Abstract
Inhibition of gene expression can be achieved with RNA interference (RNAi) or U1 small nuclear RNA—snRNA—interference (U1i). U1i is based on U1 inhibitors (U1in), U1 snRNA molecules modified to inhibit polyadenylation of a target pre-mRNA. In culture, we have shown that the combination of RNAi and U1i results in stronger inhibition of reporter or endogenous genes than that obtained using either of the techniques alone. We have now used these techniques to inhibit gene expression in mice. We show that U1ins can induce strong inhibition of the expression of target genes in vivo. Furthermore, combining U1i and RNAi results in synergistic inhibitions also in mice. This is shown for the inhibition of hepatitis B virus (HBV) sequences or endogenous Notch1. Surprisingly, inhibition obtained by combining a U1in and a RNAi mediator is higher than that obtained by combining two U1ins or two RNAi mediators. Our results suggest that RNAi and U1i cooperate by unknown mechanisms to result in synergistic inhibitions. Analysis of toxicity and specificity indicates that expression of U1i inhibitors is safe. Therefore, we believe that the combination of RNAi and U1i will be a good option to block damaging endogenous genes, HBV and other infectious agents in vivo.
INTRODUCTION
Inhibition of the expression of toxic genes has great therapeutic potential. For most applications, gene expression inhibition has been obtained using RNA interference (RNAi). The inhibitors that mediate RNAi are double-stranded small RNA molecules called small interfering RNAs (siRNAs). siRNAs can be synthetic molecules or can be produced in the cell from precursors called small hairpin RNAs (shRNAs). For RNAi, exogenous siRNAs are directed to the RNA-induced silencing complex (RISC), bind the target with perfect complementarity and induce target mRNA cleavage and as a result, target gene expression is inhibited (1). Inhibition of gene expression can also be performed using U1 small nuclear RNA—snRNA—interference (U1i) (2). U1i is based on a natural property of U1 snRNA ribonucleoprotein (U1 snRNP). When nucleotides 2–11 of U1 snRNA bind to a target pre-mRNA, U1 snRNP inhibits pre-mRNA polyadenylation (3) (Figure 1a). Without a polyA tail, the pre-mRNA fails to mature and is rapidly degraded leading to reduced expression. U1i requires expression of an exogenous 5′-end mutated U1 snRNA designed to base pair to the 3′-terminal exon or, in case of unspliced transcripts, to any position of a target gene (4,5). This U1 snRNA inhibitor (U1in) is expressed from a plasmid that includes U1 snRNA promoter and terminator sequences. Upon plasmid transfection, U1in is expressed, binds target mRNA and blocks expression by hindering polyadenylation (4,5). When target sites are located outside the 3′-terminal exon of a spliced transcript or when they form secondary structures, inhibition is lost (5,6). As in many cases, prediction of the secondary structure of long RNAs is not accurate and selection of good target sites is a challenge that generally decides the success of the technique. When appropriate targets are chosen, U1in can inhibit the expression of reporter or endogenous genes after transient or stable transfections, suggesting a low toxicity of this technique (5).
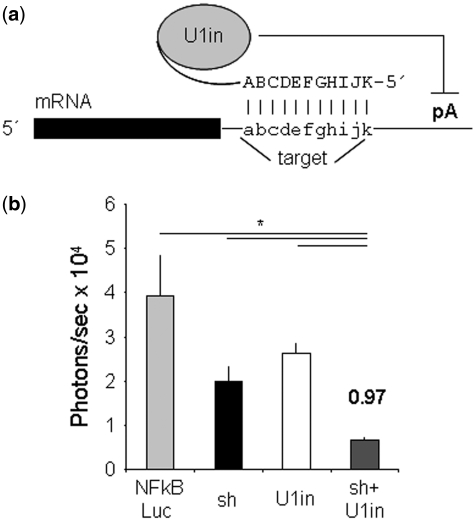
Figure 1.
Schematic of U1i and inhibition of Notch1 in vivo. (a) Schematic of U1-mediated inhibition. U1in are exogenous 5′-end modified U1 snRNPs designed to base pair to the 3′-terminal exon of a target gene. In this study, the U1in binds Notch1 or HBV mRNA and blocks expression by hindering polyadenylation (pA). (b) C57BL/6 mice were injected with pNFκβ-Luc plasmid (NFkBLuc) combined or not with plasmids expressing shαNotch1 (sh), U1inαNotch1 (U1in) or the combination of both. Luciferase activity was measured in living mice with a CCD camera at 4 days post-injection. Luciferase activity was quantified and plotted. The SI and the statistical significance are indicated.
In tissue culture, the combination of RNAi and U1i results in a stable inhibition of reporter or endogenous genes, that is stronger than that obtained using either of the techniques alone (2). The combination allows functional inhibition with decreased doses of the inhibitors and therefore it should serve to decrease unwanted effects. This is therapeutically relevant and consequently, we wanted to know whether the combination of RNAi and U1i also leads to increased inhibitions in animal models. In this work, we have evaluated the functionality of U1i in vivo to target sequences of therapeutic relevance such as those of hepatitis B virus (HBV) or endogenous Notch1. To this aim, we have designed novel shRNAs and U1ins against HBV expression. We show for the first time that U1i inhibits HBV expression after hydrodynamic injection in mice. Besides, we show that a previously validated U1in inhibits the expression of endogenous Notch1 gene in mouse liver. Furthermore, the combination of U1in and shRNA results in synergistic inhibition in mice. Surprisingly, inhibitions obtained by the combination of U1in and shRNA are higher than those obtained by combination of two shRNAs or two U1ins. This suggests that RNAi and U1i cooperate by an unknown mechanism to result in synergistic inhibitions. We believe that the combination of RNAi and U1i could serve as the basis for a novel antiviral therapy against HBV and other infectious agents and to obtain increased inhibition of the expression of endogenous genes.
MATERIALS AND METHODS
Cell lines and DNA constructs
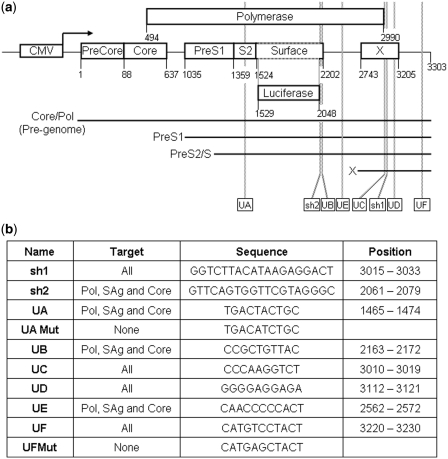
HuH7 cell line was obtained from the American Type Culture Collection (ATCC) and cultured in Dulbecco's Modified Eagle Medium (DMEM), supplemented with 10% FBS and 1% penicillin-streptomycin, at 37°C in a 5% CO2 atmosphere. All cell culture reagents were obtained from Gibco BRL/Life Technologies. The pCH Firefly Luc vector (pCH-Fluc) was constructed by replacing the preS2/S ORF region of pCH-9/3091 HBV replication competent plasmid with Firefly luciferase-encoding DNA (7). pNFκβ-Luc (pNFκβ 3xLuc; Clontech Co) was used to express Firefly luciferase under pNFκβ promoter. Plasmid pRL-SV40 (Promega) was used as Renilla luciferase transfection control. Plasmids expressing U1inαNotch1 and shαNotch1 targeting Notch1 have been described (2). pGemU1inαHBV plasmids, expressing U1ins that target HBV genome (U1inαHBV) or mutant controls, were cloned by ligation of base paired oligonucleotides with the U1inαHBV sequences into the BclI–BglII site of pGEMU1inWT (2) (Figure 2b). The U1 snRNA gene expressed from this plasmid contains four point mutations, but the resulting U1 snRNA is identical in functionality to endogenous U1 snRNA. Plasmids expressing shRNAs that target the HBV genome (shαHBV) were cloned by ligation of base paired oligonucleotides with the shαHBV sequences into the HingIII–BglII sites of pSuper (8) (Figure 2b). The 5′-end of the shRNA starts with the sense strand and is followed by a TTCAAGAGA loop, the antisense strand and UU. The sense and antisense strands have perfect complementarity and are 19 nt long.
Figure 2.
Schematic of the pCH-Fluc with the HBV genome expressing luciferase and the inhibitors that target HBV. (a) HBV genome was cloned after a CMV promoter. The boxes represent the ORFs for Pre-core and core, polymerase (pol), X protein and PreS1, S2 and surface (S) antigen, which has been replaced by Firefly luciferase. The numbers show the position of the nucleotides that mark the start and the stop of each ORF of HBV, starting at the ATG of Pre-core protein. The position where the luciferase sequence was inserted is also indicated. The last number indicates the position of the cleavage and polyadenylation. The parallel lines indicate the four HBV transcripts. All transcripts share the same polyadenylation sequences and therefore the polyA tail is initiated at the same position. Note that luciferase is probably translated from an RNA transcribed by the S promoter (PreS2 and S proteins). However the upstream PreS1 promoter should generate a longer RNA which may encode for a PreS1/Luciferase fusion protein that could show luciferase activity. The CMV promoter generates the longest RNA from which luciferase is unlikely to be translated. The position of the inhibitors is shown at the bottom of the figure. (b) List of inhibitors used in this study. Position and sequence of the target is also indicated.
Design of U1in target sites
The target sites for the U1ins were 10–11 nt-long sequences chosen from conserved sequences in the HBV genome. Besides, they fulfill at least two of the following criteria. Firstly, they are accessible sequences according to mfold (9). Note that mfold only predicts 2D structures and not the potential occupancy of the target sites by protein factors. UA, UB or UD accomplish this criterion. Secondly, they are repetitive sequences in the HBV genome according to SRF and therefore, in theory, they could represent accessible sites susceptible to be bound by cell regulators (10). Such a transient accessibility may be advantageous as U1 snRNA binds pre-mRNA co-transcriptionally, before other cellular factors may interact with the target. UA, UC, UD or UE are repetitive. Thirdly, they include putative target sites for liver miRNAs according to several prediction programs or they are targeted by functional siRNAs. This last criterion indirectly measures accessibility of the target and has proven useful in the design of U1in targeting Notch1 (2). UA, UB, UC and UF fulfill this criterion. Fourthly, they contain sequence motifs significantly associated with antisense activity according to the rules described by McQuisten and Peek (11). UB, UE and UF comply with this condition. After this analysis, target sites found in the 3′-terminal exon of unrelated genes were discarded to reduce secondary effects. Redundancy was evaluated using Basic Local Alignment Search Tool (BLAST) set up to an Expect of 1000 and a word of 7 (12). Finally, we rejected target sites with A at position 2 and therefore susceptible to being bound by U5 snRNP, and G at position 8 and T at position 9 which could allow binding to U6 snRNP. Therefore the chosen target sites have decreased potential to be removed by splicing after binding to the U1in.
Cell transfections
Cells were transfected with calcium phosphate as described (13). The conditions used were set up as described (2). In brief, 150 000 cells were transfected with 0.75 µg of pCH-Fluc, 0.25 µg of pRL-SV40 and 1 µg of the plasmid expressing the control inhibitor, a combination of the plasmid expressing the control inhibitor and the plasmid expressing the inhibitor or a combination of plasmids expressing different inhibitors (0.5 µg each). Mock transfected cells express a U1in of irrelevant sequence. The total amount of transfected DNA was maintained constant by adding a control plasmid. Cells were harvested 72-h post-transfection.
Hydrodynamic tail vein injection and animal studies
This study was performed following the regulations of the Animal Care Ethical Committee of the University of Navarra. Six-to-eight-week-old male C57BL/6 mice were injected with a total of 25 µg of plasmids by hydrodynamic tail vein injection. Animals received in 3 s a final volume of 2 ml of physiological saline solution containing 10 µg of plasmid expressing secreted alkaline phosphatase (pSEAP, Clontech) or 5 µg of pCH-Fluc plasmid or pNFκβ-Luc combined or not with plasmids expressing U1in and/or shRNA inhibitors. The only exception to this rule was the injection of a total of 50 µg that included 30 µg of pNFκβ-Luc plasmid, where indicated. The amount of plasmids expressing the inhibitors used was generally 10 µg of each plasmid unless otherwise indicated. The total amount of injected DNA was maintained constant by adding an irrelevant carrier plasmid.
Luciferase activity was measured at the indicated time points in living mice with a CCD Camera (Xenogen, Alameda, CA) 5 min after intraperitoneal injection of 3 mg of D-luciferin (Promega) and anesthetics (14). Bioluminescence was measured for 5 min until the luciferase signal started to decrease and light intensity was quantified as photons/s with Living Image software. Blood samples were collected at the indicated time points by retro-orbital bleed and animals were subsequently sacrificed. SEAP was measured with Phospha-Light System following manufacturer’s instructions (Applied Biosystems). Serum transaminases (alanine aminotransferase and aspartate) were measured in blood serum (ABX Diagnostics, Montpellier, France) in a Cobas Integra autoanalyzer (Roche, Mannheim, Germany) (15). Liver samples were paraffin-embedded and processed for hematoxylin–eosin staining or snap-frozen in liquid nitrogen and stored at −80°C.
Liver RNA analysis
Total RNA was isolated from liver samples using TRIReagent following the manufacturer’s instructions (Sigma). Two microgram of total RNA were DNAse treated (Invitrogen) and reverse-transcribed with MMLV reverse transcriptase (Promega) (16). U1 snRNA expression was quantified by real-time (RT)-PCR (BioRad) with primers designed to detect exogenous U1 snRNA (U1Fw: GATCTCATAGTTCCATGGCAGGGGAGATACCAT and U1Rev: CGAGTTTGGCACATTTGGCC) and normalized to mouse γ-actin expression (ActFw ACTGCGCTTCTTGCCGC and mActRev CATGACGCCCTGGTGTC). PreS2/S Luc mRNA expression was quantified from pA+ liver RNA isolated with PolyATract mRNA Isolation System (Promega) by RT-PCR (BioRad) with primers SFw: GGAAAACTCGACGCAAGAAA and SRev: TACAGACTTGGCCCCCAATA. Oligo hybridization was performed at 60°C. Primer extension analysis was performed with oligonucleotides designed to detect sh1 (AAGGTCTTACATAAGAGG), sh2 (AAGTTCAGTGGTTCGT) or U6 snRNA (TGCTAATCTTCTCTGTATCGT). Oligonucleotides were labeled with T4 polynucleotide kinase (New England Biolabs) in the presence of [γ-32P]ATP. Primer extension conditions were set up to work in the linear range (17). Six picomoles of labeled oligonucleotides were incubated with 2 µg of total RNA. Only 1 µg of RNA was used in the primer extensions with oligonucleotide U6. Samples were loaded onto a 14% polyacrylamide gel and separated by electrophoresis. Gels were dried and exposed to a screen that was developed in a Cyclone phosphorimager (Perkin Elmer).
In vitro luciferase activity measurements
Transfected cells and liver samples were processed for protein isolation in 1× passive lysis buffer (Promega) with 1× protease inhibitors (Roche). Luciferase activity was measured using the Dual Luciferase System (Promega) in a Berthold Luminometer (Lumat LB 9507) as previously described (13). When extracts from transfected cells were used, the values obtained for Firefly luciferase were corrected for equal transfection efficiency with Renilla luciferase activity. Firefly luciferase activity measured in liver extracts is shown in relative light units (RLU) per µg of liver tissue protein as determined by the Bradford assay.
Mathematical formulations
For the studies performed in vitro, the fold inhibition (FI) for each inhibitor was calculated as the ratio of the luciferase activity obtained in controls with no inhibitors versus the luciferase activity obtained in cells expressing the inhibitors studied. By definition, the control has an inhibitory activity set to 1.0. For calculations with values obtained from living animals, we made use of the luciferase activity measured at Day 1 post-injection to calculate the FI. Thus, the FI for each inhibitor was calculated as the ratio of the luciferase activity obtained in Day 1 versus the luciferase activity obtained in any other time point. This helps to correct for small differences in the efficiency of the hydrodynamic injection given that inhibition was not detected at Day 1 with any of the inhibitors studied. Only small differences were observed between the FI obtained with the two methods of calculation. The synergy index (SI) for the combination of RNAi and U1 inhibition was calculated as previously described (2). The critical value of SI is zero and indicates an additive effect. When S is positive it shows synergism and when negative it shows antagonism.
Statistical analysis
Data are expressed as mean ± standard deviation (SD). Statistical analysis was performed with ANOVA using the GraphPad Prism Software. Statistically significant differences are indicated with a star (P < 0.05), two stars (P < 0.01) or three stars (P < 0.001).
RESULTS
Effect of U1i and the combination of RNAi and U1i on the expression of an endogenous gene in mice
We have recently shown that in tissue culture the combination of RNAi and U1i results in a stable inhibition of the endogenous gene Notch1, which is stronger than that obtained using either of the techniques alone (2). We have now evaluated the inhibition of Notch1 in vivo using the previously validated U1in (U1inαNotch1), or shRNA (shαNotch1) targeting Notch1 or the combination of both (2). To express the inhibitors in mouse liver, we made use of hydrodynamic injections. Using this technique, co-transfection of several plasmids is highly efficient, but only 10–30% of the hepatocytes are transfected. Therefore, Notch1 downregulation could not be evaluated in liver extracts. Instead we introduced, together with the inhibitors, a reporter gene that quantifies Notch1 activity. Notch1 expression increases NFκβ activity by facilitating its nuclear retention (18), resulting in increased expression from NFκβ-specific promoters. Thus, as a reporter, we used a plasmid that expresses Firefly luciferase under an NFκβ specific promoter (pNFκβ-Luc). Decreased Notch1 in transfected cells results in decreased NFκβ activity and therefore in reduced luciferase activity from pNFκβ-Luc plasmid (2,18). To test the system in vivo C57BL/6 mice (n = 5) were injected with 5 µg of pNFκβ-Luc plasmid mixed or not with 10 µg of plasmids expressing U1inαNotch1, shαNotch1 or the combination of both. Luciferase activity was quantified at Days 1 and 4 post-injection. FI for each inhibitor was calculated as the ratio of the luciferase activity obtained at Day 1 versus the luciferase activity obtained at Day 4. Luciferase expression was similar 1 day after injection and decreased drastically at Day 4 in all groups (data not shown). Nevertheless, co-expression of pNFκβ-Luc with U1inαNotch1 or shαNotch1 resulted in an inhibition of luciferase expression (Figure 1b). U1inαNotch1 and shαNotch1 inhibited luciferase down to 56.5% (1.8-fold) or 68.5% (1.5-fold), respectively. Surprisingly, the combination of U1inαNotch1 and shαNotch1 resulted in a significant inhibition of luciferase down to 15.4% (6.5-fold). A SI was calculated as the ratio of the FI of the combination of techniques minus the addition of the fold inhibitions obtained with each of the inhibitors alone (2). The SI was 0.97, higher than zero, indicating synergism. Similar results were obtained when C57BL/6 mice (n = 5) were injected with 30 µg of pNFκβ-Luc plasmid mixed or not with 10 µg of plasmids expressing U1inαNotch1, shαNotch1 or the combination of both and luciferase activity was quantified at Days 1 and 3 post-injection (FI of U1inαNotch1 = 1.8, FI of shαNotch1 = 1.6, FI of the combination = 4.4, SI = 0.27). Differences in luciferase expression were not observed compared to control animals when a non-functional shRNA was used alone or in combination with the U1inαNotch1.
Design of novel inhibitors against HBV expression and analysis of the effect of the combination of RNAi and U1i on HBV expression in culture
Strong inhibitions are desirable to block the expression of viral genes for therapeutic applications. Thus, we decided to evaluate the effect of the combination of RNAi and U1i on the inhibition of expression from HBV sequences. HBV is a non-cytopathic enveloped virus with a partly double-stranded DNA genome of 3.2 kbps. HBV replicates by reverse transcription of an RNA intermediate, the pregenomic RNA. Therefore, targeting HBV RNAs does not only affect viral gene products but directly impacts on viral replication. The viral genome contains regulatory regions embedded in four open reading frames that encode core, polymerase, surface (S) and X proteins (Figure 2a). The HBV genome is compact in that viral transcripts have overlapping sequences and share the same polyadenylation signals. This facilitates the design of inhibitors that target more than one transcript. To block expression from HBV sequences, we constructed a plasmid that expresses a shRNA whose functionality against HBV expression has been established (19). This shRNA, named sh1, targets a sequence located within the X ORF (Figure 2). Following recommendations previously described (2), we also designed a novel shRNA targeting HBV, named sh2. sh2 targets a region upstream of the stop codon of the S gene. Finally, we designed six U1in that target putative accessible sites of the HBV genome (see ‘Material and Methods’ section for details). The inhibitors were named UA, UB, UC, UD, UE and UF. All the target sites are located in the S region but are shared by other viral genes due to the HBV compact genome (Figure 2). UA targets a site upstream of the ATG of the S gene, UB site is upstream of the stop codon of the S gene and the remaining U1in target sites are located downstream of the stop codon of the S gene. In fact, the sequence bound by UF is the most accessible region closest to the polyadenylation signal according to mfold. This could be relevant for U1i functionality as exogenous U1in inhibits polyA tail addition.
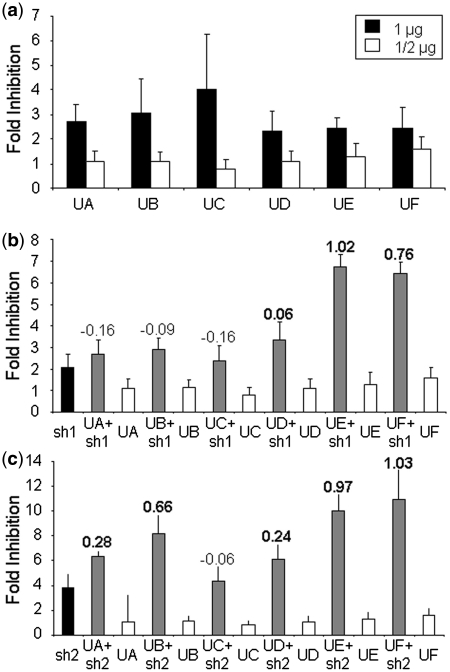
The effect of the combination of the U1in and the shRNAs targeting HBV was evaluated in cells expressing pCH-FLuc. pCH-FLuc contains all the sequences from HBV pCH-9/3091 with the S sequence replaced by the Firefly luciferase ORF (7) (Figure 2a). Expression from pCH-FLuc has been shown previously to correlate well with expression from the complete HBV genome and allows a quantitative read out important for calculating the strength of inhibitions (7,20). To evaluate expression from HBV in the presence of the U1in, HuH7 cells were co-transfected with pCH-FLuc and a plasmid expressing a non-functional U1in control or the plasmids that express each of the U1in αHBV. A plasmid that expresses Renilla luciferase was also co-transfected as an internal control. Cells were collected at 72-h post-transfection and luciferase activity was evaluated in cell extracts. Renilla luciferase activity was similar in all cases, suggesting comparable transfection efficiency and specificity of the inhibitor for pCH-FLuc. The FI of luciferase exerted by the U1in αHBV expression was calculated as the ratio of the luciferase activity obtained in cells transfected with a plasmid expressing a non-functional U1in control versus the luciferase activity obtained in cells expressing each of the U1in αHBV inhibitors. Luciferase activity was similar in mock transfected cells and in cells expressing a non-functional U1in. However, all U1in αHBV inhibited expression from HBV sequences by 50–75% (2–4-fold, Figure 3a). As described previously (2), inhibition of Firefly luciferase expression decreased when the amount of plasmid expressing U1in αHBV was decreased to half in the transfection mixture. Using half of the dose, UC failed to inhibit expression from HBV sequences. The most robust inhibitor was UF. There were no significant differences among UA, UB, UD or UE.
Figure 3.
Analysis of U1ins targeting HBV in culture. (a) Expression inhibition of HBV sequences with U1i. Luciferase activity was measured in HuH7 cells co-transfected with pCH-FLuc and a control plasmid or 1 µg or 1/2 µg of each plasmid expressing U1inαHBV. (b and c) Effect of the co-expression of U1inαHBV and shRNAs αHBV on the expression from HBV sequences. Luciferase activity was measured as before but in cells co-transfected with pCH-FLuc and 1/2 µg of a plasmid expressing a control U1in, each U1inαHBV, sh1 (b), sh2 (c) or a combination of 1/2 µg of a plasmid expressing an U1inαHBV and 1/2 µg of a plasmid expressing sh1 (b) or sh2 (c). The FI is indicated for each case. Note that the scale is different for each figure. The SI is indicated at the top of the corresponding bars. Synergistic inhibitions are highlighted with an SI in bold. Data are mean ± SD from a minimum of five independent experiments.
In spite of the poor functionality of the lower dose of U1in αHBV, we tested whether the inhibition increased when these inhibitors were co-expressed with shRNAs targeting HBV. To this end, cells were treated as before with the exception that pCH-FLuc was co-transfected with a combination of a control plasmid and a plasmid expressing each U1in αHBV, sh1 or sh2 or with the combination of a plasmid expressing an U1in αHBV and a plasmid expressing a shRNA αHBV in all possible combinations. Seventy-two hours post-transfection luciferase activity was measured in cell extracts and FI was calculated as described above. The combination of the shRNAs with the non-efficient UC resulted in an inhibition similar to that obtained with the shRNAs alone (Figure 3b and c). Thus, UC is not a functional inhibitor on its own or after combination with the shRNAs. However, the best inhibition of luciferase was observed when any of the other U1in αHBV were co-expressed with the shRNAs. Combination of U1inαHBV with sh2 was better than with sh1, as the former leads to synergistic increased inhibition with all functional U1in αHBV tested.
Effect of UA and the combination of RNAi and UA on HBV expression in mice
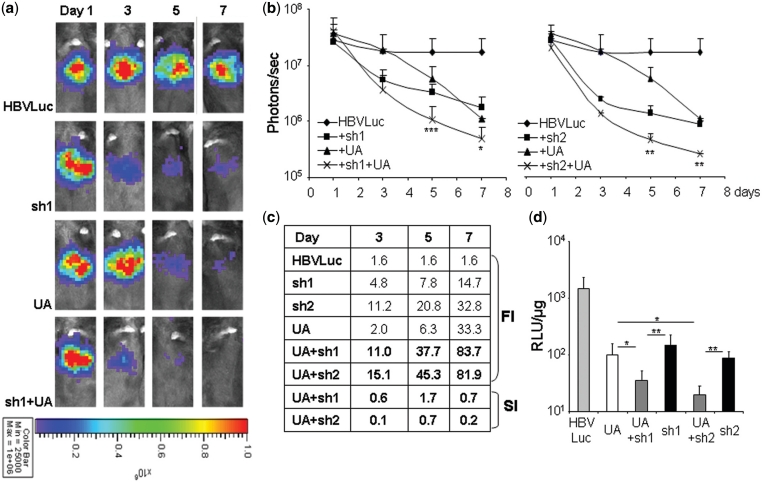
To test the system in vivo we made use of hydrodynamic injections. We first analyzed the luciferase expression from pCH-Fluc in mice. C57BL/6 mice (n = 5) were injected with pCH-Fluc and luciferase activity was quantified with a CCD camera at Days 1, 3, 5, 7, 9, 11 and 14 post-injection. Luciferase expression from pCH-Fluc was detected at Day 1 and was relatively stable until Day 7 post-injection (Supplementary Figure S1). Therefore, this time was chosen as the end-point for our experiments. We decided to evaluate first the effect in vivo of one of the U1inαHBV. Thus, C57BL/6 mice (n = 5) were injected with 5 µg of pCH-Fluc plasmid mixed or not with 10 µg of plasmids expressing UA, sh1, sh2 or the combination of UA with sh1 or sh2. Luciferase activity was quantified at Days 1, 3, 5 and 7 post-injection. FI for each inhibitor was calculated as the ratio of the luciferase activity obtained at Day 1 versus the luciferase activity obtained at any other time point. Animals were sacrificed at Day 8 for further analysis. Luciferase expression in all groups was similar 1 day after injection, indicating that inhibition is not detected at this time point (Figure 4a and b). Co-expression of HBV sequences encoding luciferase with sh1 or sh2 resulted in a dramatic inhibition of luciferase expression (sh1: inhibition of 93.2%; FI = 14.7 and sh2: inhibition of 96.9%; FI = 32.8) (Figure 4). As observed in culture, sh2 was more potent than sh1. Inhibition by both shRNAs targeting HBV was detected at 3 days post-injection and gradually increased until the end of the study. Surprisingly, in mice, inhibition with UA was as potent as with the shRNAs (inhibition of 97.0%; FI = 33.3). Furthermore, the combination of UA and sh1 or sh2 resulted in the strongest inhibition of expression from HBV sequences (UA + sh1: inhibition of 98.8%; FI = 83.7; UA + sh2: inhibition of 98.7%; FI = 81.9). Further analysis revealed that the combination of UA and sh1 or sh2 resulted in a statistically significant stronger synergistic inhibition of HBV expression as compared to that obtained using either of the inhibitors on their own (Figure 4c). Similar results were obtained when luciferase activity was evaluated in vitro using liver extracts from animals sacrificed 8 days post-hydrodynamic injection (Figure 4d). The inhibition obtained was similar in animals expressing UA, sh1 or sh2. Again, co-expression of UA and sh1 or sh2 resulted in a statistically significant stronger inhibition of HBV expression.
Figure 4.
Analysis of the inhibition of luciferase expression from HBV by RNAi and UA in mice. C57BL/6 mice were injected with pCH-Fluc plasmid (HBVLuc) combined or not with plasmids expressing UA, sh1, sh2 or the combination of UA and sh1 or UA and sh2. Luciferase activity was measured in living mice with a CCD camera at the indicated times post-injection (a–c) or in liver extracts obtained 8 days post-injection (d). Representative pictures are shown (a). The color scale used is identical for all images and is shown at the bottom. Luciferase activity was quantified and plotted (b) or used to calculate the FI and SI (c). RLU indicates relative light units (d). Data are mean ± SD from three independent experiments. Significant differences are indicated with asterisks. Note that in b significant differences shown compare luciferase activity obtained with the best inhibitor, either UA or shRNA, on its own with the luciferase activity obtained by combination of UA and the shRNA.
To set up the best conditions for inhibition, similar experiments were carried out using decreasing amounts (10, 5 and 2.5 µg) of plasmids expressing sh1, sh2 or UA (Supplementary Figure S2). A 5 or 2.5 µg of plasmids expressing sh1 or sh2, respectively, resulted in inhibitions similar to those observed with 10 µg of plasmid. Inhibition by UA was more sensitive to the use of lower doses of plasmid. Non-significant differences were observed between 10 or 5 µg of plasmids expressing UA combined with the same amount of plasmids expressing any of the shRNAs. However, inhibition obtained by combination of 10 µg of plasmid expressing UA and 10 µg of plasmid expressing any shRNA was the fastest and tended to last longer (see in vitro analysis in Supplementary Figure S2f). Therefore, we decided to use these conditions unless specifically indicated.
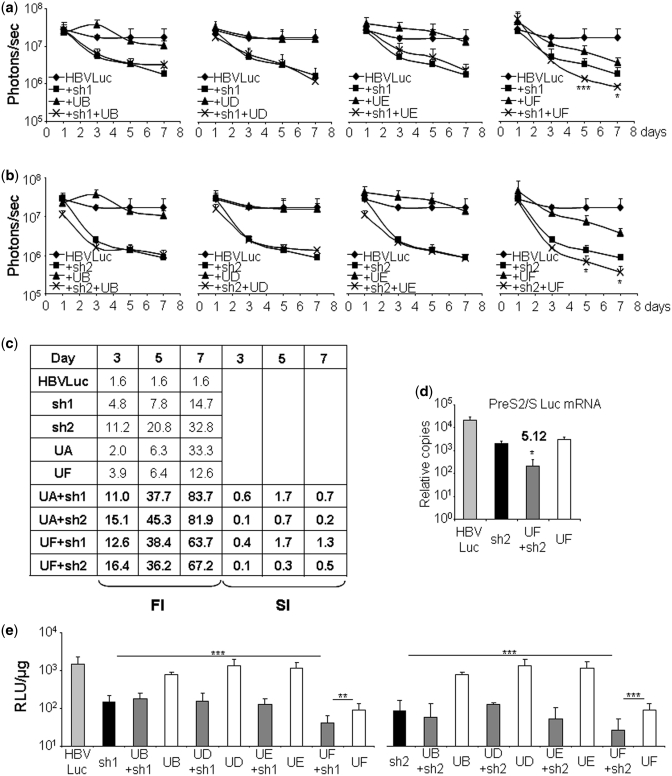
Analysis of the combination of U1ins and shRNAs in HBV expression in mice
The good performance of UA or the combination of UA and shRNAs αHBVs in vivo encouraged us to test the effect of the functional inhibitors UB, UD, UE or UF on expression from HBV sequences in mice. C57BL/6 mice (n = 5) were injected with pCH-Fluc plasmid combined or not with plasmids expressing each one of these U1in αHBV and/or each shRNA αHBV in all possible combinations and luciferase activity was evaluated. The experiments performed were similar to those described above. Luciferase expression in all groups was generally similar 1 day after injection, indicating that inhibition was not detected at this time point (Figure 5a–c). Functionality of the shRNAs αHBV has been already described. UF is also a good inhibitor of HBV expression (inhibition of 92.1%; FI = 12.6). In general, the results obtained in culture do not help to predict the outcome in mice. In culture UF was stronger than UA while in vivo UA was the strongest. In culture UA, UB, UD and UE were similar whereas in mouse UB, UD and UE were not functional. UC was not functional neither in vitro nor in vivo (data not shown). UB, UC, UD and UE neither inhibited luciferase expression on their own nor in combination with any shRNA αHBV tested. However, the combination of UA or UF with the shRNA αHBVs resulted in increased synergistic inhibitions (UF + sh1: inhibition of 98.4%; FI = 63.7; UF + sh2: inhibition of 98.5%; FI = 67.2). We also quantified the transcripts that may encode for luciferase (PreS2/SLuc mRNA and PreS1Luc mRNA) in RNA isolated from liver extracts obtained from animals sacrificed 8 days post-hydrodynamic injection. The results show a synergistic decrease in the accumulation of PreS2/SLuc and PreS1Luc mRNAs when UF and sh2 were combined compared to that observed when only UF or sh2 were used (Figure 5d and Supplementary Figure S3). These results suggest that the inhibitors alone or in combination affect the stability of the target mRNA and do not act at a translational level. Similar results were obtained when luciferase activity was evaluated in vitro using liver extracts (Figure 5e).
Figure 5.
Effect of U1ins and shRNAs targeting HBV in mice. C57BL/6 mice were injected with pCH-Fluc plasmid (HBVLuc) combined or not with plasmids expressing UB, UD, UE, UF and/or sh1 (a, c and e) or sh2 (b–e). Luciferase activity was quantified in living mice with a CCD camera at the indicated times post-injection (a–c) or in liver extracts obtained 8 days post-injection (e). PreS2/S Luc mRNA was quantified by RT–PCR from the same extracts (d). The relative number of copies of PreS2/S Luc mRNA obtained from 67 ng of pA + mRNA compared to a standard plasmid is shown. FI was calculated as described (c). The SI has been calculated for the combination of U1in and shRNA (c and d). RLU indicates relative light units (e). Data are mean ± SD from at least two independent experiments. Significant differences are indicated with asterisks. Statistical analysis shown compares animals treated with the combination of U1in and shRNA with animals treated with the best inhibitor alone (a, b and d) or with either of the inhibitors on its own (e). Statistical analysis of UB, UD and UE resulted in non-significant differences.
Analysis of the specificity of U1in mediated inhibition
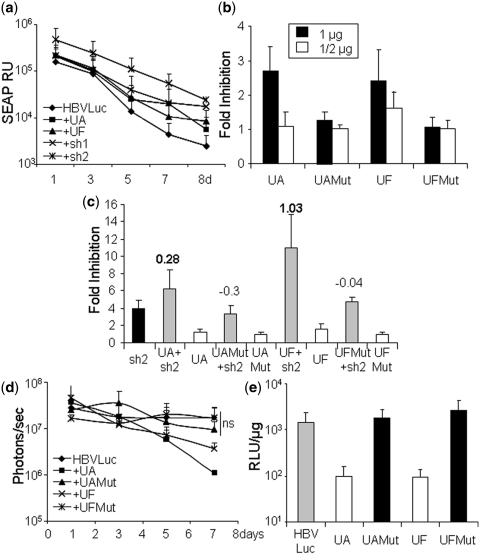
The inhibition observed with UA and UF seems to be specific since the expression of other exogenous U1ins, such as UB, UC, UD or UE did not affect luciferase expression in vivo. To further evaluate specificity, we first quantified SEAP in serum obtained from C57BL/6 mice (n = 5) at different times post-hydrodynamic injection of a plasmid expressing pSEAP together with HBVLuc, as a negative control, UA, UF, sh1 or sh2 (Figure 6a). SEAP decreased with time with similar kinetics in all cases. This indicates that SEAP expression was not affected by UA or UF. Similar results were obtained when SEAP was co-expressed with other U1in (data not shown).
Figure 6.
Analysis of the specificity of U1ins targeting HBV. (a) C57BL/6 mice were injected with pSEAP and pCH-Fluc plasmid (HBVLuc) combined or not with plasmids expressing UA, UF, sh1 or sh2. SEAP was quantified in blood extracted at the indicated times post-injection. SEAP RU indicates relative units of SEAP. (b and c) Luciferase activity was evaluated in HuH7 cells treated as described in Figure 3 with the exception that the cells were transfected with plasmids expressing inhibitors UA, UAMut, UF and UFMut alone (b) or in combination with sh1 or sh2 (c). Cells expressing sh1 or sh2 alone were also evaluated (c). The FI is indicated for each case. Note that the scale is different for each figure. The SI is indicated at the top of the corresponding bars. Synergistic inhibitions are highlighted with an SI in bold. (d and e) C57BL/6 mice were injected with pCH-Fluc plasmid (HBVLuc) combined or not with plasmids expressing UA, UAMut, UF and UFMut. Luciferase activity was quantified in living mice with a CCD camera at the indicated times post-injection (d) or in liver extracts obtained 8 days post-injection (e). RLU indicates relative light units. Data are mean ± SD from at least two independent experiments.
Finally, to determine whether the 5′-terminal sequence of UA or UF was important for inhibition of luciferase expression, we constructed mutated versions named UAMut and UFMut, in which the two central nucleotides of the target complementary sequence have been exchanged by their complementary counterparts (Figure 2b). Luciferase expression from HBVLuc was evaluated in vitro when UA, UAMut, UF and UFMut were expressed in HuH7 cells alone or in combination with plasmids expressing sh1 or sh2 (Figure 6b and c) and in vivo after hydrodynamic injection of C57BL/6 mice (n = 5) (Figure 6d and e). As expected, UAMut and UFMut failed to inhibit luciferase expression in tissue culture and in mouse liver.
Analysis of the combinations of inhibitors of the same kind on HBV expression in vivo
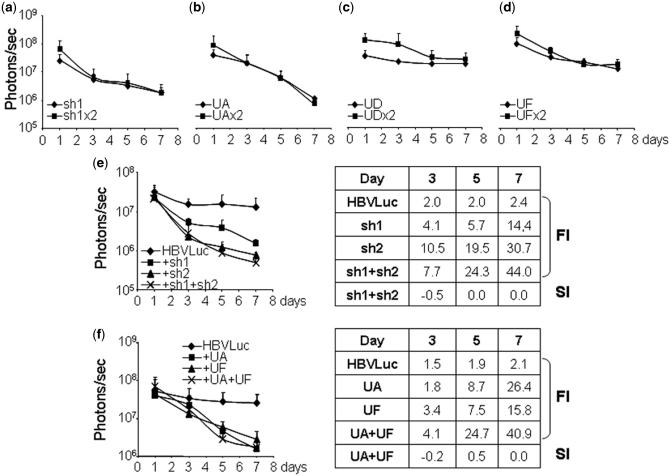
The increased inhibition observed by combination of RNAi and U1i could simply be the result of the combination of two different inhibitors or even the result of the increased dose of inhibitors. We rationalized that increasing the dose of inhibitors of the same kind should have, at most, an additive effect (i.e. SI = 0). However, we decided to verify this experimentally. First, we tested the effect of higher doses of a single inhibitor on luciferase expression from HBV sequences in vivo. To this aim, luciferase activity was evaluated in C57BL/6 mice (n = 5) injected with pCH-Fluc and 10 or 20 µg of plasmids expressing sh1, UA, UD or UF. Analysis revealed that luciferase expression was similar in animals that received 10 or 20 µg of plasmids expressing the inhibitors (Figure 7a–d).
Figure 7.
Effect of combining HBV inhibitors on luciferase expression in vivo. (a–d) Effect of increased doses of inhibitors. C57BL/6 mice were injected with pCH-Fluc plasmid (HBVLuc) combined with 10 or 20 µg (×2) of plasmids expressing sh1 (a), UA (b), UD (c) or UF (d). (e and f) Effect of combining inhibitors of the same kind. C57BL/6 mice were injected with pCH-Fluc plasmid (HBVLuc) combined with 10 µg of plasmids expressing sh1, sh2 or the combination of sh1 and sh2 (e) or UA, UF or the combination of UA and UF (f). Luciferase activity was quantified in living mice with a CCD camera at the indicated times post-injection. FI and SI were calculated as described. Data are mean ± SD from two independent experiments.
Further, we tested the effect of the combination of two inhibitors of the same kind. In this case, luciferase activity was evaluated in C57BL/6 mice (n = 5) injected with pCH-Fluc and 10 µg of plasmids expressing sh1, sh2, UA, UF or the combination of sh1 and sh2 (Figure 7e) or UA and UF (Figure 7f). The results show that the combination of inhibitors of the same kind results in non-statistically significant increased inhibition compared to that obtained using either of the inhibitors on their own. Increased inhibitions obtained with the combination of sh1 and sh2 or UA and UF generally represent additive effects (SI = 0). In all cases, the combination of U1i and RNAi resulted in stronger inhibitions than the combination of two shRNAs or two U1ins (Figures 4 and 5 and data not shown).
Analysis of the accumulation of inhibitors in vivo after combination of RNAi and U1i
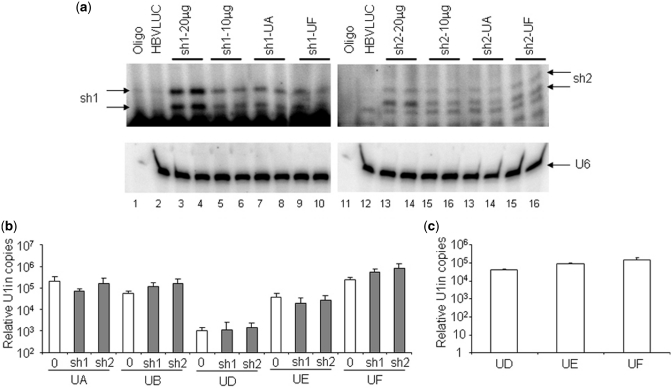
Even when the snRNA and the shRNA processing pathways seem to have little in common, one of these could activate the other by an unknown mechanism. This could result in an increased accumulation of one of the inhibitors and, therefore, in increased inhibition. To address this possibility, we measured the accumulation of the inhibitors in mouse liver. Processed sh1 and sh2 was evaluated by primer extension of RNA isolated from liver extracts obtained 8 days after hydrodynamic injection (Figure 8a). As expected, animals treated with 20 µg of plasmid expressing sh1 or sh2 showed twice the amount of inhibitor than animals treated with 10 µg of plasmid. This confirms that the primer extension is sensitive to detect increased concentration of inhibitors. However, the amount of inhibitor was similar in animals expressing the RNAi inhibitor alone or in combination with an U1in. Thus, the increased inhibition observed by combination of RNAi and U1i is not caused by an increased accumulation of shRNA.
Figure 8.
Quantification of shRNAs and U1in targeting HBV. (a) Quantification of shRNAs. sh1 and sh2-derived siRNAs were visualized by extension with a sh1 (lanes 1–10) or sh2-specific (lanes 11–20) labeled primer of RNAs isolated from the liver of C57BL/6 mice injected with pCH-Fluc plasmid (HBVLuc, lanes 2 and 12) alone or combined with 20 or 10 µg of plasmids expressing sh1 or sh2 or with 10 µg of plasmids expressing sh1 or sh2 and 10 µg of plasmids expressing UA or UF. U6 snRNA was also evaluated by primer extension as a loading control (bottom). Two shRNA-expressing animals were evaluated for each condition. Labeled primer incubated with buffer was run in parallel (Oligo). (b) Quantification of U1in in vitro and in vivo. Exogenous U1in expression was quantified by RT-PCR from liver extracts obtained as described in Figure 5 (b) or extracts from HuH7 cells transfected with plasmids expressing UD, UE or UF as described in Figure 3 (c). Actin mRNA was also quantified to allow comparison between different samples. Data are mean ± SD from five (a) or six (b) samples from two independent experiments.
To measure hepatic U1in αHBV, expression of exogenous U1 snRNA was evaluated by quantitative RT-PCR in liver extracts obtained 8 days after hydrodynamic injection. Note that the exogenous snRNAs differ from the endogenous in the 5′-end and in four internal point mutations that do not disturb functionality and allow easy quantification. A quantitative RT-PCR of actin mRNA was performed in parallel to correct for small loading differences. As expected, exogenous U1 snRNA was only detected in animals treated with plasmids expressing U1in. Interestingly, there were no significant differences in levels of exogenous U1in between animals treated with plasmids expressing U1in alone or in combination with shRNA (Figure 8b). Levels of UA, UB, UF, UAMut and UFMut were similar in all animals that expressed these small RNAs (Figure 8b and Supplementary Figure S4). Surprisingly, some non-functional U1in tended to have a lower accumulation in vivo. Levels of UD and UE were reproducibly lower than a threshold of 50 000 relative copies.
To determine whether the low expression of UD and UE was intrinsic to the constructs that express these RNAs, HuH7 cells were transfected with plasmids expressing UD, UE or UF and exogenous U1in was evaluated in cell extracts as described. The expression levels of these constructs were similar in tissue culture (Figure 8c). Finally, we evaluated DNA levels of the plasmid that express U1in by quantitative PCR in liver extracts obtained 8 days after hydrodynamic injection. All animals showed similar levels of U1in expressing plasmids (Supplementary Figure S5). This indicates that the lower accumulation of UD and UE does not result from a decreased stability of the plasmids that express these inhibitors in mouse liver.
DISCUSSION
Our study shows that expression of U1ins can result in strong inhibition of the expression of target genes in mice. Furthermore, the combination of U1i and RNAi can result in synergistic inhibition in animal models, relevant to decreasing expression from sequences of therapeutic relevance. In particular, we tested inhibition of endogenous Notch1 and inhibition of expression from HBV sequences. Notch1 downregulation was evaluated by measurement of Notch1 effect on NFκβ (2,18). Thus, we could determine the functional relevance of Notch1 downregulation. However, this method could not be used to quantify strong inhibitions of Notch1 expression, as NFκβ activity is only increased linearly when Notch1 is expressed over a relatively high threshold level (18). Therefore, we evaluated inhibition of luciferase expression from HBV sequences, which allowed quantification within a broad linear range. To this end, we used a plasmid that contains all HBV sequences except for the ORF of the S gene, which has been replaced with the ORF of Firefly luciferase (Figure 2a). This plasmid, pCH-Fluc, was previously used to analyze RNAi-mediated inhibition in culture (7,20). Here we show that pCH-Fluc can also be used in vivo. When we introduced pCH-Fluc into mice by hydrodynamic injection, luciferase activity was stable for one week and decreased slowly at later times (Supplementary Figure S1). This is different to what has been observed in mice with expression from other viral promoters such as CMV or SV40, which are inactivated very rapidly (21) (Figure 6a). Actually, pCH-Fluc also has a CMV promoter that drives the expression of the chimeric core/pol-luciferase mRNA (Figure 2a). This long chimeric mRNA was not detected by quantitative RT-PCR of pA+ RNA isolated from the liver 8 days after hydrodynamic injection of pCH-Fluc (data not shown). As relatively constant levels of luciferase activity were detected until this time point, we conclude that the chimeric core/pol-luciferase mRNA does not contribute significantly to luciferase activity. Instead, luciferase is probably translated from an RNA transcribed from the S promoter that results in translation of PreS2 and S proteins from HBV genome. The upstream PreS1 promoter should generate a longer RNA which may encode for a PreS1/Luciferase fusion protein that could also show luciferase activity. One day after hydrodynamic injection of pCH-Fluc, luciferase activity was similar in control animals and in animals co-injected with plasmids expressing functional shRNA or U1in targeting HBV (Figures 4–7 and Supplementary Figure S2). This suggests that luciferase is expressed very rapidly and that the effect of the inhibitors is only detected at later time points. We believe that measurement of luciferase from pCH-Fluc in vivo and in vitro is a good method to quantify the effect of inhibitors that target HBV sequences. However, further experiments are required to determine whether the inhibitors affect the stability of transcripts expressed from the complete viral genome and have an impact on viral viability.
In this study we evaluated the effects in tissue culture and in mice of six novel U1ins, designed following our best criteria for functionality (see ‘Materials and Methods’ section for details). We also constructed two plasmids that express shRNAs targeting HBV. In culture, the U1ins tested were worse inhibitors than the shRNAs and showed poor functionality when a lower dose was used (Figure 3), indicating that the design of functional U1in could require further development. Surprisingly, in mice, some U1ins were functional and resulted in similar inhibition to that observed with shRNAs (Figure 5c). Inhibition is specific as functional U1in did not affect expression of reporter genes that lack a target sequence (Figure 6a). Furthermore, mutant versions of functional U1in failed to inhibit luciferase expression from HBV (Figure 6).
On the other hand, some U1in were functional in tissue culture and not in mouse. UA, UB, UC, UD and UE showed similar efficacy in HuH7 cells, but only UA was functional in the liver (Figures 3–5). This was not observed with the shRNAs analyzed, which were functional both in vitro and in vivo. One of the inhibitors, UB, accumulated to normal levels in the liver where it was not functional. We speculate that UB and other U1in may be more sensitive to target accessibility than shRNAs. Some of the U1in target sites may be accessible in some cells (such as immortalized HuH7 cells) and not in others (such as primary mouse hepatocytes). In the case of UD and UE, the lack of functionality correlated with low levels of accumulation of the inhibitor. In HuH7 cells UD and UE were expressed to similar levels as UF while in liver cells they showed lower levels than any other U1in tested (Figure 8). This is particularly dramatic in the case of UD whose levels were 2 logs lower than those of UF. Transcription or stability of UD could be lower in mice for unknown reasons. Alternatively, UD expressing cells could be eliminated due to toxicity caused by UD-mediated off-target inhibition. This does not seem to be the case. UD-treated liver cells expressed similar levels of luciferase as control cells and similar levels of the U1in expressing plasmid as cells treated with the functional UF (Figure 5 and Supplementary Figure S5). Furthermore, we tested safety of the expression of UD and other U1ins, shRNAs and combinations of both and we found no signs of toxicity for the entire period of observation (data not shown). Necropsies performed in animals sacrificed 8 or 15 days after hydrodynamic injection revealed no apparent systemic abnormalities. Similar levels of ALT and AST transaminases were detected in the serum of all animals (data not shown). Liver sections were stained with hematoxylin–eosin and subjected to histopathological analysis. Liver histology confirmed the absence of malignant or premalignant lesions, lack of inflammation, necrosis or apoptosis and showed appropriate liver architecture in all animals (data not shown). Further experiments will be required to address the safety of U1i when inhibitors are ubiquitously expressed at high levels and for longer periods of time. In fact, it has been recently published that toxicity was not detected when U1ins and shRNAs were expressed from adeno-associated viral vectors for 2 months (22). This, together with our results, is encouraging for the use of the combination of U1i and RNAi for gene therapy applications. Also, more experiments are required to address why some inhibitors are functional in culture and not in mouse liver. In this work, we show that three U1in (U1inαNotch1, UA, UF) worked in vivo out of the six that show synergistic inhibitions in culture when combined with shRNAs. Until the major reasons that drive the lack of functionality in vivo of these U1ins are understood and U1in able to work in vivo can be designed, we recommend that the users of this technology test in vivo at least three U1in functional in culture.
Our results show a synergistic effect of the combination of RNAi and U1i both in culture and in mice, although synergism is not observed with all possible combinations between U1in and shRNAs. In fact, we had previously reported that poor inhibitors do not show synergism in cultured cells (2). In mice, UA and UF are strong inhibitors and when co-expressed with sh1 or sh2 result in a 60–80 FI of HBV expression (Figure 5c). Similar results were observed when UA or UF were combined with another shRNA targeting HBV (data not shown). Surprisingly, combination of two shRNAs or two U1ins resulted in lower inhibition that the combination of U1i and RNAi. The former represented an additive effect while the later showed synergistic inhibition. Therefore, synergism between U1i and RNAi indicates that these mechanisms can work together to produce higher levels of inhibition than those obtained when the techniques are used independently. Further experiments will be required to clarify the exact molecular mechanism underlying this phenomenon. Our results indicate that the combination of techniques does not lead to a higher accumulation of U1in or shRNA molecules that could account for the synergism observed.
The increased inhibitions obtained by combining U1i and RNAi could be of interest to decrease the dose of each inhibitor and still obtain functional inhibitions with lower secondary effects (23–27). Besides, the combination could be used to obtain a high inhibition which is mandatory, for instance, when targeting a replicative viral RNA. An inhibition of 97.5% (40-fold) or of 98.75% (80-fold) may have the same functional effect in the inhibition of endogenous genes such as Notch1. However, in the cases of viruses replicating via RNA, such as HCV, HIV or HBV, an inhibition of 97.5% would leave twice as many viral RNA copies than an inhibition of 98.75%. The production of few viral particles from a partially inhibited infected cell may be sufficient to sustain the viral infection in the patient. Furthermore, the combination of RNAi and U1i could serve to decrease the possibility of viral escape by selection of viruses resistant to a single inhibitor. This is important for HBV infection as HBV polymerase lacks proofreading activity resulting in rapid mutagenesis of HBV and emergence of resistant variants against current treatments. In conclusion, we believe that combination of RNAi and U1i could be the basis for a novel therapy for the treatment of HBV and, possibly, other infections.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5 and Supplementary Materials and Methods.
FUNDING
The Ministry of Science and Innovation (BIO2006-13225, BIO 2009/09295); Euskal Irrati Telebista (EITB) Maratoia 2006 (BIOEF BIO07/CA/024); through the ‘UTE project CIMA’ and by the project RNAREG (CSD2009-00080), funded by the Ministry of Science and Innovation under the programme CONSOLIDER INGENIO 2010. L.B. is the recipient of a Torres Quevedo fellowship (PTQ-09-02-02213). Funding for open access charge: UTE project CIMA.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Marina Barriocanal for technical assistance, Oscar Aparicio for cloning of shRNA expressing plasmids, Patrick Arbuthnot (University of the Witwatersrand, South Africa) for plasmid pCH-Fluc and Paul Miller for English editorial work.
REFERENCES
- 1.Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 2.Abad X, Razquin N, Abad A, Fortes P. Combination of RNA interference and U1 inhibition leads to increased inhibition of gene expression. Nucleic Acids Res. 2010;38:e136. doi: 10.1093/nar/gkq299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gunderson SI, Polycarpou-Schwarz M, Mattaj IW. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol. Cell. 1998;1:255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- 4.Beckley SA, Liu P, Stover ML, Gunderson SI, Lichtler AC, Rowe DW. Reduction of target gene expression by a modified U1 snRNA. Mol. Cell. Biol. 2001;21:2815–2825. doi: 10.1128/MCB.21.8.2815-2825.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fortes P, Cuevas Y, Guan F, Liu P, Pentlicky S, Jung SP, Martinez-Chantar ML, Prieto J, Rowe D, Gunderson SI. Inhibiting expression of specific genes in mammalian cells with 5' end-mutated U1 small nuclear RNAs targeted to terminal exons of pre-mRNA. Proc. Natl Acad. Sci. USA. 2003;100:8264–8269. doi: 10.1073/pnas.1332669100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abad X, Vera M, Jung SP, Oswald E, Romero I, Amin V, Fortes P, Gunderson SI. Requirements for gene silencing mediated by U1 snRNA binding to a target sequence. Nucleic Acids Res. 2008;36:2338–2352. doi: 10.1093/nar/gkn068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ely A, Naidoo T, Mufamadi S, Crowther C, Arbuthnot P. Expressed anti-HBV primary microRNA shuttles inhibit viral replication efficiently in vitro and in vivo. Mol. Ther. 2008;16:1105–1112. doi: 10.1038/mt.2008.82. [DOI] [PubMed] [Google Scholar]
- 8.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 9.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma D, Issac B, Raghava GP, Ramaswamy R. Spectral Repeat Finder (SRF): identification of repetitive sequences using Fourier transformation. Bioinformatics. 2004;20:1405–1412. doi: 10.1093/bioinformatics/bth103. [DOI] [PubMed] [Google Scholar]
- 11.McQuisten KA, Peek AS. Identification of sequence motifs significantly associated with antisense activity. BMC Bioinformatics. 2007;8:184. doi: 10.1186/1471-2105-8-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 13.Aparicio O, Razquin N, Zaratiegui M, Narvaiza I, Fortes P. Adenovirus virus-associated RNA is processed to functional interfering RNAs involved in virus production. J. Virol. 2006;80:1376–1384. doi: 10.1128/JVI.80.3.1376-1384.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vera M, Sobrevals L, Zaratiegui M, Martinez L, Palencia B, Rodriguez CM, Prieto J, Fortes P. Liver transduction with a simian virus 40 vector encoding insulin-like growth factor I reduces hepatic damage and the development of liver cirrhosis. Gene Ther. 2007;14:203–210. doi: 10.1038/sj.gt.3302858. [DOI] [PubMed] [Google Scholar]
- 15.Sobrevals L, Rodriguez C, Romero-Trevejo JL, Gondi G, Monreal I, Paneda A, Juanarena N, Arcelus S, Razquin N, Guembe L, et al. Insulin-like growth factor I gene transfer to cirrhotic liver induces fibrolysis and reduces fibrogenesis leading to cirrhosis reversion in rats. Hepatology. 2010;51:912–921. doi: 10.1002/hep.23412. [DOI] [PubMed] [Google Scholar]
- 16.Aparicio O, Carnero E, Abad X, Razquin N, Guruceaga E, Segura V, Fortes P. Adenovirus VA RNA-derived miRNAs target cellular genes involved in cell growth, gene expression and DNA repair. Nucleic Acids Res. 2010;38:750–763. doi: 10.1093/nar/gkp1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narvaiza I, Aparicio O, Vera M, Razquin N, Bortolanza S, Prieto J, Fortes P. Effect of adenovirus-mediated RNA interference on endogenous microRNAs in a mouse model of multidrug resistance protein 2 gene silencing. J. Virol. 2006;80:12236–12247. doi: 10.1128/JVI.01205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shin HM, Minter LM, Cho OH, Gottipati S, Fauq AH, Golde TE, Sonenshein GE, Osborne BA. Notch1 augments NF-kappaB activity by facilitating its nuclear retention. EMBO J. 2006;25:129–138. doi: 10.1038/sj.emboj.7600902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shlomai A, Shaul Y. Inhibition of hepatitis B virus expression and replication by RNA interference. Hepatology. 2003;37:764–770. doi: 10.1053/jhep.2003.50146. [DOI] [PubMed] [Google Scholar]
- 20.Ely A, Naidoo T, Arbuthnot P. Efficient silencing of gene expression with modular trimeric Pol II expression cassettes comprising microRNA shuttles. Nucleic Acids Res. 2009;37:e91. doi: 10.1093/nar/gkp446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kramer MG, Barajas M, Razquin N, Berraondo P, Rodrigo M, Wu C, Qian C, Fortes P, Prieto J. In vitro and in vivo comparative study of chimeric liver-specific promoters. Mol. Ther. 2003;7:375–385. doi: 10.1016/s1525-0016(02)00060-6. [DOI] [PubMed] [Google Scholar]
- 22.Koornneef A, van Logtenstein R, Timmermans E, Pisas L, Blits B, Abad X, Fortes P, Petry H, Konstantinova P, Ritsema T. AAV-mediated in vivo knockdown of luciferase using combinatorial RNAi and U1i. Gene Ther. 18:929–935. doi: 10.1038/gt.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 24.Persengiev SP, Zhu X, Green MR. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs) RNA. 2004;10:12–18. doi: 10.1261/rna5160904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 2003;34:263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- 26.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 27.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.