Abstract
Campylobacter coli is the most common Campylobacter species found in pig (95%), but the ability of this bacterium to survive in swine manure as well as the potential for causing human illness are poorly understood. We present here laboratory-scale experiments to investigate the effect of temperature on the survival of C. coli in spiked swine manure samples at temperatures from 4 to 52°C. The survival of C. coli during storage for 30 days was studied by three different methods: bacterial culture (plate counting), DNA qPCR, and mRNA RT-qPCR. The results indicate that C. coli could survive in swine manure up to 24 days at 4°C. At higher temperatures, this bacterium survived only 7 days (15°C) or 6 days (22°C) of storage. The survival of C. coli was extremely short (few hours) in samples incubated at 42 and 52°C. The results from the RT-qPCR method were consistent with the data from the bacterial culture method, indicating that it detected only viable C. coli cells, thus eliminating false-positive resulting from DNA from dead C. coli cells.
Keywords: RT-qPCR, Campylobacter coli, mRNA, ceuE, swine manure
Introduction
Livestock wastes such as manure or slurry from intensive animal production may contain pathogenic microorganisms including viruses, bacteria (Escherichia coli, Campylobacter spp., and Salmonella), and protozoa (Mawdsley et al., 1995; Semenov et al., 2009; Klein et al., 2011). There has been an increasing concern about which effect of pathogens in animal manure may have on human and animal health (Bicudo and Goyal, 2003). The manure is a potential source of contamination to the aquatic environment particularly where the slurry is used for fertilizing soil (Mawdsley et al., 1995; Marti et al., 2009; Klein et al., 2011). In addition, it has been reported that many farmers spread manure on the land straight after removal from the tanks, either because of inadequate storage capacity or greater convenience (Nicholson et al., 2005) which may release Campylobacters as well as other intestinal pathogens into the environment via the feces from infected animals.
Campylobacter spp. is currently the most common cause of human gastrointestinal disease worldwide. It is estimated approximately nine million human campylobacteriosis cases are reported annually in 27 countries in the EU (EU27; Andreoletti et al., 2011). The major sources of Campylobacter spp. are in animal intestinal tracts including chickens, cattle, pigs, wild-living mammals, and birds (Nielsen et al., 1997; Inglis et al., 2010; Oporto and Hurtado, 2011). Although 95% of the human campylobacteriosis cases attributed to Campylobacter jejuni, the importance of human campylobacteriosis caused by Campylobacter coli is being recognized due to an increased resistance of this pathogen to a greater number of antimicrobials (Gebreyes et al., 2005). Pigs are known to be frequently infected with Campylobacter (prevalence between 50 and 100%), to exhibit high counts of this pathogen in their feces, and to show a dominance of C. coli species (Boes et al., 2005; Jensen et al., 2006; Oporto et al., 2007).
It has been reported that soil is a source of microbial contamination for fruits and vegetables, as evidenced by the isolation of soil-residing pathogenic bacteria including Campylobacters from fresh produce. Pathogens may be transferred to the environment by application of inadequately composted or raw animal manures or sewage (Berger et al., 2010; Gardner et al., 2011; Verhoeff-Bakkenes et al., 2011). When pig feces or manures are applied to the agricultural field, the presence of C. coli could contaminate groundwater and soil either directly or indirectly after rainfalls. Although C. coli is responsible for less than 5–7% of human campylobacteriosis reported cases, the impact of this bacterium is still substantial. It is estimated that human campylobacteriosis caused by C. coli infection has an annual cost of millions of dollars but despite the economic importance of this pathogen, most Campylobacter research focuses upon C. jejuni (Humphrey et al., 2007; Sheppard et al., 2010). Furthermore, it has been reported recently that drinking water is the source of C. coli infection in grandparent breeder farms (Pérez-Boto et al., 2010). Therefore, control of the survival of this pathogen in the slurry during storage (prior to field application) is important to prevent infection in man and in animal as well as to prevent environmental contamination.
This study aimed to investigate the effect of various temperatures on the survival of C. coli in swine slurry using three different techniques: bacterial culture, DNA-based quantitative PCR (qPCR) and reverse transcription quantitative real-time PCR (RT-qPCR). Conventional bacterial culture methods for detection of Campylobacter spp. involving enrichment, isolation, and identification at the species level are labor-intensive and time-consuming, requiring 5–6 days to complete (Collette et al., 2008). While the major limitation of the DNA-based qPCR method is the potential detection of both live and dead, or non-culturable cells (Wolffs et al., 2005), RT-qPCR method in which mRNA is targeted instead of DNA has greater potential for detecting viable cells (Maurer, 2006). Five different temperatures were selected: 4°C – a temperature used to mimic the average temperature in the slurry tank during the winter time in Denmark; 15 and 22°C, representing the average temperatures in spring and summer times, respectively; 42°C is optimal growth temperature for thermophilic Campylobacters; 52°C – the temperature was chosen because it has been reported that most anaerobic digestion processes of bio-waste are operated at temperatures more than 50°C (Chen, 1983; Han and Dague, 1997; Wagner et al., 2008). A putative virulence gene, the ceuE gene of C. coli was chosen as a biomarker for C. coli detection for both qPCR and RT-qPCR assays. This gene was selected because it represents a good candidate for C. coli detection as it is present in all isolated strains described to date (Gonzalez et al., 1997; Gebreyes et al., 2005; Nayak et al., 2005). Furthermore, several ceuE DNA-based methods have been developed for detection of C. coli directly from complex biological samples such as feces with a high sensitivity and specificity (Bang et al., 2003; Hong et al., 2003).
Materials and Methods
Bacterial strains and culture conditions
Campylobacter coli reference strain CCUG-10955 isolated from swine manure (Culture Collection of University of Gothenburg) was used in this study for spiking of swine manure samples. The strain was recovered on blood agar base No. 2 (CM271; Oxoid, Greve, Denmark) supplemented with 5% (v/v) sterile defibrinated calf blood and isolated on modified charcoal cefoperazone deoxycholate agar (mCCDA CM0739; Oxoid, Greve, Denmark) with selective supplement SR0155 (Oxoid, Greve, Denmark). The medium was prepared according to the manufacturer’s instruction. A solid selective medium, Abeyta–Hunt–Bark (AHB) agar [National Veterinary Institute, Technical University of Denmark (DTU-Vet), Aarhus, Denmark] with 1% triphenyltetrazolium chloride (+TCC), was used for direct determination of colony-forming unit (CFU). All Campylobacter spp. used in this study were grown on blood agar plates at 42°C in microaerophilic conditions, whereas Salmonella, Escherichia coli, and Listeria strains were grown on blood agar plates at 37°C in aerobic conditions. Clostridium strain was grown on blood agar plates at 37°C in anaerobic conditions.
Bacterial DNA of Campylobacters (n = 9), Salmonella (n = 5), E. coli (n = 2), Listeria (n = 1), and Clostridium (n = 1; Table 1) was extracted using QIAamp® DNA Mini Kit (Qiagen, Copenhagen, Denmark). The DNA concentration was determined using a NanoDrop 1000 spectrophotometer Thermo Scientific (Saveen Werner ApS, Denmark). The bacterial DNA samples (2 ng/μl) were used to evaluate the specificity of the qPCR assays.
Table 1.
The bacterial strains used in this study.
| No. | Species | Strains | Real-time PCR |
|---|---|---|---|
| 1 | C. coli | CCUG 10955 | + |
| 2 | C. coli | CCUG 11283 | + |
| 3 | C. coli | CCUG 10951 | + |
| 4 | C. coli | CCUG 12079 | + |
| 5 | C. jejuni | SC11 | − |
| 6 | C. jejuni | CCUG 11824 | − |
| 7 | C. lari | CCUG 19512 | − |
| 8 | C. upsaliensis | CCUG 15015 | − |
| 9 | C. fetus | CCUG 6823 | − |
| 10 | Salmonella Typhimurium | NCTC 12023 | − |
| 11 | S. Typhimurium LT2 | NCTC 12416 | − |
| 12 | S. Enteritidis | NCTC 13349 | − |
| 13 | S. Enteritidis | NCTC 12694 | − |
| 14 | S. Dublin | NCTC 09676 | − |
| 15 | Escherichia coli | NCTC 9001 | − |
| 16 | E. coli CDT producing | E6468/62 D2253 (O127:H11) | − |
| 17 | Clostridium perfringens | NCTC8239 | − |
| 18 | Listeria monocytogenes | NCTC 7973 | − |
Manure samples
Liquid manure slurries used in this study were collected from seven different pig farms for three times in 2 weeks in January, 2010 in Jutland (Denmark). A total of 5 l of manure slurry were collected from two slurry tanks at each farm using a bucket after 10 min of mechanical mixing of the tank content. Subsequently, the contents of the bucket were stirred and a 200-ml sample was collected into a plastic bag. A total of 50 samples were stored in ice-boxes and immediately transported to the laboratory. On arrival, all samples were tested for the presence of Campylobacter spp. by both bacterial culture and qPCR methods as described below (see Detection of C. coli by Bacterial Culture Method and Detection and Quantification of C. coli by qPCR and RT-qPCR). Of 50 samples tested, 25 were Campylobacter-negative. All Campylobacter-negative liquid manure samples were pooled and aliquoted into 90 ml volumes and spiked with C. coli as follows. At an onset of the experiment, each manure sample (90 ml) was spiked with 10 ml of C. coli in physiological saline (0.09% NaCl) to reach a final concentration of 1 × 109 CFU/ml. The spiked samples (in triplicate) were stored in Erlenmeyer flasks (Carolina, USA) and incubated at various temperatures (4, 15, 22, 42, and 52°C) under aerobic conditions for up to 30 days. The samples incubated at high temperatures (42 and 52°C) were tested at 5 and 3 h, respectively after spiking and were not processed after day 1 until day 30. The samples incubated at 15 and 22°C were not processed after day 7. However, all samples incubated at all selected temperatures were tested by culture, qPCR, and RT-qPCR assays at day 30.
Total bacterial RNA and DNA extraction
The total bacterial nucleic acids (RNA and DNA) were extracted from manure samples using cetyltrimethylammonium bromide (CTAB) buffer and a part of the RNeasy Mini RNA isolation kit (Qiagen, Copenhagen, Denmark) according to the manufacturer’s protocol. Briefly, 1 ml of each bacterial manure suspension was transferred to a microcentrifuge tube and centrifuged at 8,000 g for 7 min. The pellets were mixed with 0.5 ml of CTAB extraction buffer, 0.5 ml of phenol–chloroform–isoamyl alcohol (25:24:1, pH 8.0) and 250 mg of zirconia/silica beads. The sample and beads was mixed by vortex for 30 s. The lysate was centrifuged at 13,000 g for 5 min. The aqueous phase was purified by chloroform–isoamyl alcohol (24:1) extraction. The mixture was centrifuged at 13,000 g for 5 min. The volume of the aqueous phase was estimated and the nucleic acids were precipitated by adding a 0.08 volume of chilled 7.5 M ammonium acetate and a 0.54 volume of chilled isopropanol. For the DNA extraction, instructions for step (a) were followed, and for the RNA extraction, instructions for step (b) were followed.
-
a)
The tube was inverted 20–30 times to mix the components and incubated on ice for 30–40 min. The precipitated DNA was collected by centrifugation at 13,000 g for 10 min at 4°C. The DNA pellet was washed once using ice-cold 70% ethanol and dried by air. The DNA pellet was suspended in 50 μl of DNase-free water. The DNA preparation was used immediately or stored at −20°C until needed.
-
b)
The mixture, including any precipitate that may have formed, was transferred to an RNeasy spin column placed in a 2-ml collection tube from the RNeasy Mini RNA isolation kit (Qiagen, Copenhagen, Denmark) and centrifuged for 15 s at 8,000 g. Washing steps were followed according to the manufacturer’s protocol. The RNA was eluted in 50 μl of RNase-free water and treated with 0.3 U ml−1 of DNase I Amplification Grade (Invitrogen, Denmark) according to the manufacturer’s instruction. The DNA-free RNA products were transcribed to complementary DNA (cDNA) using the iScript™ cDNA Synthesis Kit (Bio-Rad, USA) with pre-mixed RNase inhibitor and random hexamer primers, according to the manufacturer’s instruction.
Design of primers and standard curve for qPCR
The sequences from ceuE gene of C. coli (accession number: X88849.1) were obtained from NCBI GenBank and used for primer design. After multiple sequence alignment by using the ClustalW program (Chenna et al., 2003), a primer pair namely ceuE-F/ceuE-R with sequences flanking to the conserved regions in C. coli ceuE gene was designed using the Primer 3 program (http://frodo.wi.mit.edu/primer3/). The forward primer (ceuE-F), 5′-AAATTTCCGCTTTTGGACCT-3′ (corresponding to nucleotide position 3328–3348 in ceuE gene) and the reverse primer (ceuE-R), 5′-CCTTGTGCGCGTTCTTTATT-3′ (corresponding to nucleotide position 3504–3524 in ceuE gene) were used to amplify a 196-bp fragment.
To enable accurate quantification of C. coli, a standard curve for the qPCR assays was generated. A 24-h growth of C. coli at 42°C in microaerophilic conditions on blood agar plates was harvested in physiological saline (0.09% NaCl). Serial 10-fold dilutions of C. coli were added to each Campylobacter-negative manure sample, and the spiked materials were immediately used for DNA isolation. This experiment was carried in duplicate. The DNA extracts of 10-fold dilutions from 1 × 108 to 1 × 102 CFU/ml were used for qPCR assays to establish the standard curve and used for quantifying C. coli in swine manure.
Detection of C. coli by bacterial culture method
Duplicate 10-fold serial dilutions ranging from 100 to 10−9 of each sample were prepared and 100 μl of each dilution was spread in duplicate onto pre-dried (at 22°C for 45 min) AHB plates and incubated for 48 h at 42°C in microaerophilic conditions. The selective AHB agar plates were applied according to the recommendations of ISO 10272-1:2006 (Anonymous, 2006). Plates were inspected to detect the presence of colonies presumed to be Campylobacter because of their characteristics. The detection limit of culture method was 500 CFU/ml. Five presumptive Campylobacter colonies from each manure sample were picked and used directly for verification by a conventional PCR method described previously (Lund et al., 2003).
Detection and quantification of C. coli by qPCR and RT-qPCR
Quantitative real-time PCR and RT-qPCR were carried out in an Mx3005P thermocycler (Stratagene, Denmark) using ceuE primers. The PCR mixtures (25 μl) contained 5 μl DNA or 5 μl cDNA, 12.5 μl of 2× PCR master mix (Promega, Denmark), 400 nM of each primer and 50000× diluted SYBR green (Invitrogen, Denmark). The qPCR conditions consist of an initial heat–denaturing step at 94°C for 5 min; followed by 45 cycles of 94°C for 15 s, annealing at 56°C for 20 s, and extended at 72°C for 15 s; followed by an elongation step at 72°C for 3 min. In every qPCR analysis, the C. coli standard for absolute quantification was included. A negative control (5 μl of water) and a positive DNA control (5 μl) of C. coli DNA (2 ng/μl) were included.
Post amplification melting temperature (Tm) analysis from 60 to 95°C at 0.5°C increments was conducted to confirm specific ceuE product (Tm = 80°C). The Mx3005P detection software was used to determine threshold cycle (Ct) values, Tm, and the standard curve. Negative controls included RNase- and DNase-free water and nucleic acid extracts from un-spiked manure samples to determine any possible cross-reactivity or contamination (false-positive results). The product of ended point qPCR assays was also analyzed using agarose gel electrophoresis. Five microliters of PCR products were loaded on 2% of agarose gel (BioWhittaker, Inc., USA) containing 0.1 μg of ethidium bromide/ml and the electrophoresis was performed at 400 V for 45 min. The gel was visualized on an UV transillumination (Ultra-Violet Products, Ltd., Cambridge, UK).
Statistical analyses
The values were expressed as the average ± SD. The data were analyzed for statistical significance using one-way ANOVA (ANalysis Of VAriance, Microsoft Excel). A p-value ≤0.05 was considered to be statistically significant.
Results
Specificity and sensitivity of qPCR and RT-qPCR assays
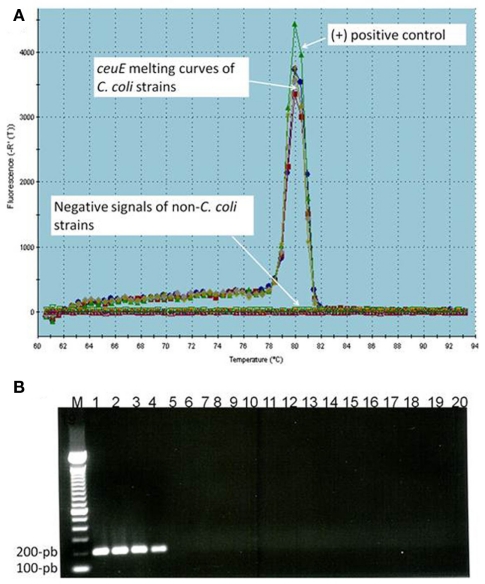
The specificity of assays was determined by qPCR assays with the DNA targets isolated from pure cultures of 18 Campylobacter and non-Campylobacter strains (Table 1). All C. coli strains (n = 4) were identified correctly. None of the five different Campylobacter species and none of the non-Campylobacter strains employed in the tests gave any positive signal (Table 1). The specificity of the PCR amplified products was determined by both melting curves (Tm) and agarose gel electrophoresis analysis. As expected, a Tm single peak at 80°C for C. coli ceuE gene amplified products (Figure 1A) and a single band of 196-bp ceuE amplified product was obtained with agarose gel electrophoresis (Figure 1B). Un-spiked manure samples and water gave the expected negative results both in the qPCR assays and in the melting curve analysis (Figure 1). In all RT-qPCR assays, the RNA samples were amplified by qPCR to test for DNA contamination. We did not obtain any peaks at 80°C or any 196-bp amplified product by gel electrophoresis (data not shown) on DNaseI-treated nucleic acid extracts, verifying that DNA was totally removed. By using serial dilutions of Campylobacter DNA and mRNA extracted as described in Section “Materials and Methods” from a known number of C. coli, the sensitivity of qPCR and RT-qPCR were tested. The sensitivity of the DNA-based qPCR assay was as low as 100 CFU/ml, whereas the sensitivity of the RT-qPCR assay was 1000 CFU/ml, respectively.
Figure 1.
(A) Melting temperature curve of ceuE qPCR products: (+) control: positive control; arrows represent ceuE melting curves of C. coli strains (4 different strains) and negative signals of non-C. coli strains (14 different strains), un-spiked samples, and water. (B) Agarose gel electrophoresis of qPCR products: Lane 1–4 (four different C. coli strains) with 196-bp band, lane M, 100-bp DNA marker; lane 5–18, 14 non-C. coli strains; lane 19, water; lane 20, un-spiked manure sample.
Standard curve for absolute quantification of qPCR assays
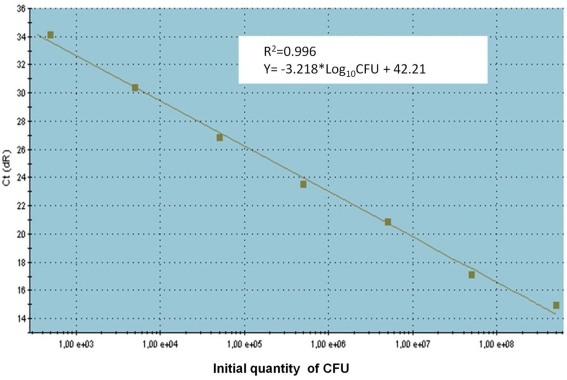
To determine absolute quantification of qPCR, nucleic acid standard was generated from genomic DNA of C. coli. The Ct-values were plotted as a function of the cell concentration and the plot showed the expected linear relationship between the log10 of Campylobacter CFU per milliliter (CFU/ml) and Ct-values (Figure 2). The standard curve slope was −3.218, which corresponded to ~100% efficiency for the PCR assay, using the formula E (efficiency) = (10−1/slope) − 1 and the calibration curve is linear with a correlation coefficient (R2) = 0.996.
Figure 2.
The standard curve for absolute quantification of C. coli in swine manure. Standard curves produced from 10-fold serial dilutions ranging from 1 × 102 to 1 × 108 CFU/ml swine manure sample of C. coli (CCUG 11283), showing the relationship between Ct-values and CFU/ml for qPCR assays. Ct, cycle threshold.
Determination of survival of C. coli in swine manure by bacterial culture and RT-qPCR methods
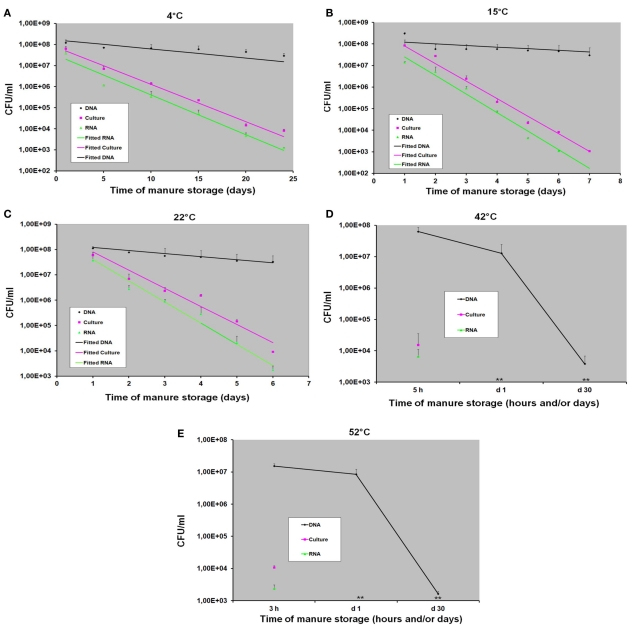
Figure 3 shows the levels of C. coli in manure samples incubated at five different temperatures: 4, 15, 22, 42, and 52°C. A decrease level of C. coli in all swine manure samples was observed throughout the experiment at all incubation temperatures by both bacterial culture and RT-qPCR methods. At 4°C, the viable C. coli cells were detected up to day 24 of storage by both methods (Figure 3A). Using bacterial culture and RT-qPCR methods, approximately 5 × 107 and 6.0 × 103 CFU/ml were obtained at day 1 and day 24, respectively (Figure 3A). At 15°C, the viable C. coli cells in manure samples were still detectable up to day 7 (approximately 1.2 × 103 CFU/ml) by bacterial culture method but could only be detected by RT-qPCR until day 6 (~1 × 103 CFU/ml; Figure 3B). At 22°C, the viable C. coli cells were detected up to day 6 with approximately 6.2 × 103 and 2 × 103 CFU/ml obtained by bacterial culture method and RT-qPCR method, respectively (Figure 3C). As shown in Figures 3D,E), a rapid decrease of the counts of viable C. coli cells was observed at 42°C (approximately 1.5 × 104 CFU/ml) and 52°C (approximately 1 × 104 CFU/ml) using the bacterial culture method after 5 and 3 h of incubation, respectively. At these high temperatures, viable C. coli cells were not detected by both methods after 24 h. It should note that all samples were incubated until day 30.
Figure 3.
The detection and quantification of C. coli in swine manure samples incubated at various temperatures (4, 15, 22, 42, and 52°C) by bacterial culture (counting), DNA-based qPCR (DNA), and RT-qPCR (RNA) methods with (A) at 4°C, (B) at 15°C, (C) at 22°C, (D) at 42°C, and (E) at 52°C. Data are means and SE of at least three independent experiments; (**): not detected.
Persistence of C. coli DNA in swine manure
As shown in Figures 3A–C, a slight decrease level of C. coli DNA was obtained using DNA-based qPCR method at 4, 15, and 22°C. At 4°C, approximately 1.2 × 108 and 2.8 × 107 CFU/ml were obtained at day 1 and day 24, respectively. At 15 and 22°C, we observed the similar amounts of C. coli DNA ranging from ~1 × 108 to 2.7 × 107 CFU/ml at day 1 and day 7, respectively (Figures 3B,C). Although none of viable C. coli cells was observed by either bacterial culture or RT-qPCR method at day 30 of storage, high levels (~2 × 107 CFU/ml) of C. coli DNA were still observed by DNA-based qPCR method in all samples at these incubation temperatures (4, 15, and 22°C; data not shown). At higher temperatures (42 and 52°C), although a slight decrease level of C. coli DNA was obtained after 24 h, it was still persistent until day 30 with approximately 1.5 × 103 CFU/ml (Figures 3D,E).
Discussion
The introduction of new molecular methods has become an especially important advance in reducing the time required for the detection of Campylobacter spp. and detecting viable bacteria in environmental samples through their DNA (Rudi et al., 2004; Ridley et al., 2008). The precise correlation of cell viability and the detected level of DNA have been shown to be poor, since bacterial DNA persists in dead cells for significant periods of time (Masters et al., 1994; Young et al., 2007). It has been demonstrated that bacterial DNA persisted in a PCR-detectable form in culture–negative environmental (Deere et al., 1996), and clinical samples (Hellyer et al., 1999). In contrast, the half-life of most bacterial mRNA has been reported to range from 0.5 to 50 min (Takayama and Kjelleberg, 2000). In addition, it has been shown that the use of bacterial mRNA for RT-qPCR could provide a more closely correlated indication of the cell viability status than DNA-based methods (Keer and Birch, 2003).
In the present study, we use mRNA as a maker for cell viability, and ceuE gene, a putative virulence gene of C. coli was selected as a biomarker for viable cells using RT-qPCR method. The ceuE gene product – a lipoprotein, plays an important role as a component of a protein-binding-dependent transport system for the siderophore enterochelin of C. coli (Richardson and Park, 1995). Our data indicated that the viable cells counts of C. coli in swine manure at all incubation temperatures determined by RT-qPCR and by culture method were almost equivalent (Figure 3). The results are in a good agreement with a previous study reported by Matsuda et al. (2006) who used RT-qPCR to enumerate bacteria in human feces and peripheral blood. Moreover, the positive signals were observed by RT-qPCR as long as viable C. coli cells were counted by bacterial culture method. In contrast, our results showed that the levels of C. coli DNA in manure obtained by DNA-based qPCR method were significantly (p < 0.001) higher than those obtained by either bacterial culture or RT-qPCR method in all manure samples at all incubation temperatures tested. Although no viable C. coli cells were detected by either bacterial culture or RT-qPCR in any manure samples stored at day 30, the significant levels of C. coli DNA were still detected by DNA-based qPCR showing that this method gave false-positive resulting from DNA from dead C. coli cells. Similar results have been found in several previous studies and the explanation for this phenomenon is the use of qPCR to detect the DNA as target could also detect the DNA from dead or non-viable cells (Lund et al., 2004; Rudi et al., 2004; Wolffs et al., 2005). It was reported that DNA from dead bacterial cells could persist for up to 3 weeks after the cell death (Josephson et al., 1993) and that persistence could lead to an overestimation of the number of viable cells and false-positive results (Wolffs et al., 2005). RT-qPCR is therefore superior to DNA-based qPCR for determining the concentration of viable bacteria.
Recently, it has been reported that propidium monoazide PCR (PMA-PCR) and ethidium monoazide PCR (EMA-PCR) could be used to detect and to quantify viable Campylobacter in complex samples (Rudi et al., 2005; Inglis et al., 2010; Josefsen et al., 2010). However, the advantage of our method presented here is that by detecting the mRNA level of a putative virulence gene, it is not only possible to detect and quantify viable C. coli but also to study the potential pathogenicity of this bacterium during the storage of manure.
Temperature has been shown to be a major factor determining pathogen inactivation during the storing and composting of animal manures (Hutchison et al., 2005; Nicholson et al., 2005; Larney and Hao, 2007). However, little is known about quantitative data on microbial inactivation rates and the influence of temperature in these materials, if not controversial (Inglis et al., 2010). In this study, the influence of temperature on the survival and fate of C. coli in swine manure stored at various temperatures was investigated. Using bacterial culture and RT-qPCR methods, a great decline of viable C. coli cells was observed at high temperatures (15, 22, 42, and 52°C). Our findings are in very good agreement with data from (Hänel and Atanassova, 2007) who showed that the number of Campylobacter on turkey meat samples incubated at 25°C was severely decreased in comparison to the same samples incubated at 4°C. Our data also showed that C. coli could survive up to 24 days in the samples incubated at 4°C in aerobic conditions. In contrast, at higher temperatures (at 42 or 52°C), no viable C. coli cells were detected after 24 h using either bacterial culture or RT-qPCR method. These findings are in agreement with data from previous study reported by Garénaux et al. (2009) who revealed that a cross protection between the cold shock response and oxidative stress response might explain the increased resistance of bacteria at low temperature. In addition, it has been shown that superoxide dismutase, as well as other oxidized stress related proteins were over-expressed at 4°C (Stintzi, 2003). Several studies have suggested that the enhanced survival of Campylobacter in various biological milieus is due to cold stress (Buswell et al., 1998; Chan et al., 2001; Moen et al., 2005). Furthermore, a number of genes involved in energy metabolism have been reported to be up-regulated at 5°C in comparison to at 25°C (Moen et al., 2005). Few data are available on survival of Campylobacter spp. under oxidative stress conditions in animal manures, especially swine manure. In this study, the swine manure samples were collected from open slurry tanks at the pig farm and the conditions for testing resembled aerobic conditions at the farm. From the data of our study, it seems that the survival of C. coli in swine manure under aerobic conditions depends on temperature. This is of particular importance because at low temperature (4°C) used allows bacterial survival longer and at higher rates (24 days), while at higher temperatures (42 and 52°C), survival of C. coli is severely affected (few hours).
Outbreaks of food-borne illness caused by food-borne pathogens associated with contaminated fruit and vegetables have recently reported and received worldwide attention (Pakalniskiene et al., 2009; Gajraj et al., 2011; Gardner et al., 2011). Vegetables can become contaminated with pathogenic organisms while growing or during harvesting and the most likely source is the application of manure or compost as fertilizer to fields where crops are grown and the fecal contamination of irrigation water (Berger et al., 2010; Oliveira et al., 2010). In addition, the storage of manure plays an important role in survival of pathogens during transmission (Kearney et al., 1993). The results of our study suggest that swine manure before application on the agricultural soil should be treated properly such as increasing the temperature up to 42°C or even more than 52°C for few hours since low temperatures allow Campylobacters survive a longer time (at least 24 days at 4°C).
In summary, this study compared, for the first time, the survival of C. coli in swine manure at various temperatures is investigated using bacterial culture method and molecular methods. The data suggest that C. coli in swine manure might be sensitive to aerobic conditions at high temperatures (15 and 22°C), especially at 42 and 52°C. Exposure to high temperatures has a stronger effect on survival of C. coli in swine manure than at low temperature (4°C). Furthermore, a good correlation was observed throughout the experiments between the number of viable C. coli cells obtained by RT-qPCR and those obtained by bacterial culture method. In contrast, greater differences between DNA C. coli levels obtained by DNA-based qPCR and CFU levels obtained by either bacterial culture or RT-qPCR method. Our findings draw an attention for the need of determining the level of contaminated pathogens at various temperatures in whole-slurry or manure before applying to the agricultural soil.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the Pathos Project funded by the Strategic Research Council of Denmark (ENV 2104-07-0015). We thank Jonas Høgberg for skilled technical assistance.
References
- Andreoletti O., Budka H., Buncic S., Collins J. D., Griffin J., Hald T., Havelaar A., Hope J., Klein G., Mclauchlin J., Müller-Graf C., Nguyen-The C., Noerrung B., Peixe L., Maradona M. P., Ricci A., Sofos J., Threlfall J., Vågsholm I., Vanopdenbosch E. (2011). Scientific opinion on Campylobacter in broiler meat production: control options and performance objectives and/or targets at different stages of the food chain. EFSA J. 9, 141 [Google Scholar]
- Anonymous (2006). Microbiology of Food and Animal Feeding Stuffs-Horizontal Method for Detection and Enumeration of Campylobacter spp. BS EN ISO 10272-1:2006. London: British Standards Institution [Google Scholar]
- Bang D. D., Nielsen E. M., Scheutz F., Pedersen K., Handberg K., Madsen M. (2003). PCR detection of seven virulence and toxin genes of Campylobacter jejuni and Campylobacter coli isolates from Danish pigs and cattle and cytolethal distending toxin production of the isolates. J. Appl. Microbiol. 94, 1003–1014 10.1046/j.1365-2672.2003.01926.x [DOI] [PubMed] [Google Scholar]
- Berger C. N., Sodha S. V., Shaw R. K., Griffin P. M., Pink D., Hand P., Frankel G. (2010). Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environ. Microbiol. 12, 2385–2397 10.1111/j.1462-2920.2010.02297.x [DOI] [PubMed] [Google Scholar]
- Bicudo J. R., Goyal S. M. (2003). Pathogens and manure management systems: a review. Environ. Technol. 24, 115–130 10.1080/09593330309385542 [DOI] [PubMed] [Google Scholar]
- Boes J., Nersting L., Nielsen E. M., Kranker S., En Oslash E. C., Wachmann H. C., Baggesen D. L. (2005). Prevalence and diversity of Campylobacter jejuni in pig herds on farms with and without cattle or poultry. J. Food Prot. 68, 722–727 [DOI] [PubMed] [Google Scholar]
- Buswell C. M., Herlihy Y. M., Lawrence L. M., Mcguiggan J. T. M., Marsh P. D., Keevil C. W., Leach S. A. (1998). Extended survival and persistence of Campylobacter spp. in water and aquatic biofilms and their detection by immunofluorescent-antibody and -rRNA staining. Appl. Environ. Microbiol. 64, 733–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K. F., Le Tran H., Kanenaka R. Y., Kathariou S. (2001). Survival of clinical and poultry-derived isolates of Campylobacter jejuni at a low temperature (4°C). Appl. Environ. Microbiol. 67, 4186–4191 10.1128/AEM.67.9.4186-4191.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. R. (1983). Kinetic analysis of anaerobic digestion of pig manure and its design implications. Agric. Wastes 8, 65–81 10.1016/0141-4607(83)90105-1 [DOI] [Google Scholar]
- Chenna R., Sugawara H., Koike T., Lopez R., Gibson T. J., Higgins D. G., Thompson J. D. (2003). Multiple sequence alignment with the clustal series of programs. Nucleic Acids Res. 31, 3497–3500 10.1093/nar/gkg500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collette F., Whichard J., Nachamkin I. (2008). “Diagnosis and antimicrobial susceptibility of Campylobacter species,” in Campylobacter, 3rd Edn, eds Nachamkin I., Szymanski C. M., Blaser M. J. (Washington, DC: ASM Press; ), 227–243 [Google Scholar]
- Deere D., Porter J., Pickup R. W., Edwards C. (1996). Survival of cells and DNA of Aeromonas salmonicida released into aquatic microcosms. J. Appl. Microbiol. 81, 309–318 10.1111/j.1365-2672.1996.tb04333.x [DOI] [PubMed] [Google Scholar]
- Gajraj R., Pooransingh S., Hawker J. I., Olowokure B. (2011). Multiple outbreaks of Salmonella braenderup associated with consumption of iceberg lettuce. Int. J. Environ. Health Res. 1–6 10.1080/09603123.2011.613114 [DOI] [PubMed] [Google Scholar]
- Gardner T. J., Fitzgerald C., Xavier C., Klein R., Pruckler J., Stroika S., Mclaughlin J. B. (2011). Outbreak of campylobacteriosis associated with consumption of raw peas. Clin. Infect. Dis. 53, 26–32 10.1093/cid/cir687 [DOI] [PubMed] [Google Scholar]
- Garénaux A., Ritz M., Jugiau F., Rama F., Federighi M., De Jonge R. (2009). Role of oxidative stress in C. jejuni inactivation during freeze-thaw treatment. Curr. Microbiol. 58, 134–138 10.1007/s00284-008-9289-3 [DOI] [PubMed] [Google Scholar]
- Gebreyes W. A., Thakur S., Morrow W. E. M. (2005). Campylobacter coli: prevalence and antimicrobial resistance in antimicrobial-free (ABF) swine production systems. J. Antimicrob. Chemother. 56, 765–768 10.1093/jac/dki305 [DOI] [PubMed] [Google Scholar]
- Gonzalez I., Grant K., Richardson P., Park S., Collins M. (1997). Specific identification of the enteropathogens Campylobacter jejuni and Campylobacter coli by using a PCR test based on the ceuE gene encoding a putative virulence determinant. J. Clin. Microbiol. 35, 759–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Dague R. R. (1997). Laboratory studies on the temperature-phased anaerobic digestion of domestic primary sludge. Water Environ. Res. 69, 1139–1143 10.2175/106143097X125885 [DOI] [Google Scholar]
- Hänel C. M., Atanassova V. (2007). Impact of different storage factors on the survivability of Campylobacter jejuni in turkey meat. FEMS Immunol. Med. Microbiol. 49, 146–148 10.1111/j.1574-695X.2006.00179.x [DOI] [PubMed] [Google Scholar]
- Hellyer T. J., Desjardin L. E., Hehman G. L., Cave M. D., Eisenach K. D. (1999). Quantitative analysis of mRNA as a marker for viability of Mycobacterium tuberculosis. J. Clin. Microbiol. 37, 290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Berrang M. E., Liu T., Hofacre C. L., Sanchez S., Wang L., Maurer J. J. (2003). Rapid detection of Campylobacter coli, C. jejuni, and Salmonella enterica on poultry carcasses by using PCR-enzyme-linked immunosorbent assay. Appl. Environ. Microbiol. 69, 3492–3499 10.1128/AEM.69.6.3492-3499.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey T., O’Brien S., Madsen M. (2007). Campylobacters as zoonotic pathogens: a food production perspective. Int. J. Food Microbiol. 117, 237–257 [DOI] [PubMed] [Google Scholar]
- Hutchison M. L., Walters L. D., Moore A., Avery S. M. (2005). Declines of zoonotic agents in liquid livestock wastes stored in batches on-farm. J. Appl. Microbiol. 99, 58–65 10.1111/j.1365-2672.2005.02591.x [DOI] [PubMed] [Google Scholar]
- Inglis G. D., Mcallister T. A., Larney F. J., Topp E. (2010). Prolonged survival of Campylobacter species in bovine manure compost. Appl. Environ. Microbiol. 76, 1110–1119 10.1128/AEM.01902-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen A. N., Dalsgaard A., Baggesen D. L., Nielsen E. M. (2006). The occurrence and characterization of Campylobacter jejuni and C. coli in organic pigs and their outdoor environment. Vet. Microbiol. 116, 96–105 10.1016/j.vetmic.2006.03.006 [DOI] [PubMed] [Google Scholar]
- Josefsen M. H., Lofstrom C., Hansen T. B., Christensen L. S., Olsen J. E., Hoorfar J. (2010). Rapid quantification of viable Campylobacter bacteria on chicken carcasses, using real-time PCR and propidium monoazide treatment, as a tool for quantitative risk assessment. Appl. Environ. Microbiol. 76, 5097–5104 10.1128/AEM.00411-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephson K. L., Gerba C. P., Pepper I. L. (1993). Polymerase chain reaction detection of nonviable bacterial pathogens. Appl. Environ. Microbiol. 59, 3513–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney T. E., Larkin M. J., Levett P. N. (1993). The effect of slurry storage and anaerobic digestion on survival of pathogenic bacteria. J. Appl. Microbiol. 74, 86–93 10.1111/j.1365-2672.1993.tb03000.x [DOI] [PubMed] [Google Scholar]
- Keer J. T., Birch L. (2003). Molecular methods for the assessment of bacterial viability. J. Microbiol. Methods 53, 175–183 10.1016/S0167-7012(03)00025-3 [DOI] [PubMed] [Google Scholar]
- Klein M., Brown L., Ashbolt N. J., Stuetz R. M., Roser D. J. (2011). Inactivation of indicators and pathogens in cattle feedlot manures and compost as determined by molecular and culture assays. FEMS Microbiol. Ecol. 77, 200–210 10.1111/j.1574-6941.2011.01098.x [DOI] [PubMed] [Google Scholar]
- Larney F. J., Hao X. (2007). A review of composting as a management alternative for beef cattle feedlot manure in southern Alberta, Canada. Bioresour. Technol. 98, 3221–3227 10.1016/j.biortech.2006.07.005 [DOI] [PubMed] [Google Scholar]
- Lund M., Nordentoft S., Pedersen K., Madsen M. (2004). Detection of Campylobacter spp. in chicken fecal samples by real-time PCR. J. Clin. Microbiol. 42, 5125–5132 10.1128/JCM.42.11.5125-5132.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund M., Wedderkopp A., Wainø M., Nordentoft S., Bang D. D., Pedersen K., Madsen M. (2003). Evaluation of PCR for detection of Campylobacter in a national broiler surveillance programme in Denmark. J. Appl. Microbiol. 94, 929–935 10.1046/j.1365-2672.2003.01934.x [DOI] [PubMed] [Google Scholar]
- Marti R., Dabert P., Pourcher A.-M. (2009). Pig manure contamination marker selection based on the influence of biological treatment on the dominant fecal microbial groups. Appl. Environ. Microbiol. 75, 4967–4974 10.1128/AEM.02791-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters C. I., Shallcross J. A., Mackey B. M. (1994). Effect of stress treatments on the detection of Listeria monocytogenes and enterotoxigenic Escherichia coli by the polymerase chain reaction. J. Appl. Microbiol. 77, 73–79 10.1111/j.1365-2672.1994.tb03047.x [DOI] [PubMed] [Google Scholar]
- Matsuda K., Tsuji H., Asahara T., Kado Y., Nomoto K. (2006). Sensitive quantitative detection of commensal bacteria by rRNA-targeted reverse transcription (RT)-PCR. Appl. Environ. Microbiol. 73, 32–39 10.1128/AEM.01224-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer J. J. (2006). “The mythology of PCR: a warning to the wise,” in PCR Methods in Foods, ed. Maurer J. (New York, NY: Springer; ), 27–40 [Google Scholar]
- Mawdsley J. L., Bardgett R. D., Merry R. J., Pain B. F., Theodorou M. K. (1995). Pathogens in livestock waste, their potential for movement through soil and environmental pollution. Appl. Soil Ecol. 2, 1–15 10.1016/0929-1393(94)00039-A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moen B., Oust A., Langsrud O., Dorrell N., Marsden G. L., Hinds J., Kohler A., Wren B. W., Rudi K. (2005). Explorative multifactor approach for investigating global survival mechanisms of Campylobacter jejuni under environmental conditions. Appl. Environ. Microbiol. 71, 2086–2094 10.1128/AEM.71.4.2086-2094.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak R., Stewart T. M., Nawaz M. S. (2005). PCR identification of Campylobacter coli and Campylobacter jejuni by partial sequencing of virulence genes. Mol. Cell. Probes 19, 187–193 10.1016/j.mcp.2004.11.005 [DOI] [PubMed] [Google Scholar]
- Nicholson F. A., Groves S. J., Chambers B. J. (2005). Pathogen survival during livestock manure storage and following land application. Bioresour. Technol. 96, 135–143 10.1016/j.biortech.2004.02.030 [DOI] [PubMed] [Google Scholar]
- Nielsen E. M., Engberg J., Madsen M. (1997). Distribution of serotypes of Campylobacter jejuni and C. coli from Danish patients, poultry, cattle and swine. FEMS Immunol. Med. Microbiol. 19, 47–56 10.1016/S0928-8244(97)00049-7 [DOI] [PubMed] [Google Scholar]
- Oliveira M., Usall J., Viñas I., Anguera M., Gatius F., Abadias M. (2010). Microbiological quality of fresh lettuce from organic and conventional production. Food Microbiol. 27, 679–684 10.1016/j.fm.2010.03.008 [DOI] [PubMed] [Google Scholar]
- Oporto B., Esteban J. I., Aduriz G., Juste R. A., Hurtado A. (2007). Prevalence and strain diversity of thermophilic Campylobacters in cattle, sheep and swine farms. J. Appl. Microbiol. 103, 977–984 10.1111/j.1365-2672.2007.03328.x [DOI] [PubMed] [Google Scholar]
- Oporto B., Hurtado A. (2011). Emerging thermotolerant Campylobacter species in healthy ruminants and swine. Foodborne Pathog. Dis. 8, 807–813 10.1089/fpd.2010.0803 [DOI] [PubMed] [Google Scholar]
- Pakalniskiene J., Falkenhorst G., Lisby M., Madsen S. B., Olsen K. E. P., Nielsen E. M., Mygh A., Boel J., Mølbak K. (2009). A foodborne outbreak of enterotoxigenic E. coli and Salmonella Anatum infection after a high-school dinner in Denmark, November 2006. Epidemiol. Infect. 137, 396–401 10.1017/S0950268808000484 [DOI] [PubMed] [Google Scholar]
- Pérez-Boto D., García-Peña F. J., Abad-Moreno J. C., Hurtado-Pizarro M. D., Pérez-Cobo I., Aurora Echeita M. (2010). Drinking water as the source of Campylobacter coli infection in grandparent heavy breeders. Avian Pathol. 39, 483–487 10.1080/03079457.2010.518138 [DOI] [PubMed] [Google Scholar]
- Richardson P. T., Park S. F. (1995). Enterochelin acquisition in Campylobacter coli: characterization of components of a binding-protein-dependent transport system. Microbiology 141, 3181–3191 10.1099/13500872-141-12-3181 [DOI] [PubMed] [Google Scholar]
- Ridley A. M., Allen V. M., Sharma M., Harris J. A., Newell D. G. (2008). Real-time PCR approach for detection of environmental sources of Campylobacter strains colonizing broiler flocks. Appl. Environ. Microbiol. 74, 2492–2504 10.1128/AEM.01242-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudi K., Hoidal H. K., Katla T., Johansen B. K., Nordal J., Jakobsen K. S. (2004). Direct real-time PCR quantification of Campylobacter jejuni in chicken fecal and cecal samples by integrated cell concentration and DNA purification. Appl. Environ. Microbiol. 70, 790–797 10.1128/AEM.70.8.5010-5018.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudi K., Moen B., Dromtorp S. M., Holck A. L. (2005). Use of ethidium monoazide and PCR in combination for quantification of viable and dead cells in complex samples. Appl. Environ. Microbiol. 71, 1018–1024 10.1128/AEM.71.2.1018-1024.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenov A. V., Van Overbeek L., Van Bruggen A. H. C. (2009). Percolation and survival of Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium in soil amended with contaminated dairy manure or slurry. Appl. Environ. Microbiol. 75, 3206–3215 10.1128/AEM.01791-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard S. K., Dallas J. F., Wilson D. J., Strachan N. J. C., Mccarthy N. D., Jolley K. A., Colles F. M., Rotariu O., Ogden I. D., Forbes K. J., Maiden M. C. J. (2010). Evolution of an agriculture-associated disease causing Campylobacter coli clade: evidence from national surveillance data in Scotland. PLoS ONE 5, e15708. 10.1371/journal.pone.0015708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A. (2003). Gene expression profile of Campylobacter jejuni in response to growth temperature variation. J. Bacteriol. 185, 2009–2016 10.1128/JB.185.6.2009-2016.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K., Kjelleberg S. (2000). The role of RNA stability during bacterial stress responses and starvation. Environ. Microbiol. 2, 355–365 10.1046/j.1462-2920.2000.00119.x [DOI] [PubMed] [Google Scholar]
- Verhoeff-Bakkenes L., Jansen H. A. P. M., In ‘T Veld P. H., Beumer R. R., Zwietering M. H., Van Leusden F. M. (2011). Consumption of raw vegetables and fruits: a risk factor for Campylobacter infections. Int. J. Food Microbiol. 144, 406–412 10.1016/j.ijfoodmicro.2010.10.027 [DOI] [PubMed] [Google Scholar]
- Wagner A. O., Gstraunthaler G., Illmer P. (2008). Survival of bacterial pathogens during the thermophilic anaerobic digestion of biowaste: laboratory experiments and in situ validation. Anaerobe 14, 181–183 10.1016/j.anaerobe.2008.03.004 [DOI] [PubMed] [Google Scholar]
- Wolffs P., Norling B., Rådström P. (2005). Risk assessment of false-positive quantitative real-time PCR results in food, due to detection of DNA originating from dead cells. J. Microbiol. Methods 60, 315–323 10.1016/j.mimet.2004.10.003 [DOI] [PubMed] [Google Scholar]
- Young G., Turner S., Davies J. K., Sundqvist G., Figdor D. (2007). Bacterial DNA persists for extended periods after cell death. J. Endod. 33, 1417–1420 10.1016/j.joen.2007.09.002 [DOI] [PubMed] [Google Scholar]