Abstract
Glioblastoma is the most malignant type of brain tumor. Despite recent advances in therapeutic modalities, the prognosis of glioblastoma remains very poor. Recent studies have indicated that RelA/nuclear factor (NF)-κB is consistently activated in human glioblastoma. In this study, we searched for a new treatment modality for glioblastoma, by examining the effects of dehydroxymethylepoxyquinomicin (DHMEQ), a unique small molecule inhibitor of NF-κB. Addition of DHMEQ to cultured human glioblastoma cells inhibited the nuclear translocation of RelA. It also reduced the growth rate of human glioblastoma cells significantly in 6 cell lines and modestly in 3 among 10 cell lines examined. Then, we performed further analyses using 3 sensitive cell lines (U87, U251, and YKG-1). The growth retardation was accompanied by G2/M arrest in vitro. Increased apoptosis was observed in U87 and YKG-1, but not U251 cells after DHMEQ treatment. Then, we tested the efficacy of DHMEQ in chemoprevention through the use of a nude mouse model. Subcutaneous tumors formed by U87 or U251 cells were reduced by ∼40% in size by intraperitoneal administration of DHMEQ started immediately after implantation of the cells. DHMEQ treatment achieved statistically significant improvements in survival curves of mice intracranially implanted with U87 or U251 cells. Histological analysis revealed increased areas of necrosis, increased numbers of collapsed microvessels, decreased nuclear immunoreactivity of RelA, and decreased immunoreactivity of urokinase-type plasminogen activator in the DHMEQ-treated U87 tumor tissues. These results suggest that the targeting of NF-κB by DHMEQ may serve as a promising treatment modality in glioblastoma.
Keywords: DHMEQ; glioblastoma; NF-κB, RelA
Glioblastoma is the most malignant form of brain tumors and is characterized by rapid growth, extensive invasiveness, and enhanced angiogenesis.1 Even with the latest combined treatment modalities, the prognosis of patients with glioblastoma remains extremely poor. Therefore, new and innovative therapies for glioblastoma are critically required. Nuclear factor-κB (NF-κB) consists of a family of transcription factors, composed of 5 proteins: RelA (p65), RelB, c-Rel, NF-κB1 (p50), and NF-κB2.2 They form a variety of hetero- and homodimers. Among them, RelA is responsible for most of the transcriptional activity of NF-κB.2 There is accumulating evidence that NF-κB is involved in a wide range of biological phenomena observed in malignant tumors, including cell growth, resistance to apoptosis, angiogenesis, invasion, and metastasis.2,3 The effects of NF-κB on these malignant phenotypes are likely to be mediated by upregulated transcription of target genes that encode, for example, cell cycle-related protein, such as cyclin D1; anti-apoptosis proteins; growth factor receptors; angiogenesis factors, such as vascular endothelial growth factor; and proteases, such as urokinase-type plasminogen activator (uPA).2,3 Of note, activated RelA/NF-κB is consistently observed in gliomas with a tendency for positive correlation with malignant progression.4,5 In vitro studies using cultured glioblastoma cells suggest that RelA/NF-κB is involved in malignant properties of glioblastoma, such as enhanced growth, resistance to apoptosis, invasiveness, and generation of angiogenesis factors.5–8 Therefore, RelA/NF-κB may be a promising therapeutic molecular target in glioblastoma.
Dehydroxymethylepoxyquinomicin (DHMEQ) is a recently synthesized small molecule inhibitor of NF-κB.9,10 It directly binds to the canonical and noncanonical NF-κB components to inhibit their DNA binding.11 It also inhibits nuclear translocation of RelA.12 In the initial in vivo study of DHMEQ, it inhibited type-2 collagen-induced arthritis in mice, a model for rheumatoid arthritis in which activation of NF-κB by tumor necrosis factor α (TNFα) plays a pivotal role.10 It also inhibited renal inflammation and cancer cachexia.10 Subsequently, it has been shown that DHMEQ could suppress the growth of various types of tumor cells.10,13–23
In our search for a new treatment modality for glioblastoma, we examined the effects of DHMEQ on glioblastoma cells, because RelA/NF-κB signaling is known to be activated in glioblastoma cells.4–8 To our knowledge, our data demonstrated, for the first time, that DHMEQ could suppress the growth of glioblastoma cells and improved the survival of mice after intracranial transplantation of glioblastoma cells.
Materials and Methods
Cell Lines and Clinical Samples
The human glioblastoma cell lines U87, YKG-1, A172, T98G, U251, KS-1, YH13, U373, NYGM, and MGM-1 were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS). NYGM and MGM-1 were established in our laboratory.24,25 U251, YKG-1, T98G, A172, and KS-1 were obtained from the RIKEN cell bank (Tsukuba, Japan). YH-13 was from the Health Science Research Resource Bank (Osaka, Japan). U87 and U373 were from Dainippon Sumitomo Pharma (Osaka, Japan). The experimental protocol for clinical samples was approved by the Human Ethics Review Committees of Miyazaki University. Tissue samples were obtained from surgically resected low-grade gliomas, anaplastic gliomas, and glioblastomas. Normal brain mRNAs were obtained from nonneoplastic portions. Tumors were assessed according to the World Health Organization classification.26
Reagents and Antibodies
DHMEQ was synthesized as described elsewhere,9 dissolved in dimethyl sulfoxide (DMSO; 10 mg/mL), and stored at −20°C. Recombinant TNFα was purchased from Boehringer Mannheim Biochemica. Antibodies used were anti-NF-κB p65 (C22B4) rabbit monoclonal (Cell Signaling Technology), anti-lamin A/C rabbit polyclonal (Cell Signaling), anti-cleaved caspase 3 (Asp175) rabbit polyclonal (Cell Signaling), anti-CD31 rabbit polyclonal (AnaSpec), and anti-uPA (R&D Systems) polyclonal antibodies.
Reverse-Transcriptase Polymerase Chain Reaction
Reverse-transcriptase polymerase chain reaction (RT-PCR) was performed as described elsewhere,27 using the following primers: RelA, forward 5′-GGGATGGCTTCTATGAGGCTGA-3′ and reverse 5′-GGTCTGGATGCGCTGACTGA-3′; uPA, forward 5′-GCCATCCCGGACTATACAGA-3′ and reverse 5′-AGGCCATTCTCTTCCTTGGT-3′; and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), forward 5′-GTGAAGGTCGGAGTCAACG-3′ and reverse, 5′-GGTGAAGACGCCAGTGGACTC-3′.
Immunoblot Analysis
Cultured cells (60%–70% confluency) were incubated with or without DHMEQ (10 µg/mL) in growth medium for 2.5 h, followed by treatment with TNFα (20 ng/mL) for 30 min in serum-free DMEM, 0.1% bovine serum albumin (BSA). After washing in phosphate-buffered saline (PBS), nuclear proteins were extracted using CelLytec NuCLEAR extraction kit (Sigma). For immunoblot, each extracted protein (4.5 µg) was separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis under reducing conditions, transferred onto Immobilon membrane (Millipore), and processed as described elsewhere.27
Cell Proliferation and Cytotoxicity Assays
Cells were seeded into 96-well plates (103 cells/well) and cultured with various concentrations of DHMEQ, and the cell numbers were evaluated using TetraColor ONE assay (SEIKAGAKU) as described elsewhere.27 Cytotoxic activity of DHMEQ was evaluated by calculating the concentration required for 50% inhibition of cellular growth (IC50).
Cell Cycle Analysis and Cell Viability Assay
For cell cycle analysis, cells (50% confluency) were starved in serum-free condition for 24 h, followed by incubation in growth medium with or without DHMEQ. At the end of the treatment, the cells were trypsinized to obtain a single-cell suspension, centrifuged, and the pellet was resuspended in PBS, followed by fixation in ice-cold 70% ethanol. The fixed cells were centrifuged at ×450 g for 5 min, washed with PBS, resuspended in Guava Cell Cycle Reagent (Millipore), incubated for 30 min at room temperature in the dark, and analyzed by Guava EasyCyte flow cytometer (Millipore) according to the manufacturer's instructions. For cell viability assay, cells were seeded into 6-well plates (1 × 105 cells/well) and cultured without or with DHMEQ. The numbers of live cells, apoptotic cells, and dead cells were calculated using Guava ViaCount assay system (Millipore) and Guava EasyCyte flow cytometer with Cytosoft software (Millipore) according to the manufacturer's instructions.
Murine Xenograft Models
All animal procedures were performed in accordance with institutional guidelines, and the protocol was approved by the Animal Care Committee of University of Miyazaki. Six-week-old male athymic nude mice (BALB/cAJc1-nu) with a mean body weight of 20 g were obtained from CLEA Japan. In all experiments, the experimental group was injected intraperitoneally with 10 mg/kg DHMEQ 3 times per week started on the day of tumor injection, and the control group was administered vehicle DMSO solutions. For subcutaneous implantation, glioblastoma cells (5 × 106) were implanted subcutaneously in the back of mice. Tumor size was measured twice per week for 4 weeks. Tumor volume was estimated by the formula A × B2 × 0.5, where A is length and B is width. When mice were sacrificed, blood samples were collected by intracardiac puncture, and hematological biochemistry tests were performed in a clinical laboratory (SRL; Tokyo, Japan). For intracranial transplantation, U87 or U251 cells (1 × 105 or 1 × 106 cells/10-µL DMEM, respectively) were stereotactically transplanted into the forebrain of mice as described elsewhere.28 The mice were carefully observed every day for 38 days to calculate the Kaplan-Meier survival curves. In another experiment, the brain specimens were prepared after euthanasia of mice 29 days after the transplantation. The brain tissues were fixed in 4% formaldehyde in PBS and sectioned coronally at the point of cellular implantation, followed by embedding in paraffin.
Immunostaining
Formalin-fixed paraffin-embedded tissue sections (4 μm) were processed for antigen retrieval (autoclaving in 10 mM citrate buffer [pH, 6.0], for RelA and CD31, and in 1 mM EDTA [pH, 8.0], for cleaved caspase 3). After blocking in 3% BSA and 10% normal goat serum in PBS, the sections were immunostained as described elsewhere.29 For immunocytochemistry, cells were seeded into Lab-Tek chamber slide (Nalge Nunc International) and incubated with or without DHMEQ (10 µg/mL) for 2.5 h. Then, the cells were treated with TNFα (20 ng/mL) for 30 min, fixed with 4% paraformaldehyde in PBS for 15 min, and washed in PBS 3 times. The cells were blocked for 1 h with 5% normal goat serum in PBS with 0.003% Triton X-100 at room temperature, followed by incubation with anti-NF-κB p65 antibody overnight at 4°C. After washing with PBS, the cells were incubated for 1 h at room temperature with Alexa Fluor 488-conjugated goat anti-rabbit IgG (Invitrogen) in PBS, washed with PBS, counterstained with 4,'6-diamino-2-phenylindole (Sigma), and investigated with Axio Imager.A2 (Carl Zeiss MicroImaging).
Quantification of Microvessel Density, Apoptotic Cells, and Nuclear RelA-Positive Cells In Vivo
To count microvessel densities, apoptotic cells, nuclear RelA-positive cells, and uPA-positive cells, tissue sections were immunostained with anti-CD31, anti-cleaved caspase 3, anti-NF-κB p65 (RelA), and anti-uPA antibodies, respectively. The stained sections were examined at low power field (×40). Then, 5 areas with the most intense labeling were selected and photographed at ×200 (CD31, cleaved caspase 3, and uPA) or ×400 (RelA) magnification. Two independent investigators (M.K. and H.K. for microvessels and apoptotic cells; K.Y. and Hir.T. for uPA; M.K. and Hir.T. for RelA) counted the positive vessels per cells, and the mean number per field was calculated. For semi-quantification of uPA immunoreactivity, the following score was used: numbers of strongly positive cells × 2 + numbers of weakly positive cells/field.
Statistical Analysis
Data were analyzed using the Statview 4.0 program (Brainpower), and the results were expressed as the mean ± standard deviation. Comparison between 2 unpaired groups was done using 2-way repeated-measures analysis of variance or Mann-Whitney U test. Survival of mice was assessed by estimating the Kaplan-Meier survival curves, which were compared by log-rank test. Significance was set at P < .05.
Results
DHMEQ Suppressed Proliferation of Glioblastoma Cells In Vitro
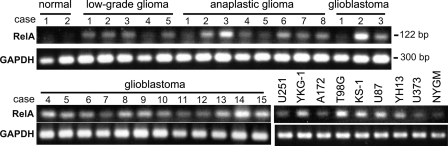
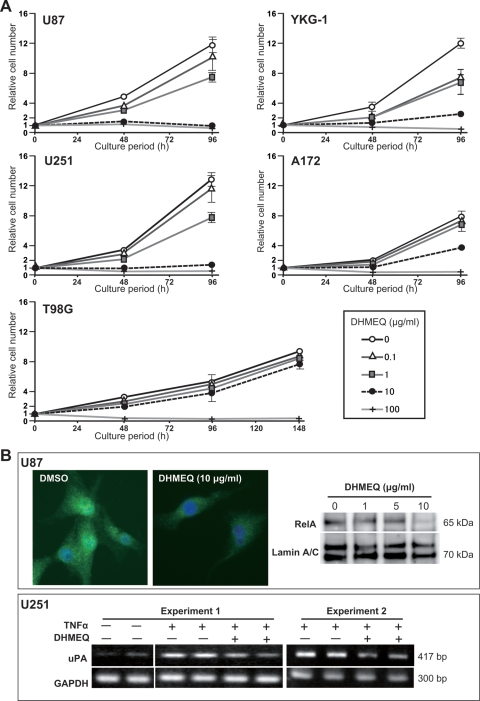
Enhanced RelA mRNA levels were consistently detected in high-grade gliomas, particularly in glioblastoma, and human glioblastoma cell lines (Fig. 1). We examined the effects of DHMEQ on the proliferation of 10 human glioblastoma cell lines. As shown in Fig. 2A, DHMEQ suppressed the growth of the glioblastoma cell lines. However, the extents of suppression varied among the cell lines (Table 1). Most (9 of 10) cell lines were sensitive to DHMEQ, in which significant suppressive effects were observed in 6 cell lines (YKG-1, U251, U87, MGM-1, NYGM, and U373). YKG-1 showed the highest sensitivity, with an IC50 value of 1.1 µg/mL followed, in descending order, by U251 (IC50, 1.4 µg/mL) and U87 (IC50, 1.8 µg/mL). T98G was resistant to the treatment (IC50, 26.7 µg/mL). As expected, the nuclear translocation of RelA was significantly suppressed by DHMEQ treatment (Fig. 2B). We also examined the effect of DHMEQ on TNFα-induced upregulation of an NF-κB target gene product, uPA. The DHMEQ treatment (6 h) suppressed TNFα-induced upregulation of uPA mRNA level in U251 (Fig. 2B). However, the basal level of uPA mRNA was not altered by DHMEQ. Similar effects were observed in YKG-1 (data not shown). On the other hand, U87 did not show TNFα-induced uPA upregulation, and the expression level of uPA in this cell line was not altered by DHMEQ (data not shown).
Fig. 1.
RT-PCR analysis for RelA expression in resected tissues of nontumor cerebrum (2 cases), low-grade gliomas (5 cases), anaplastic gliomas (8 cases), glioblastomas (15 cases), and 9 human glioblastoma cell lines. Twenty-six cycles of amplification. Corresponding GAPDH signals are also indicated.
Fig. 2.
Effects of DHMEQ on cellular growth and nuclear translocation of RelA in glioblastoma cell lines. (A) Effects of varying concentrations of DHMEQ (0–100 µg/mL) on the growth of U87, YKG-1, U251, A172, and T98G. Cells were cultured in growth medium (DMEM, 10% FBS) for indicated periods. Error bars, SD (n = 3). (B) Effects of DHMEQ (10 µg/mL) on TNFα-induced (20 ng/mL) nuclear translocation of RelA in U87 (upper panel) and uPA expression in U251 cells (lower panel). For RelA translocation, immunofluorescence study (green, RelA; blue, nuclei) and immunoblot analysis of nuclear extracts of U87 cells are shown. Nuclear translocation of RelA after 30 min of TNFα stimulation was disturbed by DHMEQ pretreatment (2.5 h). RT-PCR analysis for the expression of uPA indicated that DHMEQ pretreatment (6 h) suppressed the TNFα-induced uPA upregulation.
Table 1.
Inhibitory effect of DHMEQ on in vitro growth of human glioblastoma cell lines
| Cell lines | IC50 (μg/mL) |
|---|---|
| YKG-1 | 1.1 |
| U251 | 1.4 |
| U87 | 1.8 |
| MGM-1 | 3.2 |
| U373 | 3.4 |
| NYGM | 3.5 |
| YH13 | 8.5 |
| A172 | 11.3 |
| KS-1 | 16.7 |
| T98G | 26.7 |
DHMEQ Induced G2/M Cell Cycle Arrest and Apoptosis in Glioblastoma Cells
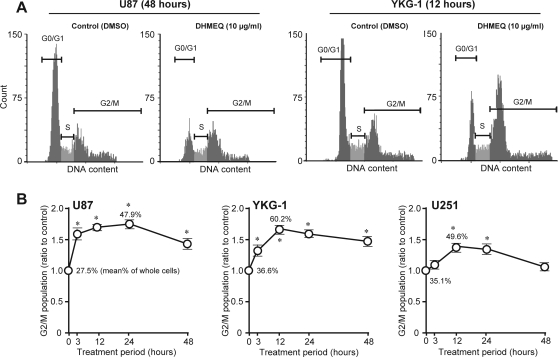
Previous studies have indicated that DHMEQ induces G1 arrest17 or G2/M arrest21 of tumor cells, possibly depending on the types of cells used. Therefore, we examined the effects of DHMEQ on cell cycle progression of U87, YKG-1, and U251 cells. After DHMEQ treatment (10 µg/mL), U87 and YKG-1 showed statistically significant increases in the sizes of the G2/M population accompanied by reductions in the G0/G1 fractions, and representative histogram patterns are shown in Fig. 3A. Peak effects were observed 24 h and 12 h after treatment in U87 and YKG-1, respectively (Fig. 3B). In U87 cells, G0/G1 fraction was reduced by 36%, compared with control cells, after 24 h of DHMEQ treatment (data not shown). In YKG-1, G0/G1 fraction was reduced by 33% after 12 h of DHMEQ treatment (Fig. 3A). A tendency of similar cell cycle arrest was observed in U251; however, the effect was modest in this cell line, compared with U87 and YKG-1.
Fig. 3.
Effects of DHMEQ on cell cycle and apoptosis in glioblastoma cell lines. (A) Representative DNA cell cycle histograms of vehicle (DMSO)–treated and DHMEQ-treated (10 µg/mL) U87 (48-h treatment) and YKG-1 (12-h treatment) cells. (B) Time course of G2/M arrest. Ratios of the proportion of G2/M subpopulation in DHMEQ-treated cells to that of vehicle-treated cells are plotted (mean ± SD of 5 independent experiments). Mean values of G2/M proportion (%) in the nontreated cells and that in the cells showing peak effect are also indicated. *P < .001, compared with control (Mann-Whitney U test).
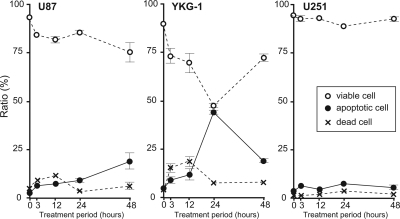
Because NF-κB is known to inhibit apoptosis through the induction of anti-apoptotic proteins and suppression of pro-apoptotic genes,3 we further examined whether apoptosis also contributed to the observed growth inhibition of glioblastoma cells. The ratios of apoptotic cells were evaluated 3, 12, 24, and 48 h after DHMEQ treatment. Peak effects were observed 48 h and 24 h after the treatment in U87 and YKG-1, respectively (Fig. 4). On the other hand, U251 was resistant to DHMEQ-induced apoptosis (Fig. 4).
Fig. 4.
Time course of apoptotic cell ratio. Proportions (%) of dead, apoptotic, and viable cells in glioblastoma cells (U87, YKG-1, and U251) treated with or without DHMEQ (10 µg/mL) are plotted. Values are mean ± SD of 3 independent experiments.
DHMEQ Treatment Suppressed the Growth of Glioblastoma Cells In Vivo
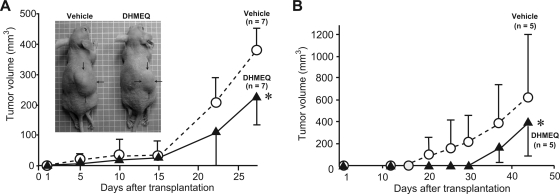
To test the effect of DHMEQ on the growth of glioblastoma cells in vivo, we analyzed the nude mouse xenotransplantation model using the highly tumorigenic glioblastoma cell line U87. Subcutaneous injection (5 × 106 cells) of U87 resulted in tumor formation in nude mice. Intraperitoneal injection of DHMEQ (10 mg/kg, 3 times per week) reduced the volume of subcutaneous tumors by 40% (P < .05) (Fig. 5A). Hepatotoxicity was not observed after intraperitoneal DHMEQ injections, as judged by measurements of alanine aminotransferase, aspirate aminotransferase, alkaline phosphatase, and total bilirubin levels in serum samples (Table 2). Similar results were obtained in U251 (39% reduction of subcutaneous tumor volume; P < .05) (Fig. 5B). YKG-1 could not form subcutaneous tumor.
Fig. 5.
Effect of DHMEQ treatment on in vivo tumor volume of U87 (A) and U251 (B) cells; 5 × 106 cells were implanted into nude mouse subcutaneously. A representative photo of subcutaneous U87 tumors (indicated by arrows) is also shown. *, P < .05, 2-way repeated-measures analysis of variance.
Table 2.
Effect of intraperitoneal administration of DHMEQ on serum parameters of mice with subcutaneous transplantation of U87 cells
| Parameters* | Vehicle treatment | DHMEQ treatment** |
|---|---|---|
| T-Bil (mg/dL) | 0.02 ± 0.01 | 0.03 ± 0.01 |
| AST (IU/L) | 245.6 ± 69.8 | 231.8 ± 127.7 |
| ALT (IU/L) | 63.6 ± 65.4 | 33.2 ± 13.2 |
| ALP (IU/L) | 274.6 ± 66.8 | 269.8 ± 50.5 |
Values are mean + SD (n = 7).
*Serum samples were collected 4 weeks after subcutaneous transplantation of U87 cells, and each parameter was measured.
**10 mg/kg, 3 times/week.
ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspirate aminotransferase; IU, international units; T-Bil, total bilirubin.
DHMEQ Improved the Survival of Mice After Intracranial Implantation of Glioblastoma Cells
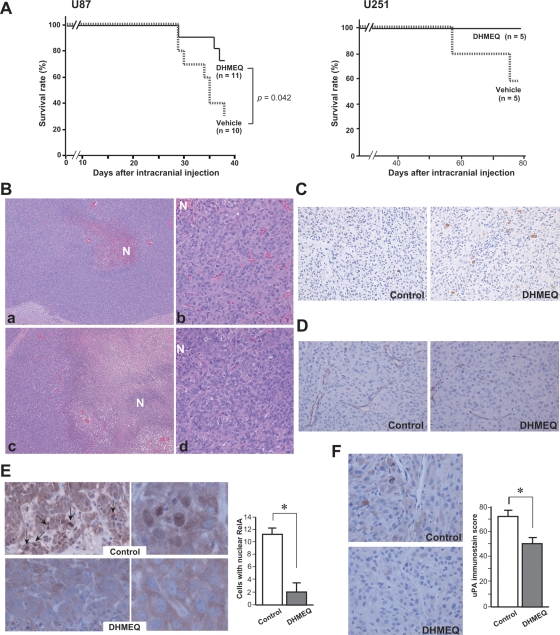
Finally, we examined the effect of DHMEQ with use of an intracranial glioblastoma model. In this model, U87 (1 × 105) or U251 (1 × 106) cells were injected into the forebrain and the mice were observed for 38 days or 77 days, respectively. YKG-1 did not show tumorigenic potential in these experimental conditions. As shown in Fig. 6A, intraperitoneal administration of DHMEQ resulted in improved survival, compared with the control group, during the observation period in both U87 and U251 cells. Tumor formation was confirmed in all mice histologically. Next, to compare the size and histology of brain tumors, we performed a similar intracranial implantation experiment and sacrificed the mice 29 days after transplantation of U87. Autopsy revealed that, although the mean tumor masses were comparable between the DHMEQ group and the control group (data not shown), the DHMEQ-treated tumors showed larger necrotic areas (Fig. 6B). Apoptotic cells, indicated by immunoreactivity for cleaved caspase 3, were increased (Fig. 6C), and the difference of labeling index (control, 3.0 ± 0.3; DHMEQ, 7.5 ± 2.6/ × 200 field) was statistically significant (P < .05, Mann-Whitney U test). In addition, although the difference of microvessel densities was not statistically significant (control, 11.4 ± 2.3; DHMEQ, 7.1 ± 2.9/ × 200 field), DHMEQ-treated tumors showed a tendency for increased collapsed microvessels (Fig. 6D). Immunohistochemical study for RelA (Fig. 6E) indicated that the number of nuclear RelA-positive glioblastoma cells was decreased by DHMEQ treatment (P < .05). Moreover, uPA immunoreactivity was also decreased in the DHMEQ-treated U87 cells (Fig. 6F).
Fig. 6.
Effect of DHMEQ treatment on intracranial implantation of glioblastoma cells. (A) Kaplan-Meier survival curves of mice treated with or without DHMEQ; 1 × 105 (U87) or 1 × 106 (U251) cells were transplanted into the forebrain of each mouse. P value, log-rank test. (B) Histology (hematoxylin and eosin stain) of the intracranial tumors derived from U87 without (a, b) or with (c, d) DHMEQ treatment. DHMEQ-treated tumor (c) shows larger necrotic area (N) than control tumor (a). Original magnification, ×40 (a, c) or ×200 (b, d). (C) Immunoreactivity of cleaved caspase 3 (U87, ×200). (D) Immunoreactivity of CD31 (U87, ×200). Microvessels in DHMEQ-treated tumors show a collapsed pattern. (E) Immunoreactivity of RelA (U87; ×400, left panel; ×1300, right panel). Arrows indicate nuclear immunoreactivity. Mean ± SD (n = 4) of positive cells/×400 field is indicated as a bar graph. *P < .05, Mann-Whitney U test. (F) Immunoreactivity of uPA in tumor cells (U87). Immunoscore was calculated and indicated as a bar graph (mean ± SD, n = 4). *P < .05, Mann-Whitney U test.
Discussion
Despite extensive studies on the molecular basis of the pathogenesis and biology of glioblastoma, the prognosis of glioblastoma remains quite poor, providing a strong incentive to develop additional treatment strategies against this disease.1,30,31 In this study, we explored an anti-proliferative effect of DHMEQ on glioblastoma cells in vitro and its possible usefulness in the treatment of glioblastoma in vivo. Growth inhibitory effects of DHMEQ on various kinds of tumors have been reported elsewhere, including prostatic, thyroid, breast, hepatocellular, urothelial and nasopharyngeal carcinomas,13–15,20–22 adult T-cell leukemia,16 multiple myeloma,17 B-cell lymphoma,19 synovial sarcoma,18 and uveal melanoma.23 Our study indicates that glioblastoma is an additional promising target of the DHMEQ therapy, and the results of our in vivo study provided supportive evidence for possible clinical application of DHMEQ.
Various molecules targeting the NF-κB pathway at different points in the signaling cascade have been developed.32 Among them, DHMEQ is unique in that it inhibits the translocation of NF-κB proteins into the nucleus without changing the amount of IκB.10,12 The inhibitory effect of DHMEQ is more potent on the NF-κB1(p50)/RelA(p65) heterodimer than on the NF-κB1(p50) homodimer. The amino acid sequence CEGRSAGSI, which appears in RelA, c-Rel, and RelB, is the target.33 Although approaches to the targeting of RelA/NF-κB for the treatment of glioblastoma require further analysis, several studies have suggested the possibility that RelA/NF-κB could be a molecular target in glioblastoma therapy. In human gliomas, both the expression levels and the activation status of RelA/NF-κB were correlated with tumor grade.4,5 Inhibition of RelA by curcumin or antisense oligonucleotide suppressed cellular migration and invasion in vitro.5 Similarly, knockdown of RelA by short hairpin RNA suppressed the growth of glioblastoma cells in vitro.7 Furthermore, transfection of anti-RelA/NF-κB intracellular antibody into glioblastoma cells reduced invasive and angiogenic capabilities of the cells in vitro and, more importantly, suppressed intracranial growth in vivo.6 In our current study, targeting RelA/NF-κB with DHMEQ induced growth suppression, G2/M cell cycle arrest, and apoptosis of glioblastoma cells. Therefore, our data confirm the usefulness of targeting RelA/NF-κB in glioblastoma. Moreover, in vivo survival of mice transplanted intracranially with U87 or U251 glioblastoma cells was improved after intraperitoneal administration of DHMEQ, suggesting a clinical application of this small molecule inhibitor of RelA/NF-κB for the treatment of intracranial tumors. However, responses to DHMEQ treatment were not always the same in the glioblastoma cell lines used. For example, the T98G cell line was resistant to DHMEQ-induced growth suppression. The U251 cell line showed significant growth suppression after DHMEQ treatment, but was resistant to DHMEQ-induced apoptosis. It is probable that a dominant intracellular signaling cascade may be different between glioblastoma cell lines, which may result in different responses to DHMEQ treatment. On this point, a recent report reveals that phospatidylinositol-3-kinase/Akt survival signaling is functioning in T98G, which harbors PTEN (phosphatase and tension homolog located on chromosome 10), whereas NF-κB survival signaling is functioning in PTEN-deficient U87.34 The p53 status may also influence the response to DHMEQ, and U251 and T98G have mutant p53 status.27
In summary, our study showed that DHMEQ, a small molecule inhibitor of NF-κB, induced growth arrest and apoptosis of glioblastoma cells. Furthermore, in vivo study showed improved survival in the mouse intracranial glioblastoma model, suggesting that the administration of DHMEQ may have clinical use in the chemoprevention of glioblastoma growth. On the other hand, our data also suggest that DHMEQ treatment alone is not enough to induce complete remission of the glioblastoma tumor in vivo. Moreover, not all glioblastoma cell lines showed significant DHMEQ sensitivity in vitro. Taken together, we propose that DHMEQ may serve as a promising tool for adjuvant therapy for enhancing the efficacy of conventional treatment modalities in glioblastoma. Further study will be required to explore the usefulness of DHMEQ in the treatment of glioblastoma, particularly in combination with other treatment modalities, such as radiation therapy or chemotherapy using temozolomide.35
Funding
This work was supported in part by Grant-in-Aid for Scientific Research (20390114 to H.K.) and for Young Scientists (22790384 to T.F.) from the Ministry of Education, Science, Sports and Culture, Japan.
Acknowledgment
We thank Ms. Y. Tobayashi for her help in immunostain.
Conflict of interest statement. None declared.
References
- 1.Khasraw M, Lassman AB. Advances in the treatment of malignant gliomas. Curr Oncol Rep. 2010;12:26–33. doi: 10.1007/s11912-009-0077-4. doi:10.1007/s11912-009-0077-4. [DOI] [PubMed] [Google Scholar]
- 2.Patrick A.B. IκB–NF-κB Structures: At the interface of inflammation control. Cell. 1998;95:729–731. doi: 10.1016/s0092-8674(00)81694-3. doi:10.1016/S0092-8674(00)81694-3. [DOI] [PubMed] [Google Scholar]
- 3.Rayet B, Gélinas C. Aberrant rel/nfkb genes and activity in human cancer. Oncogene. 1999;18:6938–6947. doi: 10.1038/sj.onc.1203221. doi:10.1038/sj.onc.1203221. [DOI] [PubMed] [Google Scholar]
- 4.Wang H, Wang H, Zhang W, Huang HJ, Liao WS, Fuller GN. Analysis of the activation status of Akt, NFkappaB, and Stat3 in human diffuse gliomas. Lab Invest. 2004;84:941–951. doi: 10.1038/labinvest.3700123. doi:10.1038/labinvest.3700123. [DOI] [PubMed] [Google Scholar]
- 5.Tsunoda K, Kitange G, Anda T. Expression of the constitutively activated RelA/NF-kappaB in human astrocytic tumors and the in vitro implication in the regulation of urokinase-type plasminogen activator, migration, and invasion. Brain Tumor Pathol. 2005;22:79–87. doi: 10.1007/s10014-005-0186-1. doi:10.1007/s10014-005-0186-1. [DOI] [PubMed] [Google Scholar]
- 6.Li L, Gondi CS, Dinh DH, Olivero WC, Gujrati M, Rao JS. Transfectionwith anti-p65 intrabody suppresses invasion and angiogenesis in glioma cells by blocking nuclear factor-κB transcriptional activity. Clin Cancer Res. 2007;13:2178–2190. doi: 10.1158/1078-0432.CCR-06-1711. doi:10.1158/1078-0432.CCR-06-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith D, Shimamura T, Barbera S, Bejcek BE. NF-kappaB controls growth of glioblastomas/astrocytomas. Mol Cell Biochem. 2008;307:141–147. doi: 10.1007/s11010-007-9593-4. doi:10.1007/s11010-007-9593-4. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Gong LY, Song LB, et al. Oncoprotein Bmi-1 renders apoptotic resistance to glioma cells through activation of the IKK-nuclear factor-kappaB Pathway. Am J Pathol. 2010;176:699–709. doi: 10.2353/ajpath.2010.090502. doi:10.2353/ajpath.2010.090502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsumoto N, Ariga A, To-e S, et al. Synthesis of NF-kappaB activation inhibitors derived from epoxyquinomicin C. Bioorg Med Chem Lett. 2000;10:865–869. doi: 10.1016/s0960-894x(00)00114-1. doi:10.1016/S0960-894X(00)00114-1. [DOI] [PubMed] [Google Scholar]
- 10.Umezawa K. Inhibition of tumor growth by NF-kappaB inhibitors. Cancer Sci. 2006;97:990–995. doi: 10.1111/j.1349-7006.2006.00285.x. doi:10.1111/j.1349-7006.2006.00285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto M, Horie R, Takeiri M, Kozawa I, Umezawa K. Inactivation of nuclear kappa B components by covalent bincing of (-)-dehydroxymethylepoxyquinomicin to specific cysteine residues. J Mod Chem. 2008;51:5780–5788. doi: 10.1021/jm8006245. doi:10.1021/jm8006245. [DOI] [PubMed] [Google Scholar]
- 12.Ariga A, Mamekawa J, Matsumoto N, Inoue J, Umezawa K. Inhibition of tumor necrosis factor-α -induced nuclear translocation and activation of NF-κB by dehydroxymethylepoxyquinomicin. J Biol Chem. 2002;277:27625–27630. doi: 10.1074/jbc.M112063200. [DOI] [PubMed] [Google Scholar]
- 13.Kikuchi E, Horiguchi Y, Nakashima J, et al. Suppression of hormone-refractory prostate cancer by a novel nuclear factor kappaB inhibitor in nude mice. Cancer Res. 2003;63:107–110. [PubMed] [Google Scholar]
- 14.Starenki DV, Namba H, Saenko VA. Induction of thyroid cancer cell apoptosis by a novel nuclear factor kappaB inhibitor, dehydroxymethylepoxyquinomicin. Clin Cancer Res. 2004;10:6821–6829. doi: 10.1158/1078-0432.CCR-04-0463. doi:10.1158/1078-0432.CCR-04-0463. [DOI] [PubMed] [Google Scholar]
- 15.Matsumoto G, Namekawa J, Muta M. Targeting of nuclear factor kappaB Pathways by dehydroxymethylepoxyquinomicin, a novel inhibitor of breast carcinomas: antitumor and antiangiogenic potential in vivo. Clin Cancer Res. 2005;11:1287–1293. [PubMed] [Google Scholar]
- 16.Watanabe M, Ohsugi T, Shoda M, et al. Dual targeting of transformed and untransformed HTLV-1-infected T cells by DHMEQ, a potent and selective inhibitor of NF-kappaB, as a strategy for chemoprevention and therapy of adult T-cell leukemia. Blood. 2005;106:2462–2471. doi: 10.1182/blood-2004-09-3646. doi:10.1182/blood-2004-09-3646. [DOI] [PubMed] [Google Scholar]
- 17.Watanabe M, Dewan MZ, Okamura T, et al. A novel NF-kappaB inhibitor DHMEQ selectively targets constitutive NF-kappaB activity and induces apoptosis of multiple myeloma cells in vitro and in vivo. Int J Cancer. 2005;114:32–38. doi: 10.1002/ijc.20688. doi:10.1002/ijc.20688. [DOI] [PubMed] [Google Scholar]
- 18.Horiuchi K, Morioka H, Nishimoto K, et al. Growth suppression and apoptosis induction in synovial sarcoma cell lines by a novel NF-kappaB inhibitor, dehydroxymethylepoxyquinomicin (DHMEQ) Cancer Lett. 2008;272:336–344. doi: 10.1016/j.canlet.2008.07.021. doi:10.1016/j.canlet.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 19.Dabaghmanesh N, Matsubara A, Miyake A, et al. Transient inhibition of NF-kappaB by DHMEQ induces cell death of primary effusion lymphoma without HHV-8 reactivation. Cancer Sci. 2009;100:737–746. doi: 10.1111/j.1349-7006.2009.01083.x. doi:10.1111/j.1349-7006.2009.01083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lampiasi N, Azzolina A, D'Alessandro N, et al. Antitumor effects of dehydroxymethylepoxyquinomicin, a novel nuclear factor-kappaB inhibitor, in human liver cancer cells are mediated through a reactive oxygen species-dependent mechanism. Mol Pharmacol. 2009;76:290–300. doi: 10.1124/mol.109.055418. doi:10.1124/mol.109.055418. [DOI] [PubMed] [Google Scholar]
- 21.Wong JH, Lui VW, Umezawa K, et al. A small molecule inhibitor of NF-kappaB, dehydroxymethylepoxyquinomicin (DHMEQ), suppresses growth and invasion of nasopharyngeal carcinoma (NPC) cells. Cancer Lett. 2010;287:23–32. doi: 10.1016/j.canlet.2009.05.022. doi:10.1016/j.canlet.2009.05.022. [DOI] [PubMed] [Google Scholar]
- 22.Kodaira K, Kikuchi E, Kosugi M, et al. Potent cytotoxic effect of a novel nuclear factor-kappaB inhibitor dehydroxymethylepoxyquinomicin on human bladder cancer cells producing various cytokines. Urology. 2010;75:805–812. doi: 10.1016/j.urology.2009.11.048. doi:10.1016/j.urology.2009.11.048. [DOI] [PubMed] [Google Scholar]
- 23.Dror R, Lederman M, Umezawa K, Barak V, Pe'er J, Chowers I. Characterizing the involvement of the nuclear factor-kappa B (NF-κB) transcription factor in uveal melanoma. Invest. Ophthalmol. Vis Sci. 2010;51:1811–1816. doi: 10.1167/iovs.09-3392. doi:10.1167/iovs.09-3392. [DOI] [PubMed] [Google Scholar]
- 24.Nuki Y, Uchinokura S, Miyata S, et al. Establishment and characterization of a new human glioblastoma cell line, NYGM. Hum Cell. 2004;17:145–150. doi: 10.1111/j.1749-0774.2004.tb00031.x. doi:10.1111/j.1749-0774.2004.tb00031.x. [DOI] [PubMed] [Google Scholar]
- 25.Moriyama T, Kataoka H, Seguchi K, et al. Establishment and characterization of a new human glioblastoma cell line (MGM-1) with highly motile phenotype. Hum Cell. 1997;10:105–109. doi: [PubMed] [Google Scholar]
- 26.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. WHO Classification of Tumours of the Central Nervous System. 4th edn. Lyon, France: International Agency for Research on Cancer (IARC); 2007. [Google Scholar]
- 27.Fukushima T, Tezuka T, Shimomura T, Nakano S, Kataoka H. Silencing of insulin-like growth factor-binding protein-2 in human glioblastoma cells reduces both invasiveness and expression of progression-associated gene CD24. J Biol Chem. 2007;282:18634–18644. doi: 10.1074/jbc.M609567200. doi:10.1074/jbc.M609567200. [DOI] [PubMed] [Google Scholar]
- 28.Uchinokura S, Miyata S, Fukushima T, et al. Role of hepatocyte growth factor activator (HGF activator) in invasive growth of human glioblastoma cells in vivo. Int J Cancer. 2006;118:583–592. doi: 10.1002/ijc.21362. doi:10.1002/ijc.21362. [DOI] [PubMed] [Google Scholar]
- 29.Nagaike K, Kawaguchi M, Naoki Takeda N, et al. Defect of hepatocyte growth factor activator inhibitor type 1/serine protease inhibitor, Kunitz type 1 (Hai-1/Spint1) leads to ichthyosis-like condition and abnormal hair development in mice. Am J Pathol. 2008;173:1464–1475. doi: 10.2353/ajpath.2008.071142. doi:10.2353/ajpath.2008.071142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. doi:10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shirahata M, Oba S, Iwao-Koizumi K, et al. Using gene expression profiling to identify a prognostic molecular spectrum in gliomas. Cancer Sci. 2009;100:165–172. doi: 10.1111/j.1349-7006.2008.01002.x. doi:10.1111/j.1349-7006.2008.01002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Epinat JC, Gilmore TD. Diverse agents act at multiple levels to inhibit the Rel/NF-kappaB signal transduction pathway. Oncogene. 1999;18:6896–6909. doi: 10.1038/sj.onc.1203218. doi:10.1038/sj.onc.1203218. [DOI] [PubMed] [Google Scholar]
- 33.Watanabe M, Nakashima M, Togano T, et al. Identification of the RelA domain responsible for action of a new NF-kappaB inhibitor DHMEQ. Biochem Biophys Res Commun. 2008;376:310–314. doi: 10.1016/j.bbrc.2008.08.148. doi:10.1016/j.bbrc.2008.08.148. [DOI] [PubMed] [Google Scholar]
- 34.Zhang R, Banik NL, Ray SK. Combination of all-trans retinoic acid and interferon-gamma suppressed PI3K/Akt survival pathway in glioblastoma T98G cells whereas NF-kB survival signaling in glioblastoma U87MG cells for induction of apoptosis. Neurochem Res. 2007;32:2194–2202. doi: 10.1007/s11064-007-9417-7. doi:10.1007/s11064-007-9417-7. [DOI] [PubMed] [Google Scholar]
- 35.Fukushima T, Takeshima H, Kataoka H. Anti-glioma therapy with temozolomide and status of the DNA-repair gene MGMT. Anticancer Res. 2009;29:4845–4854. [PubMed] [Google Scholar]