-dependent DNA synthesis was determined using two different template/primer (T/P) substrates, as described below:
RT DNA synthesis was evaluated using a 214-nt RNA or DNA template that corresponds to the HIV-1 sequence used for (−) strong stop DNA synthesis (Figure 2). The RNA and DNA templates were prepared as described previously (3). Both the RNA- and DNA-dependent DNA synthesis reactions were primed with the same 5′-end-32P-labeled 18-nt DNA primer (5′-GTCCCTGTTCGGGCGCCA-3′) that corresponds to the HIV-1 primer binding site. DNA synthesis reactions (20 µl) were carried out in a 50-mM Tris–HCl (pH 7.5) buffer that contained 50 mM KCl, 10 mM MgCl2, 20 nM T/P, 0.5 µM of each dNTP and varying concentrations of the 6-modified-3′-azido-ddGTP analogs. Reactions were initiated by the addition of 200 nM WT HIV-1 RT, incubated at 37°C for 30 min (DNA-dependent DNA synthesis) or 60 min (RNA-dependent DNA synthesis) and then quenched by addition of 20 µl of gel loading buffer (98% deionized formamide containing 1 mg/ml each of bromophenol blue and xylene cyanol). Samples were then denatured at 95°C for 10 min and polymerization products were separated from substrates by denaturing gel electrophoresis using 14% acrylamide gels containing 7 M urea. DNA synthesis was analyzed by phosphorimaging using a GS-525 Molecular Imager and Quantity One Software (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Inhibition of HIV-1 RT DNA synthesis was also evaluated to determine IC50 values using a 5′-end-32P-labeled 19-nt DNA primer (5′-TTGTAGCACCATCCAAAGG-3′) that was annealed to one of three 36-nt DNA templates that contained five consecutive thymine, cytosine or adenosine bases. In this regard, inhibition of HIV-1 RT DNA synthesis by analogs with adenine, guanine or thymine bases were evaluated using the templates (T1, 5′-CAGACTTTTTCAGACCTTTGGATGGTGCTACAAGCT-3′), (T2, 5′-CAGAGCCCCCGAGACCTTTGGATGGTGCTACAAGCT-3′), or (T3, 5′-CTGCTAAAAACTGCCCTTTGGATGGTGCTACAAGCT-3′), respectively. The T1 template was used for inhibition of WT and mutant HIV-1 RT DNA synthesis by each of the 6-modified-3′-azido-ddGTP analogs. Reaction conditions and gel electrophoresis were identical to those described above except 100 nM of WT or mutant HIV-1 RT was used to initiate the reaction. Reactions were quenched following incubation at 37°C for 10 min. The amount of final product on the denaturing polyacrylamide gel electrophoresis (PAGE) gels was quantified by densiometric analysis using Quantity One Software. The concentration of each 6-modified-3′-azido-ddGTP analog required to inhibit the formation of final product by 50% was calculated using non-linear regression analyses (SigmaPlot Software Version 11, Systat Software, Inc., San Jose, CA, USA) from at least three independent experiments. Two-tailed homoscedastic t-tests were used to calculate the reported P-values.
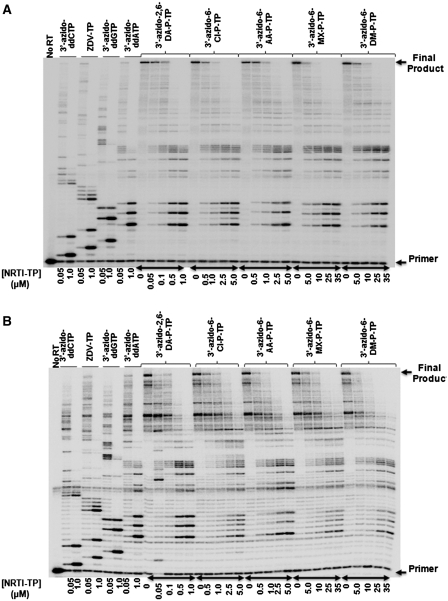
Figure 2.
6-Modified-3′-azido-ddGTPs are recognized and incorporated by WT HIV-1 as adenosine analogs. (A) Representative denaturing polyacrylamide gel showing chain-termination of HIV-1 RT DNA synthesis by each of the 3′-azido-ddGTPs on a heteropolymeric DNA/DNA T/P under steady-state assay conditions. (B) Representative denaturing polyacrylamide gel showing chain termination of WT HIV-1 RT DNA synthesis by each of the 3′-azido-ddGTPs on a heteropolymeric RNA/DNA T/P under steady-state assay conditions. Reaction conditions are described in the ‘Materials and Methods’ section.
Pre-steady-state incorporation of 6-modified-3′-azido-ddGTP analogs
A rapid quench instrument (Kintek RQF-3 instrument, Kintek Corporation, Clarence, PA, USA) was used for pre-steady-state experiments. The typical experiment was performed at 37°C in 50 mM Tris–HCl (pH 7.5) containing 50 mM KCl, 10 mM MgCl2 and varying concentrations of nucleotide. All concentrations reported refer to the final concentrations after mixing. HIV-1 RT (200 nM) was pre-incubated with 20 nM T/P substrate, prior to rapid mixing with nucleotide and divalent metal ions to initiate the reaction that was quenched with 50 mM EDTA. The sequences of the template and 5′-radiolabeled primer are shown in Table 1. The quenched samples were then mixed with an equal volume of gel loading buffer and products were separated from substrates as described above. The disappearance of substrate (20mer) and the formation of product (21-mer) were quantified using a Bio-Rad GS525 Molecular Imager (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Data were fitted by nonlinear regression with Sigma Plot software (Systat Software, Inc., San Jose, CA, USA) using the appropriate equations (9). The apparent burst rate constant (kobs) for each particular concentration of dNTP was determined by fitting the time courses for the formation of product (21mer) using the following equation: [21mer] = A[1 − exp(−kobst)], where A represents the burst amplitude. The turnover number (kpol) and apparent dissociation constant for dNTP (Kd) were then obtained by plotting the apparent catalytic rates (kobs) against dNTP concentrations and fitting the data with the following hyperbolic equation: kobs = (kpol[dNTP])/([dNTP] + Kd). Catalytic efficiency was calculated as the ratio of turnover number over dissociation constant ([kpol/Kd]). Selectivity for natural dNTP versus 6-modified-3′-azido-ddGTP analog was calculated as the ratio of catalytic efficiency of dNTP over that of the 6-modified-3′-azido-ddGTP analog ([(kpol/Kd)dNTP/(kpol/Kd)Analog]).
Table 1.
Pre-steady-state kinetic values for incorporation of 6-modified-3′-azido-ddGTP analogs by WT HIV-1 RT
| Incorporation as an ‘A’ analoga | ||||
|---|---|---|---|---|
| Nucleotide analog | kpol (s−1) | Kd (µM) | kpol/Kd (µM−1 s−1) | Selectivityb versus dATP |
| dATP | 17.0 ± 2.1c | 0.33 ± 0.05 | 52 | – |
| 3′-azido-ddATP | 14.0 ± 5.6 | 0.32 ± 0.13 | 44 | 1.2 |
| 3′-azido-2,6-DA-P-TP | 14.0 ± 4.9 | 0.29 ± 0.05 | 48 | 1.1 |
| 3′-azido-6-Cl-P-TP | 2.7 ± 0.5 | 1.8 ± 1.0 | 1.5 | 35 |
| 3′-azido-6-AA-P-TP | 2.1 ± 0.2 | 4.8 ± 2.3 | 0.44 | 118 |
| 3′-azido-6-MX-P-TP | 2.8 ± 0.2 | 12.0 ± 1.2 | 0.23 | 226 |
| Incorporation as a ‘G’ analogd | ||||
|---|---|---|---|---|
| Nucleotide analog | kpol (s−1) | Kd (µM) | kpol/Kd (µM−1 s−1) | Selectivity versus dGTP |
| dGTP | 17.3 ± 1.4 | 0.17 ± 0.05 | 102 | – |
| 3′-azido-2,6-DA-P-TP | 0.16 ± 0.02 | 34 ± 14 | 0.005 | 20 400 |
| 3′-azido-6-Cl-P-TP | 0.37 ± 0.04 | 0.92 ± 0.02 | 0.402 | 254 |
| 3′-azido-6-AA-P-TP | n.m.e | >50 | n.m. | n.m. |
| 3′-azido-6-MX-P-TP | 0.22 ± 0.09 | 16.3 ± 4.3 | 0.013 | 7846 |
a3′-ACAGGGACAAGCCCGCGGTGACGATCTCTTAAAGGTAAGACTGATTTTCCCAGACTC-5′; 5′-TCGGGCGCCACTGCTAGAGA: Sequence of the T/P substrate used to assess the 6-modified-3′-azido-ddGTP nucleotides as ‘A’ analogs.
b Selectivity is (kpol/Kd)dNTP/(kpol/Kd)3′-azido-ddNTP
cValues represent the mean ± standard deviation of three to four independent experiments
d3′-ACAGGGACAAGCCCGCGGTGACGATCTCTCAAAGGTAAGACTGATTTTCCCAGACTC-5′; 5′-TCGGGCGCCACTGCTAGAGA: Sequence of the T/P substrate used to assess the 6-modified-3′-azido-ddGTP nucleotides as ‘G’ analogs
en.m. Not measurable.
The rate of incorporation was so inefficient that accurate kinetic parameters could not be determined.
Excision of the 6-modified-3′-azido-ddG monophosphate analogs by WT or TAM-containing HIV-1 RTs
The ability of WT or TAM-containing HIV-1 RT to facilitate ATP-mediated excision of the 6-modified-3′-azido-ddG-MP analogs from a DNA/DNA chain-terminated T/P was assessed as described previously (3). For excision of 3′-azido-ddAMP and each 6-modified-3′-azido-ddGMP analog the T/P pair was T1/pr23A (pr23A: 5′-TTGTAGCACCATCCAAAGGTCTG-3′). The 23-nt DNA primer was 5′-radiolabeled with [γ-32P]ATP, annealed to T1, and then chain-terminated with the appropriate nucleotide analog. Reactions were carried out in a 50-mM Tris–HCl (pH 7.5) buffer that contained 50 mM KCl, 10 mM MgCl2, 3 mM ATP and 1 μM dATP/10 μM ddGTP. Reactions were initiated by the addition of 200 nM WT or mutant RT. Aliquots (5 µl) were removed at defined times, quenched with gel loading buffer, denatured at 95°C for 10 min, and product was then resolved from substrate by denaturing PAGE and analyzed, as described above.
Molecular modeling
Molecular models of incorporation were constructed using the X-ray crystallographic coordinates (PDB entry 1 RTD) for the RT-T/P-TTP ternary complex (10). The 3′-OH of the primer strand as well as the 6-modified-3′-azido-2′,3-ddGTP analogs and complementary bases in the template strand were built into the model to generate a pre-initiation complex, as described previously (11). Tautomerization and protonation potential of the entire system was calculated using the Generalized Born/Volume Integral (GB/VI) electrostatics method of Labute (12). Energy gradient minimization was carried out using MMFF94x force field in the Molecular Operating Environment (MOE 2008.02; Chemical Computing Group, Montreal, Quebec, Canada). Ligand interactions were quantified and images generated using the LigX interaction function in MOE. Structural models comparing incorporation and excision of specific nucleotide analogs used the PDB co-ordinates 3KLF and 3KLE that contain the excision product AZT adenosine dinucleoside tetraphosphate (AZTppppA) (13). To eliminate the potential force field bias from alignment of bases, empirically derived angle relations were used for all analogs tested. 3KLF and 3KLE were fit to the p66 subunit of 1RTD using the Matchmaker function in Chimera version 1.51 (UCSF Chimera, Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco). Aligned structures were opened in MOE and dihedral angles were measured from bases relative to the ribose ring O and C1 of both template and bound nucleotides for each complex. MOE's builder function was used to modify each template/bound nucleotide base pair retaining the experimental dihedrals. Hydrogen-bonding potential between the aligned base pairs was quantified using the ‘FindHBond’ function in Chimera with default settings. Parts of modeling workflow was automated using Pipeline Pilot 7.5 (Accelrys Corporation, San Diego, CA, USA)
RESULTS
Steady-state incorporation of 6-modified 3′-azido-ddGTP nucleotides by HIV-1 RT
To characterize the ability of the WT HIV-1 RT to incorporate the 6-modified-3′-azido-ddGTP analogs we first conducted steady-state DNA synthesis reactions using heteropolymeric DNA/DNA and RNA/DNA T/P substrates (Figure 2). The sequences of the RNA and DNA templates correspond to the HIV-1 sequence used for (−) strong stop DNA synthesis, and both reactions were primed with the same 18-nt DNA primer that was complementary to the HIV-1 primer binding site (3). 3′-Azido-ddGTP, 3′-azido-ddATP, 3′-azido-ddCTP and ZDV-TP were included as controls in these experiments. Unexpectedly, we found that HIV-1 RT recognized all of the 6-modified-3′-azido-ddGTP nucleotides as adenosine analogs and incorporated them opposite thymine in DNA (Figure 2A) or uracil in RNA (Figure 2B). As anticipated, their incorporation resulted in chain-termination of DNA synthesis. Under the assay conditions described in Figure 2, the 6-modified-3′-azido-ddGTP nucleotides were not incorporated into the nascent DNA opposite cytosine. However, if the dNTP concentration was lowered to 0.1 μM, chain termination opposite cytosine was observed (data not shown). We also determined the concentration of 6-modified-3′-azido-ddGTP analog required to inhibit DNA synthesis (i.e. IC50) using a different heteropolymeric DNA/DNA T/P substrate, as described in the ‘Materials and Methods’ section (Table 2). Their order of potency against WT RT was determined to be: 3′-azido-ddATP, 3′-azido-ddGTP >3′-azido-2,6-DA-P-TP > 3′-azido-6-Cl-P-TP, 3′-azido-6-AA-P-TP > 3′-azido-6-DM-P-TP, 3′-azido-6-MX-P-TP.
Table 2.
Inhibition of steady-state WT and mutant HIV-1 RT DNA synthesis by 6-modified-3′-azido-ddGTP analogs
| Nucleotide analog | HIV-1 RT |
||||
|---|---|---|---|---|---|
| WT | K65R | L74V | M184V | Q151Ma | |
| ZDV-TP | |||||
| IC50 (µM)b | 0.13 ± 0.04 | 0.66 ± 0.20 | 0.18 ± 0.10 | 0.12 ± 0.03 | 2.7 ± 1.0 |
| Fold-Rc | – | 5.2 | 1.4 | 0.9 | 21.0 |
| 3′-azido-ddGTP | |||||
| IC50 (µM) | 0.15 ± 0.01 | 0.36 ± 0.02 | 0.25 ± 0.10 | 0.06 ± 0.02 | 0.30 ± 0.20 |
| Fold-R | – | 2.5 | 1.7 | 0.4 | 2.1 |
| 3′-azido-ddATP | |||||
| IC50 (µM) | 0.16 ± 0.10 | 0.70 ± 0.10 | 0.42 ± 0.10 | 0.19 ± 0.10 | 1.44 ± 1.00 |
| Fold-R | – | 4.3 | 2.5 | 1.2 | 8.8 |
| 3′-azido-2,6-DA-P-TP | |||||
| IC50 (µM) | 0.80 ± 0.10 | 2.5 ± 1.6 | 1.3 ± 0.4 | 0.34 ± 0.10 | 4.2 ± 3.6 |
| Fold-R | – | 3.1 | 1.6 | 0.4 | 5.3 |
| P-valued | – | 0.36 | 0.08 | 0.02 | 0.45 |
| 3′-azido-6-Cl-P-TP | |||||
| IC50 (µM) | 3.2 ± 0.8 | 9.1 ± 3.8 | 11.8 ± 3.9 | 2.5 ± 1.0 | 30.8 ± 6.1 |
| Fold-R | – | 2.9 | 3.7 | 0.8 | 9.7 |
| P-value | – | 0.12 | 0.15 | 0.22 | 0.81 |
| 3′-azido-6-AA-P-TP | |||||
| IC50 (µM) | 4.10 ± 0.8 | 8.8 ± 0.8 | 10.3 ± 1.9 | 4.8 ± 0.6 | 21.8 ± 12.0 |
| Fold-R | – | 2.2 | 2.5 | 1.2 | 5.3 |
| P-value | – | 0.0006 | 0.97 | 0.94 | 0.43 |
| 3′-azido-6-DM-P-TP | |||||
| IC50 (µM) | 23.4 ± 13.0 | >50 | >50 | N.D.e | >50 |
| Fold-R | – | >2.0 | >2.0 | >2.0 | |
| P-value | – | – | – | – | – |
| 3′-azido-6-MX-P-TP | |||||
| IC50 (µM) | 24.4 ± 7.0 | 80.1 ± 6.1 | >100 | 32.9 ± 23.0 | 78.0 ± 34.0 |
| Fold-R | – | 3.3 | >4.0 | 1.3 | 3.2 |
| P-value | – | 0.10 | – | 0.71 | 0.31 |
aThe Q151M RT contained the A62V, V75I, F77L, F116Y and Q151M mutations.
bIC50 values are the concentration of drug required to inhibit 50% of DNA synthesis under steady-state assay conditions.
Data are shown as the mean ± standard deviation of at least three independent experiments.
cFold-resistance (Fold-R) values are calculated by IC50mutant RT/IC50WT RT.
dP-value compares the Fold-R values determined for the 6-modified 3′-azido-ddGTP analogs to the Fold-R value determined for 3′-azido-ddATP.
eNot determined.
Pre-steady-state incorporation of 6-modified 3′-azido-ddGTP nucleotides by HIV-1 RT
Pre-steady-state kinetic analyses were carried out to elucidate, in detail, the interactions between the 6-modified-3′-azido-ddGTP analogs and the polymerase active site of WT HIV-1 RT (Table 1). Because the 6-modified-3′-azido-ddGTP analogs behave as adenine nucleotide analogs, we included 3′-azido-ddATP as a control. The results (Table 1) show that the catalytic efficiency of incorporation (kpol/Kd) of 3′-azido-2,6-DA-P-TP was similar to that of dATP and 3′-azido-ddATP. Consistent with the steady-state kinetic experiments, 3′-azido-6-Cl-P-TP, 3′-azido-6-AA-P-TP and 3′-azido-6-MX-P-TP were all less efficiently incorporated by WT HIV-1 compared to dATP. The observed decreases in catalytic efficiency for each of these substrates was driven by both a decrease in the rate of nucleotide incorporation (i.e. kpol) and a decrease in the affinity of the nucleotide for the polymerase active site (i.e. Kd). Due to limited quantities of the 3′-azido-6-DM-P-TP, we were unable to obtain pre-steady-state kinetic data for this analog. We also assessed the ability of the 6-modified-3′-azido-ddGTP analogs to be incorporated opposite cytosine (Table 1). The data show that, in comparison to dGTP, the 6-modified-3′-azido-ddGTP analogs were inefficiently incorporated by HIV-1 RT. The only compound with reasonable activity as a G-analog was 3′-azido-6-Cl-P-TP; however, its selectivity value versus dGTP (254) was ~7-fold less than its selectivity value versus dATP (35).
Activity of the 6-modified-3′-azido-ddGTP analogs against HIV-1 RT containing NRTI discrimination mutations
The amino acid substitutions K65R, L74V, Q151M (in complex with A62V, V75I, F77L and F116Y) or M184V in HIV-1 RT improve the enzyme's ability to discriminate between the natural dNTP substrate and an NRTI-TP. These substitutions are typically referred to as NRTI discrimination mutations. To determine whether NRTI discrimination mutations impacted RTs’ ability to recognize and incorporate the 6-modified-ddGTP analogs, we determined the concentration of nucleotide analog required to inhibit 50% of the DNA polymerase activity (i.e. IC50) under steady-state assay conditions using the heteropolymeric DNA/DNA T/P substrates as described in the ‘Materials and Methods’ section (Table 2). 3′-Azido-ddATP, 3′-azido-ddGTP and ZDV-TP were again included as controls. The 6-modified-3′-azido-ddGTP analogs were also incorporated as adenosine analogs by the mutant HIV-1 RTs (data not shown). In general, the mutant HIV-1 RTs exhibited fold-resistance values to each of the 6-modified-3′-azido-ddGTP analogs that were similar (i.e. <2-fold) to the fold change in resistance values determined for 3′-azido-ddATP. The only statistically significant exceptions (P < 0.05) included 3′-azido-6-AA-P-TP and 3′-azido-2,6-DA-P-TP which exhibited better activity against K65R HIV-1 RT and M184V HIV-1 RT, respectively. Of interest, the fold-resistance values for all of the 3′-azido-dideoxypurines against HIV-1 RT containing A62V/V75I/F77L/Y116F/Q151M were significantly less than the fold-resistance value that was determined for ZDV-TP.
Excision of 6-modified-3′-azido-ddG-5′-MP analogs by WT and TAM containing HIV-1 RT
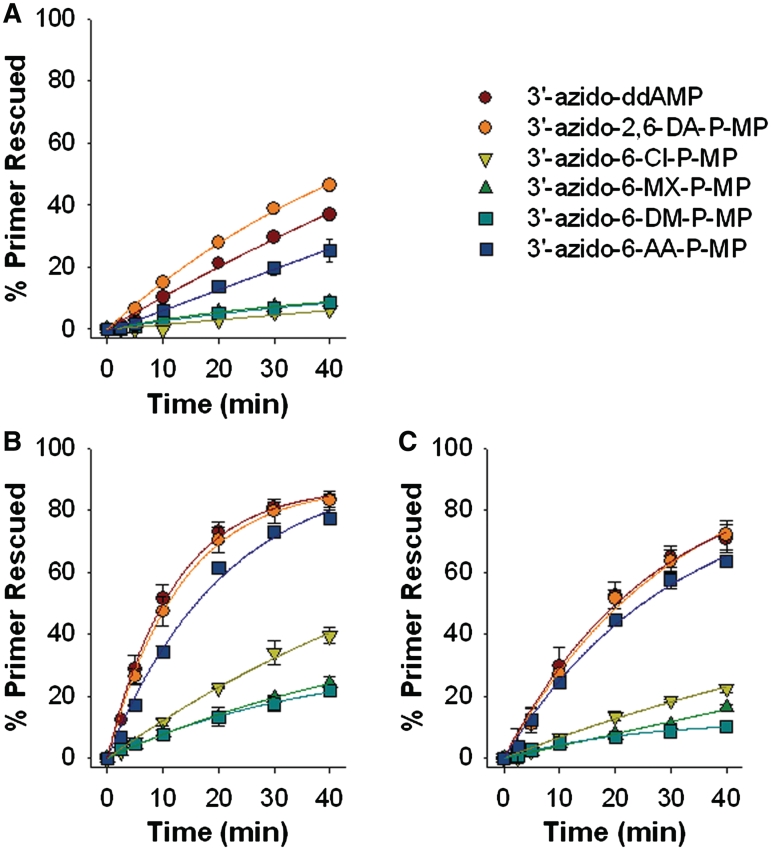
HIV-1 RT has the intrinsic ability to rescue DNA synthesis from an NRTI-MP blocked primer, using ATP as a phosphate donor (14). This ATP-mediated excision activity of HIV-1 RT is selectively increased by TAMs (15). Accordingly, we investigated the ability of WT and TAM-containing RT to excise the 6-modified-3′-azido-ddGMP analogs from a DNA chain-terminated T/P. Two RT enzymes that contained different patterns of TAMs [e.g. D67N/K70R/T215F/K219Q (TAM67) and M41L/L210W/T215Y (TAM41)] were studied. Our results (Figure 3) showed that the WT, TAM67, and TAM41 RTs could unblock T/Ps chain-terminated with 3′-azido-ddAMP, 3′-azido-2,6-DA-P-MP and 3′-azido-6-AA-P-MP, although TAM41 RT and TAM67 RT were more efficient in this regard than was the WT enzyme. By contrast, both the WT and TAM-containing RTs could not efficiently unblock T/P substrates chain-terminated with 3′-azido-6-MX-P-MP, 3′-azido-6-Cl-P-MP or 3′-azido-6-DM-P-MP.
Figure 3.
ATP-mediated excision of 6-modified-3′-azido-ddGMP analogs by WT (A), TAM67 (B) or TAM41 (C) HIV-1 RT. Data are the mean ± standard deviation from at least three independent experiments. Reaction conditions are described in the ‘Materials and Methods’ section.
Molecular models of 3′-azido-2,6-DA-P-TP and 3′-azido-6-Cl-P-TP in the active site of HIV-1 RT
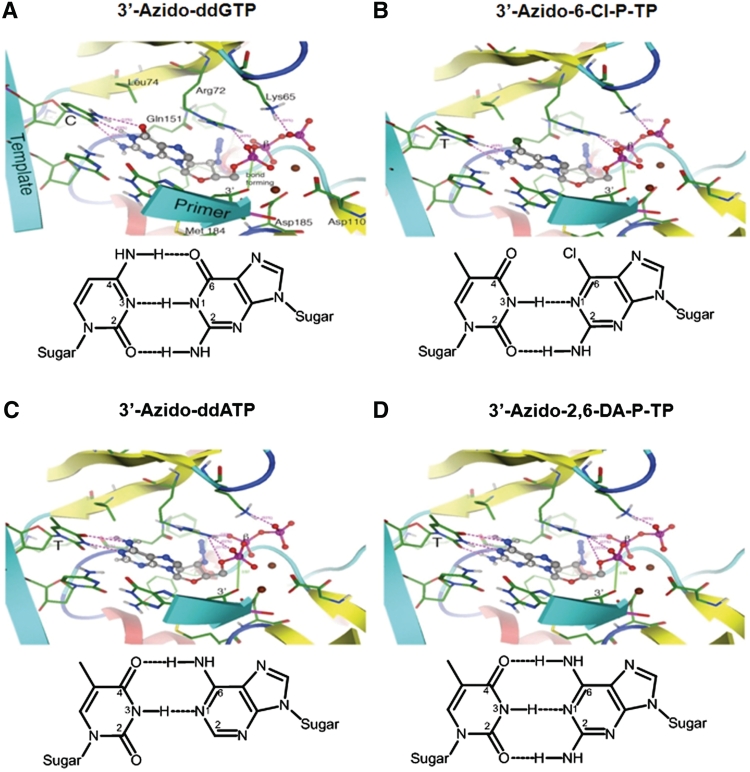
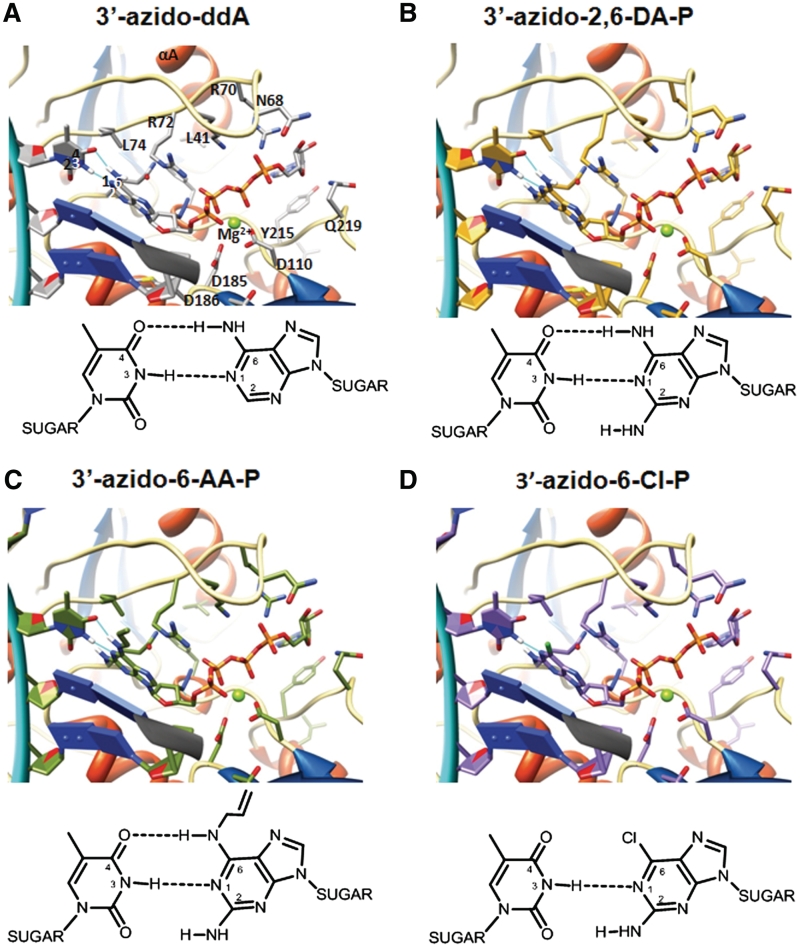
We used molecular modeling to gain structural insight into the binding interaction between the 6-modified-3′-azido-ddGTP analogs and the thymine base in the DNA template in the active site of HIV-1 RT. Specifically we modeled 3′-azido-2,6-DA-P-TP and 3′-azido-6-Cl-P-TP into the active site of HIV-1 RT using the co-ordinates (1RTD) from the RT-T/P-TTP complex (10). Both 3′-azido-2,6-DA-P-TP and 3′-azido-6-Cl-P-TP were found to fit comfortably into the active site of HIV-1 RT and formed hydrogen-bond interactions with the thymine base in the DNA template (Figure 4). However, the base hydrogen-bonding characteristics differed for each analog. The base of 3′-azido-6-Cl-P-TP forms two hydrogen-bond interactions through the nitrogen (NH) at position 1 and the amine (NH2) at position 2 with the template thymine (Figure 4B). In addition to these two hydrogen-bonding interactions, 3′-azido-2,6-DA-P-TP can form an additional hydrogen bond via the amino group (NH2) at the 6-position of the purine base (Figure 4D). Therefore, the 2,6-diaminopurine:thymine base-pairing interaction mimics the canonical Watson–Crick guanine:cytidine interaction.
Figure 4.
Molecular models of 3′-azido-ddGTP (A), 3′-azido-6-Cl-P-TP (B), 3′-azido-ddATP (C) and 3′-azido-2,6-DA-P-TP (D) in the DNA polymerase active site of HIV-1 RT. Each of the analogs was modeled into the ternary RT-T/P-TTP complex (pdb co-ordinates 1RTD). A schematic representation of the hydrogen-bonding patterns between the incoming nucleotide analog and the template base is depicted below each model.
Molecular models for ATP-mediated excision of the 6-modified-3′-azido-ddG-5′-MP analogs
To gain structural insight into why the 6-modified-3′-azido-ddG analogs are differentially excised by HIV-1 RT containing TAMs, we modeled their excision products (6-modified-3′-azido-ddG adenosine dinucleoside tetraphosphate) in the enzyme's active site. These models reveal a hydrogen-bonding network that is different from those observed for the TP forms. 3′-Azido-ddA forms hydrogen bonds from the 1 and 6 atoms to the 3 and 4 atoms of the complementary thymine template in both the incorporation (Figure 4C) and excision (Figure 5A) models. By contrast, the hydrogen bond observed between the 2-amino substitution and the 2-carbonyl of the thymine template observed for the nucleotide triphosphate analogs (Figure 4B and D) is lost in all related excision complexes (Figure 5B–D). 3′-azido-2,6-DA-P (Figure 5B) and 3′-azido-6-AA-P (Figure 5C) retain an H-bond from the 6 atom position of the purine ring and the 4 atom position on the template thymine, whereas this interaction is not maintained for 3′-azido-6-Cl-P (Figure 5D), 3′-azido-6-DM-P and 3′-azido-6-MX-P (data not shown). Taken together, these different hydrogen-bonding patterns may explain why 3′-azido-2,6-DA-P and 3′-azido-6-AA-P are more efficiently excised by HIV-1 RT than are 3′-azido-6-Cl-P, 3′-azido-6-DM-P and 3′-azido-6-MX-P.
Figure 5.
Molecular models of the excision products of 3′-azido-ddA (A), 3′-azido-2,6-DA-P (B), 3′-azido-6-AA-P (C) and 3′-azido-6-Cl-P (D) in the DNA polymerase active site of HIV-1 RT containing TAMs. Each of the analogs was modeled into the ternary RT-T/P-AZTppppA complex (pdb co-ordinates 3KLE). A schematic representation of the hydrogen-bonding patterns between the excised nucleotide analog and the template base is depicted below each model.
DISCUSSION
In this study, we demonstrated that 6-modified-3′-azido-ddGTP nucleotides act as adenosine mimetics for DNA synthesis carried out by HIV-1 RT. Of the five analogs studied, 3′-azido-2,6-DA-P-TP, which has a 2,6-diaminopurine base, was found to be the most efficient substrate for incorporation by HIV-1 RT. In pre-steady-state kinetic experiments, 3′-azido-2,6-DA-P-TP was recognized and incorporated by WT HIV-1 RT as efficiently as dATP and 3′-azido-ddATP. The analogs that contain the 2-amino-6-N,N-dimethylaminopurine (3′-azido-6-DM-P-TP) or 2-amino-6-methoxypurine (3′-azido-6-MX-P-TP) bases, however, were poor substrates for incorporation by HIV-1 RT. Our molecular modeling studies suggest that 6-modifications with a branched side-chain (e.g. 3′-azido-6-DM-P-TP) or one with electronic incompatibility (e.g. 3′-azido-6-MX-P-TP) may alter the alignment of the bases and move their α-phosphate away from the primer 3′-OH, thus impairing nucleotide incorporation. In contrast, 3′-azido-2,6-DA-P-TP, 3′-azido-6-CL-P-TP and 3′-azido-6-AA-P-TP form relatively planar base-pairing interactions with the template thymine that enhance their α-phosphate interaction with the 3′-OH of the primer. Compared to the canonical A:T base pair, the additional hydrogen bond achieved by the 3′-azido-2,6-DA-P:thymine base pair may further stabilize the α-phosphate, facilitating the favorable catalytic efficiency observed in pre-steady-state assays. In contrast, the 6-modified-3′-azido-ddGTP nucleotide analogs are inefficiently incorporated by HIV-1 RT opposite cytosine bases. Taken together, these findings are consistent with those of Cheong et al. who reported that oligodeoxyribonucleotide duplexes containing 2,6-diaminopurine formed more stable base pairs opposite thymine than cytosine (16).
The mutations K65R, L74V, Q151M (in complex with A62V, V75I, F77L and F116Y) or M184V in HIV-1 RT allow the enzyme to discriminate between the natural dNTP substrate and modified nucleotide analogs. In Table 2, we show that the K65R, L74V and Q151M mutations confer low levels of resistance to both 3′-azido-ddATP and 3′-azido-ddGTP at the enzyme level. Modification at the 6-position of the purine base does not appear to enhance this discrimination phenotype: the K65R, L74V or A62V/V75I/F77L/F116Y/Q151M HIV-1 RTs all exhibited fold-resistance changes to each of the 6-modified-3′-azido-ddGTP analogs that were largely similar (i.e. <2-fold) to the fold change in resistance values determined for 3′-azido-ddATP. Interestingly, the structure–activity relationship for the ATP-mediated excision of the 6-modified-3′-azido-ddGMP analogs by WT or TAM-containing HIV-1 RT differed from the structure–activity relationship for their incorporation. For example, whereas 3′-azido-6-CL-P-TP is a relatively good substrate for incorporation, it is not efficiently excised by either WT or TAM-containing HIV-1 RT. In this regard, our modeling data show that the hydrogen-bonding patterns observed for this analog differs from that of 3′-azido-2,6-DA-P and 3′-azido-6-AA-P (Figure 5). As such, it is possible that the change in transition state geometry required for excision is not sufficiently stabilized by the 6-Cl substituted guanine analog base-paired with thymine. Taken together, these data suggest that the optimal conformation of the HIV-1 RT active site differs for incorporation and excision and further suggests that 6-position modifications differentially effect these two reactions. This finding provides ‘proof of concept’ that modified base analogs can be identified that are efficiently incorporated by HIV-1 RT but serve as a poor substrate for HIV-1 ATP-mediated excision reactions. Further optimization of these divergent but favorable properties of purine analogs is in progress.
We previously demonstrated that 3′-azido-2,6-DA-P, 3′-azido-6-CL-P, 3′-azido-6-DM-P, 3′-azido-6-MX-P and 3′-azido-6-AA-P showed comparable or superior antiviral activities to 3′-azido-ddG in primary human cells (5, Figure 1). Intracellular pharmacology analyses, however, revealed that each of these compounds was efficiently metabolized by adenosine deaminase to 3′-azido-ddG in cells (5). Therefore, the observed anti-HIV-1 activity of each of these nucleoside analogs was not primarily due to incorporation of the ambiguous purine nucleotides described in this study. However, we recently identified RS-788, a 5′-monophosphate prodrug of 3′-azido-2,6-DA-P, as a potent and selective inhibitor of HIV-1 replication (Schinazi et al., 16th Conference on Retroviruses and Opportunistic Infections (2009) Abstract 557). In peripheral blood mononuclear cells, RS-788 is metabolized ~1:1 to both 3′-azido-2,6-DA-P-TP and 3′-azido-ddGTP. Consequently, RS-788 delivers two chemically distinct metabolites each of which are potent HIV-1 RT chain terminators that are incorporated opposite different complementary bases (cytosine for 3′-azido-ddGTP and thymine for 3′-azido-2,6-DA-P-TP). Therefore, our studies provide a promising new approach for the design and development of novel ambiguous NRTI that exert profound antiviral activity against WT and drug-resistant HIV-1.
FUNDING
The National Institutes of Health (R01-AI-071846 to J.W.M., 5P30-AI-050409 to R.F.S., 5R37-AI-041980 to R.F.S., 5R37-AI-025899 to R.F.S.) and the Department of Veterans Affairs (R.F.S.). Funding for open access charge: National Institutes of Health (grant R01 - AI - 071846).
Conflict of interest statement. Professor R.F.S. is a founder and major shareholder of RFS Pharma LLC. J.W.M. is a consultant to RFS Pharma and owns options in RFS Pharma.
ACKNOWLEDGEMENTS
J.H.N. thanks Sun Microsystems and Accelrys Corporation for Academic Achievement awards of modeling hardware and software.
REFERENCES
- 1.Goody RS, Müller B, Restle T. Factors contributing to the inhibition of HIV reverse transcriptase by chain-terminating nucleotides in vitro and in vivo. FEBS Lett. 1991;291:1–5. doi: 10.1016/0014-5793(91)81089-q. [DOI] [PubMed] [Google Scholar]
- 2.Parikh UM, Koontz DL, Chu CK, Schinazi RF, Mellors JW. In vitro activity of structurally diverse nucleoside analogs against human immunodeficiency virus type 1 with the K65R mutation in reverse transcriptase. Antimicrob. Agents Chemother. 2005;49:1139–1144. doi: 10.1128/AAC.49.3.1139-1144.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sluis-Cremer N, Arion D, Parikh U, Koontz D, Schinazi RF, Mellors JW, Parniak MA. The 3′-azido group is not the primary determinant of 3′-azido-3′-deoxythymidine (AZT) responsible for the excision phenotype of AZT-resistant HIV-1. J. Biol. Chem. 2005;280:29047–29052. doi: 10.1074/jbc.M503166200. [DOI] [PubMed] [Google Scholar]
- 4.Sluis-Cremer N, Koontz D, Bassit L, Hernandez-Santiago BI, Detorio M, Rapp KL, Amblard F, Bondada L, Grier J, Coats SJ, et al. Anti-human immunodeficiency virus activity, cross-resistance, cytotoxicity, and intracellular pharmacology of the 3′-azido-2′,3′-dideoxypurine nucleosides. Antimicrob. Agents Chemother. 2009;53:3715–3719. doi: 10.1128/AAC.00392-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang HW, Coats SJ, Bondada L, Amblard F, Detorio M, Asif G, Fromentin E, Solomon S, Obikhod A, Whitaker T, et al. Synthesis and evaluation of 3′-azido-2′,3′-dideoxypurine nucleosides as inhibitors of human immunodeficiency virus. Bioorg. Med. Chem. Lett. 2010;20:60–64. doi: 10.1016/j.bmcl.2009.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le Grice SF, Grüninger-Leitch F. Rapid purification of homodimer and heterodimer HIV-1 reverse transcriptase by metal chelate affinity chromatography. Eur. J. Biochem. 1990;187:307–314. doi: 10.1111/j.1432-1033.1990.tb15306.x. [DOI] [PubMed] [Google Scholar]
- 7.Le Grice SF, Cameron CE, Benkovic SJ. Purification and characterization of human immunodeficiency virus type 1 reverse transcriptase. Methods Enzymol. 1995;262:130–144. doi: 10.1016/0076-6879(95)62015-x. [DOI] [PubMed] [Google Scholar]
- 8.Ludwig J, Eckstein F. Rapid and efficient synthesis of nucleoside 5′-O-(1-thiotriphosphates), 5′-triphosphates and 2′,3′-cyclophosphorothioates using 2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one. J. Org. Chem. 1989;54:631–635. [Google Scholar]
- 9.Johnson KA. Rapid quench kinetic analysis of polymerases, adenosinetriphosphatases, and enzyme intermediates. Methods Enzymol. 1995;249:38–61. doi: 10.1016/0076-6879(95)49030-2. [DOI] [PubMed] [Google Scholar]
- 10.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 11.Sarafianos SG, Clark AD, Jr, Das K, Tuske S, Birktoft JJ, Ilankumaran P, Ramesha AR, Sayer JM, Jerina DM, Boyer PL, et al. Structures of HIV-1 reverse transcriptase with pre- and post-translocation AZTMP-terminated DNA. EMBO J. 2002;21:6614–6624. doi: 10.1093/emboj/cdf637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Labute P. Protonate3D: assignment of ionization states and hydrogen coordinates to macromolecular structures. Proteins. 2009;75:187–205. doi: 10.1002/prot.22234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tu X, Das K, Han Q, Bauman JD, Clark AD, Jr, Hou X, Frenkel YV, Gaffney BL, Jones RA, Boyer PL, et al. Structural basis of HIV-1 resistance to AZT by excision. Nat. Struct. Mol. Biol. 2010;17:1202–1209. doi: 10.1038/nsmb.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer PR, Matsuura SE, So AG, Scott WA. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc. Natl Acad. Sci. USA. 1998;95:13471–13476. doi: 10.1073/pnas.95.23.13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer PR, Matsuura SE, Mian AM, So AG, Scott WA. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol. Cell. 1999;4:35–43. doi: 10.1016/s1097-2765(00)80185-9. [DOI] [PubMed] [Google Scholar]
- 16.Cheong C, Tinoco I, Jr, Chollet A. Thermodynamic studies of base pairing involving 2,6-diaminopurine. Nucleic Acids Res. 1988;16:5115–5122. doi: 10.1093/nar/16.11.5115. [DOI] [PMC free article] [PubMed] [Google Scholar]