Abstract
1p/19q codeletion is a favorable prognostic marker of oligodendrogliomas. While fluorescence in situ hybridization (FISH) and microsatellite-based polymerase chain reaction (PCR) for loss of heterozygosity (LOH) are common methods to test for 1p/19q codeletion, it is unclear which test is better at prognostic stratification. This study analyzed outcomes of 111 oligodendrogliomas with both 1p/19q FISH and LOH done at the time of diagnosis. Overall concordance between the 2 assays was 81.1%. In grade III oligodendrogliomas, LOH was better than FISH at survival stratification (p < 0.0001 for LOH vs. p = 0.02 for FISH), although increasing the stringency of FISH interpretation criteria improved concordance and prognostic power. Oligodendrogliomas that were 1p/19q-codeleted by FISH but also had 10q LOH were negative for 1p/19q codeletion by PCR analysis in over 70% of cases, with very poor survival in the grade III subset. Thus, although PCR-based LOH is a better stratifier of 1p/19q status, FISH still has clinical and prognostic utility, especially if 10q data can be incorporated.
Keywords: 1p/19q, 10q, Epidermal growth factor receptor (EGFR), Fluorescence in situ hybridization (FISH), Loss of heterozygosity (LOH), Microsatellite, Oligodendroglioma
INTRODUCTION
Codeletion of the short arm of chromosome 1 and the long arm of chromosome 19 (1p/19q) has long been known to be a prognostic marker of oligodendroglial tumors (1, 2), specifically correlating with improved response to adjuvant radiochemotherapy and longer survival (3–9). This codeletion is the result of an unbalanced translocation between chromosomes 1 and 19, with loss of the der(1;19)(p10;q10) (10, 11). 1p/19q codeletion is associated with a proneural expression profile, MGMT promoter methylation, and IDH1/2 mutations (12–14), and is mutually exclusive with EGFR amplification (15). On a histologic level, the codeletion is more commonly seen in oligodendrogliomas with classic oligodendroglial morphology (e.g. round nuclei, delicate branching vasculature, and microcalcifications) compared to tumors with only some “oligo-like” features (16, 17). Detection of 1p/19q codeletion can help clarify whether a hybrid oligoastrocytoma will behave more like an oligodendroglioma or an astrocytoma (18, 19), although not in the pediatric population in which even classic-appearing oligodendrogliomas rarely have this genetic feature (20, 21).
Because of its diagnostic and prognostic power, testing for 1p/19q codeletion has been a mainstay of the neuropathologic armamentarium for some time. Several methods have been described to detect this biomarker but 2 of the most commonly used are fluorescence in situ hybridization (FISH) and polymerase chain reaction (PCR)-based microsatellite loss of heterozygosity (LOH). FISH is based on fluorophore-labeled DNA probes that selectively bind to regions of interest in the target nuclei. PCR-based LOH detects allelic loss via DNA repeats known as microsatellites. When 2 alleles have nearby microsatellites of varying lengths, that locus is said to be informative. This is because there would be 2 different sizes of PCR products if both alleles are intact but only one size if an allele is lost. Both FISH and PCR-based LOH can be performed on formalin-fixed, paraffin-embedded tissues and neither requires a large amount of tumor tissue; therefore, they are particularly well-suited to neurosurgical specimens.
FISH is more popular than the PCR technique because it can target specific areas of interest in biopsy material when there is a matched hematoxylin and eosin-stained slide available for comparison. However, the length of DNA that FISH probes can cover is limited, i.e. there is a risk of false-positive results if the tumor has partial/interstitial deletions rather than whole-arm losses. Prior studies suggested that 1p36.32 and 19q13.3 were minimally deleted regions in oligodendroglial tumors (22, 23), forming the rationale for selecting those areas to target by FISH.
LOH on 10q has long been known to correlate with higher World Health Organization (WHO) grade in gliomas (7, 24, 25). Several tumor suppressors have been identified on 10q, most notably PTEN, which normally serves as a brake on the oncogenic receptor tyrosine kinase pathways (26). Some studies have shown 10q deletion to be an adverse prognostic marker in oligodendrogliomas (7, 27, 28), but not all studies have shown this correlation (9). Recent work in particular has suggested that 10q LOH is more important than 1p/19q status insofar as oligodendrogliomas with codeletion still behave much worse if 10q is also deleted (28). It is possible, however, that some cases with 10q LOH may have only small losses on both chromosomal arms, as opposed to the extensive losses that correlate with better prognosis.
Prior studies have shown that there is generally good concordance between FISH and PCR-based LOH (23, 29–31), but outcome-based studies directly comparing these tests, using results generated and interpreted at the time of diagnosis, are rare. It is, therefore, of interest to know whether one test has greater prognostic power, i.e. which method is superior at identifying tumors with a better prognosis, and whether 10q status can help refine 1p/19q FISH results and prognosis in such tumors. Herein we describe outcomes from a cohort of 111 oligodendrogliomas, including 79 WHO grade II oligodendrogliomas and 32 grade III anaplastic oligodendrogliomas, in which all cases were tested for 1p/19q codeletion via both FISH and PCR-based LOH analysis at the time of initial biopsy and diagnosis.
MATERIALS AND METHODS
Cohort
Between 2002 and 2010, 111 oligodendrogliomas (WHO grades II–III) were prospectively analyzed for 1p/19q codeletion by FISH and LOH at the University of Pittsburgh (Table 1). Cases of recurrent and/or treated gliomas were excluded, as were pediatric cases (i.e. patient age under 18 years). Diagnoses were made according to standard WHO criteria at the time of initial biopsy. The diagnosis “oligoastrocytoma” was used sparingly due to its well-known low interobserver reproducibility. Instead, only gliomas that had strong oligodendroglial morphology (e.g. round nuclei, little to no angulated nuclei, branching vasculature, and absence of pseudopalisading necrosis) were classified as oligodendrogliomas; all others were classified as astrocytomas, except 4 cases that had equally prominent oligodendroglial and astrocytic components and were excluded from analysis. Postsurgical follow-up data was obtained by the University of Pittsburgh Hillman Cancer Registry. All data collection and analyses were done in accordance with the University of Pittsburgh and the University of Kentucky committees on human subjects.
Table 1.
Clinical Characteristics of the Oligodendroglioma Cohort
| Parameter | Grade II oligodendroglioma | Grade III oligodendroglioma |
|---|---|---|
| No. male (%) | 41 (51.9) | 22 (68.8) |
| Median age (range) | 42 y (19—79 y) | 49 y (25—80 y) |
| No. biopsy only (%) | 21 (29.2) | 7 (26.9) |
| No. GTR (%) | 22 (30.6) | 6 (23.1) |
| No. STR (%) | 29 (40.3) | 13 (50.0) |
| No. temporal lobe (%) | 14 (23.7) | 3 (15.8) |
| No. non-temporal lobe (%) | 45 (76.3) | 16 (84.2) |
| No. with no adjuvant therapy (%) | 26 (36.6) | 8 (30.8) |
| No. with radiation only (%) | 13 (18.3) | 4 (15.4) |
| No. with chemo only (%) | 24 (33.8) | 3 (11.5) |
| No. with radiochemotherapy (%) | 8 (11.3) | 11 (42.3) |
| Mean follow-up interval | 3.4 y | 3.4 y |
| Median follow-up interval | 2.9 y | 1.9 y |
| Follow-up range | 0.3—17.5 y | 0.1—18.9 y |
| No. died (%) | 11 (15.5) | 19 (59.4) |
| No. 1p/19q intact by FISH and LOH (%) | 22 (27.8) | 4 (12.5) |
| No. 1p/19q codeleted by FISH and LOH (%) | 43 (54.4) | 22 (68.8) |
| No. codeleted by FISH not LOH (%) | 14 (17.7) | 6 (18.8) |
| No. codeleted by LOH not FISH (%) | 0 (0) | 0 (0) |
| No. with 10q LOH (%) | 17/72 (23.6) | 9/26 (34.6) |
Seventy-nine grade II oligodendrogliomas and 32 grade III oligodendrogliomas were subjected to prospective molecular testing at the time of diagnosis. The type of surgery was known in 72 grade II and 26 grade III tumors; postoperative treatment information was available for 71 grade II and 26 grade III tumors; precise tumor location was retrievable for 59 grade II and 19 grade III tumors. Overall 1p/19q concordance between FISH and LOH was 81.1%. FISH = fluorescence in situ hybridization; GTR = gross total resection; STR = subtotal resection; LOH = loss of heterozygosity.
FISH
Formalin-fixed paraffin-embedded blocks were analyzed via FISH using probes for 1p36 and 19q13 (Abbott Molecular, Des Plaines, IL), as previously described (32). For ploidy control, locus-specific probes were used for chromosomes 1 (1q25) and 19 (19p13) (Supplemental Fig. 1). At least 60 cells were analyzed in the targeted region per case. Deletion was scored for 1p36 and 19q13 if the target:ploidy control ratio was less than 0.87, with at least 20% of nuclei showing deletion. These cutoff points exceeded 3 standard deviations from the mean of 20 non-neoplastic autopsy brain tissue specimens.
PCR-Based Microsatellite LOH Analysis
DNA from formalin-fixed paraffin-embedded tissue was probed as described previously (32) with 7 microsatellite markers on chromosome 1p22.2—36.32 (D1S1172, D1S226, D1S162, D1S1161, D1S199, D1S407, D1S171), 2 on 19q13.32 and 19q13.41 (D19S112 and D19S206, respectively), and 2 on 10q (D10S1173 and D10S520) (Supplemental Fig. 1). PCR was performed and the products were analyzed using capillary gel electrophoresis on GeneMapper ABI 3730 (Applied Biosystems, Foster City, CA). When available, patient-matched germline DNA from a peripheral blood sample was used as a control. When normal tissue was not available, peak height ratios falling outside of 2 standard deviations beyond the mean of previously validated normal values for each polymorphic allele paring were assessed as showing LOH. LOH regions were defined by bracketing only with informative markers. Non-informative loci were mapped and included within preserved or lost regions. At least half of all informative microsatellite loci on both 1p and 19q had to show LOH in order to be designated as having 1p/19q codeletion. Any loss on 10q was counted as 10q LOH.
Statistics
The Fisher exact test was used to compare relative risk of adverse outcome between 2 groups. Means were compared between multiple groups by Kruskal-Wallis test (nonparametric ANOVA and Dunn’s post hoc) or between 2 groups by Mann-Whitney test where appropriate. Univariate survival rates were compared via log rank tests on Kaplan-Meier curves; multivariate analyses were done via Cox proportional hazards survival regression. Statistical analyses were performed using GraphPad software (La Jolla, CA), Microsoft Excel (Redmond, WA), and http://statpages.org/prophaz.html. Differences were considered significant when p < 0.05.
RESULTS
Cohort Characteristics
A total of 111 oligodendrogliomas were prospectively analyzed for 1p/19q codeletion by both FISH and LOH techniques (Table 1; Supplemental Fig. 1). Median patient age was 42 years for grade II tumors and 49 years for grade III tumors. The majority of patients were male (56.8%). Of the 30 who had died at the time of analysis, median survival was 1.8 years, ranging from 47 days to 16.8 years. Of the 238 who were still alive at the time of analysis, median follow-up time was 2.8 years (range: 7.8 months to 19 years). Cases with radiotherapy all received external beam radiation; specific data on the types of chemotherapy used in each case was not retrievable.
1p/19q FISH Improves with More Stringent Cutoff Criteria
Overall, there was an 81.1% concordance between FISH and LOH analyses in all oligodendrogliomas (Table 1) (2-tailed p < 0.0001 via Spearman rank correlation). Twenty cases were positive for 1p/19q codeletion by FISH but negative by LOH analysis, whereas only 1 case was codeleted by LOH but not by FISH.
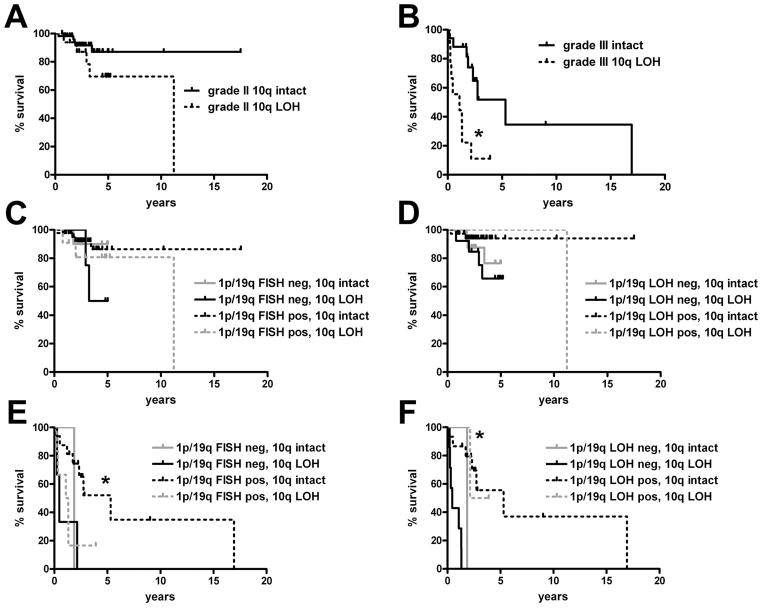
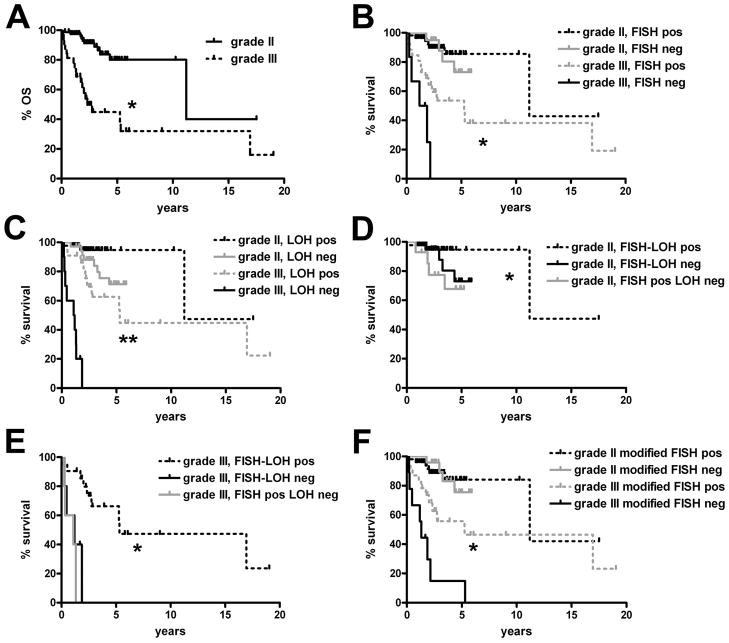
As expected, grade III oligodendrogliomas had worse survival than grade II tumors (Fig. 1A, p < 0.0001). Neither 1p/19q assay was able to stratify WHO grade II oligodendrogliomas by survival but the grade II curves generated by FISH were nearly identical whereas PCR-based analysis showed a trend toward improved survival when 1p/19q LOH was detected (p = 0.64 for FISH vs. 0.09 for LOH) (Fig. 1B, C). Further survival splitting according to FISH-LOH concordance showed that there was a significant difference between cases that were codeleted by both FISH and LOH vs. those that were positive by just FISH but not LOH (p = 0.04) (Fig. 1D).
Figure 1.
Survival according to 1p/19q fluorescence in situ hybridization (FISH) and loss of heterozygosity (LOH) analyses. (A) Grade III oligodendrogliomas had worse survival than grade II tumors (*p < 0.0001). (B, C) 1p/19q FISH (B) was weaker than PCR-based LOH (C) in differentiating longer-term from shorter-term survivors. This difference was most prominent in grade III anaplastic oligodendroglial tumors, in which prognostic stratification was better by LOH (**p < 0.0001) than by FISH (*p = 0.02). FISH could not stratify grade II oligodendrogliomas (p = 0.64), survival differences trended toward significance by LOH (p = 0.09). (D) Grade II oligodendrogliomas with 1p/19q codeletion by both modalities had longer survival than those that were positive by FISH but not by LOH (*p = 0.04; p = 0.26 vs. tumors that were negative by both tests). (E) Grade III anaplastic oligodendrogliomas that were codeleted by FISH and LOH showed longer survival compared to tumors that were negative by both tests (*p = 0.0006), or positive only by FISH but not LOH (*p < 0.0001). (F) Increasing the stringency of FISH codeletion criteria to a cutoff 1p36/1q25 and 19q13/19p13 ratio of 0.75 for each probe pair (from the original cutoff of 0.87) improved statistical power in stratifying grade III anaplastic oligodendrogliomas (*p = 0.007).
The difference between the 2 tests was greater in grade III anaplastic oligodendrogliomas (Fig. 1B, C) because although FISH reached significance in its stratification of grade III oligodendrogliomas (Fig. 1B) (p = 0.02), LOH analysis was more powerful (Fig. 1C) (p < 0.0001). This improvement was achieved by “avoiding” tumors that ultimately behaved aggressively; of the 5 patients whose tumors were diagnosed as codeleted by FISH but not by LOH, median survival was only 1.1 years and none lived beyond 1.3 years after their diagnosis (Fig. 1E). This dismal survival curve was statistically indistinguishable from the cases that were 1p/19q-intact by both FISH and LOH, in which median survival was 1.2 years. In contrast, patients whose grade III oligodendrogliomas were codeleted by both FISH and LOH had a median survival of 5.3 years. In the single anaplastic oligodendroglioma where LOH showed codeletion but not FISH, the patient died 2.2 years after diagnosis (not graphed in Fig. 1E).
Original criteria for deletion of either 1p36 or 19q13 were derived from nonneoplastic autopsy control brain tissues (see Materials and Methods). Although 1p36/1q25 and 19q13/19p13 cutoff ratios of 0.87 were 3 standard deviations below control means, we sought to determine whether even more stringent cutoff criteria would improve the statistical power of 1p/19q FISH. Supporting this hypothesis was the finding that mean ratios for 1p36/1q25 and 19q13/19p13 were significantly higher in gliomas that were positive for 1p/19q codeletion by FISH but not by LOH (p < 0.001) (Supplemental Fig. 2). Visual inspection of the ratio scatterplots suggested that lowering 1p36/1q25 and 19q13/19p13 cutoffs to 0.75 might improve specificity without excessive loss of sensitivity. Indeed, setting the cutoff ratios at 0.75 slightly increased overall FISH-LOH concordance to 82.9% and improved specificity and PPV relative to 1p/19q LOH (Table 2). Although some sensitivity and NPV was lost, this modification still produced a slightly improved Spearman rank correlation coefficient between FISH and LOH in all oligodendrogliomas (0.65 for modified FISH vs 0.63 with standard FISH cutoffs). Most importantly, the modification improved stratification power, as the modified FISH cutoff criteria were usually better at separating less aggressive from more aggressive anaplastic oligodendrogliomas (Fig. 1F) (p = 0.007) compared to standard FISH criteria (Fig. 1B). Stratification of grade II oligodendrogliomas remained non-significant even with the modified FISH criteria (Fig. 1F) (p = 0.92).
Table 2.
Comparison Between 2 Different Fluorescence In Situ Hybridization Interpretation Criteria and Loss of Heterozygosity Methods of 1p/19q Testing
| 1p/19q FISH criteria | 1p/19q FISH relative to 1p/19q LOH | |||
|---|---|---|---|---|
| % Sensitivity (95% CI) | % Specificity (95% CI) | % PPV (95% CI) | % NPV (95% CI) | |
| Original FISH criteria | 98.5 (91.7—100.0) | 56.5 (41.1—71.0) | 76.2 (65.6—84.8) | 96.3 (81.0—99.9) |
| Modified FISH criteria | 93.9 (85.0—98.3) | 67.4 (51.9—80.5) | 80.3 (69.6—88.5) | 88.6 (73.3—96.8) |
One hundred eleven grade II and III oligodendrogliomas were analyzed for 1p/19q codeletion by both FISH and LOH assays. Relative to 1p/19q LOH testing, increasing the stringency of FISH ratio cutoff criteria from < 0.87 (Original) to < 0.75 (Modified) improved the specificity and positive predictive value (PPV) of FISH, at the cost of reduced sensitivity and negative predictive value (NPV). CI = confidence interval; FISH = fluorescence in situ hybridization; LOH = loss of heterozygosity.
10q Status Further Refines 1p/19q Data in Oligodendrogliomas
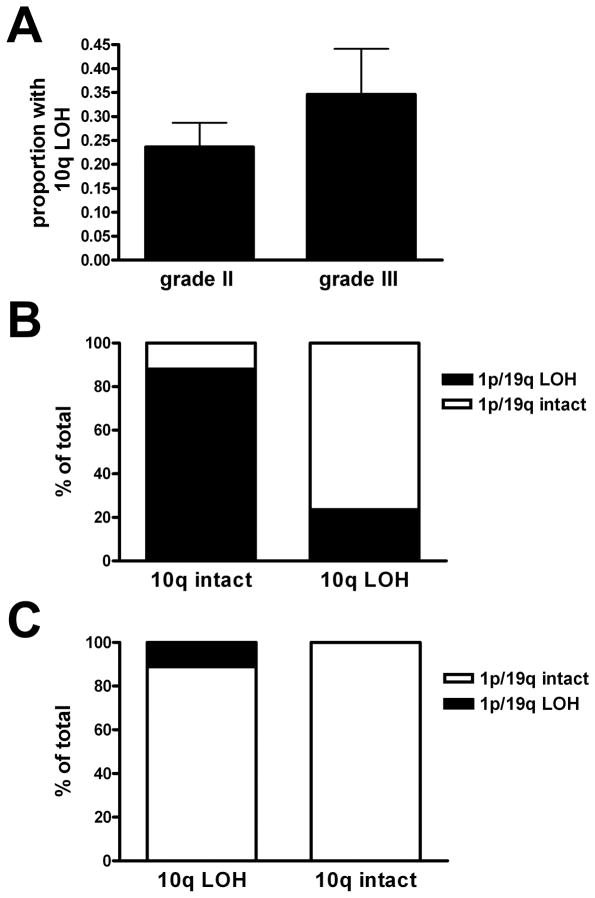
10q status had been determined at the time of diagnosis in 72 of 79 grade II oligodendrogliomas and in 26 of 32 grade III oligodendrogliomas; the remainder were either noninformative or were consult cases not subjected to 10q testing. 10q LOH was detected in 17 grade II and 9 grade III oligodendrogliomas (23.6% and 34.6%, respectively); this difference was not significant (p = 0.40) (Fig. 2A). Of the 11 grade II oligodendrogliomas that had 1p/19q codeletion by original FISH criteria plus 10q LOH, only 3 were also codeleted by PCR-based LOH analysis. Likewise, of the 6 grade III oligodendrogliomas with 1p/19q codeletion and 10q LOH, only 1 was also codeleted by LOH analysis. Overall, oligodendrogliomas codeleted by FISH and intact for 10q were also codeleted for 1p/19q via PCR-based microsatellite analysis in 52 of 59 cases (88.1%) (Fig. 2B, Table 3). In contrast, oligodendrogliomas that were 1p/19q-codeleted by FISH but had 10q LOH also showed 1p/19q codeletion via PCR-based LOH analysis in only 4 of 17 cases (23.5%) (p < 0.0001 via Fisher exact test). When FISH called an oligodendroglioma 1p/19q-intact, 10q status had no significant association with PCR-based LOH concordance, as the tumor was likely to be intact by microsatellite analysis regardless of 10q (Fig. 2C, p = 0.41).
Figure 2.
10q loss of heterozygosity (LOH) in gliomas. (A) 23.6% of grade II oligodendrogliomas had 10q LOH vs. 34.6% of grade III oligodendrogliomas (p = 0.40 by Mann-Whitney test). (B) When either a grade II or grade III oligodendroglioma was called 1p/19q-codeleted by fluorescence in situ hybridization (FISH), concordance with PCR-based 1p/19q microsatellite LOH analysis was much less likely if 10q was also deleted (p < 0.0001, n = 76). (C) If an oligodendroglioma was 1p/19q-intact by FISH, 10q status had no bearing on whether the FISH result would be concordant with PCR-based 1p/19q microsatellite analysis (p = 0.41, n = 22).
Table 3.
Concordance Between 1p/19q Fluorescence In Situ Hybridization and PCR-Based Microsatellite Loss of Heterozygosity Data Is Further Refined by 10q Status
| 1p/19q FISH | 10q status | 1p/19q PCR microsatellite-based LOH | % FISH-PCR concordance | ||
|---|---|---|---|---|---|
| codeleted | Intact | ||||
| original criteria | codeleted | 10q intact | 52 | 7 | 88.1 |
| 10q LOH | 4 | 13 | 23.5 | ||
| intact | 10q intact | 0 | 13 | 100.0 | |
| 10q LOH | 1 | 8 | 88.9 | ||
| modified criteria | codeleted | 10q intact | 49 | 5 | 90.7 |
| 10q LOH | 4 | 10 | 28.6 | ||
| intact | 10q intact | 3 | 15 | 83.3 | |
| 10q LOH | 1 | 11 | 91.7 | ||
Ninety-eight World Health Organization grade II and III oligodendrogliomas were tested for 1p/19q codeletion by FISH and PCR-based LOH analysis and for 10q deletion. Both by original and more stringent modified criteria, if a case was 1p/19q-codeleted and intact for 10q, PCR analysis also called the case codeleted about 90% of the time. If a case was called 1p/19q-codeleted by FISH yet had 10q LOH, PCR analysis called the same tumor negative for codeletion over 70% of the time. If a tumor was 1p/19q intact by either original or modified FISH criteria and also 10q intact, PCR analysis also called it 1p/19q intact 80–100% of the time; the presence of 10q LOH did not appreciably change that proportion (about 90%). FISH = fluorescence in situ hybridization; LOH = loss of heterozygosity; PCR = polymerase chain reaction.
Re-interpretation of oligodendrogliomas by modified FISH criteria produced similar results (Table 3). In sum, if cases called 1p/19q-codeleted by original FISH criteria but with 10q LOH had been interpreted as 1p/19q-intact, concordance with PCR-based analysis would have improved from 75% to 88%. Likewise, 10q data improved modified FISH-LOH concordance from 81% to 87%.
From an outcomes-based perspective, the survival of 10q-deleted oligodendrogliomas was shorter compared to their grade-matched 10q-intact counterparts (Fig. 3A, B), although this was significant only in grade III tumors (p = 0.002). 10q LOH suggested shorter overall survival in grade II oligodendrogliomas when 1p/19q was intact, though this difference was not significant either by FISH (Fig. 3C) or LOH (Fig. 3D. The only grade III oligodendrogliomas in which longer-term survival occurred was when 1p/19q was codeleted and 10q was intact (Fig. 3E, p = 0.03). The same pattern was seen via 1p/19q LOH analysis (Fig. 3F). Because of the strong inverse relationship between 1p/19q LOH and 10q LOH (Fig. 2), subgroup statistical power became problematic. For example, when stratified by 1p/19q PCR-based LOH analysis, there were only 2 grade III oligodendrogliomas that were intact for both 1p/19q and 10q, and 2 other cases with concomitant 1p/19q LOH and 10q LOH. The survival of 1p/19q-intact grade III oligodendrogliomas was slightly but significantly longer when 10q was also intact (p = 0.03 for 1p/19q LOH-negative 10q-intact vs. 1p/19q LOH-negative 10q LOH), though the subgroup sizes are small. Modified FISH criteria showed similar results (not shown).
Figure 3.
Impact of 10q loss of heterozygosity (LOH) on survival in oligodendrogliomas. (A, B) 10q LOH did not significantly change overall survival in grade II gliomas (A, p = 0.12) though it did in grade III gliomas (B, *p = 0.002). (C, D) Factoring in 1p/19q status, 10q LOH still did not significantly alter survival in grade II oligodendrogliomas as assessed by 1p/19q FISH (C, p = 0.39) or PCR-based LOH (D, p = 0.27), although the number of cases with 1p/19q codeletion and 10q LOH was very small (n = 3). (E) In grade III oligodendrogliomas, 1p/19q-codeleted cases by fluorescence in situ hybridization (FISH) had better survival only if 10q was also intact (*p = 0.03 vs. all other groups). (F) 10q status did not significantly refine survival when grade III oligodendrogliomas were 1p/19q-codeleted via PCR-based LOH (p = 0.86). Survival of 1p/19q-intact grade III oligodendrogliomas was slightly but significantly longer when 10q was also intact (*p = 0.03 for 1p/19q LOH-negative (neg.) 10q-intact vs. 1p/19q LOH-neg 10q LOH).
Treatment variables, 1p/19q, and 10q
Grade III oligodendrogliomas with either gross total resection or subtotal resection showed longer overall survival (OS) compared to those that were only biopsied (p = 0.03; Supplemental Fig. 3A), as well as a trend for longer progression-free survival (PFS) (p = 0.08, not shown). In grade II oligodendrogliomas, surgery vs. biopsy showed a trend towards longer OS (p = 0.08) but not PFS (p = 0.13) (not shown). Treatment with radiation and/or chemotherapy showed a strong trend toward better OS in grade III oligodendrogliomas (p = 0.05; Supplemental Fig. 3B), as well as significantly longer PFS (p = 0.009, not shown). However, in grade II oligodendrogliomas, adjuvant therapy showed no significant correlation with PFS (p = 0.82) or OS (p = 0.63) (not shown).
Focusing only on the 18 grade III oligodendrogliomas known to have received adjuvant radiation and/or chemotherapy, 1p/19q LOH data significantly stratified for OS (p < 0.0001) and PFS (p = 0.0003), whereas original 1p/19q FISH criteria did not (OS p = 0.23, PFS p = 0.39). Modified FISH criteria improved PFS stratification (p = 0.04) but still could not quite reach significance for OS (p = 0.14). Grade III oligodendrogliomas subjected to adjuvant therapy still had worse PFS (p = 0.0007) and OS (p = 0.001) if 10q was deleted. None of the FISH criteria, 1p/19q LOH, or 10q LOH achieved statistical significance in adjuvant-treated grade II oligodendrogliomas (not shown).
Multivariate Analyses
On multivariate analysis using original 1p/19q FISH criteria, the most significant variables for longer OS and PFS were patient ages <45 years, WHO grade II, and intact 10q (Table 4). All 3 variables were also independent OS and PFS prognostic factors when applying the modified 1p/19q FISH criteria (not shown). Patient age and WHO grade showed the same power when using 1p/19q PCR-based LOH data instead of FISH, with 10q status weakening slightly to a trend toward significance. Neither resection vs. biopsy or adjuvant therapy were independent prognostic factors. Surprisingly, 1p/19q status also was not, unless 10q was excluded from the modeling, whereupon 1p/19q PCR microsatellite LOH reached significance for OS (0.04) and a trend toward improved PFS (p = 0.08). However, even after excluding 10q data, 1p/19q FISH never approached independent prognostic significance by either original (p = 0.50 and 0.98) or modified criteria (p = 0.61 and 0.37) for PFS or OS, respectively. Our prior work showed that grade III oligodendrogliomas with moderate-to strong expression of epidermal growth factor receptor (EGFR) by immunohistochemistry had better overall survival than cases in which EGFR immunohistochemistry was negative or weak (33). In 26 grade III oligodendrogliomas with known 1p/19q status, 10q status, polysomy status (according to the criteria established by Snuderl et al [34]) and EGFR expression, younger patient age, intact 10q, and moderate-to-strong EGFR staining were independent favorable prognostic variables for OS when incorporating either original or modified 1p/19q FISH criteria (Table 5). Modeling with 1p/19q PCR-based LOH data increased 1p/19q prognostic strength (though not to the point of significance) at the expense of all other variables. (Of note, there were not enough cases with these data plus treatment and progression-free survival information to expand multivariate modeling.)
Table 4.
Multivariate Analysis of Overall Survival and Progression-Free Survival in Oligodendrogliomas
| Overall survival with 1p/19q FISH criteria | Progression-free survival with 1p/19q FISH criteria | ||||
|---|---|---|---|---|---|
| Variable | Relative Risk (95% CI) | p | Variable | Relative Risk (95% CI) | p |
| age < 45 y | 0.12 (0.042—0.37) | 0.0002 | age < 45 y | 0.25 (0.11—0.60) | 0.0017 |
| GTR or STR | 0.51 (0.16—1.6) | 0.24 | GTR or STR | 0.52 (0.17—1.5) | 0.24 |
| adjuvant Rx | 0.62 (0.26—1.5) | 0.29 | adjuvant Rx | 0.58 (0.26—1.3) | 0.18 |
| WHO grade II | 0.16 (0.065—0.39) | 0.0001 | WHO grade II | 0.14 (0.060—0.34) | 0.00001 |
| 1p19q codeleted | 1.68 (0.47—6.1) | 0.42 | 1p19q codeleted | 1.8 (0.58—5.8) | 0.31 |
| 10q intact | 0.36 (0.15—0.88) | 0.026 | 10q intact | 0.41 (0.18—0.95) | 0.037 |
| Overall survival with 1p/19q PCR-based LOH | Progression-free survival with 1p/19q PCR-based LOH | ||||
| Variable | Relative Risk (95% CI) | p | Variable | Relative Risk (95% CI) | p |
| age < 45 y | 0.15 (0.048—0.44) | 0.0007 | age < 45 y | 0.27 (0.11—0.63) | 0.0025 |
| GTR or STR | 0.75 (0.30—1.9) | 0.55 | GTR or STR | 0.79 (0.32—1.9_ | 0.61 |
| adjuvant Rx | 0.75 (0.31—1.8) | 0.52 | adjuvant Rx | 0.69 (0.30—1.6) | 0.39 |
| WHO grade II | 0.13 (0.048—0.34) | 0.00001 | WHO grade II | 0.13 (0.050—0.32) | 0.00001 |
| 1p19q LOH | 0.44 (0.15—1.3) | 0.13 | 1p19q LOH | 0.59 (0.22—1.6) | 0.30 |
| 10q intact | 0.46 (0.19—1.1) | 0.09 | 10q intact | 0.51 (0.22—1.2) | 0.12 |
Sixty-four grade II and 20 grade III oligodendrogliomas were modeled by Cox Proportional Hazards according to key clinical, histologic, and molecular variables. The most powerful factors for favorable progression-free and overall survival were age <45 years, WHO grade II histology, and intact 10q (underlined). When incorporating 10q status with the multivariate analyses, 1p/19q status was not an independently prognostic variable. CI = confidence interval; FISH = fluorescence in situ hybridization; GTR = gross total resection; LOH = loss of heterozygosity; STR = subtotal resection. (Relative to the 1p/19q FISH data, similar p values were seen when using the modified 1p/19q FISH criteria, not shown.)
Table 5.
Multivariate Analysis of Overall Survival Based on Age and Molecular Variables in Grade III Oligodendrogliomas
| variable | original 1p/19q FISH criteria | modified 1p/19q FISH criteria | 1p/19q PCR LOH | |||
|---|---|---|---|---|---|---|
| Relative Risk (95% CI) | p | Relative Risk (95% CI) | p | Relative Risk (95% CI) | p | |
| age <45 years | 0.07 (0.01—0.51) | 0.009 | 0.06 (0.008—0.5) | 0.009 | 0.12 (0.02—0.9) | 0.04 |
| 10q intact | 0.27 (0.08—0.85) | 0.025 | 0.28 (0.09—0.9) | 0.03 | 0.36 (0.1—1.3) | 0.12 |
| no polysomy | 0.70 (0.19—2.5) | 0.58 | 0.61 (0.16—2.3) | 0.46 | 0.72 (0.2—2.2) | 0.56 |
| EGFR IHC moderate-to-strong | 0.27 (0.08—0.88) | 0.03 | 0.27 (0.08—0.9) | 0.03 | 0.42 (0.12—1.4) | 0.17 |
| 1p/19q deleted | 0.72 (0.17—3.0) | 0.65 | 0.53 (0.16—1.8) | 0.31 | 0.30 (0.05—1.7) | 0.18 |
Analysis of 26 grade III anaplastic oligodendrogliomas showed that younger patient age, intact 10q, and moderate-to-strong EGFR expression on IHC were independent favorable prognostic variables for overall survival when incorporating either original or modified 1p/19q FISH criteria. When using 1p/19q PCR-based LOH data instead, the only variable that retained significance was patient age. CI = confidence interval; EGFR = epidermal growth factor; FISH = fluorescence in situ hybridization; IHC = immunohistochemistry. LOH = loss of heterozygosity.
DISCUSSION
Despite its status as the most widely accepted biomarker in the workup of gliomas, 1p/19q codeletion has never been shown to be a truly predictive marker in the sense that HER2 is, wherein amplification predicts a specific response to trastuzumab in breast and upper gastrointestinal cancers. Thus, there has not been a broad-based initiative to standardize 1p/19q testing, which has resulted in remarkable interinstitutional heterogeneity of assays, probes, and cutoff criteria. For example, even when using FISH (specifically the commercially-available 1p36/1q25 and 19q13/19p13 probes from Vysis, Abbott Park, IL), many laboratories focus on 1p/1q and 19q/19p ratios in their evaluation whereas others rely solely on the percent of tumor nuclei showing relative 1p and 19q loss, with a cutoff generally around 40% (35, 36). Our original ratio cutoffs were established as 3 standard deviations beyond the ratios normally seen in non-neoplastic control tissues. To enhance the rigor of the assay, 20 non-neoplastic control samples were used. Applying hindsight, it is clear that increasing the number had the inevitable effect of decreasing standard deviations, i.e. the cutoffs were too inclusive, producing lower concordance with PCR-based LOH data and less prognostic power. Lowering the ratio cutoffs improved concordance with LOH and the prognostic power of FISH (Fig. 1).
From a biological perspective a more stringent pair of ratio cutoffs makes sense, because the unbalanced translocation that produces 1p/19q codeletion is believed to occur at a relatively early stage of gliomagenesis and should therefore be present in most if not all glioma cells. Thus, “true” 1p36/1q25 and 19q13/19p13 ratios ought to be closer to the hypothetical “pure” ratio of 0.5. In contrast, random interstitial deletions could occur at any point during tumor development and would thus be expected to show more cell-to-cell variation, with some cells in a tumor containing partial deletions but not others. It is therefore not surprising that 1p36/1q25 and 19q13/19p13 ratios tended to be higher in gliomas that had apparent codeletion by FISH but not by LOH vs. tumors with codeletion by both modalities (Supplemental Fig. 2).
There are no perfect criteria or cutoffs that produce 100% sensitivity and specificity for 1p/19q FISH (as in any diagnostic test). For example, applying the modified FISH criteria produced a better p value in survival stratification of grade III oligodendrogliomas because it avoided calling 2 cases as codeleted that were intact by LOH analysis; those patients died 2 months and 1.3 years after diagnosis. Yet it also excluded a case that was actually codeleted by original FISH criteria and by LOH analysis; that patient survived 5.3 years. Eschewing ratios altogether, instead using cutoffs of at least 40% tumor cells with relative 1p and 19q deletion, produced a comparable degree of prognostic stratification to the 0.75 ratio cutoffs in anaplastic oligodendrogliomas (p = 0.009 for % deleted vs. 0.007 for ratios, data not shown). Furthermore, both the modified ratios and ≥40% relative deletion criteria had 95% concordance with each other in all oligodendrogliomas. Thus, it probably does not matter whether one employs ratios or % deletion because they produce essentially the same overall results. What seems more important is that these cutoffs have been determined in this study using prognostic stratification and whole-arm assays as the primary litmus tests rather than relying on non-neoplastic tissues.
Although in most cases there was good concordance between FISH and PCR-based LOH, in this cohort 1p/19q PCR-based LOH showed stronger prognostic value in grade III anaplastic oligodendrogliomas. That neither test was able to significantly stratify grade II oligodendrogliomas (Fig. 1B, C) is not surprising because 1p/19q codeletion appears to have less prognostic impact in low-grade gliomas and may only show significance with prolonged follow-up intervals (8, 37, 38). Indeed, an analysis of our grade II subset suggested insufficient power (0.67) to stratify by 1p/19q status given our cohort size, median follow-up interval and the relatively longer survival in this subgroup. On the other hand, because grade III tumors have shorter survival overall and a much sharper survival difference by 1p/19q (or 10q) status, the subset of 32 cases was adequately powered (0.91), Still, testing for codeletion at least has diagnostic value, particularly in gliomas in which morphologic features are equivocal between astrocytoma and oligodendroglioma (19).
Oligodendrogliomas with true 1p/19q codeletion but 10q LOH are rare to begin with and sometimes have fewer classic features of oligodendrogliomas, suggesting that the diagnosis of oligodendroglioma in the face of 10q deletion may be reconsidered (7, 25, 27, 39–42). Our present results support and extend these findings, specifically making the critical point that most cases called “codeleted” by FISH are false-positives if 10q is also deleted. Whereas rare gliomas do indeed have concomitant whole-arm 1p/19q codeletion and 10q LOH, many so-called 1p/19q-codeleted gliomas are likely not truly whole-arm codeleted when 10q is also lost (Fig. 2; Table 3).
Prior work showed that increased EGFR expression is paradoxically correlated with improved survival in anaplastic oligodendrogliomas (33, 43). The current data reconfirm this on multivariate analysis (Table 5). The reasons for this are not clear but it is not due to nonrandom segregation of 1p/19q LOH (p = 0.46), 10q LOH (p = 0.54), or younger patient age (p = 0.41).
Although PCR-based LOH is more powerful than FISH, and assays such as single nucleotide polymorphism, array comparative genomic hybridization, and multiplex ligation-dependent probe amplification show great promise (44–47), our results indicate that it is likely that FISH will remain a widely-used technique in the workup of gliomas. Standardized protocols for 1p/19q FISH have recently been recommended (36), which should be of great assistance to laboratories worldwide; however, universal standardization of 1p/19q FISH probe loci, interpretation criteria and reporting criteria are also needed. Based on our data, we suggest the following scoring and reporting parameters for 1p/19q FISH: 1) If using 1p36/1q25 and 19q13/19p13 ratios, they should each be <0.75. 2) If using relative deletion, both 1p36 and 19q13 should be deleted in at least 40% of cells. 3)10q interrogation, either by PCR analysis or FISH (e.g. testing for PTEN deletion) should be done in cases where 1p/19q FISH is positive or equivocal for codeletion, and if 10q is deleted, the 1p/19q data should be interpreted with great caution; Indeed, it may even be advisable to test 10q status in oligodendrogliomas regardless of the 1p/19q results, as it may be an independent adverse prognostic factor (Tables 4, 5).
These are useful parameters to serve as a starting point for the development of universal scoring and reporting criteria for 1p/19q FISH. Once such standards are in place, they will enhance the quality of prognostic information provided to clinicians and patients. They will also improve inter-institutional reproducibility of glioma research, especially work that focuses on 1p/19q codeletion, 10q deletion, and their roles in glioma biology.
Supplementary Material
Acknowledgments
Craig Horbinski was supported by a University of Kentucky College of Medicine Physician-Scientist Fellowship and NIH 1K08CA155764-01A1.
The authors thank the In Situ Hybridization Laboratory at the University of Pittsburgh for their assistance with the FISH data and the Molecular Anatomic Pathology Laboratory at the University of Pittsburgh for their assistance with the LOH data. The authors also acknowledge the assistance of Sharon Winters of the Hillman Cancer Registry in providing clinical follow-up data. Portions of this work were presented at the 87th Annual Meeting of the American Association of Neuropathologists, Seattle, WA on June 25, 2011.
References
- 1.Reifenberger J, Reifenberger G, Liu L, et al. Molecular genetic analysis of oligodendroglial tumors shows preferential allelic deletions on 19q and 1p. Am J Pathol. 1994;145:1175–90. [PMC free article] [PubMed] [Google Scholar]
- 2.Kraus JA, Koopmann J, Kaskel P, et al. Shared allelic losses on chromosomes 1p and 19q suggest a common origin of oligodendroglioma and oligoastrocytoma. J Neuropathol Exp Neurol. 1995;54:91–5. doi: 10.1097/00005072-199501000-00011. [DOI] [PubMed] [Google Scholar]
- 3.Cairncross JG, Ueki K, Zlatescu MC, et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst. 1998;90:1473–9. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- 4.Smith JS, Perry A, Borell TJ, et al. Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J Clin Oncol. 2000;18:636–45. doi: 10.1200/JCO.2000.18.3.636. [DOI] [PubMed] [Google Scholar]
- 5.Perry A, Fuller CE, Banerjee R, et al. Ancillary FISH analysis for 1p and 19q status: preliminary observations in 287 gliomas and oligodendroglioma mimics. Front Biosci. 2003;8:1–9. doi: 10.2741/896. [DOI] [PubMed] [Google Scholar]
- 6.van den Bent MJ, Looijenga LH, Langenberg K, et al. Chromosomal anomalies in oligodendroglial tumors are correlated with clinical features. Cancer. 2003;97:1276–84. doi: 10.1002/cncr.11187. [DOI] [PubMed] [Google Scholar]
- 7.Fallon KB. Prognostic value of 1p, 19q, 9p, 10q, and EGFR-FISH analyses in recurrent oligodendrogliomas. J Neuropathol Exp Neurol. 2004;63:314–22. doi: 10.1093/jnen/63.4.314. [DOI] [PubMed] [Google Scholar]
- 8.Felsberg J, Erkwoh A, Sabel MC, et al. Oligodendroglial tumors: refinement of candidate regions on chromosome arm 1p and correlation of 1p/19q status with survival. Brain Pathol. 2004;14:121–30. doi: 10.1111/j.1750-3639.2004.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kouwenhoven MC, Gorlia T, Kros JM, et al. Molecular analysis of anaplastic oligodendroglial tumors in a prospective randomized study: A report from EORTC study 26951. Neuro Oncol. 2009;11:737–46. doi: 10.1215/15228517-2009-011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jenkins RB. A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res. 2006;66:9852–61. doi: 10.1158/0008-5472.CAN-06-1796. [DOI] [PubMed] [Google Scholar]
- 11.Griffin CA. Identification of der(1;19)(q10;p10) in five oligodendrogliomas suggests mechanism of concurrent 1p and 19q loss. J Neuropathol Exp Neurol. 2006;65:988–94. doi: 10.1097/01.jnen.0000235122.98052.8f. [DOI] [PubMed] [Google Scholar]
- 12.Labussiere M, Idbaih A, Wang XW, et al. All the 1p19q codeleted gliomas are mutated on IDH1 or IDH2. Neurology. 2010;74:1886–90. doi: 10.1212/WNL.0b013e3181e1cf3a. [DOI] [PubMed] [Google Scholar]
- 13.van den Bent MJ, Dubbink HJ, Marie Y, et al. IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tumors: a report of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Clin Cancer Res. 2010;16:1597–604. doi: 10.1158/1078-0432.CCR-09-2902. [DOI] [PubMed] [Google Scholar]
- 14.Ducray F, Idbaih A, de Reynies A, et al. Anaplastic oligodendrogliomas with 1p19q codeletion have a proneural gene expression profile. Mol Cancer. 2008;7:41. doi: 10.1186/1476-4598-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Idbaih A, Marie Y, Lucchesi C, et al. BAC array CGH distinguishes mutually exclusive alterations that define clinicogenetic subtypes of gliomas. Int J Cancer. 2008;122:1778–86. doi: 10.1002/ijc.23270. [DOI] [PubMed] [Google Scholar]
- 16.Giannini C, Burger PC, Berkey BA, et al. Anaplastic oligodendroglial tumors: refining the correlation among histopathology, 1p 19q deletion and clinical outcome in Intergroup Radiation Therapy Oncology Group Trial 9402. Brain Pathol. 2008;18:360–9. doi: 10.1111/j.1750-3639.2008.00129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDonald JM, See SJ, Tremont IW, et al. The prognostic impact of histology and 1p/19q status in anaplastic oligodendroglial tumors. Cancer. 2005;104:1468–77. doi: 10.1002/cncr.21338. [DOI] [PubMed] [Google Scholar]
- 18.Eoli M, Bissola L, Bruzzone MG, et al. Reclassification of oligoastrocytomas by loss of heterozygosity studies. Int J Cancer. 2006;119:84–90. doi: 10.1002/ijc.21759. [DOI] [PubMed] [Google Scholar]
- 19.Fuller CE, Schmidt RE, Roth KA, et al. Clinical utility of fluorescence in situ hybridization (FISH) in morphologically ambiguous gliomas with hybrid oligodendroglial/astrocytic features. J Neuropathol Exp Neurol. 2003;62:1118–28. doi: 10.1093/jnen/62.11.1118. [DOI] [PubMed] [Google Scholar]
- 20.Raghavan R, Balani J, Perry A, et al. Pediatric oligodendrogliomas: a study of molecular alterations on 1p and 19q using fluorescence in situ hybridization. J Neuropathol Exp Neurol. 2003;62:530–7. doi: 10.1093/jnen/62.5.530. [DOI] [PubMed] [Google Scholar]
- 21.Kreiger PA, Okada Y, Simon S, et al. Losses of chromosomes 1p and 19q are rare in pediatric oligodendrogliomas. Acta Neuropathol. 2005;109:387–92. doi: 10.1007/s00401-004-0976-2. [DOI] [PubMed] [Google Scholar]
- 22.Barbashina V, Salazar P, Holland EC, et al. Allelic losses at 1p36 and 19q13 in gliomas: correlation with histologic classification, definition of a 150-kb minimal deleted region on 1p36, and evaluation of CAMTA1 as a candidate tumor suppressor gene. Clin Cancer Res. 2005;11:1119–28. [PubMed] [Google Scholar]
- 23.Smith JS, Alderete B, Minn Y, et al. Localization of common deletion regions on 1p and 19q in human gliomas and their association with histological subtype. Oncogene. 1999;18:4144–52. doi: 10.1038/sj.onc.1202759. [DOI] [PubMed] [Google Scholar]
- 24.Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J Neuropathol Exp Neurol. 2005;64:479–89. doi: 10.1093/jnen/64.6.479. [DOI] [PubMed] [Google Scholar]
- 25.Miller CR, Dunham CP, Scheithauer BW, et al. Significance of necrosis in grading of oligodendroglial neoplasms: a clinicopathologic and genetic study of newly diagnosed high-grade gliomas. J Clin Oncol. 2006;24:5419–26. doi: 10.1200/JCO.2006.08.1497. [DOI] [PubMed] [Google Scholar]
- 26.Abounader R. Interactions between PTEN and receptor tyrosine kinase pathways and their implications for glioma therapy. Expert Rev Anticancer Ther. 2009;9:235–45. doi: 10.1586/14737140.9.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sasaki H, Zlatescu MC, Betensky RA, et al. PTEN is a target of chromosome 10q loss in anaplastic oligodendrogliomas and PTEN alterations are associated with poor prognosis. Am J Pathol. 2001;159:359–67. doi: 10.1016/S0002-9440(10)61702-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramirez C, Bowman C, Maurage CA, et al. Loss of 1p, 19q, and 10q heterozygosity prospectively predicts prognosis of oligodendroglial tumors--towards individualized tumor treatment? Neuro Oncol. 2010;12:490–9. doi: 10.1093/neuonc/nop071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jha P, Sarkar C, Pathak P, et al. Detection of allelic status of 1p and 19q by microsatellite-based PCR versus FISH: limitations and advantages in application to patient management. Diagn Mol Pathol. 2011;20:40–7. doi: 10.1097/PDM.0b013e3181e961e9. [DOI] [PubMed] [Google Scholar]
- 30.Broholm H, Born PW, Guterbaum D, et al. Detecting chromosomal alterations at 1p and 19q by FISH and DNA fragment analysis--a comparative study in human gliomas. Clin Neuropathol. 2008;27:378–87. doi: 10.5414/npp27378. [DOI] [PubMed] [Google Scholar]
- 31.Horbinski C. Practical molecular diagnostics in neuropathology: making a tough job a little easier. Semin Diagn Pathol. 2010;27:105–13. doi: 10.1053/j.semdp.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 32.Horbinski C, Hamilton RL, Nikiforov Y, et al. Association of molecular alterations, including BRAF, with biology and outcome in pilocytic astrocytomas. Acta Neuropathol. 2010;119:641–9. doi: 10.1007/s00401-009-0634-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horbinski C, Hobbs J, Cieply K, et al. EGFR expression modulates oligodendroglioma behavior. Am J Pathol. 2011 Oct;179:1638–44. doi: 10.1016/j.ajpath.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snuderl M, Eichler AF, Ligon Kl, et al. Polysomy for chromosomes 1 and 19 predicts earlier recurrence in anaplastic oligodendrogliomas with concurrent 1p/19q loss. Clin Cancer Res. 2009;15:6430–7. doi: 10.1158/1078-0432.CCR-09-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horbinski C, Miller CR, Perry A. Gone FISHing: clinical lessons learned in brain tumor molecular diagnostics over the last decade. Brain Pathol. 2010;21:57–73. doi: 10.1111/j.1750-3639.2010.00453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woehrer A, Sander P, Haberler C, et al. FISH-based detection of 1p 19q codeletion in oligodendroglial tumors: procedures and protocols for neuropathological practice - a publication under the auspices of the Research Committee of the European Confederation of Neuropathological Societies. Clin Neuropathol. 2011;30:47–55. doi: 10.5414/npp30047. [DOI] [PubMed] [Google Scholar]
- 37.Hashimoto N. Correlation between genetic alteration and long-term clinical outcome of patients with oligodendroglial tumors, with identification of a consistent region of deletion on chromosome arm 1p. Cancer. 2003;97:2254–61. doi: 10.1002/cncr.11322. [DOI] [PubMed] [Google Scholar]
- 38.Okamoto Y. Population-based study on incidence, survival rates, and genetic alterations of low-grade diffuse astrocytomas and oligodendrogliomas. Acta Neuropathol. 2004;108:49–56. doi: 10.1007/s00401-004-0861-z. [DOI] [PubMed] [Google Scholar]
- 39.Ueki K, Nishikawa R, Nakazato Y, et al. Correlation of histology and molecular genetic analysis of 1p, 19q, 10q, TP53, EGFR, CDK4, and CDKN2A in 91 astrocytic and oligodendroglial tumors. Clin Cancer Res. 2002;8:196–201. [PubMed] [Google Scholar]
- 40.Perry A, Aldape KD, George DH, et al. Small cell astrocytoma: an aggressive variant that is clinicopathologically and genetically distinct from anaplastic oligodendroglioma. Cancer. 2004;101:2318–26. doi: 10.1002/cncr.20625. [DOI] [PubMed] [Google Scholar]
- 41.Kitange G, Misra A, Law M, et al. Chromosomal imbalances detected by array comparative genomic hybridization in human oligodendrogliomas and mixed oligoastrocytomas. Genes Chromosomes Cancer. 2005;42:68–77. doi: 10.1002/gcc.20108. [DOI] [PubMed] [Google Scholar]
- 42.Lavon I. Longitudinal assessment of genetic and epigenetic markers in oligodendrogliomas. Clin Cancer Res. 2007;13:1429–37. doi: 10.1158/1078-0432.CCR-06-2050. [DOI] [PubMed] [Google Scholar]
- 43.Zhou Y-H, Hess KR, Raj VR, et al. Establishment of prognostic models for astrocytic and oligodendroglial brain tumors with standardized quantification of marker gene expression and clinical variables. Biomarker Insights. 2010;5:153–68. doi: 10.4137/BMI.S6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cowell JK, Barnett GH, Nowak NJ. Characterization of the 1p/19q chromosomal loss in oligodendrogliomas using comparative genomic hybridization arrays (CGHa) J Neuropathol Exp Neurol. 2004;63:151–8. doi: 10.1093/jnen/63.2.151. [DOI] [PubMed] [Google Scholar]
- 45.Alvarez K, Kash SF, Lyons-Weiler MA, et al. Reproducibility and performance of virtual karyotyping with SNP microarrays for the detection of chromosomal imbalances in formalin-fixed paraffin-embedded tissues. Diagn Mol Pathol. 2010;19:127–34. doi: 10.1097/PDM.0b013e3181d527c5. [DOI] [PubMed] [Google Scholar]
- 46.Buckley PG, Alcock L, Heffernan J, et al. Loss of chromosome 1p/19q in oligodendroglial tumors: refinement of chromosomal critical regions and evaluation of internexin immunostaining as a surrogate marker. J Neuropathol Exp Neurol. 2011;70:177–82. doi: 10.1097/NEN.0b013e31820c765b. [DOI] [PubMed] [Google Scholar]
- 47.Franco-Hernandez C, Martinez-Glez V, de Campos JM, et al. Allelic status of 1p and 19q in oligodendrogliomas and glioblastomas: multiplex ligation-dependent probe amplification versus loss of heterozygosity. Cancer Genet Cytogenet. 2009;190:93–6. doi: 10.1016/j.cancergencyto.2008.09.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.