Transient and moderate affinity protein-protein interactions (PPIs) play a critical role in the regulation of essential cellular processes including protein folding, ubiquitylation, and transcription. A number of disease states are believed to be the result of aberrations within these protein networks; therefore, a longstanding therapeutic goal has been to design small molecules that can tunably modulate the constituent interactions (1–8). However, the discovery of small molecule modulators has been hindered by lack of structural and mechanistic information, in part due to the limitations of the approaches currently available for studying transient PPIs in their native environments. Techniques such as co-crystallization and co-purification in vitro and two-hybrid studies in vivo are best suited for probing stably associated proteins, but are less ideal for studying proteins that engage in modest-affinity and/or transient multi-protein binding interactions (9–12). Here we demonstrate the in vivo covalent capture of such binding partners of the prototypical activator VP16, focusing on the chromatin-modifying coactivator complex Swi/Snf. Through these in vivo photocrosslinking experiments we find that one region of VP16 contacts the Snf2 ATPase and a second relies upon the Snf5 scaffolding component for Swi/Snf binding, suggesting a cooperative recruitment mechanism for this complex at individual promoters. A similar in vivo analysis of the mechanistically related activator Gal4 reveals Snf2 to be a shared target, suggesting that the ATPase may be a viable target for small molecule intervention in the expanding roster of disease states that exhibit mis-regulated Swi/Snf (13–15). The success of using a genetically encoded photo-activatable amino acid for characterizing activator-coactivator complexes in vivo indicates that this strategy can be implemented more broadly for the capture and discovery of transient protein-protein interactions in their native contexts.
Transcriptional activators are signal responsive regulatory proteins that assemble the transcriptional machinery at the promoter of a gene through dynamic binding interactions with a variety of coactivator complexes, including chromatin-modifying, helicase, and scaffolding complexes (19, 23). Activators are modular in architecture and are minimally composed of a DNA binding domain (DBD) that localizes the activator to its cognate DNA binding site and a transcriptional activation domain (TAD) that mediates the majority of contacts with transcriptional complexes. Although the interactions between activators and suppressor proteins can be high affinity and specific in nature, activator-coactivator interactions are mediated through lower affinity, transient contacts (Figure 1a) (16–21). In vivo co-localization studies have defined the complexes that are recruited by activators during transcription, but they have not readily provided information on the direct coactivator targets within these complexes (24–26). For example, the well-characterized amphipathic activator VP16 has been shown to recruit the Swi/Snf chromatin-remodeling complex early in transcription initiation, as evidenced by both in vivo and in vitro co-localization studies (27–32). In vitro assays have identified several subunits within this complex as possible targets of VP16 but in vivo interaction studies have not distinguished which of the components are the relevant binding partner(s) (17, 33, 34). Thus there is a clear need for in vivo methodologies that can capture transient activator-coactivator interactions in their native environment.
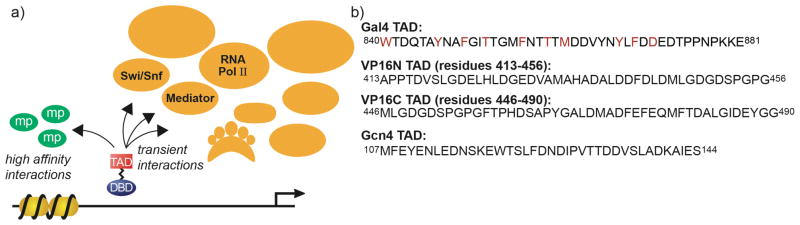
Figure 1.
(a) The transcriptional activation domain (TAD) of amphipathic activators can engage in high affinity interactions, such as those with masking proteins (mp), but the interactions between the TAD and coactivator complexes are more moderate in affinity and transient in nature (16–21). (b) Amphipathic activators share little sequence homology but do share binding targets, at least in vitro. The photocrosslinking amino acid, Bpa, has been incorporated within the Gal4 TAD (positions of incorporation in red) with little impact on the function and binding profile of that TAD (22).
In vivo crosslinking with the genetically incorporated, photo-labile amino acid p-benzoyl-L-phenylalanine (Bpa) has been demonstrated previously as a useful method for capturing direct, high affinity protein-protein interactions (22, 35–37). More specifically, Bpa placement within the TAD of the activator Gal4 did not impair function of the protein and photoactivation lead to covalent capture of its high affinity (low nanomolar KD) suppressor protein Gal80 (22, 38). However, while successful in the case of a very tight interaction, this method has not been employed in the case of moderate-affinity transient interactions such as those between activators and coactivators. In this study, we test the utility of in vivo Bpa crosslinking for capturing VP16-coactivator interactions and for resolving the identity of the Swi/Snf components targeted by this activator.
RESULTS AND DISCUSSION
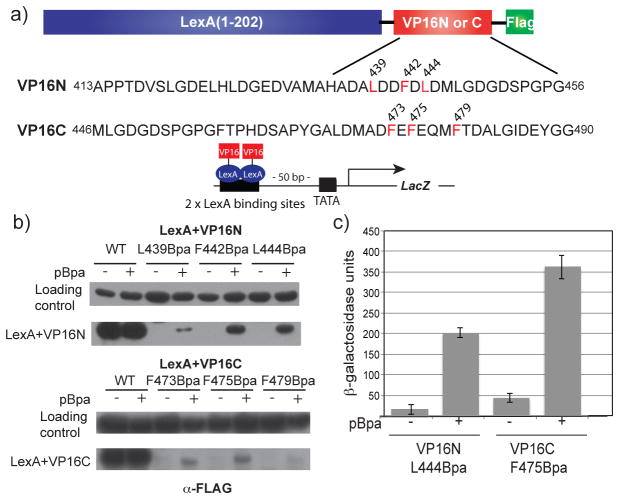
The VP16 TAD is comprised of two sub-domains that can function independently from one another, an amino terminal VP16N (residues 413–456) and a carboxy terminal VP16C (residues 446–490) (Figure 1b); therefore, we incorporated Bpa within regions of each subdomain shown to be involved in forming protein interactions. Further, because Bpa crosslinking is affected by its intrinsic reactivity and by positioning (22, 39), we examined six distinct mutants (VP16N: L439, F442, L444; VP16C: F473, F475, F479) (40, 41)(42). Bpa incorporation into VP16 is dependent on an enhanced nonsense suppression method that has been described previously (22, 43, 44). Each Bpa-containing construct was expressed in Saccharomyces cerevisiae as a fusion protein bearing the bacterial LexA DBD and a carboxy-terminal FLAG tag for detection (Figure 2a). All six Bpa mutants were assayed for Bpa incorporation and activation potential in a yeast strain with an integrated LacZ reporter gene under the control of a GAL1 promoter bearing two LexA binding sites (Figure 2b,c). The LexA+VP16C F479Bpa mutant was poorly expressed and was therefore removed from further testing. The remaining mutants displayed good incorporation and activity profiles (Figure 2d and Figure S7).
Figure 2.
Incorporation of Bpa within the VP16 TAD. (a) Plasmids encoding the DNA binding domain (DBD) of LexA fused to either the N- or C-terminal VP16 TAD as well as a FLAG tag were constructed. The LexA DBD was utilized to exclusively examine transcriptional activation at the 2 unique LexA binding sites upstream of the LacZ reporter in S. cerevisiae. Positions at which Bpa mutagenesis was carried out are within regions of the VP16N or VP16C subdomains known to participate in coactivator binding (sites of incorporation highlighted in red). The loading control is an approximately 71 kDa, FLAG-detected yeast protein whose expression level does not vary with activator identity.(22) (b) Yeast cells bearing plasmids encoding the various LexA+VP16 constructs and the Bpa specific tRNA/synthetase pair expressed by pSNRtRNA-pBpaRS were grown in the presence or absence of 1 mM Bpa and analyzed by Western blot (c) LexA+VP16N L444Bpa and LexA+VP16C F475Bpa were assessed for their ability to upregulate transcription of an integrated LacZ reporter gene in S. cerevisiae as measured by liquid β-galactosidase assays. Each activity is the average of values from at least three independent experiments with the indicated error (SDOM). See Supporting Information Figure S7 for activity assays of the remaining mutants.
A strong body of in vitro evidence exists to support the Mediator protein and coactivator Med15 as a target of VP16 and this model is supported by in vivo deletion and mutagenesis experiments (16, 39). The interaction between Med15 and activators such as VP16 is moderate in affinity, with dissociation constants ≥2 orders of magnitude weaker than the Gal4-Gal80 interaction (Figure 3a) (16, 22, 38, 39). Thus, this appeared to be an excellent test case of the effectiveness of the in vivo crosslinking strategy for capturing moderate affinity binding interactions. We first tested the ability of each VP16 subdomain to crosslink to the coactivator Med15 in vivo by co-expressing myc-tagged Med15(1–416) alongside either LexA+VP16N L444Bpa or LexA+VP16C F475Bpa and irradiating live yeast with 365 nm UV light. The covalent adducts were isolated from the yeast lysate and analyzed by Western blot (Figure 3b). A direct contact between each subdomain of VP16 and Med15 was observed and this was dependent upon irradiation, thus validating the utility of this method in capturing a moderate affinity, in vivo interaction of a transcriptional activator.
Figure 3.
In vivo photocrosslinking captures the moderate affinity interaction between LexA+VP16 and the Mediator protein, Med15. a) VP16 has been shown to interact transiently with the coactivator Med15, as determined by measured kinetic rate constants. Equilibrium binding measurements place the affinity of the TAD for Med15 in the moderate category, with DNA-bound homodimers exhibiting the highest affinity (0.1 μM) and isolated TADs in the low to mid-micromolar range (16, 39). (b) Live yeast cells bearing plasmids expressing LexA+VP16N L444Bpa or LexA+VP16C F475Bpa fusion proteins, in addition to a plasmid expressing myc-Med15(1–416) were irradiated with UV light (365 nm) for 30 minutes. Subsequently, cell lysates were immunoprecipitated with αLexA and analyzed by Western blot (αmyc). For both constructs, a crosslink with Med15 is observed. Supporting Figure S1 shows expression of myc-Med15(1–416) and Supporting Figure S2 shows the full Western blot, complete with molecular weight references.
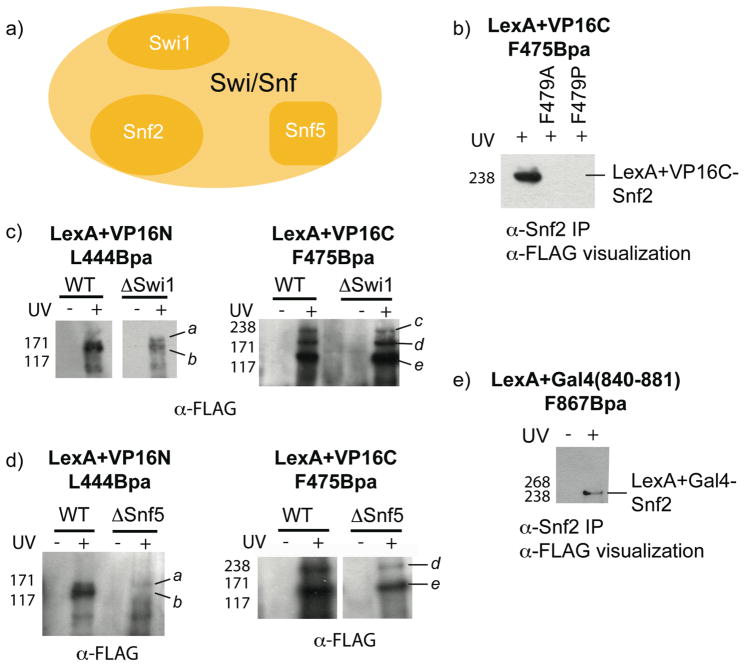
As outlined earlier, the Swi/Snf chromatin-modifying complex has also been proposed to be a direct binding partner of activators such as VP16 but there is conflicting evidence as to which subunit(s) serves as the activator-binding motif in vivo. VP16 enhances Swi/Snf recruitment to promoters such as the GAL1 promoter used in our studies and structural studies of Swi/Snf in complex with the nucleosome suggest that the catalytic subunit Snf2 is positioned close to the activator (27–32, 45–47). However, in vitro binding studies have shown several additional subunits can serve as binding partners (Swi1, Snf5) (17, 33, 34). We hypothesized that the in vivo crosslinking strategy could be used to test if the Swi/Snf complex is directly bound by VP16 in the cell and, if so, to identify Swi/Snf subunits that are directly bound by VP16 in the native complex environment. In the case of both the LexA+VP16N L444Bpa and the LexA+VP16C F475Bpa activators, irradiation of the live yeast cells expressing the activators followed by visualization of all crosslinked products via immunodetection of the FLAG tag revealed several bands in the 130–220 kDa molecular weight range, consistent with the size range expected for covalent complexes with the Snf2, Swi1 and Snf5 subunits (Figure 4 and Supporting Information Figure S3). To test this, immunoprecipitation of whole-cell extracts from irradiated cells with an antibody to Snf2 was carried out. In these experiments, no detectable LexA+VP16N-Snf2 product was observed for any of the three LexA+VP16N Bpa mutants, but, as seen in Figure 4b, the LexA+VP16C F475Bpa mutant crosslinks directly to Snf2. Consistent with this result, point mutations (F479A and F479P) known to decrease VP16 coactivator binding in vitro were introduced and, in line with these earlier biochemical experiments, abrogation of VP16 crosslinking to Snf2 in vivo is observed (48–50). These results are consistent with the recent structural model proposed by Dechassa et al that places Snf2 proximal to the transcriptional activator in the context of a Swi/Snf-nucleosome-activator complex (32). Further, the data suggests that it is the C-terminus of the VP16 TAD that is responsible for the bulk of the Snf2 recruitment activity (51).
Figure 4.
Analysis of VP16 crosslinking to the Swi/Snf coactivators, Snf2, Swi1 and Snf5. (a) The recruitment of the Swi/Snf chromatin remodeling complex by VP16 has been proposed to occur through interactions with the Snf2, Swi1 and Snf5 subunits although the direct binding partners in vivo have not been determined (17, 33, 34). (b) Live yeast cells expressing LexA+VP16C F475Bpa were irradiated with 365 nm light (30 minutes) and subsequently the cell lysates were immunoprecipitated with an antibody to Snf2 and resolved by Western blot (αFLAG), revealing a direct interaction between VP16C and endogenous Snf2. In line with previous biochemical experiments, when phenylalanine 479 in VP16C was mutated to either alanine or proline, crosslinking to Snf2 was abolished. (c,d) LexA+VP16C F475Bpa and LexA+VP16N L444Bpa were expressed in yeast strains lacking either Swi1 or Snf5 and the live yeast cells were irradiated with 365 nm light. Subsequently, cell lysates were immunoprecipitated (αLexA) and resolved by Western blot (αFLAG). In the individual blots for LexA+VP16N, the marks a and b denote crosslinked protein bands at the appropriate size for Swi1 and Snf5, respectively. In the individual blots for LexA+VP16C, the marks c, d, and e indicate bands at the appropriate size for Snf2, Swi1 and Snf5, respectively. (e) To test if Gal4 also contacts Snf2, crosslinking experiments were carried out with live yeast cells expressing LexA+Gal4 F867Bpa as in (b). The full Western blot of b) and e) can be found in Supplemental Figures S4 and S6, respectively.
In contrast to the Snf2 immunoprecipitation experiments, enrichment with either a Swi1 or Snf5 antibody did not result in any detectible crosslinked product (52). To probe these interactions further, we generated yeast strains lacking either Swi1 or Snf5 and carried out crosslinking experiments. No differences in crosslinked product formation between the WT strain and Swi1 delete strain were observed with either VP16-derived activator, suggesting that Swi1 is not a direct target of VP16 (Figure 4c). In contrast, deletion of Snf5 disrupts the normal binding pattern of LexA+VP16N L444Bpa, consistent with Snf5 interacting with VP16N (Figure 4d). However, upon deletion of Snf5, LexA+VP16C F475Bpa displays no change in binding pattern, suggesting that the VP16C TAD does not interact with Snf5. Together with the results of Figure 4b, these data support a model in which the subdomains of VP16 work cooperatively to recruit the Swi/Snf complex, with VP16C directly contacting Snf2 and VP16N depending on Snf5 during transcription initiation.
Snf2 is an ATPase that is essential for Swi/Snf function and is highly conserved among eukaryotes, making it a likely shared target among other transcriptional activators (53). In fact, in addition to VP16, the amphipathic activators Gal4 and Gcn4 have been shown to recruit Swi/Snf to a variety of promoters in vivo and in vitro binding studies have suggested that these activators contact a conserved set of targets within this complex (17, 31, 33, 34, 54–58). To test if Snf2 is a shared target of these activators, Gal4 and Gcn4 were modified to contain pBpa within regions of each TAD implicated in coactivator binding and then tested for their ability to crosslink to Snf2. As shown in Figure 4e, Gal4 directly interacts with Snf2, whereas Gcn4 does not for any position tested (Supporting Information Figure S5). These data suggest that Snf2 (Brg1/Brm in metazoans) could be a key target for small molecule probe development in order to characterize the role of the conserved Swi/Snf complexes that are associated with pathophysiological processes (13–15, 59). However, further studies will be needed to dissect if VP16 and Gal4, as well as other activators, interact with the same binding site within Snf2. In addition, experiments with crosslinking moieties of distinct chemical reactivities will enable further refinement of the binding models.
Taken together, these data demonstrate that genetically encoded photocrosslinkers are a viable and perhaps indispensible tool for capturing moderate affinity and transient protein-protein interactions in vivo. In this instance, employment of the in vivo photocrosslinking strategy revealed an interaction model for the cooperative recruitment of the chromatin-modifying complex Swi/Snf to gene promoters and, further, identified the Snf2 coactivator and ATPase to be a direct target of the prototypical transcriptional activators VP16 and Gal4. These data represent a significant step towards the development of a complete interaction map of the direct binding partners of transcriptional activators, long an elusive goal. Successful implementation of the in vivo crosslinking methodology for this class of moderate affinity, transient interactions sets the stage for the dissection of the complex interactions of the many other cellular mechanisms that function through similar binding networks.
Supplementary Material
Acknowledgments
We are grateful to the NIH (GM 2RO106553) for support of this work. We thank Dr. L. Lee for construction of the Gcn4 expression plasmids and the Matzger laboratory for use of a photocrosslinking apparatus.
Literature Cited
- 1.Lee LW, Mapp AK. Transcriptional switches: chemical approaches to gene regulation. J Biol Chem. 2010;285:11033–11038. doi: 10.1074/jbc.R109.075044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koehler AN. A complex task? Direct modulation of transcription factors with small molecules. Curr Opin Chem Biol. 2010;14:331–340. doi: 10.1016/j.cbpa.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arkin MR, Whitty A. The road less traveled: modulating signal transduction enzymes by inhibiting their protein-protein interactions. Curr Opin Chem Biol. 2009;13:284–290. doi: 10.1016/j.cbpa.2009.05.125. [DOI] [PubMed] [Google Scholar]
- 4.Evans CG, Chang L, Gestwicki JE. Heat shock protein 70 (hsp70) as an emerging drug target. J Med Chem. 2010;53:4585–4602. doi: 10.1021/jm100054f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ong DS, Kelly JW. Chemical and/or biological therapeutic strategies to ameliorate protein misfolding diseases. Curr Opin Cell Biol. 2011;23:231–238. doi: 10.1016/j.ceb.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berg T. Inhibition of transcription factors with small organic molecules. Curr Opin Chem Biol. 2008;12:464–471. doi: 10.1016/j.cbpa.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 7.Arndt HD. Small molecule modulators of transcription. Angew Chem Int Ed Engl. 2006;45:4552–4560. doi: 10.1002/anie.200600285. [DOI] [PubMed] [Google Scholar]
- 8.Blazer LL, Neubig RR. Small molecule protein-protein interaction inhibitors as CNS therapeutic agents: current progress and future hurdles. Neuropsychopharmacology. 2009;34:126–141. doi: 10.1038/npp.2008.151. [DOI] [PubMed] [Google Scholar]
- 9.Berggård T, Linse S, James P. Methods for the detection and analysis of protein-protein interactions. Proteomics. 2007;7:2833–2842. doi: 10.1002/pmic.200700131. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka Y, Bond MR, Kohler JJ. Photocrosslinkers illuminate interactions in living cells. Mol Biosyst. 2008;4:473–480. doi: 10.1039/b803218a. [DOI] [PubMed] [Google Scholar]
- 11.Melcher K. New chemical crosslinking methods for the identification of transient protein-protein interactions with multiprotein complexes. Curr Protein Pept Sci. 2004;5:287–296. doi: 10.2174/1389203043379701. [DOI] [PubMed] [Google Scholar]
- 12.Perkins JR, Diboun I, Dessailly BH, Lees JG, Orengo C. Transient protein-protein interactions: structural, functional, and network properties. Structure. 2010;18:1233–1243. doi: 10.1016/j.str.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 13.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 14.Hang CT, Yang J, Han P, Cheng HL, Shang C, Ashley E, Zhou B, Chang CP. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466:62–67. doi: 10.1038/nature09130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wands AM, Wang N, Lum JK, Hsieh J, Fierke CA, Mapp AK. Transient-state kinetic analysis of transcriptional activator.DNA complexes interacting with a key coactivator. J Biol Chem. 2011;286:16238–16245. doi: 10.1074/jbc.M110.207589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferreira ME, Hermann S, Prochasson P, Workman JL, Berndt KD, Wright APH. Mechanism of transcription factor recruitment by acidic activators. J Biol Chem. 2005;280:21779–21784. doi: 10.1074/jbc.M502627200. [DOI] [PubMed] [Google Scholar]
- 18.Fuxreiter M, Tompa P, Simon I, Uversky VN, Hansen JC, Asturias FJ. Malleable machines take shape in eukaryotic transcriptional regulation. Nat Chem Biol. 2008;4:728–737. doi: 10.1038/nchembio.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mapp AK, Ansari AZ. A TAD further: exogenous control of gene activation. ACS Chem Biol. 2007;2:62–75. doi: 10.1021/cb600463w. [DOI] [PubMed] [Google Scholar]
- 20.Herbig E, Warfield L, Fish L, Fishburn J, Knutson Ba, Moorefield B, Pacheco D, Hahn S. Mechanism of Mediator recruitment by tandem Gcn4 activation domains and three Gal11 activator-binding domains. Mol Cell Biol. 2010;30:2376–2390. doi: 10.1128/MCB.01046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melcher K. The strength of acidic activation domains correlates with their affinity for both transcriptional and non-transcriptional proteins. J Mol Biol. 2000;301:1097–1112. doi: 10.1006/jmbi.2000.4034. [DOI] [PubMed] [Google Scholar]
- 22.Majmudar CY, Lee LW, Lancia JK, Nwokoye A, Wang Q, Wands AM, Wang L, Mapp AK. Impact of nonnatural amino acid mutagenesis on the in vivo function and binding modes of a transcriptional activator. J Am Chem Soc. 2009;131:14240–14242. doi: 10.1021/ja904378z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ptashne M, Gann A. Genes & Signals. Cold Spring Harbor Laboratory; New York: 2001. [Google Scholar]
- 24.Bryant GO, Ptashne M. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol Cell. 2003;11:1301–1309. doi: 10.1016/s1097-2765(03)00144-8. [DOI] [PubMed] [Google Scholar]
- 25.Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 26.Hall DB, Struhl K. The VP16 activation domain interacts with multiple transcriptional components as determined by protein-protein cross-linking in vivo. J Biol Chem. 2002;277:46043–46050. doi: 10.1074/jbc.M208911200. [DOI] [PubMed] [Google Scholar]
- 27.Memedula S, Belmont AS. Sequential recruitment of HAT and SWI/SNF components to condensed chromatin by VP16. Curr Biol. 2003;13:241–246. doi: 10.1016/s0960-9822(03)00048-4. [DOI] [PubMed] [Google Scholar]
- 28.Yudkovsky N, Logie C, Hahn S, Peterson CL. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 1999;13:2369–2374. doi: 10.1101/gad.13.18.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herrera FJ, Triezenberg SJ. VP16-dependent association of chromatin-modifying coactivators and underrepresentation of histones at immediate-early gene promoters during herpes simplex virus infection. J Virol. 2004;78:9689–9696. doi: 10.1128/JVI.78.18.9689-9696.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hassan AH, Neely KE, Workman JL. Histone acetyltransferase complexes stabilize SWI/SNF binding to promoter nucleosomes. Cell. 2001;104:817–827. doi: 10.1016/s0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- 31.Neely KE, Hassan AH, Wallberg AE, Steger DJ, Cairns BR, Wright AP, Workman JL. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol Cell. 1999;4:649–655. doi: 10.1016/s1097-2765(00)80216-6. [DOI] [PubMed] [Google Scholar]
- 32.Dechassa ML, Zhang B, Horowitz-Scherer R, Persinger J, Woodcock CL, Peterson CL, Bartholomew B. Architecture of the SWI/SNF-nucleosome complex. Mol Cell Biol. 2008;28:6010–6021. doi: 10.1128/MCB.00693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prochasson P, Neely KE, Hassan AH, Li B, Workman JL. Targeting activity is required for SWI/SNF function in vivo and is accomplished through two partially redundant activator-interaction domains. Mol Cell. 2003;12:983–990. doi: 10.1016/s1097-2765(03)00366-6. [DOI] [PubMed] [Google Scholar]
- 34.Neely KE, Hassan AH, Brown CE, Howe L, Workman JL. Transcription activator interactions with multiple SWI/SNF subunits. Mol Cell Biol. 2002;22:1615–1625. doi: 10.1128/MCB.22.6.1615-1625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chin JW, Schultz PG. In vivo photocrosslinking with unnatural amino acid mutagenesis. Chembiochem. 2002;3:1135–1137. doi: 10.1002/1439-7633(20021104)3:11<1135::AID-CBIC1135>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 36.Mohibullah N, Hahn S. Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes Dev. 2008;22:2994–3006. doi: 10.1101/gad.1724408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hino N, Okazaki Y, Kobayashi T, Hayashi A. Protein photo-cross-linking in mammalian cells by site-specific incorporation of a photoreactive amino acid. Nat Methods. 2005;2:3–8. doi: 10.1038/nmeth739. [DOI] [PubMed] [Google Scholar]
- 38.Thoden JB, Ryan LA, Reece RJ, Holden HM. The interaction between an acidic transcriptional activator and its inhibitor. The molecular basis of Gal4p recognition by Gal80p. J Biol Chem. 2008;283:30266–30272. doi: 10.1074/jbc.M805200200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Majmudar CY, Wang B, Lum JK, Hakansson K, Mapp AK. A highresolution interaction map of three transcriptional activation domains with a key coactivator from photo-cross-linking and multiplexed mass spectrometry. Angew Chem Int Ed Engl. 2009;48:7021–7024. doi: 10.1002/anie.200902669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Triezenberg SJ, Kingsbury RC, McKnight SL. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 1988;2:718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]
- 41.Regier JL, Shen F, Triezenberg SJ. Pattern of aromatic and hydrophobic amino acids critical for one of two subdomains of the VP16 transcriptional activator. Proc Natl Acad Sci USA. 1993;90:883–887. doi: 10.1073/pnas.90.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Consistent with other reports, expression of the full length VP16 TAD proved to be toxic in yeast. Therefore, we chose to use individual subdomains of the VP16 TAD which did not result in cell death.
- 43.Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG. An expanded eukaryotic genetic code. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 44.Wang Q, Wang L. New methods enabling efficient incorporation of unnatural amino acids in yeast. J Am Chem Soc. 2008;130:6066–6067. doi: 10.1021/ja800894n. [DOI] [PubMed] [Google Scholar]
- 45.Côté J, Quinn J, Workman JL, Peterson CL. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 46.Lemieux K, Gaudreau L. Targeting of Swi/Snf to the yeast GAL1 UAS G requires the Mediator, TAF IIs, and RNA polymerase II. EMBO J. 2004;23:4040–4050. doi: 10.1038/sj.emboj.7600416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peterson CL, Workman JL. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 48.Jonker HR, Wechselberger RW, Boelens R, Folkers GE, Kaptein R. Structural properties of the promiscuous VP16 activation domain. Biochemistry. 2005;44:827–839. doi: 10.1021/bi0482912. [DOI] [PubMed] [Google Scholar]
- 49.Uesugi M, Nyanguile O, Lu H, Levine AJ, Verdine GL. Induced alpha helix in the VP16 activation domain upon binding to a human TAF. Science. 1997;277:1310–1313. doi: 10.1126/science.277.5330.1310. [DOI] [PubMed] [Google Scholar]
- 50.Yang F, DeBeaumont R, Zhou S, Naar AM. The activator-recruited cofactor/Mediator coactivator subunit ARC92 is a functionally important target of the VP16 transcriptional activator. Proc Natl Acad Sci U S A. 2004;101:2339–2344. doi: 10.1073/pnas.0308676100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Although this hypothesis is supported by the lack of Snf2 crosslinking with three different VP16N Bpa mutants and by biochemical binding data from other laboratories(34), an alternative hypothesis is that none of the VP16N Bpa mutants in our study are properly positioned for Bpa crosslinking with Snf2.
- 52.Using these antibodies for detection of endogenous Swi1 and Snf5, we have observed bands corresponding to the molecular weight of free Swi1 and Snf5 but no crosslinked complex was detected. It is possible that a covalent crosslink formed as a result of these experiments may be interfering with epitope recognition by the antibody, resulting in an inability to detect a crosslinked product. Moreover, one of the challenges associated with this effort is that the Swi/Snf complex is present at very low concentrations, about 100– 500 copies per cell. Due to the low expression levels of these coactivators in combination with the instability of the activator-coactivator cross-linked products, and, presumably, modest cross-linking yields, mass spectrometric identification of any of the activator cross-linked bands have not been successful. (Sinz A. Investigation of protein-protein interactions in living cells by chemical crosslinking and mass spectrometry. Anal Bioanal Chem. 2010;8:3433–3440. doi: 10.1007/s00216-009-3405-5.) (Soutourina J, Wydau S, Ambroise Y, Boschiero C, Werner M. Direct interaction of RNA polymerase II and mediator required for transcription in vivo. Science. 2011;331:1451–1454. doi: 10.1126/science.1200188.)
- 53.Laurent BC, Treich I, Carlson M. The yeast SNF2/SWI2 protein has DNAstimulated ATPase activity required for transcriptional activation. Genes Dev. 1993;7:583–591. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- 54.Ferreira ME, Prochasson P, Berndt KD, Workman JL, Wright APH. Activator-binding domains of the SWI/SNF chromatin remodeling complex characterized in vitro are required for its recruitment to promoters in vivo. FEBS J. 2009;276:2557–2565. doi: 10.1111/j.1742-4658.2009.06979.x. [DOI] [PubMed] [Google Scholar]
- 55.Schwabish MA, Struhl K. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol Cell Biol. 2007;27:6987–6995. doi: 10.1128/MCB.00717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bryant GO, Prabhu V, Floer M, Wang X, Spagna D, Schreiber D, Ptashne M. Activator control of nucleosome occupancy in activation and repression of transcription. PLoS Biol. 2008;6:2928–2939. doi: 10.1371/journal.pbio.0060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Govind CK, Yoon S, Qiu H, Govind S, Hinnebusch AG. Simultaneous recruitment of coactivators by Gcn4p stimulates multiple steps of transcription in vivo. Mol Cell Biol. 2005;25:5626–5638. doi: 10.1128/MCB.25.13.5626-5638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swanson MJ, Qiu H, Sumibcay L, Krueger A, Kim S, Natarajan K, Yoon S, Hinnebusch AG. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol Cell Biol. 2003;23:2800–2820. doi: 10.1128/MCB.23.8.2800-2820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.