Abstract
Due to the increasing throughput of current DNA sequencing instruments, sample multiplexing is necessary for making economical use of available sequencing capacities. A widely used multiplexing strategy for the Illumina Genome Analyzer utilizes sample-specific indexes, which are embedded in one of the library adapters. However, this and similar multiplex approaches come with a risk of sample misidentification. By introducing indexes into both library adapters (double indexing), we have developed a method that reveals the rate of sample misidentification within current multiplex sequencing experiments. With ~0.3% these rates are orders of magnitude higher than expected and may severely confound applications in cancer genomics and other fields requiring accurate detection of rare variants. We identified the occurrence of mixed clusters on the flow as the predominant source of error. The accuracy of sample identification is further impaired if indexed oligonucleotides are cross-contaminated or if indexed libraries are amplified in bulk. Double-indexing eliminates these problems and increases both the scope and accuracy of multiplex sequencing on the Illumina platform.
INTRODUCTION
Over the last decade, new sequencing technologies have become available (1–5), which greatly outperform the older Sanger technology in terms of throughput and cost. Due to the large number of sequences generated with these technologies, there is a growing interest in sequencing multiple samples in parallel. Using different library construction protocols, sample-specific index sequences (also called ‘barcodes’) can be attached to the sample molecules during sequencing library preparation [e.g. (6–9)]. Subsequently, multiple libraries can be pooled and sequenced in the same region, and later computationally separated based on their index sequence. This facilitates highly parallel sequencing of a large number of samples (96 and more).
On the Illumina sequencing platform, the perhaps most widely used multiplexing strategy (the vendor's protocol) uses indexes, which are embedded within one of the adapters (8–10), separated from the actual template (Figure 1A). Thus, for a typical Illumina multiplex library, the index is sequenced after the forward read in a separate ‘index read’, for which a new sequencing primer is annealed. Although there are alternative indexing approaches, where the index is attached adjacent to the insert [e.g. (7)], this strategy has several benefits. Decoupling the actual template read and the index read allows the index read to be left out if not required and also keeps the sequencing error rate low, because phasing (3,11,12), one of the main sources of sequencing error on the Illumina platform, is reset with the annealing of a new sequencing primer. In addition, image analysis and the estimation of base calling parameters are not affected by the frequently unbalanced base composition of indexes.
Figure 1.
(A) Regular Illumina multiplex library design. The grafting sequences (P5 and P7) are used for template immobilization and amplification. Three distinct sequence reads (forward read, index read, reverse read) are primed from different adapter sites. (B) Double-index library design with an additional index incorporated into the second adapter. Here, four distinct sequence reads are performed.
While sample multiplexing greatly increases experimental scalability, it also introduces the danger of falsely assigning sequences to their original samples. Some applications, however, require highly accurate genotyping, particularly if conclusions are drawn from the occurrence of rare sequence variants. These could for example be rare transcripts or somatic mutations. In cancer research, for example, low-frequency somatic mutations can harbor important biological insights (13). The current throughput of Illumina sequencers is sufficiently high for sequencing exomes of several tumor/normal pairs to high coverage. It can therefore be anticipated that multiplex sequencing will soon be a common tool in many biomedical studies. Another very sensitive application—and the initial motivation behind this study—is ancient DNA research. Here, the observation of a single sequence may be taken as evidence for DNA survival or presence of contamination (14,15).
To avoid false-assignments of sequences to samples, previous studies have focused on generating highly distinguishable index designs (6,8,9,16), i.e. requiring several sequence changes before one index sequence is converted into another valid index sequence. This should efficiently reduce the number of index conversions due to errors in sequencing, library amplification or oligonucleotide synthesis. For example, assuming an error rate of 0.5% per position, an edit distance of three would correspond to a false-assignment probability of 4.31E-06 for 7-nt indexes (~172 per 40 million sequences in a lane of an Illumina Genome Analyzer IIx flow cell) under a simple binomial error distribution model. This is much lower than required for most applications. Another possible source of sample misidentification is cross-contamination of indexes by the oligonucleotide manufacturer or during later handling. This can be caused, for example, by sequential purification of different indexing primer/adapter oligonucleotides on the same high performance liquid chromatography (HPLC) column after synthesis. Even though columns are washed between oligonucleotides, low levels of carry-over contamination may be difficult to prevent.
In many cases, sequencing libraries need to be enriched for certain targets of interest, often by means of hybridization capture, before they are subjected to multiplex sequencing. To minimize efforts, it can be desirable to pool samples prior to target capture to perform both capture and sequencing in a multiplex setup. Since it is usually inevitable to amplify libraries after capture, this strategy introduces a step where libraries from different samples are amplified in a single reaction. Unfortunately, PCR can produce chimeras by recombining different templates molecules (a process often referred to as ‘jumping PCR’) (17–20). Consequently, multiplex capture may introduce significant levels of sample cross-contamination.
In order to better quantify and improve the accuracy of multiplexing sequencing on the Illumina platform, we have devised a new double-indexing method, which places indexes into both of the universal adapter sequences (Figure 1B), thereby extending the current system from three to four sequencing reads. Using Neandertal DNA extracts, we constructed double-indexed libraries with unique index combinations and performed three experiments. First, libraries were pooled only for sequencing (experiment no-CAP). Second, the same libraries were enriched individually for mitochondrial DNA using a recently published hybridization capture method (21) and then pooled for sequencing (experiment SP-CAP). Third, libraries were pooled prior to target enrichment, and hence capture, amplification and sequencing were all performed in a multiplex setup (MP-CAP), allowing for possible cross-contamination, caused by jumping PCR, to be quantified.
MATERIALS AND METHODS
Library preparation, amplification and target enrichment
Fifteen DNA extracts (L1–L15) were prepared using 100–200 mg of bone powder from two ~30 000-year-old Neandertal bones (Vi33.25 and Vi33.26 from Vindija Cave, Croatia) following the protocol of Rohland et al. (22). Two negative controls (L16 and L17) were carried through the extraction process. Sequencing libraries were prepared from the extracts using a previously published protocol (9) with the following modifications: (i) All SPRI purification steps were substituted by spin column purification (MinElute PCR purification kit, Qiagen). (ii) For L11-L15 and L17, USER enzyme mix (New England Biolabs) was added to the blunt-end repair reaction to remove uracils (23). (iii) Adapter concentration in the ligation reaction was reduced to 0.25 µM of each adapter. (iv) No purification step was performed after adapter fill-in with Bst polymerase. Instead, the enzyme was heat inactivated at 80°C for 20 min. The reaction mix was then used directly as template for PCR.
All libraries were amplified twice by PCR, using a polymerase that is capable of copying across deoxyuracils for the first, and a proof-reading polymerase for the second amplification. Using 5′-tailed primers (‘indexing primers’; see Supplementary Table S1 for all primer sequences), indexes were added to both ends of the library molecules during the first amplification. The entire library volumes were used as templates in 100 µl PCR reactions containing 1× Thermopol buffer (NEB), 5 U AmpliTaq Gold (Applied Biosystems), 250 μM each dNTP and 400 nM each indexing primer. Cycling conditions were comprised of an activation step lasting 12 min at 95°C, followed by 10 cycles of denaturation at 95°C for 20 s, annealing at 60°C for 30 s and elongation at 72°C for 40 s, with a final extension step at 72°C for 5 min. The index combinations used for each library are listed in Supplementary Table S2. PCR products were purified using the MinElute PCR purification kit and eluted in 20 µl EB. An amount of 5 µl of the eluates were used as template for the second round of amplification, which was performed in 100 µl reactions containing 1× Phusion High Fidelity Mastermix (NEB) and the primers IS5 and IS6 (9) at a concentration of 400 nM each. Cycling conditions were comprised of an activation step lasting 30 s at 98°C, followed by 10 cycles of denaturation at 98°C for 20 s, annealing at 60°C for 30 s and elongation at 72°C for 40 s, with a final extension step at 72°C for 5 min. PCR products were purified using the MinElute PCR purification kit and eluted in 10 µl EB. The concentrations of all libraries were determined on a Bioanalyzer 2100 (Agilent) using DNA 1000 chips.
Libraries were either directly pooled and sequenced (no-CAP experiment) or enriched for mitochondrial DNA. Enrichment was performed either individually (experiment SP-CAP) or in bulk (experiment MP-CAP) using a protocol detailed in Maricic et al. (21). After enrichment, the libraries in the SP-CAP and MP-CAP experiments were amplified for 24 cycles using Phusion polymerase under the conditions described above. Libraries were purified using the MinElute PCR purification kit, quantified on a Bioanalyzer 2100 and pooled in equimolar ratios.
Sequencing
Libraries were sequenced in three lanes of an Illumina Genome Analyzer IIx run (v4 chemistry, v2 cluster generation kit). Deviating from the manufacturer's instruction for a 2 × 101 + 7 cycles multiplexed paired end run, a φX174 control library was spiked into all lanes, contributing to on average about 1% of the reads in each lane. Furthermore, an additional seven-cycle index read was performed by repeating the chemistry steps of the first index (without commands marking this part of the read as index) at the end of the run recipe. This second index read used the custom sequencing primer shown in Figure 1B.
Data processing
The sequencing data was analyzed three times, once starting from QSEQ sequence files and CIF intensity files obtained from Illumina's Genome Analyzer SCS 2.6/RTA 1.6 software, and twice starting from raw images using OLB 1.8 and OLB 1.9. In all cases, the QSEQ raw reads obtained from Illumina's base caller Bustard were aligned to the φX174 reference sequence to obtain a training data set for the base caller Ibis 1.1.2 (12), which was then used to call bases and quality scores. The PF flag for each cluster was extracted from the QSEQ files of the Illumina pipeline output. Index pairs were analyzed starting from raw sequences and considering only perfect matches to the index sequences.
Sequences from OLB 1.8 intensity files (Ibis called) have been deposited in the ENA with accession number ERP000829.
RESULTS
Quantifying false index pairings
In contrast to current single-indexed multiplex sequencing, double-indexing allows for determining the sample origin of each sequence twice independently. Considering only perfect matches to the designated index sequences, we first compared the two index sequences to estimate the fraction of sequences with conflicting information on sample origin. From the previous considerations—based on a 0.5% sequencing error rate as the only source of error—we would expect to find approximately 172 false index pairs in 20 million reads (<0.001%). Using the sequencing error rate and other parameters of the actual experiments, we expect even fewer false index pairs (4 in 20 million reads, see Supplementary Methods section). However, in stark contrast to these expectations, we found 0.582% in no-CAP, 0.509% in SP-CAP and 0.760% in MP-CAP of the index pairings to be wrong (Table 1). Interestingly, we observed extremely high fractions of false index pairs irrespective of whether libraries were only sequenced together (no-CAP, SP-CAP) or also amplified together (MP-CAP), indicating that factors other than jumping PCR must contribute substantially to the fraction of false pairings.
Table 1.
Numerical summary of the false-assignment rates and fractions of false index pairs observed for the three different experiments no-CAP, SP-CAP and MP-CAP
| no-CAP | SP-CAP | MP-CAP | |
|---|---|---|---|
| Total number of raw reads | 34 241 955 | 48 546 372 | 34 684 183 |
| Index pairs in raw data | |||
| Correct pairs (*) (%) | 89.14 | 78.83 | 89.38 |
| False pairs (%) | 0.582 | 0.509 | 0.760 |
| False index pairs after PF-filtering of raw reads | |||
| Total number of PF-filtered reads | 27 466 817 | 37 586 292 | 27 220 161 |
| False pairs (%) | 0.523 | 0.387 | 0.691 |
| False index pairs after quality score based filtering of index reads | |||
| Average index quality filter (~PF) (%) | 0.059 | 0.192 | 0.423 |
| Mininum index quality filter (~PF) (%) | 0.060 | 0.177 | 0.422 |
| Minimum index quality score of 15 (%) | 0.060 | 0.138 | 0.428 |
| False index pairs after quality score based filtering of template reads | |||
| Read quality filter on both reads (~PF) (%) | 0.362 | 0.439 | 0.614 |
| Read quality filter on the forward read (~PF) (%) | 0.389 | 0.394 | 0.593 |
| Read quality filter on the reverse read (~PF) (%) | 0.298 | – | – |
| Quantifying cross-contamination, mixed clusters and jumping PCR | |||
| False pairs due to contamination (%) | 0.042 | 0.104 | 0.038 |
| False pairs due to mixed clusters / jump. PCR (%) | 0.018 | 0.034 | 0.390 |
~PF indicates values for a quality score cutoff that removes fewer raw reads than Illumina's Pass Filter (PF) flag.
*The fraction of correct index pairs is strongly affected by loading density. Denser loading in experiment SP-CAP led to a higher sequencing error rate and hence reduced the fraction of correct index pairs.
Removing mixed clusters with signal purity filters
To elucidate the major source of false index pairings, we first checked whether false pairs accumulate at the edges or in specific regions of the flow cell image tiles, but could not see any spatial pattern when overlaying the X,Y-coordinates for correct and false index pairs (data not shown). We then applied Illumina's Pass Filter (PF) flag to the raw sequence data, which is a widely used filter based on the signal purity of each cluster over the first 25 bases of the sequencing run. This filter is supposed to reduce the number of sequences from mixed clusters (i.e. PCR product colonies derived from more than one template molecule). In our experiments, ~80% of the sequences passed this filter (Table 1). Albeit unsatisfactory, we in fact observed a reduction in the fraction of falsely paired indexes (e.g. from 0.582% to 0.523% in no-CAP), suggesting that mixed clusters could be the source of these falsely paired indexes. To test this hypothesis, we manually checked the raw intensity signals from a few clusters with conflicting index reads. In all cases, we detected overlaying signals from at least two different sequence populations (see Supplementary Figure S1 and Supplementary Methods section). In addition, we analyzed sequence reads with conflicting index information from very short molecules, which are present in libraries constructed from ancient DNA. There, sequencing proceeds through the insert into the adapter, providing yet another independent observation of the index sequences. In almost all cases the template read generated a congruent index pair (311 of 311 in experiment no-CAP, 141 of 149 in SP-CAP, 99 of 109 in MP-CAP; see Supplementary Methods section), providing further evidence for the occurrence of mixed clusters on the flow cell.
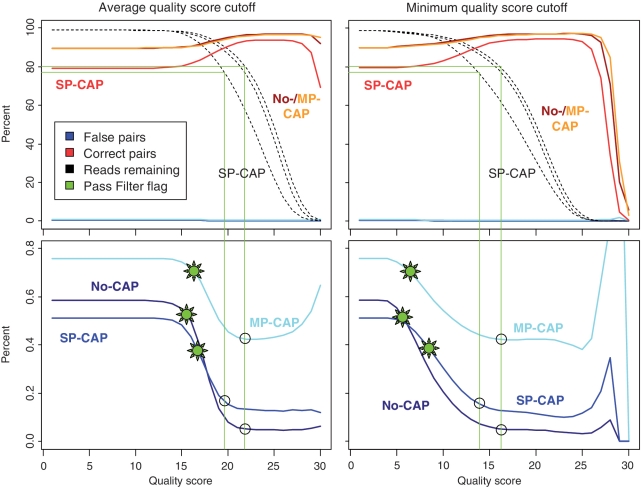
To efficiently eliminate sequences from mixed clusters, we explored the effect of applying a base quality score filter specifically to the index reads. Base quality scores are highly correlated with signal purity, but also incorporate signal strength. We considered a filter based on the average base quality score across the two index reads and another filter based on the minimum base quality score observed in the index reads. Using cut-offs that remove just a little less raw data than the Pass Filter flag, both filters remove considerably more false pairs than the Pass Filter flag (e.g. 0.059%/0.060% versus 0.523% false pairs remaining in no-CAP; Table 1). While all three experiments show similar trajectories for the different filter cut-offs (Figure 2), we note that the fraction of false pairs is always much higher for the MP-CAP experiment and that higher cluster densities in the SP-CAP experiment seem to negatively affect quality scores, as more data is removed using the same score cut-off (dashed black lines, Figure 2). To check whether the quality scores in the forward and reverse template reads also correlate with the fraction of false index pairs, we applied a minimum quality filter on those reads as well (Table 1 and Supplementary Figure S2). Although this filter is also more efficient than the PF flag, it removes fewer false pairs than a quality filter on the index reads. We therefore used a fixed minimum quality score filter of 15 on the index reads for all subsequent analyses.
Figure 2.
Changes in the fraction of false (blue) and correct (red) index pairs when applying two different types of base quality filters on the index reads (minimum accepted quality score and average quality score). The fraction remaining after PF is indicated by a green ‘sun’ symbol. Black circles denote the fraction of reads remaining when considering quality score cutoffs that remove just a little bit less raw data than the Pass Filter flag (green lines, ~20% of the data). Both filter criteria remove considerably more false pairs. While no-CAP, SP-CAP and MP-CAP show similar trajectories, the fraction of false pairs is always considerably higher for the MP-CAP experiment, in which samples have been enriched and amplified in a multiplex setup. Quality score cutoffs for the SP-CAP experiment are lower than for the other two experiments due to the 40% higher cluster density of this experiment.
Since we had stored the raw image data from the sequencing experiments, we were able to explore whether the occurrence of false index pairs changes if different versions of Illumina's image analysis software (RTA/OLB 1.6, 1.8 and 1.9) are used. We found that the newer image analysis software identified a larger number of clusters, but also increased the fraction of falsely paired indexes (Supplementary Table S3). We also compared the performance of Illumina's base caller Bustard to the base caller Ibis (12), both of which directly operate on cluster intensity files. For both base callers, the fraction of false pairs increases if a larger number of correct pairs is identified, indicating an improved performance of the newer image analysis software in extracting signals from low quality clusters.
Disentangling the factors causing false index pairs
Oligonucleotide synthesis, amplification and sequencing errors are expected to create false index pairs at comparatively low frequencies. It can therefore be assumed that the vast majority of false index pairs remaining after quality filtering (0.060% in no-CAP, 0.138% in SP-CAP and 0.428% in MP-CAP) are caused by (i) remaining sequences from mixed clusters, (ii) cross-contamination of oligonucleotides or indexed libraries, or (iii) chimera formation during bulk amplification of libraries from different samples (MP-CAP only). These factors are expected to generate different patterns of false pairings, which may be used to further disentangle the underlying causes. The conversion of indexes due to mixed clusters and jumping PCR can be assumed to occur uniformly across all index pairs. In contrast, cross-contamination of indexes is expected to be a sporadic process and its effect size may be assessed from unusually frequent false pairs.
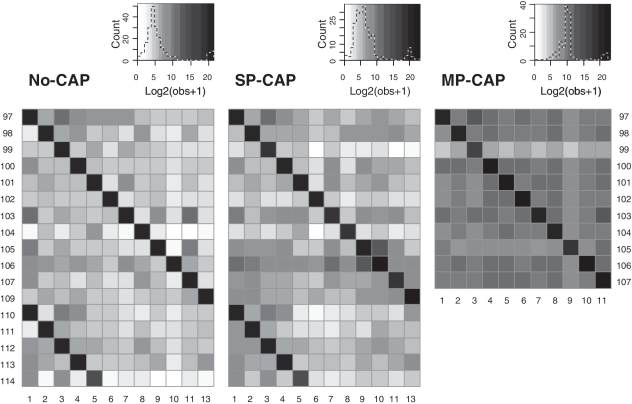
We therefore counted the occurrence of all possible index pairs in the three experiments (Figure 3). For identifying putative cross-contamination, we may identify false pairs with an overrepresentation of counts directly from the figure. Figure 3 clearly shows individual pairs, e.g. 11/103 and 11/105 in no-CAP, 97/3 and 10/105 SP-CAP, and 97/3 in MP-CAP, which are overrepresented compared to background. In addition, overrepresentation of complete rows/columns is also observed, e.g. the reverse index 1 in no-CAP and the forward index 106 in SP-CAP. Quantitatively, we checked for an overrepresentation of false index combinations compared to a background value calculated for each experiment (see Supplementary Methods section). When estimating cross-contamination from index pairs that are observed five times more frequently than the background, thus considering only the higher frequency false index pairs, we estimate that 0.042% (no-CAP), 0.104% (SP-CAP) and 0.038% (MP-CAP) of the false pairs are due to cross-contamination. The comparatively high contamination estimate for the SP-CAP experiment is also supported by another—albeit less powerful—analysis, which can be performed by counting the number of sequences derived from unused indexes (24) (see Supplementary Methods section).
Figure 3.
Heat map with the counts observed for all index combinations in the experiments no-CAP, SP-CAP and MP-CAP after applying a minimum quality score filter of 15 to the index reads. Only indexes that were actually used in the experiments are plotted; forward indexes on the horizontal and reverse indexes on the vertical axis. Color frequencies are provided for each of the experiments in the top right graphs.
Subtracting the number of false pairs derived from cross-contamination from the total number of false pairs provides an estimate for the fraction of false pairs that are caused either by remaining mixed clusters or jumping PCR. Interestingly, these numbers are low for no-CAP (0.018%) and SP-CAP (0.034%) and more than ten times higher for MP-CAP (0.390%). The low numbers in the first two experiments can be attributed to mixed cluster that could not be eliminated by quality filtering. The third experiment differs from the others in that libraries were amplified in bulk after target capture. We therefore conclude that jumping PCR generated ~0.36% chimeric reads with false index pairs in MP-CAP. If recombination happens predominantly along the adapter sequences, which are the regions of the sequencing library with the highest sequence similarity, half of the chimeric reads (0.18%) would be assigned to a false sample if only a single index read was used.
Quantifying false-assignment rates in single-indexed experiments
Assuming that both index reads are equally informative about sample origin, the false-assignment rate in experiment no-CAP would be 0.29% if only the first index was used to sort the sequences (this is the fraction of sequences with falsely paired indexes divided by two). This number can almost exclusively be attributed to sequences from mixed clusters, demonstrating that false-assignments of sequences to samples occur at high frequency in single-indexed multiplex sequencing experiments, particularly if no quality filter is imposed on the index read. Although double-indexing is the most powerful approach for identifying and excluding sequences from mixed clusters, false-assignment rates may also be inferred from existing single-indexed data, if samples with a large evolutionary distance are sequenced together. In this case, sample identification based on the index read can be compared to sample identification based on alignments to the respective reference genomes.
In our three experiments—as in all our recent sequencing runs—we spiked a φX174 control library into each lane of the flow cell (yielding ~1% of total reads). Sequences from this library are usually used to train the Ibis base caller (25) and to obtain measures of run quality for comparing different experiments. Unfortunately, φX174 and Neandertal library sequences are not sufficiently distinct for this analysis, since bacteriophage sequences may also derive from microbial contaminants in the bone. We therefore analyzed raw data from seven human genomes we sequenced recently (26). These samples were indexed, but sequenced on separate lanes of one run. Based on the sequences identified as φX174, we determined false-assignment rates in the range of 0.09%–0.22% (see Supplementary Table S4 and Supplementary Methods section). However, these numbers are very likely underestimates, because the requirement of successful alignments implicitly acts as a quality filter. When applying the stringent minimum quality score filter of 15, false assignment rates reduce to 0.01%–0.03%. Finally, we also analyzed sequencing data from mRNA libraries, which were constructed using a very different library preparation protocol (Illumina's TruSeq RNA Sample Prep Kit, FC-122-100x), and found similar results. Between 0.14% and 0.17% of φX174 reads erroneously occur with one of the sample indexes if no quality filter is applied to the index read (see Supplementary Table S5 and Supplementary Methods section). Thus, the high false-assignment rates we report do not represent artifacts of library preparation, but must be caused by the occurrence of mixed clusters on the flow cell.
DISCUSSION
Multiplex sequencing strategies have become indispensable for exploiting the capacities of high-throughput sequencing technologies in a cost- and time-efficient manner. However, little emphasis has been placed on directly assessing the level of confidence at which sequences are assigned to their source samples, probably because these strategies are believed to be sufficiently accurate for most applications based on theoretical considerations. Sequences generated with our new double-indexing method reveal actual false-assignment of up to 0.3%, orders of magnitude higher than expected. The overwhelming majority of false assignments can be explained by mixed clusters, i.e. clusters originating from two different template molecules or clusters growing into each other. False assignment occurs if the dominance of signals changes in different reads or if different signals are tracked. Although we do not understand the exact underlying processes, we independently verified the existence of this effect in data from single-indexed multiplex sequencing experiments. Thus, it neither represents an artifact of the double-indexing method nor the library preparation protocols used.
In many cases, false-assignment rates in the order of 3 in 1000 sequences can be highly problematic. For example, rare somatic mutations are used as biomarkers for cancer (27–29) or for studying mitochondrial heteroplasmy (30,31). Gene expression studies may be confounded by the bleeding-over of sequences from one sample to another. High false-assignment rates may even be problematic for accurate genotyping in studies using targeted re-sequencing. If, for instance, enrichment success in hybridization capture varies among samples, sequences bleeding over from a highly enriched sample (e.g. a positive control) may eventually constitute several percent of the target sequences in a weekly enriched sample. Apart from mixed clusters, we identified two other major sources of error that lead to false assignments of sequences to samples. The first is sporadic cross-contamination of oligonucleotides carrying different indexes, which may be introduced during synthesis or subsequent handling step. Despite being cautious, we were not able to completely avoid this type of contamination in our experiments. The second, PCR jumping, occurs only in experiments where sequencing libraries from multiple samples are amplified in bulk, leading to a significant fraction of chimeric molecules (~0.4% in our experiment). Relative to these errors, amplification and sequencing errors, which were often focused on in previous studies, occur at negligible levels.
We developed two strategies to improve the accuracy of multiplex genotyping on the Illumina platform. The first and most powerful strategy is using our double-indexing method. Here, sample identification is performed twice for each template molecule, enabling an exponential decrease of the false-assignment rates. For example, by identifying and removing false index pairs in experiment MP-CAP, the false-assignment rate drops from ~2 in 1000 to less than 1 in 100 000. Thus, using double-indexing, accurate genotyping becomes possible even if libraries from different samples are amplified in bulk or if cross-contamination is present among indexed oligonucleotides. Moreover, double-indexing greatly reduces the costs of highly multiplexed sequencing. If only 50 indexed oligonucleotides are synthesized for each of the two adapters, 2500 index combinations are theoretically available. Since this level of multiplexing is hardly ever required, the majority of index combinations will remain unused. This allows for determining false-assignment rates with nearly the same precision as if each index is only used once. Although double-indexing is recommended for all applications requiring bulk amplification of indexed libraries (e.g. multiplex target capture) or extraordinary levels of accuracy, we suggest a second—not mutually exclusive—strategy for reducing false-assignment rates also in single-indexed experiments, which is applying a quality filter on the index read. However, with this strategy it remains impossible to estimate false-assignment rates in single-indexed experiments, unless samples with a large evolutionary distance are sequenced together. Spiking in a φX174 control library will often be suitable for this purpose.
Alternative indexing strategies for the Illumina platform have been developed where an index is attached immediately adjacent to the template molecule. Using these strategies, index and template are sequenced simultaneously in the forward read. In consequence, the confounding effect of mixed clusters can be expected to be much smaller. However, as with all single-indexed approaches, other sources of sample misidentification cannot be prevented, most notably oligonucleotide cross-contamination and jumping PCR. We conclude that incorporating indexes into both ends of library molecules is a very powerful approach for improving the accuracy of multiplex sequencing. This approach can in principle also be extended to other high-throughput sequencing platforms to reduce the errors common to multiplex sequencing in general and uncover problems inherent to the specific technology.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online: Supplementary Tables 1–5, Supplementary Figures 1–2, Supplementary Methods, Supplementary Reference (32).
FUNDING
Max Planck Society. Funding for open access charge: Max Planck Society.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the members of the Department of Evolutionary Anthropology, in particular Janet Kelso and Svante Pääbo, for providing interesting discussions and useful insights.
REFERENCES
- 1.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 3.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris TD, Buzby PR, Babcock H, Beer E, Bowers J, Braslavsky I, Causey M, Colonell J, Dimeo J, Efcavitch JW, et al. Single-molecule DNA sequencing of a viral genome. Science. 2008;320:106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 5.Korlach J, Marks PJ, Cicero RL, Gray JJ, Murphy DL, Roitman DB, Pham TT, Otto GA, Foquet M, Turner SW. Selective aluminum passivation for targeted immobilization of single DNA polymerase molecules in zero-mode waveguide nanostructures. Proc. Natl Acad. Sci. USA. 2008;105:1176–1181. doi: 10.1073/pnas.0710982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer M, Stenzel U, Myles S, Prufer K, Hofreiter M. Targeted high-throughput sequencing of tagged nucleic acid samples. Nucleic Acids Res. 2007;35:e97. doi: 10.1093/nar/gkm566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craig DW, Pearson JV, Szelinger S, Sekar A, Redman M, Corneveaux JJ, Pawlowski TL, Laub T, Nunn G, Stephan DA, et al. Identification of genetic variants using bar-coded multiplexed sequencing. Nat. Methods. 2008;5:887–893. doi: 10.1038/nmeth.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mamanova L, Coffey AJ, Scott CE, Kozarewa I, Turner EH, Kumar A, Howard E, Shendure J, Turner DJ. Target-enrichment strategies for next-generation sequencing. Nat. Methods. 2010;7:111–118. doi: 10.1038/nmeth.1419. [DOI] [PubMed] [Google Scholar]
- 9.Meyer M, Kircher M. Illumina Sequencing Library Preparation for Highly Multiplexed Target Capture and Sequencing. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5448. pdb prot5448. [DOI] [PubMed] [Google Scholar]
- 10. Illumina Inc. (2008). 770-2008-011 ed http://www.illumina.com/Documents/products/datasheets/datasheet_sequencing_multiplex.pdf (11 September 2011, date last accessed)
- 11.Erlich Y, Mitra PP, delaBastide M, McCombie WR, Hannon GJ. Alta-Cyclic: a self-optimizing base caller for next-generation sequencing. Nat. Methods. 2008;5:679–682. doi: 10.1038/nmeth.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kircher M, Stenzel U, Kelso J. Improved base calling for the Illumina Genome Analyzer using machine learning strategies. Genome Biol. 2009;10:R83. doi: 10.1186/gb-2009-10-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green RE, Briggs AW, Krause J, Prufer K, Burbano HA, Siebauer M, Lachmann M, Paabo S. The Neandertal genome and ancient DNA authenticity. EMBO J. 2009;28:2494–2502. doi: 10.1038/emboj.2009.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stiller M, Knapp M, Stenzel U, Hofreiter M, Meyer M. Direct multiplex sequencing (DMPS)–a novel method for targeted high-throughput sequencing of ancient and highly degraded DNA. Genome Res. 2009;19:1843–1848. doi: 10.1101/gr.095760.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyerhans A, Vartanian JP, Wain-Hobson S. DNA recombination during PCR. Nucleic Acids Res. 1990;18:1687–1691. doi: 10.1093/nar/18.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paabo S, Irwin DM, Wilson AC. DNA damage promotes jumping between templates during enzymatic amplification. J. Biol. Chem. 1990;265:4718–4721. [PubMed] [Google Scholar]
- 19.Odelberg SJ, Weiss RB, Hata A, White R. Template-switching during DNA synthesis by Thermus aquaticus DNA polymerase I. Nucleic Acids Res. 1995;23:2049–2057. doi: 10.1093/nar/23.11.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lahr DJ, Katz LA. Reducing the impact of PCR-mediated recombination in molecular evolution and environmental studies using a new-generation high-fidelity DNA polymerase. Biotechniques. 2009;47:857–866. doi: 10.2144/000113219. [DOI] [PubMed] [Google Scholar]
- 21.Maricic T, Whitten M, Paabo S. Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLoS One. 2010;5:e14004. doi: 10.1371/journal.pone.0014004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rohland N, Hofreiter M. Comparison and optimization of ancient DNA extraction. Biotechniques. 2007;42:343–352. doi: 10.2144/000112383. [DOI] [PubMed] [Google Scholar]
- 23.Briggs AW, Stenzel U, Meyer M, Krause J, Kircher M, Paabo S. Removal of deaminated cytosines and detection of in vivo methylation in ancient DNA. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp1163. doi:10.1093/nar/gkp1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer M, Stenzel U, Hofreiter M. Parallel tagged sequencing on the 454 platform. Nat. Protoc. 2008;3:267–278. doi: 10.1038/nprot.2007.520. [DOI] [PubMed] [Google Scholar]
- 25.Kircher M, Kelso J. High-throughput DNA sequencing–concepts and limitations. Bioessays. 2010;32:524–536. doi: 10.1002/bies.200900181. [DOI] [PubMed] [Google Scholar]
- 26.Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY, Viola B, Briggs AW, Stenzel U, Johnson PL, et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, Morsberger LA, Latimer C, McLaren S, Lin ML, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell PJ, Stephens PJ, Pleasance ED, O'Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat. Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M, Schonberg A, Schaefer M, Schroeder R, Nasidze I, Stoneking M. Detecting heteroplasmy from high-throughput sequencing of complete human mitochondrial DNA genomes. Am. J. Hum. Genet. 2010;87:237–249. doi: 10.1016/j.ajhg.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He Y, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, Velculescu VE, Diaz LA, Jr, Kinzler KW, Vogelstein B, Papadopoulos N. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010;464:610–614. doi: 10.1038/nature08802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.