Abstract
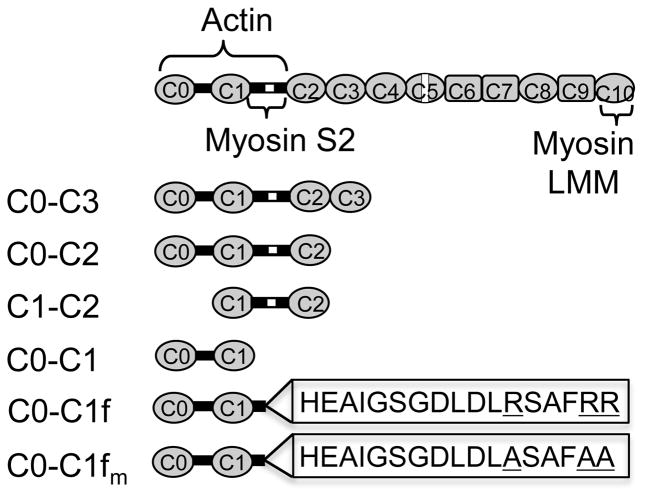
Cardiac myosin binding protein-C (cMyBP-C) has 11 immunoglobulin or fibronectin-like domains, C0 through C10, which bind sarcomeric proteins, including titin, myosin and actin. Using bacterial expressed mouse N-terminal fragments (C0 through C3) in an in vitro motility assay of myosin-generated actin movement and the laser trap assay to assess single molecule actin-binding capacity, we determined that the first N-terminal 17 amino acids of the cMyBP-C motif (the linker between C1 and C2) contain a strong, stereospecific actin-binding site that depends on positive charge due to a cluster of arginines. Phosphorylation of 4 serines within the motif decreases the fragments’ actin-binding capacity and actomyosin inhibition. Using the laser trap assay, we observed individual cMyBP-C fragments transiently binding to a single actin filament with both short (~20ms) and long (~300ms) attached lifetimes, similar to that of a known actin-binding protein, α-actinin. These experiments suggest that cMyBP-C N-terminal domains containing the cMyBP-C motif tether actin filaments and provide one mechanism by which cMyBP-C modulates actomyosin motion generation, i.e. by imposing an effective viscous load within the sarcomere.
Keywords: single molecule biophysics, laser trap, PKA phosphorylation, contractile proteins, contractility, heart
Introduction
Myosin binding protein-C (MyBP-C) is a striated muscle, thick filament associated protein [1]. Its importance is emphasized by the development of familial hypertrophic or dilated cardiomyopathy caused by mutations in the cardiac isoform (cMyBP-C) [2–4]. MyBP-C exists in a repeating pattern of 7–9 bands within the C-zones of the sarcomere [5] with each cMyBP-C containing 11 immunoglobulin or fibronectin-like domains, C0 through C10 (Figure 1) [6]. Functional roles have been proposed for distinct regions within the protein based on its multiple contractile protein binding partners: myosin [7], myosin regulatory light chain [8], titin [9], and actin [10]. However, the physiological importance of its in vitro binding to actin has been called into question with recent evidence that this binding is non-specific and that cMyBP-C phosphorylation, a potential regulator of cMyBP-C function in vivo, has no effect on such binding [11]. Here we used single molecule biophysical techniques (i.e. in vitro motility and laser trap assays) to directly confirm that the N-terminus of cMyBP-C binds actin stereospecifically, as proposed by others [12, 13], and that this binding is modulated by cMyBP-C phosphorylation.
Figure 1.
Schematic of whole cMyBP-C and expressed N-terminal fragments. Immunoglobulin I-like domains are oval, while fibronectin-3 domains are rectangular. cMyBP-C contains several cardiac specific sequences: the C0 domain, and inserts within the MyBP-C motif and the C5 domain (represented as white boxes). C0-C1f contains the first N-terminal 17 amino acids of the cMyBP-C motif, whose sequence is represented in the box. C0-C1fm is identical to C0-C1f, except for three mutations (R266A, R270A and R271A).
The impact of MyBP-C on muscle function was derived from early studies in skinned skeletal muscle fibers where MyBP-C had been chemically extracted [14, 15]. Upon its removal, faster shortening velocities and elevated force production at low calcium levels were observed, which was interpreted as MyBP-C imposing an internal load and limiting myosin head mobility. This effect could be a result of cMyBP-C’s capacity to form a link between the myosin head and thick filament backbone through its N-terminal binding to the myosin S2 region and its C-terminal binding to the myosin LMM [4]. With the N-terminus of cMyBP-C also capable of binding actin [16, 17], an equally plausible model is that cMyBP-C forms a tether between the thick and thin filaments. Such a link could modulate actomyosin activity, by competing with myosin for actin-binding sites, by altering muscle activation by modulating tropomyosin movement on the thin filament, or by simply imposing an internal load to muscle shortening.
Phosphorylation of cMyBP-C in response to β-adrenergic stimulation is critically important to the dynamic contractile regulation of the heart [4]. Phosphorylation occurs within the linker between domains C1 and C2 (i.e. MyBP-C motif) at 4 cardiac isoform-specific serines [18, 19]. Recent studies suggest that cMyBP-C phosphorylation is reduced in humans with hypertrophic cardiomyopathy or end-stage heart failure [20–22]. In fact, transgenic mice expressing non-phosphorylatable alanine substitutions for serines in the cMyBP-C motif in a cMyBP-C null background, exhibited a hypertrophied phenotype [23, 24], whereas substitution with aspartic acids as phosphomimetics protected the heart from ischemic reperfusion injury [25]. These data underscore the physiologic importance of cMyBP-C phosphorylation. Phosphorylation by protein kinase A (PKA) in vitro diminishes the binding affinity of the motif for actin as well as myosin [17, 26], although the effect of phosphorylation on actin-binding was recently challenged [11]. Therefore, phosphorylation might eliminate cMyBP-C’s role as a tether and partially account for the enhanced cardiac contractility observed with adrenergic stimulation.
Using a combination of expressed mouse cMyBP-C fragments and single molecule biophysical approaches, we identified potential site(s) within the first 4 N-terminal domains (C0-C3) responsible for actin-binding and the regulatory effect phosphorylation has on this interaction. Through structural deletions and amino acid substitutions, we identified an actin-binding site within the first 17 amino acids of the cMyBP-C motif following the C1 domain that may interact with actin in a stereospecific manner. All N-terminal fragments studied that contain these 17 amino acids inhibit actomyosin motility to the same extent as we previously reported for purified full length cMyBP-C [27]. Based on direct evidence that the N-terminal fragments transiently bind to a single actin filament in the laser trap assay, we propose that one mode by which cMyBP-C exerts its mechanical effects on actomyosin power generation is by creating a physical link between the thick and thin filaments that imposes an internal viscous load within the sarcomere.
Materials and Methods
Proteins
Actin and myosin were prepared from chicken pectoralis [28, 29]. Actin filaments were fluorescently labeled with rhodamine phalloidin [30]. cMyBP-C fragments were bacterially expressed from mouse cardiac cDNA using a pET expression system (Novagen, Madison, WI) [23]. Six expressed N-terminal fragments were studied (Fig. 1): C0-C1 (amino acids 1–254), C0-C2 (1–448), C0-C3 (1–539), C1-C2 (151–448), C0-C1f (1–271) and C0-C1fm (1–271, with three mutations: R266A, R270A, and R271A). In a recent report, many of these same fragments were similarly expressed, characterized for purity, and then their ability to decorate isolated actin filaments determined [13]. Fragments were phosphorylated by incubating with 1 unit/μl PKA (Sigma) at 30°C for 20 min then on ice overnight in 2 mM DTT, 50 mM MOPS, 1 mM MgCl2 and 2 mM ATP- pH 7.0. Phosphorylation was qualitatively verified by Pro-Q Diamond phosphostaining (Invitrogen) and gels stained using Simply Blue to confirm equivalent peptide loads. Phosphorylated amino acids were identified quantitatively by electron ionization (ESI) liquid chromatography-mass spectrometry (LC-MS) (see Supplementary Material for details) [31].
In vitro motility assay
The in vitro motility assay was described previously [27]. In brief, myosin (100 μg/ml) was added to a nitrocellulose-coated flowcell and subsequently blocked with bovine serum albumin (BSA). Then unlabeled actin was infused with a 1 mM ATP buffer, eliminating damaged myosin that avidly binds actin in an ATP-insensitive manner [32]. Next fluorescently labeled actin was incubated for 1 min and then exchanged with the final motility buffer (1 mM ATP, 25 mM Imidazole, 1 mM EGTA, 4 mM MgCl2, 10 mM DTT, 25 mM KCl and 0.5% methylcellulose, pH 7.4 - unless noted) containing an N-terminal fragment. Actin filament motility was imaged with a Nikon Eclipse TE 2000 epifluorescent microscope at 30° C. Motility was recorded and analyzed using the Motion Analysis System VP110. In general, a minimum of 300 tracks were analyzed per experiment with tracks discarded if appearing in fewer than 10 frames or deemed non-continuous, as defined by the velocity’s ratio of standard deviation/mean > 0.5.
Actin-binding assays
To visualize cMyBP-C’s ability to bind actin, we adhered N-terminal fragments to the motility surface by two different approaches in order to answer two different but related questions. The first question relates to whether actin-binding was modulated by phosphorylation. In this assay, fragments were adhered directly to the nitrocellulose-coated coverslips. C0-C3 or phosphorylated C0-C3 was added to the flowcell first for 2 min, followed by a 5 min BSA incubation. Vortexed, shredded fluorescent actin (10 nM) was added for 5 min followed by 6 actin buffer washes. Actin-binding to the cMyBP-C fragment-coated surface was determined by image analysis (ImageJ, NIH) as follows. The intensity threshold command was used to make the first frame from each recording binary, such that filaments were white and the background black. The images were thresholded so that the ratio between the actin fluorescence signal and background was at least 3:1. Then the number of spots was counted using the ImageJ Analyze Particles command. The Analyze Particles settings were as follows: minimum pixel size of 4 to screen out noise, and circularity between 0.7 and 1 to remove large aggregates of actin.
The second question relates to the fragments’ ability to inhibit actomyosin motility. In the motility assay, fragments were added to the final buffer after the nitrocellulose-coated coverslip was blocked with BSA. If fragments inhibit actomyosin motility by acting as a tether between the actin filament and the BSA-coated surface independent of myosin being present on the motility surface, then fragments must be able to bind simultaneously at one end to BSA and the other end to an actin filament. To demonstrate such binding, nitrocellulose-coated coverslips were incubated with 0.5 mg/ml BSA for 30 seconds followed by a 3 min fragment incubation. The flowcell was then washed 3 times with actin buffer (25 mM KCl, 25 mM Imidazole, 1 mM EGTA, 4 mM MgCl2, 10 mM DTT) to remove unbound cMyBP-C fragment. Then 10 nM fragmented (vortexed and sonicated), fluorescent actin was added in actin buffer for 3 min followed by 6 actin buffer rinses to remove unbound actin. Actin-binding to the cMyBP-C fragment-coated surface was determined by image analysis (ImageJ, NIH) as described above. To ensure that the BSA incubation time provided complete blocking of the nitrocellulose surface, both longer incubation times (5 min) and higher BSA concentrations (2 mg/ml) were characterized with no effect on actin-binding observed (data not shown), confirming complete BSA surface coverage under the conditions reported here.
Laser trap assay
The laser trap assay was performed as described previously [33]. Briefly, cMyBP-C fragments (1–2 nM C0-C3, 10–15 nM C0-C1f, 12–16 nM C0-C1fm: concentrations were chosen such that data were obtained from approximately one in 10 surface beads in order to ensure single molecule interactions) were incubated for 2 min in a nitrocellulose-coated flowcell with 3 μm beads as pedestals. The actin-binding protein, α-actinin (Cytoskeleton, Inc, Denver, CO), was examined at 10 pM for comparison to cMyBP-C fragments. The flowcell was then blocked with BSA for 10–15 min. The C0-C1 data were obtained slightly differently. When C0-C1 was attached directly to the nitrocellulose-coated surface before blocking with BSA, the fragment no longer interacted with actin. Knowing that actin-binding does occur in the motility assay when C0-C1 is adhered to a BSA-coated surface (Fig. S4), we coated the laser trap assay surface with BSA before 0.2 μM C0-C1 was added to the flowcell. The flowcell was washed afterwards to remove any C0-C1 from solution.
To capture actin filaments, 1 μm silica beads were incubated overnight in 10 mg/ml neutravidin, then washed, spun at 8200 g, and resuspended 6 times in actin buffer. Biotinylated actin was made by incubating 1 μM filamentous actin with 0.5 μM TRITC-phalloidin and 0.5 μM biotin-phalloidin (Invitrogen, Carlsbad, CA). Biotinylated actin was strung between two trapped neutravidin-coated beads and lowered to the pedestal surface. Actin position data were acquired with a quadrant photodiode detector at room temperature.
The data were analyzed by Mean-Variance (MV) analysis (see Fig. 4) to determine attachment time (ton), the number of events, and the binding event amplitude [33]. The short event ton was determined by MV analysis on sections of the data traces with no long (>200 ms) events. Then an average of the short ton from each trace was calculated. The number and duration of long events were measured by eye, as events were too infrequent for MV analysis [34]. The long event ton was determined from a single exponential fit to an event duration histogram. Event frequency was determined by dividing the number of events within a data trace by the total trace time.
Figure 4.
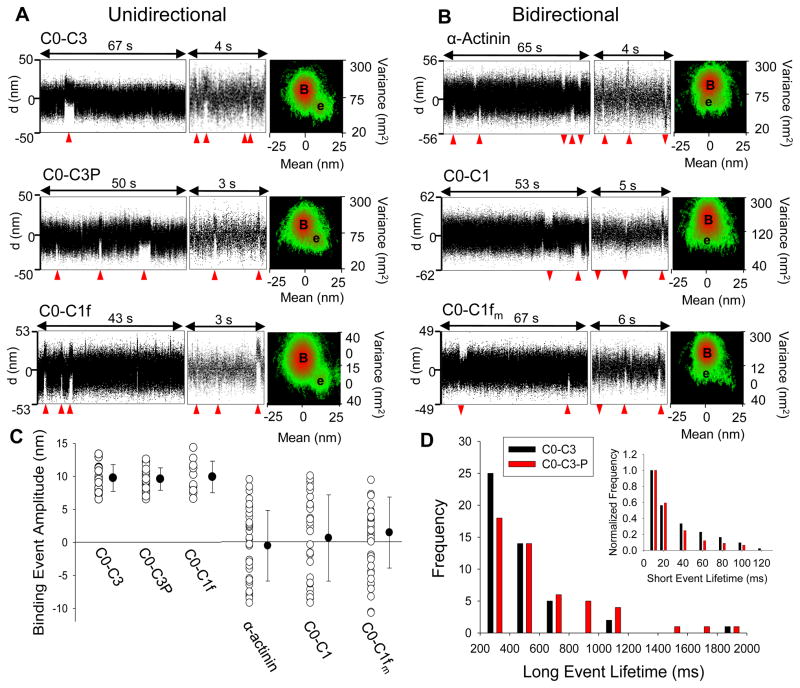
Single molecule cMyBP-C fragment laser trap data. A) Sample C0-C3, C0-C3P, and C0-C1f actin binding traces- all of which display unidirectional events. The leftmost images contain the entire data trace, in which long timescale events (denoted by red triangles) are noticeable. At a shorter time scale (center image), many short lifetime events are observed. At right is a mean-variance (MV) histogram generated from the entire data trace. In the MV, B denotes the center of the baseline population with “e” for the event population which is shifted to lower variance and centered at 10nm. B) Sample α-actinin, C0-C1, and C0-C1fm actin-binding traces. Note both the long and short bidirectional events, which are displayed in the MV as a broadly distributed event amplitude population centered at 0 nm, in contrast to the unidirectional events in A. C) Binding event amplitudes for C0-C3, C0-C3P, C0-C1f, α-actinin, C0-C1, and C0-C1fm. Open circles are mean amplitudes for individual data traces and closed circles represent mean ± standard deviation for all experiments. D) Histogram of long event lifetimes (≥200 ms) for all C0-C3 and phosphorylated C0-C3 (C0-C3-P) data traces. The inset is a histogram of short events (≤200 ms) from two sample C0-C3 or C0-C3-P data traces, normalized so that the frequency of the first bin is equal to 1 for comparison.
Statistics
Significance was determined using either a Student’s t-test for two data sets or a one-way ANOVA with Bonferroni correction for multiple comparisons. A rank sum test was used to compare data sets that were not normally distributed (Sigmastat). Data were significantly different for p ≤ 0.05. Data are reported as mean ± standard error. Experiments were repeated a minimum of 3 times.
Results
cMyBP-C and its N-terminal fragments in the in vitro motility assay
N-terminal domains are responsible for cMyBP-C’s inhibition of actin filament motility
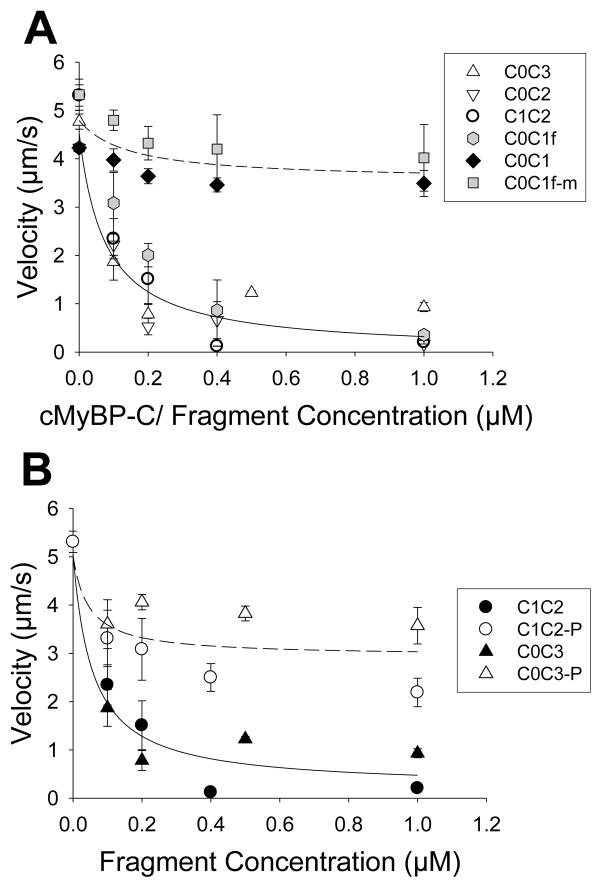
Previously, we demonstrated that whole cMyBP-C inhibited actin filament sliding velocity in a concentration-dependent manner in the in vitro motility assay, a model system for unloaded shortening velocity in muscle [27]. To define the minimal structural unit within the N-terminus of cMyBP-C that is responsible for this inhibition, we characterized several fragments with differing combinations of domains and linkers between C0 and C3 (Fig. 1, 2A). The two longest fragments, C0-C3 and C0-C2, reduced actin sliding velocities in a concentration-dependent manner to nearly the same extent (i.e. 80±5% and 94±1% at 1 μM, respectively) as we reported previously for whole cMyBP-C (i.e. 100% at 0.8 μM) [27]. This inhibition was characterized by a reduced number of moving filaments as velocity decreased and those that did move, moved slowly (data not shown). These data suggest that these N-terminal domains contain the cMyBP-C elements necessary to inhibit actin filament motility. However, further structural deletion of the fragments to C0-C1, by removal of the motif and the C2 domain, resulted in only modest inhibition (i.e. 17±6% at 1 μM), suggesting that an essential structural element was eliminated in the C0-C1 fragment. This element appears to be the first 17 N-terminal amino acids of the cMyBP-C motif, since the C0-C1f fragment, which contains these additional 17 amino acids, restored the inhibitory effects, reducing actin velocity 91±2% at 1 μM. Therefore, it was not surprising that the C1-C2 fragment, which contains these 17 amino acids was equally capable of inhibiting motility (Fig. 2A).
Figure 2.
Actin sliding velocity inhibition by N-terminal fragments (mean ± standard error, n=3). A) The solid line is a hyperbolic decay fit to all data excepting C0-C1 and C0-C1fm, and the dotted line the fit only to C0-C1 and C0-C1fm. The fits are presented as visual guides. B) Effect of N-terminal fragment phosphorylation on these fragments’ inhibition of actomyosin motility. The solid line is a hyperobolic decay fit to unphosphorylated C0-C3 and C1-C2, while the dotted line is also a hyperbolic decay fit to phosphorylated C0-C3 and C1-C2. The fits are presented as visual guides.
Closer inspection of the 17 amino acid sequence (HEAIGSGDLDLRSAFRR) reveals a patch of positive charge associated with three arginines at the C-terminus. If fragment binding to either actin and/or myosin requires these three charged residues to inhibit motility, then mutating these to alanines should have a profound effect. In fact, such a mutant of C0-C1f (C0-C1fm) was expressed and exhibited hardly any inhibition as was the case for C0-C1, which is devoid of the 17 amino acids (Fig. 2A).
Phosphorylation modulates cMyBP-C fragments’ inhibition of actomyosin motility
To determine if phosphorylation within the motif can modulate the N-terminal fragment’s inhibitory capacity in the in vitro motility assay, we added PKA-phosphorylated C0-C3 and C1-C2. Phosphorylation was confirmed by Pro-Q Diamond phosphostained gels of C0-C3 (Fig. S3) and C1-C2 (data not shown). Phosphorylated amino acids were identified and quantified at several sites within the PKA-treated C0-C3 by label-free liquid chromatography-mass spectrometry (see Supplementary Material for details) [31]. PKA treatment resulted in phosphorylation of serines 273 (100±0%), 282 (99±0%), 302 (100±0%), and 307 (84±1%) (n=4) (see Table 1). Based on combinatorial statistics using the determined percent phosphorylation at each site, 83% of the PKA-treated C0-C3 fragments are predicted to be 100% phosphorylated at all four serines.
Table 1.
Tryptic phosphopeptides observed and percent phosphorylation in PKA-treated C0-C3 protein fragment.
| Phosphorylated Amino Acid | Phosphopeptide Observed | m/z | Phosphorylation (% ± SD, n=4) |
|---|---|---|---|
| Serine 273 | 272TSpLAGAGR279 | 406.7 | 100 ± 0 |
| 271RTSpLAGAGR279 | 484.7 | ||
| Serine 282 | 281TSpDSHEDAGTLDFSSLLK298 | 1002.0 | 99 ± 0 |
| 280RTSpDSHEDAGTLDFSSLLK298 | 1080.0 | ||
| Serine 302 | 300RDSpFR304 | 760.3 | 100 ± 0 |
| 299KRDSpFR304 | 444.7 | ||
| Serine 307 | 306DSpKLEAPAEEDVWEILR322 | 1040.4 | 84 ± 1 |
| 305RDSpKLEAPAEEDVWEILR322 | 1118.5 |
Phosphorylation reduced these fragment’s ability to inhibit actin velocities approximately 3–4-fold, resulting in a level of inhibition similar to that of C0-C1 and C0-C1fm (Fig. 2A,B).
N-terminal fragments bind actin
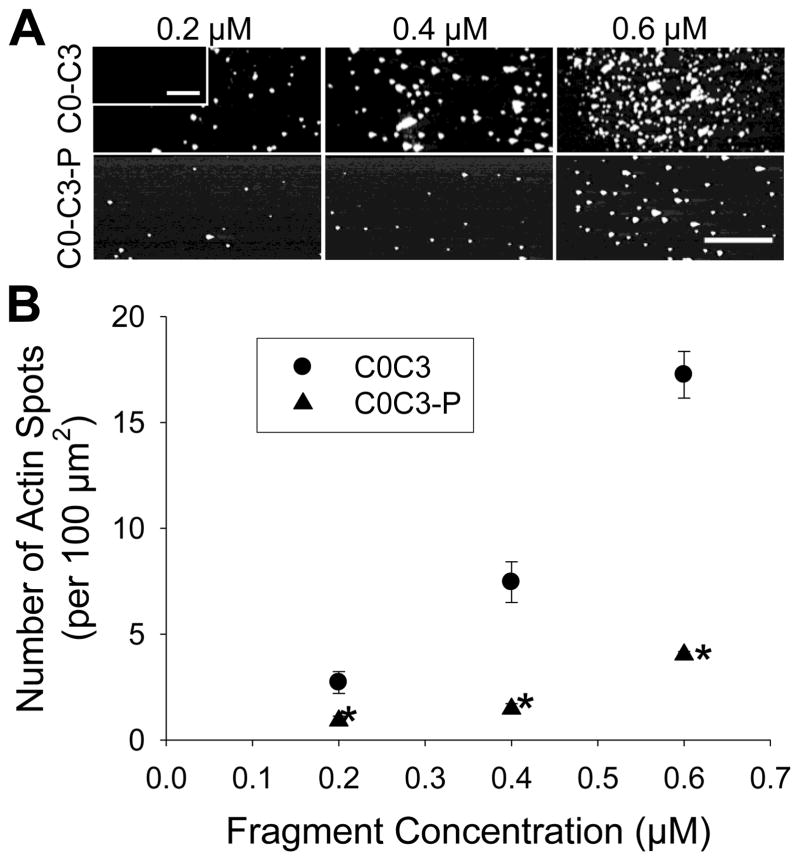
In the absence of cMyBP-C, actin filaments do not bind to a BSA blocked surface (see Fig. S4). If actin filament sliding inhibition results from cMyBP-C fragments tethering actin filaments to the motility surface, then fragments must bind actin at one end while attached to a BSA-coated surface and at its other end, given our motility assay protocol (see Material and Methods). To observe this capacity, actin was introduced to a flowcell following the incubation of 0.5–2 μM C0-C1, C0-C1f or C0-C3 to a BSA-coated coverslip surface in the absence of myosin (Figs. S4). All fragments were capable of actin-binding, although C0-C1 bound 4 times less actin than C0-C3 or C0-C1f at the same concentrations. Since phosphorylated C0-C3’s lack of inhibition of velocity resembles that of C0-C1, we examined whether phosphorylation causes a parallel reduction in actin-binding. As expected, phosphorylation of C0-C3 reduced the amount of actin bound to the surface by 4-fold (Fig. 3).
Figure 3.
Actin-binding to nitrocellulose surfaces coated with non-phosphorylated or phosphorylated C0-C3. A) Images of actin shards bound to the surface at 3 different C0-C3 (-P) concentrations. Inset in top left corner is a control, showing a BSA blocked surface with no cMyBP-C fragments. Scale bars for both inset and panels are 25 μm. B) Quantification of the number of actin shards (i.e. spots) per 100 μm2 of coverslip surface (mean ± standard error, n=3). There is a four-fold average decrease in actin bound to the surface after C0-C3 phosphorylation. The * indicates significant difference from unphosphorylated C0-C3.
cMyBP-C fragments bind actin transiently in the laser trap assay
Although the inhibition of actin filament velocity and actin-binding observed in the motility assay data suggest that cMyBP-C and its N-terminal fragments impart their effects through actin-binding, the laser trap assay is the most direct assay to characterize cMyBP-C-actin interactions at the molecular level. In fact, individual binding events were observed when a single actin filament suspended between two trapped beads was brought into contact with a sparsely coated surface of N-terminal cMyBP-C fragments. Examples of raw actin filament displacement traces and their Mean-Variance (MV) histograms are shown in Fig. 4A, B. Binding events are detected as a shift in the mean actin filament position and a reduction in the positional noise (i.e. variance), as cMyBP-C fragment binding reduces the Brownian motion of the bead-actin-bead complex (Fig. 4A). For all fragments tested (C0-C3, C0-C1f, C0-C1fm, C0-C1), the data traces are composed of frequent events (4 s−1) of short duration (<30 ms) interspersed with much less frequent (0.06 s−1) longer duration (>200 ms) events (Fig. 4, Table 2). These data offer direct evidence that the N-terminus of cMyBP-C is capable of actin-binding.
Table 2.
Laser trap cMyB-C fragment actin binding data
| cMyBP-C Fragment/α-Actinin | Binding Event Amplitude (nm) | Short lifetime (ms) | Long lifetime (ms) |
|---|---|---|---|
| C0-C3 | 9.8 ± 0.5 | 22 ± 6 | 294 ± 86 |
| C0-C3P | 9.6 ± 0.4 | 25 ± 4 | 467 ± 32 |
| C0-C1f | 9.9 ± 0.8 | 19 ± 3 | 197 ± 48 |
| α-Actinin | 0.6 ± 1.3a | 23 ± 5 | 189 ± 46 |
| C0-C1 | 0.7 ± 1.3a | 21 ± 3 | 370 ± 70 |
| C0-C1fm | 0.4 ± 1.1a,b | 26 ± 5 | 347 ± 35 |
Data are mean ± standard error.
= significantly different than C0-C3 and
= significantly different than C0-C1f.
N-terminal fragments bind with distinct event amplitudes
The structure-function relations characterized in the motility assay suggest that the inhibition of velocity with the longest N-terminal fragment (C0-C3) is maintained in the shorter C0-C1f fragment (Fig. 2A). On closer analysis, the binding events associated with these two fragments in the laser trap assay were similar for the most part (see below) in that binding to actin was described by events with mean population amplitudes of ~10 nm (Fig. 4A, C). This is apparent in the MV histogram as a distinct event population density (e in Fig. 4A) with a mean amplitude and variance offset from the baseline density (B in Fig. 4A) by ~10 nm. Since cMyBP-C or its fragments should have no intrinsic motion generating capacity, a unidirectional displacement of the actin filament, which reflects displacement of the bead-actin-bead complex in a consistent direction and amplitude as normally observed with myosin, was surprising. Although the directionality was maintained within a trace for a given actin filament, between actin filaments there was an ~ 50% probability of the events being either positive or negative in direction, suggesting that the polarity of the actin filament, which can’t be controlled in our assay, dictates the event direction.
Two trivial explanations for the unidirectional binding events were controlled for: 1) myosin contamination; 2) non-specific binding of actin to the surface. If myosin were present, then eliminating ATP should cause the contaminant myosin to form long-lasting rigor bonds with actin, which was not observed. Non-specific surface interactions were ruled out, since no events were detected when actin was lowered to a BSA-blocked surface (data not shown). As an additional control, we characterized the binding of α-actinin, a known actin-binding protein. The α-actinin binding events are similar to that of the cMyBP-C fragments, having both short (23 ms) and long (189 ms) lifetime events (Fig. 4B, Table 2). As with the cMyBP-C fragments, long lived events could be more than one molecule interacting with the actin or a property common to acting-binding proteins. In contrast to C0-C3 and C0-C1f, α-actinin events are bidirectional and variable in amplitude as evident in the MV histogram by the event population density (e in Fig. 4B) being uniformly distributed beneath the baseline density with an average event amplitude of ~0 nm (Fig. 4B, Table 2). Thus, the unidirectional events observed for both C0-C3 and C0-C1f are not a characteristic common to all actin-binding proteins.
The minimal inhibition observed for C0-C1 and C0-C1fm suggest a different mode of interaction for these two fragments with actin. This, too, was the case in the laser trap assay, where the binding events were always bidirectional, resembling α-actinin, and centered on 0 nm (Fig. 4B,C, Table 2). The difference is most striking given that C0-C1f and C0-C1fm differ by the substitution of 3 alanines for arginines, which was sufficient to alter the binding mode for the C0-C1fm. Although the C0-C1f shows unidirectional behavior comparable to the C0-C3, occasional C0-C1f data traces displayed bidirectional events, suggesting that in these instances the C0-C1f landed on the surface in such a way that the 17 amino acids of the motif were unable to properly interact with actin to generate unidirectional binding events (see Fig. S5).
Binding events of phosphorylated C0-C3 are similar to unphosphorylated C0-C3
When phosphorylated C0-C3 was added to the laser trap assay, its binding to actin was indistinguishable from unphosphorylated C0-C3 in terms of the binding event lifetimes and amplitudes (Fig. 4A,C,D, Table 2). However, to obtain a similar frequency of events, twice as much phosphorylated C0-C3 was added to the flowcell.
Discussion
A significant body of literature demonstrates actin-binding by cMyBP-C [4]. However, the physiological relevance of such binding is not uniformly accepted [11]. Therefore, we sought to determine directly whether cMyBP-C, through the use of mouse N-terminal domain fragments, was capable of binding actin at the single molecule level. In fact, the unidirectionality of cMyBP-C’s actin-binding in the laser trap assay requires a structural element localized to the initial 17 amino acids of the motif. These same 17 amino acids confer complete inhibitory capacity to a small (27 kD) cMyBP-C N-terminal fragment (C0-C1f) comparable to that previously published using full length bovine cMyBP-C in the motility assay [27]. We also confirm that phosphorylation decreases cMyBP-C’s actin-binding affinity [17] and directly decreases its inhibition of actomyosin motility. These data support a model in which cMyBP-C’s ability to bind reversibly to actin, at least in part provides an internal load to myosin power generation in muscle.
cMyBP-C’s inhibition of actomyosin motility
Full length cMyBP-C and various N-terminal fragments have profound effects on muscle fiber force and motion generation [15, 35–39], which have been recapitulated in the motility assay [27, 40–42]. These effects could arise from cMyBP-C interacting with either actin and/or myosin. For example, the cMyBP-C motif binds to myosin-S2 [43], thus potentially limiting myosin head mobility and its ability to interact with actin [44, 45]. Additionally, recent structural and binding data suggest that the C0 domain can simultaneously interact with the myosin regulatory light chain, potentially forming a strut between the myosin neck and the myosin-S2 domain, further hindering myosin’s power stroke and/or head mobility [8]. However, equally compelling data support a role for cMyBP-C’s actin-binding. Electron microscopy (EM) and neutron scattering experiments have demonstrated that cMyBP-C fragments decorate actin in a helical pattern that implies stereospecific binding [12, 13, 46]. Recent, EM tomographs of frog sartorious muscle indicate that the mass associated with MyBP-C in the C-zone extends from the thick filament to within reach of the thin filaments, suggestive of actin-binding in vivo [47]. It is important to note that actin- and myosin-based models are not mutually exclusive. In fact, the N-terminus of cMyBP-C may exist in a dynamic equilibrium between actin and myosin binding, which could be modulated by phosphorylation.
While we cannot distinguish if the primary basis for actin filament motility inhibition reported here is due to cMyBP-C’s binding to actin or to myosin, we do demonstrate that the N-terminal cMyBP-C fragments alone, when adhered to a nitrocellulose-coated coverslip directly (Fig. 3) or subsequent to a BSA coating (Fig. S4) are capable of binding and tethering actin to the surface. Although Harris and coworkers [42] previously demonstrated actin-binding to C1-C2 in the motility assay, they saw no such binding when the surface was pre-blocked with BSA in contrast to our observation (Fig. 3). Since the present study and that of Razumova et al. [42] use virtually the same bacterially expressed, N-terminal fragments that lead to similar inhibition of motility, we are hard pressed to offer any obvious reasons for the discrepancy.
Evidence that cMyBP-C’s actin-binding could inhibit motility is supported by the similarity in the actin-binding lifetimes in the laser trap assay for the N-terminal fragments compared to a known actin-binding protein, α-actinin (Fig. 4A,B, Table 2). α-actinin is routinely used as means of imposing a load in the motility assay to estimate myosin’s force generating capacity [48]. The basis for α-actinin’s slowing of actin filament velocity has been modeled as a viscous load [49], which assumes that α-actinin binds transiently to actin, as confirmed by our studies. Although C0-C1 also binds transiently to actin in the laser trap assay, actin-binding alone is not sufficient to slow actin filament velocities since C0-C1 is far less inhibitory in the motility assay (Fig. 2A). In contrast, only those fragments that contained the C1 domain and the first 17 amino acids of the motif (termed “f”) inhibited motility (see Fig. 2A). These fragments may contain an additional actin-binding site that is better able to resist the strain of myosin’s actin translocation by creating a viscous load against which myosin must operate. However, we previously argued, based on indirect evidence, that full length cMyBP-C did not act as a load in the motility assay, given that the inhibition by cMyBP-C was not affected by ionic strength [27]. In our earlier study, changes in KCl concentration were over a limited range (25–100mM), which may not have been sufficient to disrupt cMyBP-C’s electrostatic binding, since N-terminal fragments can stereospecifically decorate actin filaments even at 180mM KCl [13]. Therefore, the results presented here argue strongly for cMyBP-C acting as a viscous load through direct evidence of actin-binding. Interestingly, the PEVK region of titin with its ability to bind actin has also been proposed to act as a viscous load, slowing actomyosin motility [50]. Since this region of titin interacts with actin in the I-band and cMyBP-C may bind actin in the A-band, these two proteins could combine to act as a spatially disperse viscous element in the sarcomere to modulate cardiac contractility.
C0-C3 and C0-C1f interact stereospecifically with actin
Through structural mutagenesis as described above, we partitioned the inhibitory capacity of the cMyBP-C’s N-terminus to the C0-C1f fragment. Therefore, in the laser trap assay we characterized and compared the molecular mechanics of C0-C3 and its equally inhibitory, but smaller C0-C1f subfragment to both the C0-C1 and C0-C1fm fragments, which were less inhibitory. The molecular mechanics of C0-C3 and C0-C1f were indistinguishable, and upon binding each displaced the actin filament unidirectionally by ~10 nm. This was surprising, since neither fragment is an active motion generator. These binding events were in sharp contrast to C0-C1, which had bidirectional binding events with the average displacement centered on 0 nm; the result we had expected for all of the N-terminal fragments for the following reason. As the actin filament/bead complex undergoes ±30 nm excursions due to Brownian motion (Figs. 4A, B), fragment binding should trap the actin filament at random distances from the actin/bead complex’s equilibrium position (i.e. 0 nm). These effective displacements should be bidirectional with both positive and negative events centered at 0 nm. This accurately describes C0-C1’s binding characteristics and that of α-actinin, an extremely flexible actin-crosslinking protein (Fig. 4B, C) [51, 52]. The C0-C1 fragment is the shortest fragment tested in the laser trap, thus its bidirectional behavior might stem from it being too small to reach the actin properly. However C0-C1f is only slightly longer, yet it interacts unidirectionally with actin.
The simplest explanation for the unidirectional, 10 nm displacements for the C0-C3 and C0-C1f fragments is that at least two actin-binding sites exist, one of which is stereospecific. Thus, following the initial collision complex with actin, the fragment must rapidly bind stereospecifically to a second site with the binding energy sufficient to move the actin filament 10 nm within the trap, analogous to zipping of a zipper (Fig. 5). This motion cannot generate useful work and thus at the end of its bound lifetime the fragment must undergo a reversal of this process to effectively unzip from actin on a timescale faster than our detection capacity (~2ms). The stereospecific nature of these N-terminal fragments’ binding to actin filaments was recently confirmed by our collaborators through EM helical reconstructions [13].
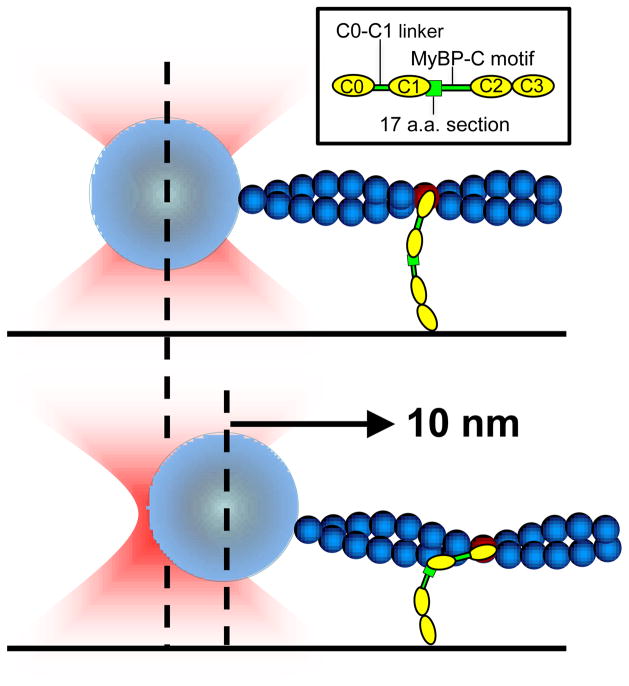
Figure 5.
Model of C0-C3 actin-binding in the laser trap assay. C0-C3 is assumed to contain two actin-binding sites. Binding of the first site in the distal N-terminus positions the C0-C3 molecule to undergo a rapid, stereospecific binding of its second, higher affinity site to actin. We assume the second binding site is located within the first 17 amino acids of the cMyBP-C motif (green box, see inset). The sequential binding of cMyBP-C’s two actin-binding sites causes a 10 nm displacement of the bead.
Multiple N-terminal actin-binding sites have been proposed in the literature based on structural, biochemical, and in vitro actin-binding/-bundling data [16, 17, 39]. Although high affinity sites within the C1 and motif domains have been proposed by Harris and coworkers [17], the exact residues were not identified. Our data point to the first 17 N-terminal amino acids of the motif and more specifically a cluster of positively charged arginines (see Fig. 1). The importance of the three arginines at residues 266, 270, and 271 in the mouse sequence is emphasized by substitution of these residues to alanines (i.e. C0-C1fm) leading to loss of inhibition of actin filament motility and converting C0-C1f into an actin-binding fragment that was indistinguishable from C0-C1 in the laser trap assay (Fig. 4A–C). Because actin’s N-terminus is negatively charged [53], this cluster of positively charged arginines (absolutely conserved in species ranging from zebrafish to human) is likely a critical element within one of cMyBP-C’s actin-binding sites. It would also explain the reduction in motif binding to actin observed by Harris and coworkers with elevated pH and ionic strength [17, 42].
Phosphorylation reduces actin binding
β-adrenergic enhancement of cardiac contractility is mediated by PKA phosphorylation of key sarcomeric proteins (e.g. cMyBP-C, cardiac troponin I, myosin regulatory light chain) [4, 54, 55]. With four cardiac-specific phosphorylation sites confirmed within the motif between the C1 and C2 domains (Table 1), we fully phosphorylated at least 83% of the molecules at all four sites of the C0-C3 fragment with PKA. Once phosphorylated, the inhibitory capacities of the C0-C3 and C1-C2 fragments in the in vitro motility assay were significantly reduced (Fig. 2B) to that of C0-C1, which is devoid of the motif. Phosphorylation clearly reduces the fragments’ ability to bind actin as demonstrated by diminished actin tethering to the motility surface (Fig. 3), in agreement with cosedimentation data from Harris and coworkers [17]. However, this is in stark contrast to a recent study by Rybakova et al [11], where phosphorylation had no effect on the actin-binding of full-length, expressed cMyBP-C. In their study phosphorylation was confirmed by Pro-Q Diamond staining, which only provides a qualitative assessment of phosphorylation without information on which of the 4 specific sites were phosphorylated. Given that the functional hierarchy resulting from the phosphorylation of these various sites is still unknown, phosphorylation of the sites needed for modulating cMyBP-C’s actin-binding may not have reached the necessary levels in that study.
Phosphorylation of the motif could affect the fragment’s actin-binding capacity by affecting the motif’s putative contact to a neighboring actin monomer in the actin filament, as proposed by Craig and coworkers [13]. More specifically, phosphorylation could affect the N-terminal 17 amino acids identified here, as the serine at position 273, once phosphorylated, would present negative counteracting charges only 2 residues away from the arginine cluster. In support of this, the phosphorylated C0-C3 and C1-C2 are indistinguishable from the C0-C1fm fragment in terms of motility inhibition.
If cMyBP-C fragment phosphorylation profoundly affects actin-binding and velocity inhibition, then why were unphosphorylated and phosphorylated C0-C3 (C0-C3-P) indistinguishable in the laser trap assay in terms of lifetimes and directionality of binding events? Twice as much C0-C3-P was required to observe a similar frequency of binding events, confirming a reduction in the fragment’s overall actin-binding. This implies that while phosphorylation of C0-C3 reduces its ability to bind actin, once the fragment interacts with actin, it binds specifically with both binding sites. This behavior contrasts with C0-C1, which has a similar lack of inhibition in the motility assay but binds actin bidirectionally in the laser trap. The dichotomy stems from the fact that in an ensemble assay, C0-C3-P’s and C0-C1’s reduced binding to actin manifest similarly, but on a molecular level C0-C1 lacks a second actin binding site, so it cannot bind actin unidirectionally like C0-C3-P. If C0-C3-P still binds actin stereospecifically, then its lack of inhibition in the in vitro motility assay can be explained if its binding lifetime to actin is force-dependent. Phosphorylation could alter the sensitivity of cMyBP-C’s binding lifetime to force so that cMyBP-C would be more likely to detach under the forces generated by myosins translocating actin filaments. Thus, we propose that phosphorylating cMyBP-C diminishes its capacity to inhibit actomyosin motility, leading to increased power generation in fibers. In fact, Sadayappan et al. showed that a mouse in which three phosphorylatable serines were mutated to aspartic acids to mimic phosphorylation had increased maximum power compared to control mice [56].
cMyBP-C effects within the sarcomere
Our data suggest a potential role of cMyBP-C in modulating cardiac contractility by forming a link between its C-terminal binding to the myosin thick filament (i.e. through its LMM binding) and the actin thin filaments. A caveat in these studies involves the use of cardiac cMyBP-C in conjunction with skeletal myosin and actin. While the mixing of isoforms is not ideal, actin is highly conserved throughout muscle, and the S2 portion of myosin that binds cMyBP-C is identical between vertebrate muscle isoforms [57]. Also, our in vitro approach, which controls protein stoichiometry, lacks the sarcomere’s nearly crystalline spatial relationships between contractile proteins. In fact, given that cMyBP-C’s location within the sarcomere is restricted to the C-zone, how are its mechanical effects manifested through its interaction with only a limited population of crossbridges? Through its actin attachment, cMyBP-C’s effect would be transmitted along the entire filament, thus potentially affecting all crossbridges that interact with that actin filament.
Supplementary Material
Highlights.
Actin-binding properties of cardiac myosin binding protein-C (cMyBP-C) defined
Single molecule binding of cMyBP-C N-terminal fragments to actin in laser trap
First 17 amino acids of the MyBP-C motif define strong actin binding site
Phosphorylation of 4 serines within the MyBP-C motif reduces actin binding
cMyBP-C may act as a viscous load in the myocardium
Acknowledgments
We thank S. Previs and K. Begin for superb technical assistance, G. Kennedy from the University of Vermont Instrumentation and Model Facility for optomechanical support, and Warshaw Lab members for numerous discussions. The LC-MS sample analysis was performed at the Proteomics Facility of the Vermont Genetics Network at the University of Vermont (NIH RR16462). This work was supported by the National Institutes of Health (HL086728, HL059408 to DMW; HL007944 to AW; HL07647 to MP) and the American Heart Association (Scientist Development Grant 0830311N to SS).
Footnotes
Disclosures: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Offer G, Moos C, Starr R. A new protein of the thick filaments of vertebrate skeletal myofibrils. Extractions, purification and characterization. J Mol Biol. 1973;74:653–76. doi: 10.1016/0022-2836(73)90055-7. [DOI] [PubMed] [Google Scholar]
- 2.Chang AN, Potter JD. Sarcomeric protein mutations in dilated cardiomyopathy. Heart Fail Rev. 2005;10:225–35. doi: 10.1007/s10741-005-5252-6. [DOI] [PubMed] [Google Scholar]
- 3.Harris SP, Lyons RG, Bezold KL. In the thick of it HCM-causing mutations in myosin binding proteins of the thick filament. Circ Res. 2011;108:751–64. doi: 10.1161/CIRCRESAHA.110.231670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barefield D, Sadayappan S. Phosphorylation and function of cardiac myosin binding protein-C in health and disease. J Mol Cell Cardiol. 2010;48:866–75. doi: 10.1016/j.yjmcc.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett P, Craig R, Starr R, Offer G. The ultrastructural location of C-protein, X-protein and H-protein in rabbit muscle. J Muscle Res Cell Motil. 1986;7:550–67. doi: 10.1007/BF01753571. [DOI] [PubMed] [Google Scholar]
- 6.Einheber S, Fischman DA. Isolation and characterization of a cDNA clone encoding avian skeletal muscle C-protein: an intracellular member of the immunoglobulin superfamily. Proc Natl Acad Sci U S A. 1990;87:2157–61. doi: 10.1073/pnas.87.6.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moos C, Offer G, Starr R, Bennett P. Interaction of C-protein with myosin, myosin rod and light meromyosin. J Mol Biol. 1975;74:653–76. doi: 10.1016/s0022-2836(75)80017-9. [DOI] [PubMed] [Google Scholar]
- 8.Ratti J, Rostkova E, Gautel M, Pfuhl M. Structure and interactions of myosin-binding protein C domain C0: cardiac-specific regulation of myosin at its neck? J Biol Chem. 2011:286. doi: 10.1074/jbc.M110.156646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furst DO, Vinkemeier U, Weber K. Mammalian skeletal-muscle C-protein - purification from bovine muscle, binding to titin and the characterization of a full-length human cDNA. J Cell Sci. 1992;102:769–78. doi: 10.1242/jcs.102.4.769. [DOI] [PubMed] [Google Scholar]
- 10.Moos C, Mason CM, Besterman JM, Feng IM, Dubin JH. The binding of skeletal muscle C-protein to F-actin, and its relation to the interaction of actin with myosin subfragment-1. J Mol Biol. 1978;124:571–86. doi: 10.1016/0022-2836(78)90172-9. [DOI] [PubMed] [Google Scholar]
- 11.Rybakova IN, Greaser ML, Moss RL. Myosin binding protein C interaction with actin: characterization and mapping of the binding site. Journal of Biological Chemistry. 2011;286:2008–16. doi: 10.1074/jbc.M110.170605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitten AE, Jeffries CM, Harris SP, Trewhella J. Cardiac myosin-binding protein C decorates F-actin: implications for cardiac function. Proc Natl Acad Sci U S A. 2008;105:18360–5. doi: 10.1073/pnas.0808903105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mun JY, Gulick J, Robbins J, Woodhead J, Lehman W, Craig R. Electron microscopy and 3D reconstruction of F-actin decorated with cardiac myosin-binding protein C (cMyBP-C) J Mol Biol. 2011;410:214–25. doi: 10.1016/j.jmb.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofmann PA, Hartzell HC, Moss RL. Alterations in Ca2+ sensitive tension due to partial extraction of C-protein from rat skinned cardiac myocytes and rabbit skeletal-muscle fibers. J Gen Physiol. 1991;97:1141–63. doi: 10.1085/jgp.97.6.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofmann PA, Greaser ML, Moss RL. C-protein limits shortening velocity of rabbit skeletal-muscle fibers at low-levels of Ca2+ activation. J Physiol. 1991;439:701–15. doi: 10.1113/jphysiol.1991.sp018689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Squire JM, Luther PK, Knupp C. Structural evidence for the interaction of C-protein (MyBP-C) with actin and sequence identification of a possible actin-binding domain. J Mol Biol. 2003;331:713–24. doi: 10.1016/s0022-2836(03)00781-2. [DOI] [PubMed] [Google Scholar]
- 17.Shaffer JF, Kensler RW, Harris SP. The myosin-binding protein C motif binds to F-actin in a phosphorylation-sensitive manner. J Biol Chem. 2009;284:12318–27. doi: 10.1074/jbc.M808850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gautel M, Zuffardi O, Freiburg A, Labeit S. Phosphorylation switches specific for the cardiac isoform of myosin binding protein-C: a modulator of cardiac contraction? EMBO J. 1995;14:1952–60. doi: 10.1002/j.1460-2075.1995.tb07187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia W, Shaffer JF, Harris SP, Leary JA. Identification of novel protein kinase A phosphorylation sites in the M-domain of human and murine cardiac myosin binding protein-C using mass spectrometry analysis. J Proteome Res. 2010;9:1843–53. doi: 10.1021/pr901006h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacques AM, Copeland O, Messer AE, Gallon CE, King K, McKenna WJ, et al. Myosin binding protein C phosphorylation in normal, hypertrophic and failing human heart muscle. J Mol Cell Cardiol. 2008;45:209–16. doi: 10.1016/j.yjmcc.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 21.El-Armouche A, Pohlmann L, Schlossarek S, Starbatty J, Yeh YH, Nattel S, et al. Decreased phosphorylation levels of cardiac myosin-binding protein-C in human and experimental heart failure. J Mol Cell Cardiol. 2007;43:223–9. doi: 10.1016/j.yjmcc.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Copeland O, Sadayappan S, Messer AE, Steinen GJ, van der Velden J, Marston SB. Analysis of cardiac myosin binding protein-C phosphorylation in human heart muscle. J Mol Cell Cardiol. 2010;49:1003–11. doi: 10.1016/j.yjmcc.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 23.Sadayappan S, Gulick J, Osinska H, Martin LA, Hahn HS, Dorn GW, 2nd, et al. Cardiac myosin-binding protein-C phosphorylation and cardiac function. Circ Res. 2005;97:1156–63. doi: 10.1161/01.RES.0000190605.79013.4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tong CW, Stelzer JE, Greaser ML, Powers PA, Moss RL. Acceleration of Crossbridge Kinetics by Protein Kinase A Phosphorylation of Cardiac Myosin Binding Protein C Modulates Cardiac Function. Circ Res. 2008;103:974–82. doi: 10.1161/CIRCRESAHA.108.177683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sadayappan S, Osinska H, Klevitsky R, Lorenz JN, Sargent M, Molkentin JD, et al. Cardiac myosin binding protein C phosphorylation is cardioprotective. Proc Natl Acad Sci U S A. 2006;103:16918–23. doi: 10.1073/pnas.0607069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gruen M, Prinz H, Gautel M. cAPK-phosphorylation controls the interaction of the regulatory domain of cardiac myosin binding protein C with myosin-S2 in an on-off fashion. FEBS Lett. 1999;453:254–9. doi: 10.1016/s0014-5793(99)00727-9. [DOI] [PubMed] [Google Scholar]
- 27.Saber W, Begin KJ, Warshaw DM, VanBuren P. Cardiac myosin binding protein-C modulates actomyosin binding and kinetics in the in vitro motility assay. J Mol Cell Cardiol. 2008;44:1053–61. doi: 10.1016/j.yjmcc.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Margossian SS, Lowey S. Preparation of myosin and its subfragments from rabbit skeletal muscle. Methods Enzymol. 1982;85(Pt B):55–71. doi: 10.1016/0076-6879(82)85009-x. [DOI] [PubMed] [Google Scholar]
- 29.Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85(Pt B):164–81. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- 30.Warshaw DM, Desrosiers JM, Work SS, Trybus KM. Smooth muscle myosin cross-bridge interactions modulate actin filament sliding velocity in vitro. J Cell Biol. 1990;111:453–63. doi: 10.1083/jcb.111.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Previs MJ, VanBuren P, Begin KJ, Vigoreaux JO, LeWinter MM, Matthews DE. Quantification of protein phosphorylation by liquid chromatography-mass spectrometry. Anal Chem. 2008;80:5864–72. doi: 10.1021/ac800337v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmiter KA, Tyska MJ, Haeberle JR, Alpert NR, Fananapazir L, Warshaw DM. R403Q and L908V mutant beta-cardiac myosin from patients with familial hypertrophic cardiomyopathy exhibit enhanced mechanical performance at the single molecule level. J Muscle Res Cell Motil. 2000;21:609–20. doi: 10.1023/a:1005678905119. [DOI] [PubMed] [Google Scholar]
- 33.Guilford WH, Dupuis DE, Kennedy G, Wu J, Patlak JB, Warshaw DM. Smooth muscle and skeletal muscle myosins produce similar unitary forces and displacements in the laser trap. Biophys J. 1997;72:1006–21. doi: 10.1016/S0006-3495(97)78753-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baker JE, Brosseau C, Joel PB, Warshaw DM. The biochemical kinetics underlying actin movement generated by one and many skeletal muscle myosin molecules. Biophys J. 2002;82:2134–47. doi: 10.1016/S0006-3495(02)75560-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herron TJ, Rostkova E, Kunst G, Chaturvedi R, Gautel M, Kentish JC. Activation of myocardial contraction by the N-terminal domains of myosin binding protein-C. Circ Res. 2006;98:1290–8. doi: 10.1161/01.RES.0000222059.54917.ef. [DOI] [PubMed] [Google Scholar]
- 36.Korte FS, McDonald KS, Harris SP, Moss RL. Loaded shortening, power output, and rate of force redevelopment are increased with knockout of cardiac myosin binding protein-C. Circ Res. 2003;93:752–8. doi: 10.1161/01.RES.0000096363.85588.9A. [DOI] [PubMed] [Google Scholar]
- 37.Harris SP, Rostkova E, Gautel M, Moss RL. Binding of myosin binding protein-C to myosin subfragment S2 affects contractility independent of a tether mechanism. Circ Res. 2004;95:930–6. doi: 10.1161/01.RES.0000147312.02673.56. [DOI] [PubMed] [Google Scholar]
- 38.Razumova MV, Bezold KL, Tu AY, Regnier M, Harris SP. Contribution of the myosin binding protein C motif to functional effects in permeabilized rat trabeculae. J Gen Physiol. 2008;132:575–85. doi: 10.1085/jgp.200810013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kulikovskaya I, McClellan G, Flavigny J, Carrier L, Winegrad S. Effect of MyBP-C binding to actin on contractility in heart muscle. J Gen Physiol. 2003;122:761–74. doi: 10.1085/jgp.200308941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shchepkin DV, Kopylova GV, Nikitina LV, Katsnelson LB, Bershitsky SY. Effects of cardiac myosin binding protein-C on the regulation of interaction of cardiac myosin with thin filament in an in vitro motility assay. Biochem Biophys Res Comm. 2010;401:159–63. doi: 10.1016/j.bbrc.2010.09.040. [DOI] [PubMed] [Google Scholar]
- 41.Lecarpentier Y, Vignier N, Oliviero P, Guellich A, Carrier L, Coirault C. Cardiac myosin-binding protein C modulates the tuning of the molecular motor in the heart. Biophys J. 2008;95:720–8. doi: 10.1529/biophysj.107.127787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Razumova MV, Shaffer JF, Tu AY, Flint GV, Regnier M, Harris SP. Effects of the N-terminal domains of myosin binding protein-C in an in vitro motility assay - Evidence for long-lived cross-bridges. J Biol Chem. 2006;281:35846–54. doi: 10.1074/jbc.M606949200. [DOI] [PubMed] [Google Scholar]
- 43.Gruen M, Gautel M. Mutations in beta-myosin S2 that cause familial hypertrophic cardiomyopathy (FHC) abolish the interaction with the regulatory domain of myosin-binding protein-C. J Mol Biol. 1999;286:933–49. doi: 10.1006/jmbi.1998.2522. [DOI] [PubMed] [Google Scholar]
- 44.Starr R, Offer G. The interaction of C-protein with heavy meromyosin and subfragment-2. Biochem J. 1978;171:813–6. doi: 10.1042/bj1710813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Colson BA, Bekyarova T, Fitzsimons DP, Irving TC, Moss RL. Radial displacement of myosin cross-bridges in mouse myocardium due to ablation of myosin binding protein-C. J Mol Biol. 2007;367:36–41. doi: 10.1016/j.jmb.2006.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kensler RW, Shaffer JF, Harris SP. Binding of the N-terminal fragment C0-C2 of cardiac MyBP-C to cardiac F-actin. J Struct Biol. 2011;174:44–51. doi: 10.1016/j.jsb.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luther P, Winkler H, Taylor K, Zoghbi ME, Craig R, Padrón R. Myosin binding protein C bridges myosin and actin filaments in intact muscle. Proc Natl Acad Sci U S A. 2011;108:11423–1142. doi: 10.1073/pnas.1103216108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.VanBuren P, Alix SL, Gorga JA, Begin KJ, LeWinter MM, Alpert NR. Cardiac troponin T isoforms demonstrate similar effects on mechanical performance in a regulated contractile system. Am J Physiol Heart Circ Physiol. 2002;282:H1665–71. doi: 10.1152/ajpheart.00938.2001. [DOI] [PubMed] [Google Scholar]
- 49.Greenberg MJ, Moore JR. The molecular basis of frictional loads in the in vitro motility assay with applications to the study of the loaded mechanochemistry of molecular motors. Cytoskeleton. 2010;67:273–85. doi: 10.1002/cm.20441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kulke M, Fujita-Becker S, Rostkova E, Neagoe C, Labeit D, Manstein DJ, et al. Interaction between PEVK-titin and actin filaments - Origin of a viscous force component in cardiac myofibrils. Circ Res. 2001;89:874–81. doi: 10.1161/hh2201.099453. [DOI] [PubMed] [Google Scholar]
- 51.Hampton CM, Taylor DW, Taylor KA. Novel structures for alpha-actinin:F-actin interactions and their implications for actin-membrane attachment and tension sensing in the cytoskeleton. J Mol Biol. 2007;368:92–104. doi: 10.1016/j.jmb.2007.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Courson DS, Rock RS. Actin cross-link assembly and disassembly mechanics for alpha-Actinin and fascin. J Biol Chem. 2010;285:26350–7. doi: 10.1074/jbc.M110.123117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kabsch W, Holmes KC. The actin fold. FASEB J. 1995;9:167–74. doi: 10.1096/fasebj.9.2.7781919. [DOI] [PubMed] [Google Scholar]
- 54.Kobayashi T, Jin L, de Tombe PP. Cardiac thin filament regulation. Pflugers Arch. 2008;457:37–46. doi: 10.1007/s00424-008-0511-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Szczesna D. Regulatory light chains of striated muscle myosin. Structure, function and malfunction. Curr Drug Targets Cardiovasc Haematol Disord. 2003;3:187–97. doi: 10.2174/1568006033481474. [DOI] [PubMed] [Google Scholar]
- 56.Sadayappan S, Gulick J, Klevitsky R, Lorenz JN, Sargent M, Molkentin JD, et al. Cardiac Myosin Binding Protein-C Phosphorylation in a beta-Myosin Heavy Chain Background. Circ. 2009;119:1253–U40. doi: 10.1161/CIRCULATIONAHA.108.798983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kunst G, Kress KR, Gruen M, Uttenweiler D, Gautel M, Fink RHA. Myosin binding protein C, a phosphorylation-dependent force regulator in muscle that controls the attachment of myosin heads by its interaction with myosin S2. Circ Res. 2000;86:51–8. doi: 10.1161/01.res.86.1.51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.