Abstract
We have developed a cost-effective, highly parallel method for purification and functionalization of 5′-labeled oligonucleotides. The approach is based on 5′-hexa-His phase tag purification, followed by exchange of the hexa-His tag for a functional group using reversible reaction chemistry. These methods are suitable for large-scale (micromole to millimole) production of oligonucleotides and are amenable to highly parallel processing of many oligonucleotides individually or in high complexity pools. Examples of the preparation of 5′-biotin, 95-mer, oligonucleotide pools of >40K complexity at micromole scale are shown. These pools are prepared in up to ~16% yield and 90–99% purity. Approaches for using this method in other applications are also discussed.
INTRODUCTION
Purification and modification of synthetic oligonucleotides are fundamental technologies required for many applications in the life sciences industry. The development of efficient techniques for large-scale oligonucleotide functionalization and purification is of great interest due to the increased demand for labeled oligonucleotides in many genomics applications, including genomic assays, gene synthesis and oligonucleotide-based drugs/therapeutics (1). Purification strategies perform the following functions: (i) separation of full-length oligonucleotides from incomplete products; (ii) removal of products that are not completely deprotected, or that are depurinated or dimerized; (iii) desalting; (iv) and removal of cleaved blocking groups. Common purification methods include alcohol-based precipitation, thin-layer chromatography (TLC), ion-exchange and reverse-phase Fast Protein Liquid Chromatography (FPLC), purification cartridges (oPC, etc.), PolyAcrylamide Gel Electrophoresis (PAGE) gels, purification by polymerization (2–4), final ‘Trityl-ON’ option followed by HPLC purification or phase tags such as Fluorous or biotin (5,6). The general strategy of phase tags is based on traditional solid-phase oligonucleotide synthesis, followed by purification using a 5′-terminal phase tag. Each step of the synthesis is followed by a capping step in order to prevent deletion sequences from accumulating. The phase-tag is added at the end of the synthesis, resulting in a mixture of full-length, phase-tagged oligonucleotide and non-tagged deletion sequences (4,7,8). Subsequently, the tagged oligonucleotide is bound to a solid phase (e.g. Fluorous or polymer), undesired capped sequences are removed by washing and the purified, full-length, tagged oligonucleotide is released. The appropriate purification method or combination of purification methods depends on its application, the size of the oligonucleotide, the degree of purity required, the quantity and number of oligonucleotides to be purified, time available and the cost of the method (http:/www.abrf.org/ABRFNews/1992/June1992/jun92oligo.html) (9).
We recently described an oligonucleotide synthesis technology that can produce >40 K oligonucleotides (10–120-mers) per day with an average yield of 10 nmol per oligonucleotide (10). These oligonucleotides are used in DNA microarray production (11), genomic-based assays (12), targeted sequencing (TS) applications (13), decoding arrays (14), Infinium® and GoldenGate® genotyping assays (15–20) and many other applications. For many of these, a high degree of purification and functionalization is required. For example, building a fixed-set, targeted exome sequencing product requires a pool of >300K oligonucleotides in large quantities (micromole to millimole) with >95% biotin-labeling. Although many purification methods can deliver highly pure and functionalized oligonucleotides, current approaches cannot purify large quantities of pools of 5′-functionalized oligonucleotides at low cost and in a fast, highly parallel fashion. The need is especially great for oligonucleotides >80-mers, as purification becomes more challenging due to smaller size differences between desired and truncated products.
In this article, we describe novel large-scale purification and preparation methods for individual or highly complex pools of 5′-biotinylated oligonucleotides. The methods are based on (i) 5′-hexahistidine phase tag (5′-hexa-His) oligonucleotide purification and (ii) exchanging the hexa-His tag for a biotin group (21). We demonstrate for the first time that the purification scale can be matched with the oligonucleotide synthesis scale. Additionally, we demonstrate that pools of >40K oligonucleotides can be purified and derivatized at the 5′ end simultaneously. The method is highly automatable, scalable, cost-effective and relatively independent of oligonucleotide length.
MATERIALS AND METHODS
Oligonucleotides with a 5′-aldehyde functional group were synthesized on a custom-built continuous oligonucleotide synthesizer at Illumina, Inc. at an average yield of 10 nmol per oligonucleotide (10). Aldehyde modifiers such as 5′ benzaldehyde-modifier-C2 phosphoramidite and 5-formylindole-CE phosphoramidite were suitable in the described synthesis scheme. The oligonucleotides were concentration-normalized and pooled to 42K complexity using a Microlab Star Hamilton robot. The oligonucleotides were received in 0.1× TE buffer. A single oligonucleotide with a 5′-aldehyde functional group was purchased from TriLink, Inc. and delivered dry. This oligonucleotide (5′ALD/GGAATACAGTTGGCTGATGGTGGATGGCTCATACCCTCCAATGATGGAAAGGCTGGAAAAGAAGAATTTTATAGGGCTCTGTGTGACACTCCAGG) was used to characterize the chemical transformations and was processed in a similar manner as the pools of oligonucleotides. Plasticware and other miscellaneous laboratory equipment were purchased from Pierce, VWR, Millipore or Cole-Parmer. All other reagents were purchased from Sigma-Aldrich, Dojindo Molecular Technologies, Solulink, Clontech Laboratories and Life Technologies.
Synthesis of 5′-hexa-His labeled oligonucleotides
300 ml of the 42 K pool of 5′-aldehyde-labeled oligonucleotides (100 µM) was transferred to two aliquots in 225 ml centrifuge tubes and precipitated with cold ethanol/sodium acetate. The pellet was washed once with 25 ml of cold 70% ethanol and dried overnight in a vacuum oven set at room temperature. The pellet was redissolved in 150 ml of 1 M citrate buffer at pH 6.0, and an aliquot was quantified by optical absorbance spectroscopy on a NanoDrop ND-1000 UV-Vis spectrophotometer. Next, 150 ml of 10 M urea solution was added, bringing the total concentration of oligonucleotides to 100 µM in the solution. Subsequently, a 20-to-1 molar excess of HyNic/PEG2/Hexa-His (Solulink PN SP-E001-005) was added. 0.66 g of HyNic/PEG2/Hexa-His was dissolved in 1 ml of 1 M citrate buffer at pH 6.0, and added to the reagent mix. Next, 2.1 g of neat aniline was added for a final aniline concentration of 100 mM in the solution. The tube was capped and placed on a rotisserie for 1 h at room temperature. Next, the oligonucleotides were precipitated with cold ethanol/sodium acetate (dividing the reaction into 6 × 50 ml aliquots, and adding 150 ml of cold ethanol to each aliquot), and washed three times with 25 ml of cold 70% ethanol and then dried in a vacuum oven set at room temperature for 4 h. The pellets of each aliquot were redissolved in 50 ml of pre-warmed, HPLC-grade water and pooled.
Purification of 5′-hexa-His labeled oligonucleotides
Nickel–resin was obtained from Clontech (His60 Ni Superflow Resin, Clontech PN 635659). Next, 200 ml of pre-homogenized Ni–resin slurry was added to each of the two separate 225 ml centrifuge tubes. Each tube was centrifuged at 1200 rcf on an Eppendorf 5810 centrifuge for 5 min. The liquid was removed by aspiration and pellets were washed with 100 ml of a solution of 5× Phosphate Buffered Saline (PBS) buffer (pH 7.4) containing 8 M urea. The washing was repeated three times to equilibrate the resin. In all, 25 ml of the 50 ml of the post-His conversion reaction was placed into each centrifuge tube. An additional 75 ml of the 5× PBS buffer containing 8 M urea was added into each centrifuge tube of equilibrated slurry. The tubes were capped, and placed on the rotisserie at room temperature for a minimum of 4 h.
The tubes were then centrifuged at 1200 rcf for 5 min at room temperature. The liquid was removed by aspiration and discarded. The resin was transferred to a plastic Buchner vacuum filter flask (Nalgene, from VWR) and washed with 500 ml of 1× PBS (pH 7.4) containing 20 mM imidazole. Next, the resin was washed with 200 ml of 0.01 N NaOH. The resin was then washed with 1 L of 1× PBS (pH 7.4) containing 20 mM imidazole. The resin was split into two aliquots and transferred to new 225 ml centrifuge tubes. To each tube was added 100 ml of 500 mM imidazole and 1× PBS buffer. The tubes were placed on the rotisserie for 5 min and centrifuged for 5 min at 1200 rcf. The liquid was aspirated and collected. The resin was then transferred to a Buchner vacuum filter flask and washed, in parts, with 500 ml of 500 mM imidazole solution and 1× PBS buffer. The collected washes were concentrated down to ~15 ml using a Centricon 70 (10k MWCO, Millipore PN UFC701008) centrifugal dialysis assembly. The solution was washed with 4 × 50 ml HPLC-grade water, concentrating after each wash with the Centricon centrifugal dialysis assembly. The final solution was concentrated to ~20 ml and isolated.
A 10 µl aliquot of the concentrated solution was quantified using the NanoDrop instrument, and 20 µl of the solution at 10 µM concentration was sent to Novatia LLC (www.enovatia.com) for LC–MS analysis as a modified 95-bp oligonucleotide. Another 10 µl aliquot was used for the FPLC analysis, as described below.
Synthesis of 5′-biotin labeled oligonucleotides
The hexa-His product was transferred to a new 225 ml centrifuge tube. Subsequently, 150 ml of 1 M citrate buffer at pH 6.0 and 150 ml of 10 M urea solution were added. A 10-fold molar excess of the hydrazide-HyNic-biotin (Solulink) was used relative to the total amount of oligo in the solution. For 30 µmol of total oligonucleotide, 300 µmol of the biotin-oxo-amine was added. Subsequently, 2.7 g of aniline was added, and the tube was capped and placed on a rotisserie at room temperature for 4 h. The oligonucleotides were then precipitated out using cold ethanol and sodium acetate, the pellet was washed with 3 × 25 ml of cold 70% ethanol, and the pellet was dried in a vacuum oven overnight at room temperature. The pellet was redissolved in pre-warmed 50 ml HPLC-grade water. The 50 ml solution of biotinylated oligonucleotides was mixed with the Nickel–resin slurry as described above, except the flow-through, containing the biotinylated oligonucleotides, was collected and retained. The flow-through was concentrated using a Centricon 70 centrifugal dialysis apparatus as described above, and washed 4 × 50 ml with HPLC-grade water down to a final concentration of ~25 ml in HPLC-grade water. A 10 µl aliquot of the final purified solution was quantified with optical absorbance spectroscopy on a NanoDrop instrument, and for the LC–MS analysis and FPLC analyses. The final yields for pool 1–8 were 2.49, 2.43, 2.74, 2.48, 2.68, 3.13, 2.18, 2.16 µmol, respectively. The final process yields for the biotin oligonucleotide pools 1–8 were 8.3, 8.1, 9.2, 9.0, 9.0, 10.5, 7.3 and 7.2%, respectively. A 15.8% overall yield for pool 7 or 8 was observed when the exchange reaction was performed with biotin-oxo-amine (Dojindo PN A305-10).
FPLC analysis
FPLC analysis was performed with a GE Akta Explorer FPLC system fitted with a Nickel–resin cartridge (HisPur Ni-NTA Chromatography Cartridge, Pierce PN 90099), using a method similar to that described by the column vendor for FPLC analysis of hexa-His-labeled biomolecules. The highest quality imidazole (BioUltra, Sigma-Aldrich, 56749) gave the best performance in our hands. The signal of the oligonucleotides was monitored at 260 and 280 nm. Integration was performed using the Agilent ChemStation software using classical baseline correction, standard tangent skim mode, tail peak skim height ratio is 0, front peak skim height ration is 0, skim valley ratio is 20 and peak to valley ratio is 500. The samples were prepared as 50 µl volume in water at 20 µM concentration. The flow rate was set at 0.2 ml/min. The binary solvent profile was 100% Buffer A until after the non-hexa-His-containing biomolecules were eluted (~25 min), then 100% Buffer B to elute off the hexa-His-containing biomolecules. Buffer A consisted of 50 mM phosphate, pH 7.4, 20 mM imidazole and 300 mM NaCl. Buffer B consisted of 50 mM phosphate, pH 7.4, 300 mM imidazole and 300 mM NaCl.
RESULTS
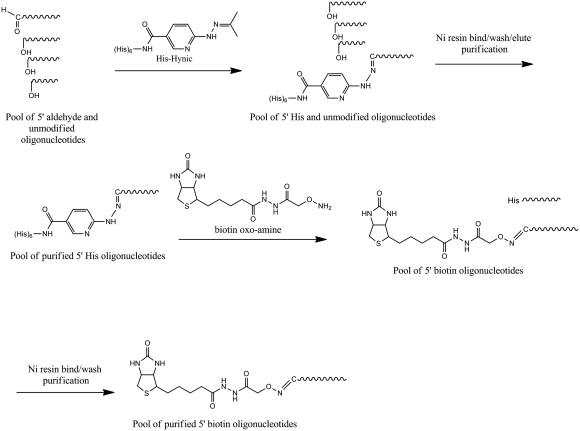
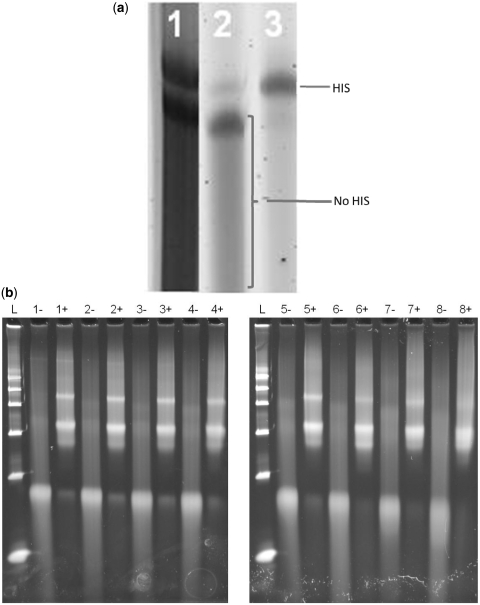
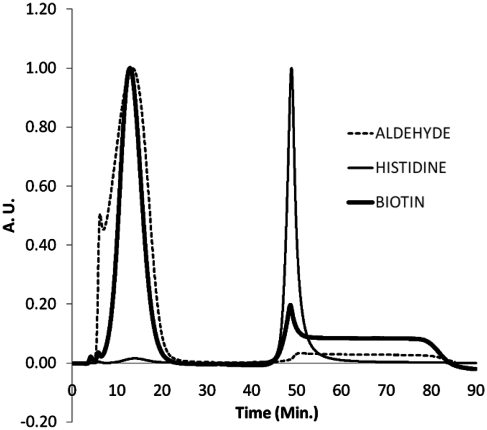
The approach of the method is depicted in Scheme 1. The method is based on synthesizing 5′-aldehyde-modified oligonucleotides, and utilizing the aldehyde group for hexa-His phase-tag attachment, followed by 5′-Biotin derivatization using an exchange reaction (21). We synthesized >300 K different 5′-aldehyde-modified oligonucleotides of 95 bp in length using a custom built continuous oligonucleotide synthesizer (10,22). The sequences of the oligonucleotides were designed against the UCSC Human Exome (NCBI/Hg19 reference genome), enabling the final biotin-oligonucleotides to be used in whole-exome TS applications (Steemers,F.J., in preparation; 23,24). The oligonucleotides were subsequently pooled into eight pools of 42 K complexity. Each pool contained oligonucleotides of a specific GC-range (Table 1), allowing specific GC-effects to be observed during the synthesis and purification. The aldehyde oligonucleotide pools were functionalized with a hexa-His tag using Hynic/Peg2/Hexa-His (25). The conversion efficiency based on FPLC and LC-MS analysis was >80% per pool. Each hexa-His-tagged oligonucleotide pool was subsequently purified using a Ni–resin (26). Oligonucleotides that lacked the hexa-His-tag, truncated oligonucleotides and synthesis failures, were removed by high-pH (0.01 N NaOH) washing. The hexa-His oligonucleotides were eluted using an imidazole elution with Dithiothreitol (DTT), (10 mM) (27). Gel analysis showed that the hexa-His product was obtained as the majority product (Figure 1a). Interestingly, we observed a small but gradual reduction in purity of the pools for the higher percent GC pools. For example, pool 1 has a purity of 95%, while pool 8 has a purity of 85% using FPLC analysis. In the subsequent step, the hexa-His tag was exchanged for a 5′-biotin moiety using biotin-hydrazide or biotin-oxo-amine (21). The conversion efficiency based on FPLC and LC-MS analysis was >80%. Unconverted hexa-His oligonucleotides were removed using simple Ni–resin filtration, and the final biotin-labeled oligonucleotides were obtained in ~7–11% yield for each of the pools using biotin-hydrazide. The purity and yield of the pools were established by capturing the FPLC trace signal (Figure 2). Overall, we obtained a purity of 97–99% (pools 1–6), 95% (pool 7), 90% (pool 8) based on 260 nm absorbance. Again, as observed with the hexa-His oligonucleotide intermediate method, the purity of the product decreased going from low to high percent GC pools. Strikingly, a doubling of the final yield to ~16% was observed for pools 7 and 8 when biotin-hydrazide was substituted for biotin-oxo-amine. These metrics were validated by using purified hexa-His oligonucleotides as input material. Additional confirmation that the final purified biotin-oligonucleotide pools were indeed biotin-labeled was obtained using a streptavidin shift assay using an agarose gel analysis (Figure 1b). Measuring the reduction in gel migration with excess streptavidin (+) versus no streptavidin (−) showed that the majority of products shifted upon addition of streptavidin, indicating that >95% of the oligonucleotides were biotinylated. Supplementary Figure 1c presents the streptavidin shift assay, except with oligonucleotides that had been biotinylated directly, without using the purification process described herein. As expected, only a very small amount of biotinylated oligonucleotide was obtained, and with very low purity. Several key steps in the protocol were identified to increase the process yield. We found that adding urea to the Ni-column/His-oligonucleotide binding step improves yield by up to 20–70%, depending on the percent GC of the pool. The higher percentage GC-pools yielded greater yield improvement with the addition of urea. In the same step, we observed yield improvements when the binding of hexa-His oligonucleotide to Ni–resin was performed as slurry instead of a traditional column format. Increased binding time up to 4 h contributed an additional ~5% to the yield improvement. The addition of urea in the exchange reaction also showed a 20–70% yield increase. Again, the higher percentage GC-pools yielded greater yield improvement with the addition of urea.
Scheme 1.
Synthesis and purification scheme for pools of 5′ biotin oligonucleotides
Table 1.
Oligonucleotide pools and their %GC range
| Pools | %GC |
|---|---|
| Pool 1 | 15–37 |
| Pool 2 | 37–42 |
| Pool 3 | 42–46 |
| Pool 4 | 46–49 |
| Pool 5 | 49–50 |
| Pool 6 | 50–55 |
| Pool 7 | 55–61 |
| Pool 8 | 61–88 |
Figure 1.
(a) Gel analysis of the purification process of a 42K oligonucleotide pool: (1) unpurified aldehyde oligonucleotides, (2) wash from Ni-column, (3) eluted hexa-His oligonucleotides. (b) Gel shift analysis of the biotinylated oligonucleotide pools 1–8 with (+) and without (−) streptavidin.
Figure 2.
Normalized FPLC traces based on maximum peak height for pool 2. Pool 2 with (i) the 5′-aldehyde functionality, (ii) purified pool 2 with the hexa-His functionality and (iii) the purified pool 2 with the biotin functionality.
To follow the chemical transformations of the oligonucleotide purification and functionalization process, mass spectroscopy was performed on a single oligonucleotide of same length and 5′-aldehyde modification. The oligonucleotide was taken through the same process steps as the oligonucleotide pools, and after each step the oligonucleotide was analyzed by LC–MS (Novatia LLC). As expected, the oligonucleotides were found to incorporate the hexa-His tag at the aldehyde position after the first reaction, and then exchange the hexa-His tag for the biotin functionality after the second reaction. Furthermore, the LC–MS of the final biotinylated product showed no starting material, no hexa-His tag intermediate products and no synthesis truncation anomalies (Supplementary Figure 3a–d). In order to determine functionality of the biotin-oligonucleotide pools, we evaluated the pools with an in-house developed TS capture method for isolating exonic regions (Truseq Exome Enrichment) in the human genome (Steemers,F.J., in preparation). For a given sequencing throughput, these targeted solutions are attractive if one desires to sequence a subset of the human genome at high coverage or sequence many samples at a lower coverage. The TS assay workflow consists of the following steps: (i) solution-phase annealing of biotin-oligonucleotides to Illumina sequencing libraries; (ii) binding of annealed biotin-oligonucleotides/libraries with magnetic streptavidin beads; (iii) stringency washing; (iv) releasing the enriched libraries of interest using denaturing conditions; and (v) sequencing the enriched libraries using the Illumina Genome Analyzer™ (28). We have evaluated the performance of the individual and combined biotinylated oligonucleotiode pools in such applications. We observed efficient and specific capture of sequencing libraries (~80% enrichment specificity), with relative uniform capture across the entire Exome (~90% of targeted bases were covered at 0.2× of the mean coverage).
DISCUSSION
A novel method is described to make individual or pools of purified 5′ biotin-functionalized oligonucleotides. The method is unique because purification is facilitated by means of a 5′-hexa-His tag, which is then exchanged for a biotin group. Hexa-His tag purification had been extensively applied previously to protein purification; however, its utility for oligonucleotide purification has not been demonstrated. The high-loading capacity of the Ni–resin is exploited for purifying large (micromole) amounts of oligonucleotides. Experiments carried out on a Ni-slurry indicated that crude oligonucleotides, loaded as high as 30 µmol per 200 µl of Ni-slurry, were readily processed, resulting in product recovery of up to 16% and purity of >90–99%. In contrast to prior work, this new approach was successfully scaled-up to high Ni–resin loadings while maintaining high purity and yields. These results demonstrate that hexa-His purification and exchange can be employed for large-scale oligonucleotide separations at high column loading. The Ni–resin solid-phase format is readily scalable and has a binding capacity that can match micromole-to-millimole quantities of oligonucleotides. Substituting biotin-hydrazide for biotin-oxo-amine in the substitution reaction doubled the yields for the final product. This effect can be attributed to the stronger nucleophilic nature of biotin-oxo-amine compared with Hydrazide-biotin, and the irreversible nature of the final product with biotin-oxo-amine.
The yields were quite respectable, given the fact that the starting full-length oligonucleotides were ~80–90% aldehyde-labeled (Smith, R. C., unpublished data), and that the theoretical maximum yield for a full-length 95-mer is ~38% based on an average stepwise coupling efficiency of 0.99 per phosphoramidite addition. Additionally, the crude aldehyde oligonucleotides contained truncated products and impurities that were included in the optical absorbance quantification to establish the amount of starting material. An interesting observation is that the yield and purity are reduced with higher percentage GC oligonucleotide pools. GC-dependent inter- and intramolecular hybridization between the pooled oligonucleotides is the most likely contributor to these phenomena. It also explains why adding urea, a denaturant, helps improve yield and purity, especially for the higher percent GC pools. The method is flexible with respect to the type of functional group or label added to the 5′ end of the oligonucleotide. Many nucleophiles in the form of oxo-amino or hydrazide can be used to introduce various chemical moieties using the exchange reaction. Moreover, this same approach can be used to purify unlabelled, full-length oligonucleotides, by using the exchange reaction with a compound such as methoxyamine. The ability to convert the hydrazone bond to a dynamic state is key to its use as an exchangable linker for the preparation of functionalized oligonucleotides (21,29). Strong nucleophiles, i.e. oxo-amine, that exchange the hexa-His group and form an irreversible bond between the oligonucleotide and the functional group are especially desirable. This results in higher yields and final product stability as compared with hydrazide-functionalized groups (30). The hexa-His and biotin derivatives were stable in our hands for at least 6 months. A significant benefit of this approach is that the overall cost of the purification method is <10% of the oligonucleotide synthesis cost. The process uses readily available reagents and equipment, allowing the method to be adopted in laboratories that are minimally equipped.
This article describes the use of hexa-His purification and functional group exchange for attaching the functional group on the 5′ end of long (>80-mers) oligonucleotides. To the best of our knowledge, this is the first reported method to demonstrate a production-scale process that can efficiently functionalize and purify pools of oligonucleotides at micromole scale. The method functionalizes only full-length oligonucleotides and efficiently removes truncated species like n − 1. However, the method does not remove depurinated, partial deprotected and modified oligonucleotides. For many applications in the genomics field, such purity is sufficient. We also demonstrate that this method effectively removes closely related impurities lacking the 5′-aldehyde functionality and other contaminants during the synthesis. Additionally, we have shown that this method scales with the binding capacity of Ni-column resin and can therefore scale to millimole quantities. Ni–resins or alternative hexa-His binding resins are readily available in various formats, including 96-well plates, columns and magnetic beads. This flexibility makes the process amenable to highly parallel formats and automation so that many individual oligonucleotides or pools of oligonucleotides can be processed. We believe that this approach is useful not only for the purification of oligonucleotides but also for other molecules such as carbohydrates, RNA, antibodies and proteins for which the researcher desires high purity and the option of introducing functional groups. Finally, we have demonstrated that the biotin–oligonucleotide pools are functional in TS assays.
In summary, the key benefits of this method are as follows: (i) purification cost is less than the oligonucleotide cost; (ii) scalability, matching purification scale with oligonucleotide synthesis scale; (iii) applicability to 95 bp and potentially longer oligonucleotides; (iv) insensitivity to various oligonucleotide synthesis impurities; (v) and applicability to pools of oligonucleotides.
We are currently exploring further purification methods by reacting the aldehyde oligonucleotide directly to a hydrazide-functionalized solid phase. Releasing the oligonucleotide into solution by a nucleophilic exchange reaction yields a purified and functionalized product. One of the benefits compared to the solution approach is that any hexa-His oligonucleotides not converted to biotinylated oligonucleotide will remain on the solid support and therefore does not need to be removed by an additional Ni-column affinity purification step. This reduces the number of steps and simplifies the workflow. An additional benefit of the solid-phase purification is that a specific amount of an individual oligonucleotide can be isolated and purified using a fixed amount of solid-phase. Such an approach would allow concentration normalization across individual oligonucleotides.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1c, 3a–d.
FUNDING
Funding for open access charge: Illumina Corporation.
Conflict of interest statement. The authors declare conflict of interest by employment and receiving stock compensation at Illumina.
Supplementary Material
ACKNOWLEDGEMENTS
The technology development summarized in this article would not have been possible without the efforts of many dedicated individuals. We would like to thank people at Illumina for their valuable contributions in Oligo Core, Custom-built continuous oligonucleotide synthesizer and Oligator™ technology, and reagent development for this technology. NgocThao Nguyen and David Canter are gratefully acknowledged for coordinating the production and pooling of the oligonucleotides. Calvin Fan is acknowledged for reviewing the manuscript. Illumina, Truseq, Infinium and GoldenGate and Oligator are registered trademarks or trademarks of Illumina, Inc. All other brands are names contained herein are the properties of their respective owners. F.S. conceived the approach and project. F.S., K.Y., R.S., M.W., R.Y., P.M. designed the experiments. K.Y., R.S., M.W., R.Y. and P.M. performed the experiments and data analysis. F.S., R.S. and K.G. wrote the manuscript.
REFERENCES
- 1.Sinha ND. Large-scale oligonucleotide synthesis using the solid-phase approach. Methods Mol. Biol. 1993;20:437–463. doi: 10.1385/0-89603-281-7:437. [DOI] [PubMed] [Google Scholar]
- 2.Ellington A, Pollard JD., Jr Synthesis and purification of oligonucleotides. Curr. Protoc. Mol. Biol. 2001 doi: 10.1002/0471142727.mb0211s42. Chapter 2, Unit2 11. [DOI] [PubMed] [Google Scholar]
- 3.Deshmukh RR, Eriksson KO, Moore P, Cole DL, Sanghvi YS. A case study: oligonucleotide purification from gram to hundred gram scale. Nucleosides Nucleotides Nucleic Acids. 2001;20:567–576. doi: 10.1081/NCN-100002333. [DOI] [PubMed] [Google Scholar]
- 4.Fang S, Fueangfung S. Scalable synthetic oligodeoxynucleotide purification with use of a catching by polymerization, washing, and releasing approach. Org. Lett. 2010;12:3720–3723. doi: 10.1021/ol101316g. [DOI] [PubMed] [Google Scholar]
- 5.Pearson WH, Berry DA, Stoy P, Jung KY, Sercel AD. Fluorous affinity purification of oligonucleotides. J. Org. Chem. 2005;70:7114–7122. doi: 10.1021/jo050795y. [DOI] [PubMed] [Google Scholar]
- 6.Olejnik J, Krzymanska-Olejnik E, Rothschild KJ. Photocleavable biotin phosphoramidite for 5'-end-labeling, affinity purification and phosphorylation of synthetic oligonucleotides. Nucleic Acids Res. 1996;24:361–366. doi: 10.1093/nar/24.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang S, Bergstrom DE. Fluoride-cleavable biotinylation phosphoramidite for 5'-end-labeling and affinity purification of synthetic oligonucleotides. Nucleic Acids Res. 2003;31:708–715. doi: 10.1093/nar/gkg130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang S, Bergstrom DE. Reversible biotinylation phosphoramidite for 5′-end-labeling, phosphorylation, and affinity purification of synthetic oligonucleotides. Bioconjug. Chem. 2003;14:80–85. doi: 10.1021/bc025626o. [DOI] [PubMed] [Google Scholar]
- 9.Buck G, et al. Strategies for purification of synthetic oligonucleotides in DNA core laboratories. 1992 [Google Scholar]
- 10.Lebl M, et al. Automatic oligonucleotide synthesizer utilizing the concept of parallele processing. Collection Symp. Ser. 2011;12:264. [Google Scholar]
- 11.Kozlov IA, Melnyk PC, Hachmann JP, Barker DL, Lebl M, Zhao C. Evaluation of different chemical strategies for conjugation of oligonucleotides to peptides. Nucleosides Nucleotides Nucleic Acids. 2007;26:1353–1357. doi: 10.1080/15257770701533909. [DOI] [PubMed] [Google Scholar]
- 12.Fan JB, Chee MS, Gunderson KL. Highly parallel genomic assays. Nat. Rev. Genet. 2006;7:632–644. doi: 10.1038/nrg1901. [DOI] [PubMed] [Google Scholar]
- 13.Steemers FJ. 2010 The Biology of Genomes Meeting, May 11–15, Cold Spring Harbor, p.296. [Google Scholar]
- 14.Gunderson KL, Kruglyak S, Graige MS, Garcia F, Kermani BG, Zhao C, Che D, Dickinson T, Wickham E, Bierle J, et al. Decoding randomly ordered DNA arrays. Genome Res. 2004;14:870–877. doi: 10.1101/gr.2255804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gunderson KL, Steemers FJ, Lee G, Mendoza LG, Chee MS. A genome-wide scalable SNP genotyping assay using microarray technology. Nat. Genet. 2005;37:549–554. doi: 10.1038/ng1547. [DOI] [PubMed] [Google Scholar]
- 16.Steemers FJ, Chang W, Lee G, Barker DL, Shen R, Gunderson KL. Whole-genome genotyping with the single-base extension assay. Nat. Methods. 2006;3:31–33. doi: 10.1038/nmeth842. [DOI] [PubMed] [Google Scholar]
- 17.Steemers FJ, Gunderson KL. Whole genome genotyping technologies on the BeadArray platform. Biotechnol. J. 2007;2:41–49. doi: 10.1002/biot.200600213. [DOI] [PubMed] [Google Scholar]
- 18.Bibikova M, Fan JB. GoldenGate assay for DNA methylation profiling. Methods Mol. Biol. 2009;507:149–163. doi: 10.1007/978-1-59745-522-0_12. [DOI] [PubMed] [Google Scholar]
- 19.Oliphant A, Barker DL, Stuelpnagel JR, Chee MS. BeadArray technology: enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques. 2002;(Suppl.32):56–61. [PubMed] [Google Scholar]
- 20.Fan JB, Oliphant A, Shen R, Kermani BG, Garcia F, Gunderson KL, Hansen M, Steemers F, Butler SL, Deloukas P, et al. Highly parallel SNP genotyping. Cold Spring Harb. Symp. Quant. Biol. 2003;68:69–78. doi: 10.1101/sqb.2003.68.69. [DOI] [PubMed] [Google Scholar]
- 21.Dirksen A, Yegneswaran S, Dawson PE. Bisaryl hydrazones as exchangeable biocompatible linkers. Angew Chem. Int. Ed. Engl. 2010;49:2023–2027. doi: 10.1002/anie.200906756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kremsky JN, Wooters JL, Dougherty JP, Meyers RE, Collins M, Brown EL. Immobilization of DNA via oligonucleotides containing an aldehyde or carboxylic acid group at the 5′ terminus. Nucleic Acids Res. 1987;15:2891–2909. doi: 10.1093/nar/15.7.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat. Genet. 42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Solulink. 9853 Pacific Heights Blvd. Suite H, San Diego, CA 92121.
- 26.Hochuli E, Dobeli H, Schacher A. New metal chelate adsorbent selective for proteins and peptides containing neighbouring histidine residues. J. Chromatogr. 1987;411:177–184. doi: 10.1016/s0021-9673(00)93969-4. [DOI] [PubMed] [Google Scholar]
- 27.Hochuli E, Bannwarth W, Dobeli H, Gentz R, Stuber D. Genetic approach to facilitate purification of recombinant proteins with a novel metal chelate adsorbent. Nat. Biotech. 1988;6:1321–1325. [Google Scholar]
- 28.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrus A, Cox S, Beavers S, Parker A, Anuskiewicz J, Mullah B. High-throughput synthesis of functionalized oligonucleotides. Nucleic Acids Symp. Ser. 1997:317–318. [PubMed] [Google Scholar]
- 30.West K, Otto S. Reversible covalent chemistry in drug delivery. Curr. Drug Discov. Technol. 2005;2:123–160. doi: 10.2174/1570163054866882. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.