Abstract
Cryopreservation of microorganisms in ancient glacial ice is possible if lethal levels of macromolecular damage are not incurred and cellular integrity is not compromised via intracellular ice formation or recrystallization. Previously, a bacterium (isolate 3519-10) recovered from a depth of 3,519 m below the surface in the Vostok ice core was shown to secrete an ice-binding protein (IBP) that inhibits the recrystallization of ice. To explore the advantage that IBPs confer to ice-entrapped cells, experiments were designed to examine the expression of 3519-10’s IBP gene and protein at different temperatures, assess the effect of the IBP on bacterial viability in ice, and determine how the IBP influences the physical structure of the ice. Total RNA isolated from cultures grown between 4 and 25°C and analyzed by reverse transcription-PCR indicated constitutive expression of the IBP gene. Sodium dodecyl sulfate-polyacrylamide gel electrophoretic analysis of 3519-10’s extracellular proteins revealed a polypeptide of the predicted size of the 54-kDa IBP at all temperatures tested. In the presence of 100 μg mL−1 of extracellular protein from 3519-10, the survival of Escherichia coli was increased by greater than 100-fold after 5 freeze-thaw cycles. Microscopic analysis of ice formed in the presence of the IBP indicated that per square millimeter field of view, there were ~5 times as many crystals as in ice formed in the presence of washed 3519-10 cells and non-IBP producing bacteria, and ~10 times as many crystals as in filtered deionized water. Presumably, the effect that the IBP has on bacterial viability and ice crystal structure is due to its activity as an inhibitor of ice recrystallization. A myriad of molecular adaptations are likely to play a role in bacterial persistence under frozen conditions, but the ability of 3519-10’s IBP to control ice crystal structure, and thus the liquid vein network within the ice, may provide one explanation for its successful survival deep within the Antarctic ice sheet for thousands of years.
Keywords: ice-binding protein, recrystallization inhibition, polycrystalline ice, freeze tolerance
Introduction
Research on the survival and distribution of microorganisms beneath the planet’s ice sheets (total surface area of ~1.5 × 107 km2) has indicated that a diverse range of habitable environments exist in the cryospheric subsurface (e.g., Skidmore, 2011). Extrapolations based on the few data which exist suggest that the ice sheets and subsurface environments of Greenland and Antarctica may contain a globally relevant, yet virtually ignored pool of microorganisms (~1029 cells) and carbon (~15 Pg of cell and dissolved organic carbon; Priscu et al., 2008; Lanoil et al., 2009). Subglacial locations harboring liquid water (e.g., subglacial lakes) have been identified as prime targets to search for viable microbial ecosystems (Fricker et al., 2011; Lukin and Bulat, 2011; Ross et al., 2011; Skidmore, 2011). There is also evidence that the physicochemical characteristics and structure of polycrystalline glacial and basal ice provide a tenable microbial habitat where cells reside either in liquid filled veins or the liquid films on the surface of entrained mineral grains (Price, 2000; Mader et al., 2006; Tung et al., 2006; Bakermans and Skidmore, 2011a). Viable bacteria and fungi have been recovered from ancient glacial ice originating from a number of geographical locations (Miteva, 2008); however, little is known about the molecular adaptations which aid these species in tolerating low water availability, osmotic stress, high hydrostatic pressure, and prolonged survival within the ice crystal matrix.
Some cold-adapted organisms mitigate the stress associated with freezing by producing ice-binding proteins (IBPs) which selectively bind to the prism facet of ice, prevent water molecules from joining along the a-axis, and influence ice structure. IBPs have been documented in a variety of cold-adapted organisms including plants (Smallwood et al., 1999), diatoms (Raymond and Knight, 2003), and fish (e.g., white flounder; Carpenter and Hansen, 1992), as well as alga and moss from cyanobacterial mats (Raymond and Fritsen, 2001). Although few bacterial IBPs have been characterized, the phenotype has been reported in isolates from Antarctic sea ice, ice-covered lakes, and glacial ice (Gilbert et al., 2005; Raymond et al., 2007, 2008), as well as cold soils (Walker et al., 2006). Analysis of the IBPs from fish (Sicheri and Yang, 1995; Davies et al., 2002), insects (Graether et al., 2000), and the Antarctic bacterium Marinomonas primoryensis (Garnham et al., 2011) have provided structural information to elucidate the mechanism by which the protein binds to the ice crystal matrix. IBPs may have thermal hysteresis activity (i.e., depression of the freezing point without a change in the melting point; also known as antifreeze activity) and/or recrystallization inhibition (RI) activity. An activity toward the inhibition of recrystallization would have particular significance to microorganisms present within an ice matrix, as the growth of large membrane-damaging ice crystals is a process known to reduce cell viability during freezing and thawing (Miller and Mazur, 1976; Gage et al., 1985; Dumont et al., 2004).
Geomicrobiological investigations of the Vostok 5G ice core have provided data on the characteristics of cells entrapped in ice for as long as 420,000 years and limnological conditions in surface waters of the largest subglacial lake in Antarctica, Subglacial Lake Vostok (Karl et al., 1999; Priscu et al., 1999; Christner et al., 2006). Ice core samples from the deepest portions of the glacial ice in the 5G bore hole (from 3,450 to 3,537 m) contain particles too large to be of aeolian origin (>30 μm) and this portion of the core is interpreted to contain entrained bedrock material (Simoes et al., 2002). During the characterization of microbes cultured from melted samples of an ice core from a depth of 3,519 m in 5G, a bacterial isolate (3519-10; Flavobacteriaceae family) was identified that possesses a 54-kDa IBP homologous to those found in some cold-adapted marine bacteria, molds, and diatoms (Raymond et al., 2008). The IBP was found to cause ice pitting and had RI activity. Further, 3519-10 was shown to have the physiological potential to maintain metabolic activity in an ice matrix at temperatures as low as −33°C (Bakermans and Skidmore, 2011b).
To investigate the phenotypic advantage that an IBP with RI activity confers to an ice-entrapped cell population, we examined the expression of 3519-10’s IBP as a function of growth temperature and its effect on cell viability during repeated freezing and thawing. Although the specific mechanism by which the IBP of 3519-10 interacts with ice crystal surfaces and protects the cells is still unclear, our results show that the presence of the IBP influences the ice crystal structure. The significance of our findings for microbial survival in polycrystalline ice is discussed.
Materials and Methods
Bacterial strains and culture conditions
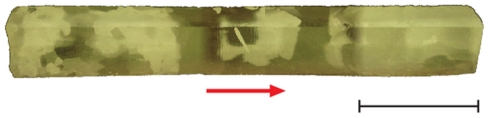
Isolate 3519-10 (family Flavobacteriaceae) was previously isolated from a Vostok 5G ice core sample recovered approximately 3,519 m below the surface (actual sampling depth of 3518.03–3518.44 m in the 5G core; Christner et al., 2006; Raymond et al., 2008; Figure 1). 3519-10 was cultured aerobically in R2 (Difco Laboratories, Inc.) liquid media and incubated with shaking at 4, 10, 15, and 25°C. Cultures were grown until the mid-logarithmic phase of growth for the isolation of RNA and early stationary phase for extracellular protein recovery. B5 was isolated from basal ice of the Taylor Glacier, Antarctica (Skidmore et al., 2009), and based on 16S rRNA identity, is a member of the genus Paenisporosarcina (Phylum Firmicutes). Paenisporosarcina isolate B5 was cultured aerobically at 15°C in liquid R2 media until the late stationary phase of growth. Escherichia coli ATCC 11775 was cultured aerobically in tryptic soy broth (TSB; Difco Laboratories, Inc.) at 37°C until the mid-logarithmic phase of growth. For crystal imaging, cells cultured at 15°C were diluted in deionized water to an approximate cell concentration of 106 colony forming units (CFU) mL−1.
Figure 1.
Composite image of ice core section 3,519 viewed under cross-polarized illumination. The arrow is “up” and indicates the ice core orientation relative to the surface. The scale bar is 10 cm.
RNA extraction, purification, and analysis
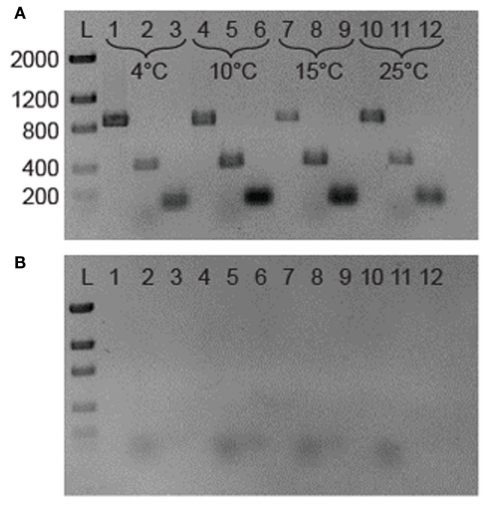
Aliquots (500 μL) of 3519-10 cell suspensions that were grown at 4, 10, 15, and 25°C were removed from culture tubes, immediately preserved in 2 volumes of RNAprotect (Qiagen), and stored at −20°C. The preserved samples were subsequently thawed on ice and lysed by the addition of 15 mg mL−1 lysozyme in 200 μL of TE buffer (10 mM Tris–Cl, 1 mM EDTA, pH 8.0), followed by incubation for 10 min at 25°C. The bulk RNA in the lysate was purified with the RNeasy Mini Kit (Qiagen) and eluted into 50 μL of RNase-free water. Genomic DNA was digested with DNase using the TURBO DNA-free kit (Ambion); its removal was confirmed with PCR (Figure 2B).
Figure 2.
(A) Reverse transcription-PCR analysis of gene expression in 3519-10. The 876-bp bands in lanes 1, 4, 7, and 10 represents the presence of 16S rRNA; the 421 and 100 bp bands are fragments of RT-PCR IBP and cspA transcripts, respectively. The template for lanes 1–3 was RNA from a 4°C grown culture of 3519-10, 4–6 from 10°C, 7–9 from 15°C, and 10–12 from 25°C cultures. (B). Identical samples lacking a RT step. Lane assignments are the same as for (A). L is a low DNA mass ladder (Invitrogen).
Specific mRNA and small subunit rRNA sequences were reverse transcribed (RT) to cDNA and subsequently amplified using a OneStep reverse transcription-PCR (RT-PCR) kit (Qiagen). Primers were designed to target a 420-bp fragment of 3519-10’s IBP gene (EU694412; IBP 421 bp F 5′-TACAAACGGCGCACTGGCCT-3′; IBP 421 bp R 5′-CAAAGCAGCTGCGCGGTTG-3′) and a 100-bp fragment of the cspA homolog (ACU07993; CspA1 F 5′-ATCCTTTGTTACCTTGCTGAACTTCGT-3′; CspA1 R 5′-AACGGAGGAGAAGACATCTTTGTTCA-3′) using sequence data available through GenBank (CP001673.1). The primers used to RT-PCR a ~900 bp region of the 16S rRNA molecule (515F 5′-GTGCCAGCAGCCGCGGTAA-3′; 1391R 5′-GACGGGCGGTGTGTRCA-3′) were those described by Reysenbach and Pace (1995). RT-PCR amplification was performed with an initial RT step at 50°C for 30 min, followed by heating at 95°C for 15 min to inactivate the reverse transcriptase. The cDNA products were subsequently amplified for 30 cycles using the following conditions: 94°C for 1 min, 50.8°C (16S rRNA primers) or 60°C (IBP and cspA primers) for 1 min, and 72°C for 1.5 min, with a terminal elongation at 72°C for 10 min. The amplified DNA products were examined after electrophoresis though a 2% (w/v) agarose gel that was stained with 1 μg mL−1 ethidium bromide.
Harvesting and preparation of extracellular protein
Extracellular proteins were captured and washed using YP-30 Microcon (Millipore) or Ultra 15 centrifugal filter devices (Amicon), both of which have reported nominal molecular weight cutoffs of 30 kDa. Cultures of 3519-10 were centrifuged at 17,000 × g for 10 min to pellet the cells, and either 1.5 or 12 mL of the supernatant was filtered through the YP-30 or Ultra 15, respectively. After centrifugation at 13,500 × g for 12 min or 4,500 × g for 20 min. (YP-30 or Ultra 15, respectively), the filters were washed in deionized H2O. The concentrated protein was recovered by inverting and centrifuging at 1,000 × g for 3 min (YP-30) or by using a pipette (Ultra 15) and was stored at 4°C until use. Samples containing deionized water were prepared in parallel to serve as controls.
The protein concentration was determined with the Coomassie (Bradford) Protein Assay kit (Pierce) and bovine serum albumin (BSA) as a standard. Absorbance at 595 nm was measured with a NanoDrop ND-1000 spectrophotometer. The extracellular proteins of 3519-10 were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The gel was buffered with tris–glycine (25 mM Tris Base, 250 mM glycine, 0.1% SDS, pH 8.3) and consisted of a 5% stacking and 12% running gel (w/v of polyacrylamide). SDS-PAGE was conducted at 70 V for 30 min and increased to 100 V for ~3 h. The separated polypeptides were visualized by staining with Coomassie stain (Weber and Osborn, 1969). The size of polypeptides was estimated by comparison to the electrophoretic migration distance of a 10- to 250-kDa molecular weight standard (New England Biolabs).
Freeze-thaw cycling viability assay
Cultures of E. coli in the logarithmic phase of growth were harvested by centrifugation at 17,000 × g for 10 min., the supernatant was removed, and cells were washed twice with an equal volume of phosphate buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4; pH 7.4). The E. coli cell suspensions (~108 CFU mL−1) were amended with the extracellular protein fraction from 3519-10 at final concentrations of 1, 10, and 100 μg mL−1. Frozen cell suspensions containing 1, 10, and 100 μg mL−1 BSA were prepared identically and served as controls. Additional controls consisted of frozen cell suspensions without amended protein and samples that remained at 25°C for the duration of the freeze-thaw cycling experiment. Each experimental data point represented a measurement from triplicate samples.
The cell suspensions were frozen rapidly by incubation at −80°C for ~45 min., and then samples were transferred to −5°C for approximately 12 h. Samples were thawed rapidly in a 25°C water bath, an aliquot was removed from each sample for serial dilution plating, and the samples were refrozen as described above. This procedure was repeated up to six times. After incubation at 37°C for 18 h, the number of E. coli colonies formed on the dilution plate series was counted to determine the number of CFU per milliliter.
Crystal structure imaging
Samples of deionized water passed though a 0.2-μm pore size filter, washed 3519-10 and Paenisporosarcina isolate B5 cells, and the extracellular proteins harvested from 3519-10 were frozen at −10°C. Samples were frozen within modified 50 mL polypropylene centrifuge tubes. The bottom of each tube was removed and the tubes were attached to a 30.5-cm2 and ~1 cm thick aluminum plate using O.C.T. embedding compound (Tissue Tek, Sakura). Twenty milliliters of liquid sample was placed in the tubes affixed to the plate and the tubes were capped. The samples were insulated on the sides and top by 2″ of extruded polystyrene foam board to minimize air space around these portions of the sample to encourage prominent crystal growth in the vertical direction (i.e., away from the aluminum plate). A thermistor connected to a HOBO® data logger (Onset Corporation) was suspended in each tube 2 cm above the aluminum plate to record the temperature of the sample during supercooling, freezing, and equilibration with the ambient temperature.
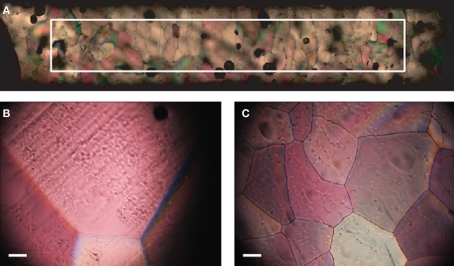
Approximately 20 h after freezing, thin sections (~1 mm) from ice in the lowest 5 mm of the ice column were prepared and imaged at −10°C under cross-polarized light using an Olympus BX51-TRF epifluorescence microscope equipped with a Linkam large area thermal stage. Eight to 15 images were collected via transect from the outside edge of the sample to the center of the sample (Figure 3A) depending on the amount of overlap between the images. The number of crystals per image was counted to calculate the average number of ice crystals per square millimeter. The field of view was calibrated using a 1-mm SPI PS8 micrometer.
Figure 3.
(A) Image transect showing ice structure from the outside edge of the sample to center (left to right in image). The superimposed rectangle delineates a 1.5-mm by 10 mm area. (B) Ice structure formed from filtered deionized water. (C) Ice structure formed in the presence of extracellular proteins from 3519-10. Scale bars in (B) and (C) are 0.1 mm.
Results
Expression of the IBP
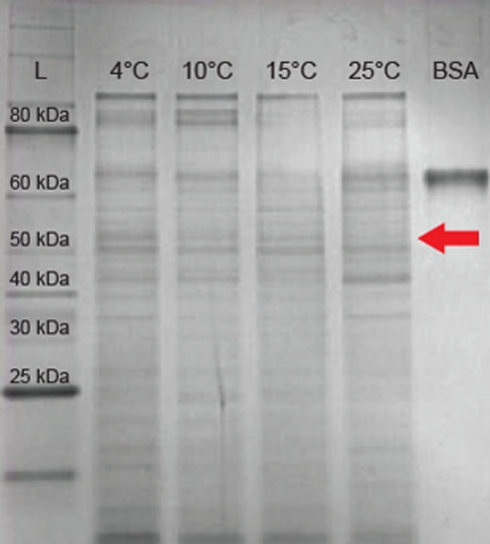
Reverse transcription-PCR amplification detected mRNA of the IBP gene at all growth temperatures tested between 4 and 25°C (Figure 2A), implying that the IBP gene is constitutively expressed during logarithmic growth. Expression of the major bacterial cold shock protein gene (cspA) and 16S rRNA were also detected under all conditions tested (Figure 2A). Amplicons were not produced in identical samples that omitted the RT step (Figure 2B), indicating that genomic DNA contamination was undetectable via PCR in the purified RNA samples. To assess if the IBP gene transcripts were translated and secreted from the cell under these conditions, preparations of the extracellular protein fraction from cultures incubated at intervals between 4 and 25°C were analyzed by SDS-PAGE (Figure 4). At all temperatures examined, there were at least 25 discernable polypeptide bands that ranged in molecular weight between 15 and 150 kDa. Denatured polypeptides smaller in size than the 30-kDa nominal molecular weight cutoff of the filtration devices are likely to represent components of multi-subunit proteins and/or hydrolyzed peptide fragments. A polypeptide that corresponded to the size of the 54-kDa IBP (Raymond et al., 2008) was observed at all growth temperatures examined (Figure 4).
Figure 4.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of extracellular proteins from 3519-10. Lane 1 contains a 10- to 250-kDa protein ladder. Lanes 2–5 contain 10 μg total extracellular protein harvested from cultures grown at 4, 10, 15, and 25°C respectively. Lane 6 contains 10 μg of BSA (66 kDa). The arrow indicates the expected migration of the IBP based on the predicted size of 54 kDa.
Bacterial viability during freezing and thawing
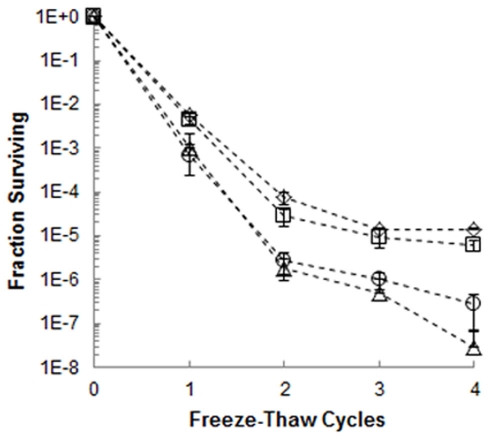
To examine the effect of the IBP on bacterial viability during the phase transition occuring between frozen incubation at −5°C and thawing, extracellular proteins harvested from 3519-10 were added to populations of E. coli at concentrations of 1, 10, and 100 μg mL−1, and the suspensions were frozen. After 4 freeze-thaw cycles, a biphasic trend was observed in the reduction of viability for all treatments (Figure 5). The first two cycles resulted in a 4–5 log reduction in the number of culturable E. coli cells, whereas only a reduction of 1–2 log was observed during cycles 2–4. However, the survival of E. coli in samples containing 100 μg mL−1 of extracellular protein from 3519-10 cultured at 4°C was significantly increased over that of the frozen control (cycles 1–4; p < 0.03; Figure 5) and had 34 and >100-fold higher recovery than frozen controls after 2 and 5 cycles, respectively. In comparison, proteins harvested from 3519-10 cultured at 25°C only significantly increased the survival of E. coli after cycles 1 and 4 (cycles 2, 3, and 5 were not statistically different from the controls p < 0.05), and in general, appeared to be less effective than proteins from the 4°C cultures in increasing the survival of E. coli under these conditions. The data from viability experiments with 1 and 10 μg mL−1 of extracellular proteins (data not shown) or 100 μg mL−1 of BSA (Figure 5) were statistically indistinguishable from the controls. After 6 freeze-thaw cycles, only samples amended with 100 μg mL−1 of extracellular protein from 3519-10 contained viable population sizes of E. coli that were detectable after dilution plating and incubation.
Figure 5.
Fraction of E. coli surviving four freeze-thaw cycles in the presence of 100 μg mL−1 of extracellular proteins harvested from a culture of 3519-10 grown at 4°C (⋄) and 25°C (□), as well as 100 μg mL−1 BSA(Δ). The bars represent the SE of triplicate samples; the frozen control (◦) is the average of six replicates from two experiments.
Ice crystal structure imaging
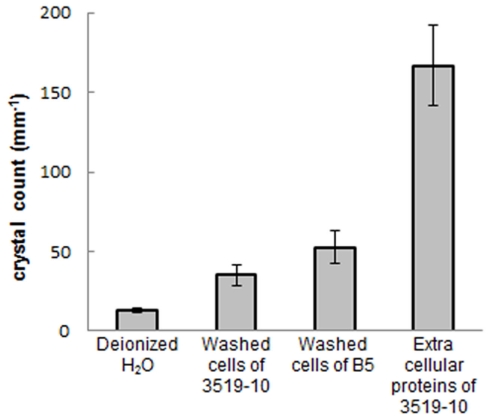
Temperature data from the probe suspended in each sample during freezing indicated that the samples supercooled and froze within ~45 min., but remained at the melting point for the subsequent ~2 h and were equilibrated with the ambient temperature (−10°C) by 3 h. Microtome-prepared thin sections of the ice were imaged approximately 20 h after freezing (Figure 3A.). Representative fields of view from the filtered deionized water sample (Figure 3B) and 3519-10 extracellular protein sample (Figure 3C) demonstrated an obvious difference in bulk ice structure. Ice samples frozen in the presence of a non-IBP producing bacteria (Paenisporosarcina isolate B5) and washed suspensions of 3519-10 contained 4.1 and 2.7-fold, respectively, more crystals per square millimeter than the ice formed from the filtered deionized water (Figure 6). Ice samples frozen in the presence of extracellular proteins from 3519-10 had 13-fold more crystals per square millimeter than filtered deionized water (Figure 6). The number of ice crystals in samples frozen in the presence of bacteria (p < 0.00001) and the extracellular proteins from 3519-10 (p < 0.00001) were statistically different from the control.
Figure 6.
Number of crystals per square millimeter for each ice sample (error bars are 1 σ). From left to right: ices made with filtered deionized water, washed 3519-10 cells, washed Paenisporosarcina isolate B5 cells, and 3519-10’s extracellular proteins.
Discussion
Microbial survival in the deep icy biosphere
The upper 3,309 m of the Vostok 5G ice core provides one of the oldest paleoclimatic ice core records examined to date (Petit et al., 1999), whereas the deepest portion of the ice core (below 3,539 m) is accreted lake water from Subglacial Lake Vostok. The latter represents the only material available to date for assessing limnological conditions in a subglacial Antarctic lake (Karl et al., 1999; Priscu et al., 1999; Christner et al., 2006). Although the surface temperature at Vostok is commonly below −55°C, the temperature of the ice at a depth of 3,519 m is approximately −8°C (Salamatin et al., 2004), and the base of the ice sheet is close to or at the pressure melting point (−2.5°C; Wüest and Carmack, 2000; Siegert et al., 2003). Analysis of ice core depths between 3,450 and 3,539 m have reported significant ice crystal deformation, evidence for basal shearing, and large particles >30 μm; characteristics which are evidence that ice in this portion of the core has interacted with the bed (Simoes et al., 2002). The large irregular crystals observed at 3,519 m (Figure 1) are consistent with these prior observations and are indicative of ice that has experienced significant shear and deformation, both of which are factors known to increase the rate of recrystallization (Paterson, 1994). Since recrystallization is damaging to biological cells and tissues (e.g., Miller and Mazur, 1976), we hypothesized that microorganisms surviving within the basal ice horizon have adaptations to mitigate cellular damage and/or influence the structure of the ice matrix they inhabit.
Basal ice in ice sheets and glaciers form when liquid water and debris at the ice-bed interface becomes entrained via freeze-on or regelation processes (Knight, 1997). Therefore, studies of basal ice can provide information on the physical, chemical, and microbiological characteristics of the subglacial environment. In some locations in East Antarctica, it has been reported that the freezing of subglacial water has formed a 1.1-km layer of basal ice, representing about half of the total ice sheet thickness in this region (Bell et al., 2011). Thus, the basal ice of glaciers and ice sheets likely represents an important transient phase for microorganisms in the subglacial environment (e.g., transport via freeze-on at the glacier bed) and may be a valuable source of microbial inocula and nutrients in downstream regions where basal melting and liquid water is widespread (Skidmore, 2011). Here we report an analysis of a bacterium isolated from basal ice from deep within the East Antarctic Ice Sheet that produces extracellular proteins, including a known IBP, that is capable of inhibiting recrystallization, conserving the ice crystal structure, and increasing the viability of bacteria entrapped within the ice matrix.
Expression of the IBP and increased tolerance to freezing and thawing
Ice-binding proteins have only been documented in cold tolerant organisms, and in bacteria, the phenotype has been observed in isolates from sea ice (Raymond and Knight, 2003; Raymond et al., 2007), glacial ice (Raymond et al., 2008), polar lakes (Raymond and Fritsen, 2000, 2001), and cold soils (Walker et al., 2006). Increased tolerance to freezing and thawing occurs when recrystallization is prevented. IBPs from a sea ice diatom (Navicula) have been shown to protect other diatoms (Raymond and Knight, 2003) and human red blood cells (Kang and Raymond, 2004) during freeze-thaw. Similarly, Walker et al. (2006) demonstrated that soil isolate “Chryseobacterium sp. strain C14” produced an ice-active substance that increased the survival of an Enterococcus sp. during freeze-thaw cycling. Consistent with prior observations, the presence of 3519-10’s IBP increased cell viability during repeated freezing and thawing (Figure 5). Since our experiments were not conducted with purified preparations of 3519-10’s IBP, it is not possible to determine the absolute concentration effect of the IBP on bacterial survival or ice crystal structure, nor are we able to discount the affect, positive or negative, that other proteins present may have had. However, in light of results from controls using BSA, our results are consistent with the activities reported for purified preparations of 3519-10’s IBP (Raymond et al., 2008). Based on the total number of extracellular polypeptides observed by SDS-PAGE (~25; Figure 4) and assuming equal weight distribution for each of the polypeptides, a crude estimate for the IBP concentration that significantly influenced bacterial viability is ~4 μg mL−1 (i.e., 100 μg mL−1/25 polypeptides = 4 μg mL−1). A concentration of 0.01–1.4 μg IBP mL−1 has been shown to inhibit ice recrystallization by microbial IBPs from Antarctic cyanobacterial mats, moss, and algae (Raymond and Fritsen, 2001), suggesting that 3519-10’s IBP has a comparable RI activity. The IBP of 3519-10 is a soluble protein that is secreted by the cell and appears capable of protecting any cell in the ice matrix (e.g., E. coli). In the habitat of polycrystalline ice, species like 3519-10 might passively protect freeze-sensitive cells or form consortial relationships with such species, providing protection from ice crystal damage in exchange for substrates or other resources.
Much of what is known about the regulation of IBP expression is restricted to studies of polar fish that seasonally experience temperatures below 0°C and induce their IBPs accordingly (Fletcher et al., 2001). In Newfoundland winter flounder, IBP mRNA is detectable in the liver at temperatures below 8°C, and the IBP accumulates to detectable levels in the blood plasma after several weeks under these conditions (Fletcher et al., 2001). In contrast, adult cod only express their IBPs at temperatures ≤1°C (Fletcher et al., 1987), while the Newfoundland ocean pout produces IBP continually independent of temperature (Fletcher et al., 1985). Based on quantitative PCR analysis, the multiple IBP isoforms of the diatom Fragilariopsis cylindrus are differentially expressed in response to osmotic shock and low temperature (−4°C; Bayer-Giraldi et al., 2010). Polypeptides and mRNA corresponding to 3519-10’s IBP were detected at all the growth temperatures tested (up to 25°C), indicating that low temperature was not required for active expression. Considering the energetic cost of protein synthesis and export, this apparent lack of regulation may hint at the importance of the IBP for ensuring survival of this bacterium in its environment. Based on the data in Figure 5, extracellular proteins containing the IBP produced by 3519-10 at 25°C did not protect E. coli viability to the same degree as those produced by cultures of 3519-10 grown at 4°C. Although the bulk protein concentration was identical between the treatments, there may have been fewer IBP molecules in the extracellular protein fraction of 25°C cultures compared to that in the 4°C cultures. Alternatively, the IBP may have a decreased activity and/or stability at warmer temperatures, providing a working hypothesis for future structural studies of this protein.
Effect of IBPs and cells on the ice crystal structure
During the phase transition to ice, particles the size of bacterial cells are rejected together with other soluble impurities into the solute-rich environment that exists at the grain boundaries (Mader et al., 2006). Under laboratory conditions, 3519-10 has been shown to be metabolically active in ice to temperatures as low as −33°C (Bakermans and Skidmore, 2011b). Price (2007) estimated that the concentration of nutrients and dissolved organic carbon in the aqueous fraction of the ice in deep portions of the Vostok core are sufficient to maintain the ambient concentration of cells in the ice for at least several hundred thousand years. In this study, the presence of 3519-10’s IBP and ~106 bacterial cells per milliliter were found to affect the ice structure (Figures 6 and 3C). Since the 3519-10 cells were washed thoroughly to remove the secreted IBP and the Paenisporosarcina isolate B5 does not produce ice-active substances, the implication is that particles the size and composition of bacterial cells influence the ice structure. Though the mechanism by which this occurs is not decipherable with the data available, it may be due to the bacteria serving as nucleation sites. These results are interesting considering that similar concentrations of cells, up to ~108 mL−1 of which at least 106 mL−1 are inferred to be viable, have been reported in basal ices (3,042–3,052 m) from the GISP2 Greenland ice core (Miteva et al., 2009). This suggests that the presence of microbial cells and their ice-interacting substances may influence the ice crystal structure of basal ices. In particular, recrystallization of ice grains via physical processes has been shown to significantly influence local strain rate (Paterson, 1994; Samyn et al., 2008). The magnitude by which microbes and their activities in a localized region of an ice mass, e.g., in the basal ice layer, could impact ice rheology is unknown and requires further investigation.
Conclusion
An IBP is a molecular adaptation that would be expected to enhance the survival and persistence of species in a diverse array of icy environments in the biosphere. Findings from the current study demonstrate that IBPs with RI activity can offer a distinct survival advantage to cell populations immured in the polycrystalline habitat of ice. IBPs are not possessed by all microorganisms inhabiting frozen environments and microorganisms like 3519-10 may use this phenotype as a basis for mutualistic interactions, similar to those mediated by diffusible compounds in other bacteria (e.g., Sher et al., 2011). A number of bacteria, including 3519-10, have demonstrated the ability to be metabolically active within the matrix of ice crystals (e.g., Amato et al., 2010; Bakermans and Skidmore, 2011b). Although expression of 3519-10’s IBP was documented at low temperature (4°C) under liquid conditions, determining if active expression of the IBP occurs in ice is territory for further study.
Proteins that affect ice crystal structure and inhibit recrystallization can be exploited to improve the texture of ice cream (Regand and Goff, 2006), cold tolerance in plants (Wen-li et al., 2005), and the cryopreservation of mammalian tissues (Bagis et al., 2006). Currently, protein structural data for bacterial IBPs are restricted to a single genetic form (Garnham et al., 2011). Unveiling the structure of 3519-10’s IBP would provide important comparative data to aid in elucidating the molecular characteristics unique to IBPs and may reveal novel properties relevant to applications in the industrial and biomedical sciences (e.g., improved cryopreservation of cell lines and tissues).
In summary, expression of 3519-10’s IBP was independent of growth temperature, affected the structure of ice crystals formed in its presence, and protected bacteria under conditions favorable to the process of recrystallization. Although the full range of molecular adaptations which play a role in bacterial persistence under frozen conditions has yet to be constrained, the ability to control ice crystal structure and preserve cellular integrity provides one possible explanation for how 3519-10 has successfully survived in deep Antarctic ice for thousands of years.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was partially supported by grants EAR-0525567, ANT-0636828, and ANT-0636770 from the National Science Foundation. Amanda Marie Achberger was also supported by funding from the NSF award ANT-0838941 and the Howard Hughes Medical Institute Biomedical Education Program. Timothy Ian Brox was also supported by scholarships from Montana Space Grant Consortium (MSGC) as part of the Montana NASA EPSCoR Program and the Montana State University (MSU) Undergraduate Scholars Program and an award to Mark Leslie Skidmore from MSGC. Mark Leslie Skidmore and Timothy Ian Brox are grateful for use of the Subzero Science and Engineering Research Facility at MSU.
References
- Amato P., Doyle S. M., Battista J. R., Christner B. C. (2010). Implications of subzero metabolic activity on long-term microbial survival in terrestrial and extraterrestrial permafrost. Astrobiology 10, 789–798 10.1089/ast.2010.0477 [DOI] [PubMed] [Google Scholar]
- Bagis H., Aktoprakligil D., Mercan H. O., Yurdusev N., Turgut G., Sekmen S., Arat S., Cetin S. (2006). Stable transmission and transcription of newfoundland ocean pout type III fish antifreeze protein (AFP) gene in transgenic mice and hypothermic storage of transgenic ovary and testis. Mol. Reprod. Dev. 73, 1404–1411 10.1002/mrd.20601 [DOI] [PubMed] [Google Scholar]
- Bakermans C., Skidmore M. L. (2011a). Microbial metabolism in ice and brine at −5°C. Environ. Microbiol. 13, 2269–2278 [DOI] [PubMed] [Google Scholar]
- Bakermans C., Skidmore M. (2011b). Microbial respiration in ice at subzero temperatures (−4 to −33°C). Environ. Microbiol. Rep. 3, 774–782 10.1111/j.1758-2229.2011.00298.x [DOI] [PubMed] [Google Scholar]
- Bayer-Giraldi M., Uhlig C., John U., Mock T., Valentin K. (2010). Antifreeze proteins in polar sea ice diatoms: diversity and gene expression in the genus Fragilariopsis. Environ. Microbiol. 12, 1041–1052 10.1111/j.1462-2920.2009.02149.x [DOI] [PubMed] [Google Scholar]
- Bell R. E., Ferraccioli F., Creyts T. T., Braaten D., Corr H., Das I., Damaske D., Frearson N., Jordan T., Rose K., Studinger M., Wolovick M. (2011). Widespread persistent thickening of the East Antarctic ice sheet by freezing from the base. Science 331, 1592–1595 10.1126/science.1200109 [DOI] [PubMed] [Google Scholar]
- Carpenter J. F., Hansen T. N. (1992). Antifreeze protein modulates cell survival during cryopreservation: mediation through influence on ice crystal growth. Proc. Natl. Acad. Sci. U.S.A. 89, 8953–8957 10.1073/pnas.89.19.8953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christner B. C., Royston-Bishop G., Foreman C. M., Arnold B. R., Tranter M., Welch K. A., Lyons W. B., Tsapin A. I., Studinger M., Priscu J. C. (2006). Limnological conditions in Subglacial Lake Vostok, Antarctica. Limnol. Oceanogr. 51, 2485–2501 10.4319/lo.2006.51.6.2485 [DOI] [Google Scholar]
- Davies P. L., Baardsnes J., Kuiper M. J., Walker V. K. (2002). Structure and function of antifreeze proteins. Phil. Trans. R. Soc. Lond. B Biol. Sci. 357, 927–935 10.1098/rstb.2002.1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont F., Marechal P. A., Gervais P. (2004). Cell size and water permeability as determining factors for cell viability after freezing at different cooling rates. Appl. Environ. Microbiol. 70, 268–272 10.1128/AEM.70.1.268-272.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher G. L., Hew C. L., Davies P. L. (2001). Antifreeze proteins of teleost fishes. Annu. Rev. Physiol. 63, 359–390 10.1146/annurev.physiol.63.1.359 [DOI] [PubMed] [Google Scholar]
- Fletcher G. L., Hew C. L., Li X., Haya K., Kao M. H. (1985). Year-round presence of high levels of plasma antifreeze peptides in a temperate fish, ocean pout (Macrozoarces americanus). Can. J. Zool. 63, 488–493 10.1139/z85-070 [DOI] [Google Scholar]
- Fletcher G. L., King M. J., Kao M. H. (1987). Low temperature regulation of antifreeze glycopeptide levels in Atlantic cod (Gadus morhua). Can. J. Zool. 65, 227–233 10.1139/z87-140 [DOI] [Google Scholar]
- Fricker H. A., Powell R., Priscu J., Tulaczyk S., Anandakrishnan S., Christner B. C., Fisher A. T., Holland D., Horgan H., Jacobel R., Mikucki J., Mitchell A., Scherer R., Severinghaus J. (2011). “Siple Coast subglacial aquatic environments: the Whillans Ice Stream Subglacial Access Research Drilling (WISSARD) project,” in Antarctic Subglacial Aquatic Environments, eds Siegert M., Kennicutt M., Bindschadler R., Geophysical Monograph 192 (Washington, DC: American Geophysical Union Press; ), 199–219 [Google Scholar]
- Gage A. A., Guest K., Montes M., Caruana J. A., Whalen D. A., Jr. (1985). The effect of varying freezing and thawing rates in experimental cryosurgery. Cryobiology 22, 175–182 10.1016/0011-2240(85)90172-5 [DOI] [PubMed] [Google Scholar]
- Garnham C. P., Campbell R. L., Davies P. L. (2011). Anchored clathrate waters bind antifreeze proteins to ice. Proc. Natl. Acad. Sci. U.S.A. 108, 7363–7367 10.1073/pnas.1100429108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert J. A., Davies P. L., Laybourn-Parry J. (2005). A hyperactive, Ca 2+-dependent antifreeze protein in an Antarctic bacterium. FEMS Microbiol. Lett. 245, 67–72 10.1016/j.femsle.2005.02.022 [DOI] [PubMed] [Google Scholar]
- Graether S. P., Kuiper M. J., Gagné S. M., Walker V. K., Jia Z., Sykes B. D., Davies P. L. (2000). β-Helix structure and ice-binding properties of a hyperactive antifreeze protein from an insect. Nature 406, 325–328 10.1038/35018610 [DOI] [PubMed] [Google Scholar]
- Kang J. S., Raymond J. A. (2004). Reduction of freeze-thaw-induced hemolysis of red blood cells by an algal ice-binding protein. Cryo Lett. 25, 307–310 [PubMed] [Google Scholar]
- Karl D. M., Bird D. F., Björkman K., Houlihan T., Shackelford R., Tupas L. (1999). Microorganisms in the accreted ice of Lake Vostok, Antarctica. Science 286, 2144–2147 10.1126/science.286.5447.2144 [DOI] [PubMed] [Google Scholar]
- Knight P. G. (1997). The basal ice layer of glaciers and ice sheets. Quat. Sci. Rev. 16, 975–993 10.1016/S0277-3791(97)00033-4 [DOI] [Google Scholar]
- Lanoil B., Skidmore M., Priscu J. C., Han S., Foo W., Vogel S. W., Tulaczyk S., Engelhardt H. (2009). Bacteria beneath the West Antarctic ice sheet. Environ. Microbiol. 11, 609–615 10.1111/j.1462-2920.2008.01831.x [DOI] [PubMed] [Google Scholar]
- Lukin V., Bulat S. (2011). “Vostok subglacial lake: details of Russian plans/activities for drilling and aampling,” in Antarctic Subglacial Aquatic Environments, eds Siegert M., Kennicutt M., Bindschadler R., Geophysical Monograph 192 (Washington, DC: American Geophysical Union Press; ), 187–197 [Google Scholar]
- Mader H. M., Pettitt M. E., Wadham J. L., Wolff E. W., Parkes R. J. (2006). Subsurface ice as a microbial habitat. Geology 34, 169–172 10.1130/G22096.1 [DOI] [Google Scholar]
- Miller R. H., Mazur P. (1976). Survival of frozen-thawed human red cells a function of cooling and warming velocities. Cryobiology 13, 404–414 10.1016/0011-2240(76)90096-1 [DOI] [PubMed] [Google Scholar]
- Miteva V. (2008). “Bacteria in snow and glacier ice,” in Psychrophiles: From Biodiversity to Biotechnology, eds Margesin R., Schinner F., Marx J.-C., Gerday C. (Berlin: Springer; ), 31–50 [Google Scholar]
- Miteva V., Teacher C., Sowers T., Brenchley J. (2009). Comparison of the microbial diversity at different depths of the GISP2 Greenland ice core in relationship to deposition climates. Environ. Microbiol. 11, 640–656 10.1111/j.1462-2920.2008.01835.x [DOI] [PubMed] [Google Scholar]
- Paterson W. S. B. (1994). The Physics of Glaciers. Oxford: Butterworth-Heinemann [Google Scholar]
- Petit J. R., Jouzel J., Raynaud D., Barkov N. I., Barnola J. M., Basile I., Bender M., Chappellaz J., Davis M., Delaygue G., Delmotte M., Kotlyakov V. M., Legrand M., Lipenkov V. Y., Lorius C., Pépin L., Ritz C., Saltzman E., Stievenard M. (1999). Climate and atmospheric history of the past 420,000 years from the Vostok ice core, Antarctica. Nature 399, 429–436 10.1038/20859 [DOI] [Google Scholar]
- Price P. B. (2000). A habitat for psychrophiles in deep Antarctic ice. Proc. Natl. Acad. Sci. U.S.A. 97, 1247–1251 10.1073/pnas.97.3.1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price P. B. (2007). Microbial life in glacial ice and implications for a cold origin of life. FEMS Microbiol. Ecol. 59, 217–231 10.1111/j.1574-6941.2006.00234.x [DOI] [PubMed] [Google Scholar]
- Priscu J. C., Adams E. E., Lyons W. B., Voytek M. A., Mogk D. W., Brown R. L., McKay C. P., Takacs C. D., Welch K. A., Wolf C. F., Kirshtein J. D., Avci R. (1999). Geomicrobiology of subglacial ice above Lake Vostok, Antarctica. Science 286, 2142–2144 10.1126/science.286.5447.2141 [DOI] [PubMed] [Google Scholar]
- Priscu J. C., Tulaczyk S., Studinger M., Kennicutt M. C., II, Christner B. C., Foreman C. M. (2008). “Antarctic subglacial water: origin, evolution and microbial ecology,” in Polar Lakes and Rivers, eds Vincent W., Laybourn-Parry J. (Oxford: Oxford University Press; ), 119–135 [Google Scholar]
- Raymond J. A., Christner B. C., Schuster S. C. (2008). A bacterial ice-binding protein from the Vostok ice core. Extremophiles 12, 713–717 10.1007/s00792-008-0178-2 [DOI] [PubMed] [Google Scholar]
- Raymond J. A., Fritsen C. H. (2000). Ice-active substances associated with Antarctic freshwater and terrestrial photosynthetic organisms. Antarct. Sci. 12, 418–424 10.1017/S0954102000000493 [DOI] [Google Scholar]
- Raymond J. A., Fritsen C. H. (2001). Semipurification and ice recrystallization inhibition activity of ice-active substances associated with Antarctic photosynthetic organisms. Cryobiology 43, 63–70 10.1006/cryo.2001.2341 [DOI] [PubMed] [Google Scholar]
- Raymond J. A., Fritsen C. H., Shen K. (2007). An ice-binding protein from an Antarctic sea ice bacterium. FEMS Microbiol. Ecol. 61, 214–221 10.1111/j.1574-6941.2007.00345.x [DOI] [PubMed] [Google Scholar]
- Raymond J. A., Knight C. A. (2003). Ice binding, recrystallization inhibition, and cryoprotective properties of ice-active substances associated with Antarctic sea ice diatoms. Cryobiology 46, 174–181 10.1016/S0011-2240(03)00026-9 [DOI] [PubMed] [Google Scholar]
- Regand A., Goff H. D. (2006). Ice recrystallization inhibition in ice cream as affected by ice structuring proteins from winter wheat grass. J. Dairy Sci. 89, 49–57 10.3168/jds.S0022-0302(06)72068-9 [DOI] [PubMed] [Google Scholar]
- Reysenbach A.-L., Pace N. R. (1995). “Reliable amplification of hyperthermophilic archaeal 16s rRNA genes by the polymerase chain reaction,” in Archaea: A Laboratory Manual: Thermophiles, eds Robb F. T., Place A. R. (New York: Cold Springs Harbor Laboratory Press; ), 101–105 [Google Scholar]
- Ross N., Siegert M. J., Rivera A., Bentley M. J., Blake D., Capper L., Clarke R., Cockell C. S., Corr H. F. J., Harris W., Hill C., Hindmarsh R. C. A., Hodgson D. A., King E. C., Lamb H., Maher B., Makinson K., Mowlem M., Parnell J., Pearce D. A., Priscu J., Smith A. M., Tait A., Tranter M., Wadham J. L., Whalley W. B., Woodward J. (2011). “Ellsworth Subglacial Lake, West Antarctica: a review of its history and recent field campagins,” in Antarctic Subglacial Aquatic Environments, eds Siegert M., Kennicutt M., Bindschadler R., Geophysical Monograph 192 (Washington, DC: American Geophysical Union Press; ), 221–233 [Google Scholar]
- Salamatin A. N., Tsyganova E. A., Lipenkov V. Y., Petit J. R. (2004). Vostok (Antarctica) ice-core time-scale from dating of different origins. Ann. Glaciol. 39, 283–292 10.3189/172756404781814023 [DOI] [Google Scholar]
- Samyn D., Svensson A., Fitzsimons S. J. (2008). Dynamic implications of discontinuous recrystallization in cold basal ice: Taylor Glacier, Antarctica. J. Geophys. Res. 113, F03S90. 10.1029/2006JF000600 [DOI] [Google Scholar]
- Sher D., Thompson J. W., Kashtan N., Croal L., Chisholm S. W. (2011). Response of Procholorococcus ecotypes to co-culture with diverse marine bacteria. ISME J. 5, 1125–1132 10.1038/ismej.2011.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicheri F., Yang D. S. C. (1995). Ice-binding structure and mechanism of an antifreeze protein from winter flounder. Nature 375, 427–431 10.1038/375427a0 [DOI] [PubMed] [Google Scholar]
- Siegert M. J., Tranter M., Ellis-Evans J. C., Priscu J. C., Lyons W. B. (2003). The hydrochemistry of Lake Vostok and the potential for life in Antarctic subglacial lakes. Hydrol. Process. 17, 795–814 10.1002/hyp.1166 [DOI] [Google Scholar]
- Simões J. C., Petit J. R., Souchez R., Lipenkov V. Y., de Angelis M., Leibao L., Jouzel J., Duval P. (2002). Evidence of glacial flour in the deepest 89 m of the Vostok ice core. Ann. Glaciol. 35, 340–346 10.3189/172756402781816816 [DOI] [Google Scholar]
- Skidmore M. (2011). “Microbial communities in Antarctic subglacial aquatic environments,” in Antarctic Subglacial Aquatic Environments, eds Siegert M., Kennicutt M., Bindschadler R., Geophysical Monograph 192 (Washington, DC: American Geophysical Union Press; ), 61–81 [Google Scholar]
- Skidmore M., Bakermans C., Brox T., Christner B. C., Montross S. (2009). Microbial respiration at sub-zero temperatures in laboratory ices. Geochim. Cosmochim. Acta 73, A1234 [Google Scholar]
- Smallwood M., Worrall D., Byass L., Elias L., Ashford D., Doucet C. J., Holt C., Telford J., Lillford P., Bowles D. J. (1999). Isolation and characterization of a novel antifreeze protein from carrot (Daucus carota). Biochem. J. 340, 385–391 10.1042/0264-6021:3400385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung H. C., Price P. B., Bramall N. E., Vrdoljak G. (2006). Microorganisms metabolizing on clay grains in 3-km-deep Greenland basal ice. Astrobiology 6, 69–86 10.1089/ast.2006.6.69 [DOI] [PubMed] [Google Scholar]
- Walker V. K., Palmer G. R., Voordouw G. (2006). Freeze-thaw tolerance and clues to the winter survival of a soil community. Appl. Environ. Microbiol. 72, 1784–1792 10.1128/AEM.72.3.1784-1792.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K., Osborn M. (1969). The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J. Biol. Chem. 244, 4406–4412 [PubMed] [Google Scholar]
- Wen-li X., Mei-qin L., Xin S., Cun-fu L. (2005). Expression of a carrot 36 kD antifreeze protein gene improves cold stress in transgenic tobacco. For. Stud. China 7, 11–15 10.1007/s11632-005-0039-3 [DOI] [Google Scholar]
- Wüest A., Carmack E. (2000). A priori estimates of mixing and circulation in the hard-to-reach water body of Lake Vostok. Ocean Model. 2, 29–43 10.1016/S1463-5003(00)00007-X [DOI] [Google Scholar]