Abstract
Transcription of ribosomal DNA by RNA polymerase I is a central feature of eukaryotic ribosome biogenesis. Since ribosome synthesis is closely linked to cell proliferation, there is a need to define the molecular mechanisms that control transcription by RNA polymerase I. To fully define the factors that control RNA polymerase I activity, biochemical analyses using purified transcription factors are essential. Although such assays exist, one limitation is the low abundance and difficult purification strategies required for some of the essential transcription factors for RNA polymerase I. Here, we describe a new method for expression and purification of the three subunit core factor complex from Escherichia coli. We demonstrate that the recombinant material is more active than yeast-derived core factor in assays for RNA polymerase I transcription in vitro. Finally, we use recombinant core factor to differentiate between two opposing models for the role of the TATA-binding protein in transcription by RNA polymerase I.
Keywords: ribosome, transcription, rRNA, gene expression, in vitro transcription
1. Introduction
Transcription of ribosomal DNA (rDNA) by RNA polymerase I (Pol I) is a central step in eukaryotic ribosome biogenesis. In Saccharomyces cerevisiae (yeast), ribosomal RNA (rRNA) accounts for approximately 80% of total RNA levels and Pol I is responsible for up to 60% of total transcription (Warner, 1999). Yeast rDNA occupies a single locus on chromosome XII as an array of approximately 200 tandem repeats of a 9.1 kb unit (Nomura et al., 2004). Only about half of the rDNA repeats is actively transcribed at any given time in a growing cell. Each repeat contains a Pol I promoter and the coding region for the 35S precursor rRNA. The 35S pre-rRNA is co- and post-transcriptionally processed into the mature 25S, 18S, and 5.8S rRNA species found in ribosomes (Osheim et al., 2004; Kos and Tollervey, 2010).
Of the three major steps in the Pol I transcription cycle (initiation, elongation and termination), transcription initiation has been studied in greatest detail. There are four essential Pol I transcription initiation factors in yeast: the upstream activation factor (UAF), TATA-binding protein (TBP), core factor, and Rrn3p [reviewed in (Schneider, 2011)]. UAF is a six subunit complex consisting of Rrn5p, Rrn9p, Rrn10p, Uaf30p, and histones H3 and H4 (Keys et al., 1996; Keener et al., 1997). UAF binds to the upstream element of the rDNA promoter (approximately −155 to −60 with respect to the transcription start site) and helps facilitate promoter-specific Pol I transcription initiation (Choe et al., 1992; Keys et al., 1996; Steffan et al., 1996; Steffan et al., 1998).
As with RNA polymerases II and III, TBP influences Pol I transcription initiation (Lin et al., 1996; Steffan et al., 1996; Keener et al., 1998; Steffan et al., 1998). In vitro studies demonstrated that TBP is required for UAF-dependent activation of Pol I transcription (Steffan et al., 1996; Keener et al., 1998). Subsequent studies in live cells and cell extracts suggested that TBP can affect Pol I transcription even in the absence of UAF (Aprikian et al., 2000). The precise role for TBP in Pol I transcription initiation is not yet clear.
Rrn3p associates directly with the A43 subunit of Pol I to render the polymerase competent for transcription initiation (Peyroche et al., 2000). The role of Rrn3p in Pol I transcription initiation may be comparable to that of prokaryotic sigma factors; however, unlike sigma factors Rrn3p has not been reported to bind DNA. Rather than direct association with DNA, Rrn3p associates with another transcription factor (core factor, see below) to recruit Pol I to the rDNA promoter.
Core factor is a heterotrimer consisting of the subunits Rrn6p, Rrn7p, and Rrn11p. All of these subunits are essential for Pol I transcription initiation both in vitro and in vivo (Lalo et al., 1996; Lin et al., 1996; Keener et al., 1998; Aprikian et al., 2000). Core factor associates with the core region of the rDNA promoter (approximately −40 to +5) in the pre-initiation complex and is thought to help recruit Pol I to the initiation complex via interactions with the Rrn3p-Pol I complex (Peyroche et al., 2000). The Rrn6p subunit of core factor has been shown to interact with both TBP and Rrn3p in vivo and in vitro (Keys et al., 1994; Keener et al., 1998; Steffan et al., 1998; Aprikian et al., 2000; Peyroche et al., 2000). Overexpression of TBP in vivo suggests that this core factor-TBP interaction can activate Pol I transcription above basal levels (Aprikian et al., 2000). Likewise, Rrn7p has been shown to interact weakly with the Rrn9p subunit of UAF in vitro, but this interaction has not been shown to be sufficient to stimulate Pol I transcription (Lalo et al., 1996; Lin et al., 1996; Steffan et al., 1996). These data suggest that core factor may act as a bridge between various activating complexes. This network of interactions is critical for the high level of rRNA synthesis required in growing cells.
Although Rrn3p and Pol I are both remarkably well conserved throughout eukaryotes, sequence conservation is low between yeast core factor and its mammalian counterpart selectivity factor 1 [SL1; (Bodem et al., 2000; Moorefield et al., 2000)]. Recently, it was shown that the TFIIB homology domains of the TAF1B subunit of SL1 could be swapped with the corresponding domains of Rrn7p and support yeast cell growth (Knutson and Hahn, 2011). Thus, SL1 and core factor are functional homologues and detailed understanding of the mechanisms by which core factor affects Pol I transcription will reveal insights into the roles for SL1 in human cells.
Core factor abundance in yeast cells is remarkably low. This challenge has rendered core factor purification and characterization laborious, though not impossible (Keener et al., 1998). Here we report the expression and purification of recombinant, transcriptionally active core factor. Previous work from the Reeder lab suggested that TBP may activate Pol I transcription independently of UAF (Aprikian et al., 2000). Transcription assays using recombinant core factor and purified transcription factors support this role for TBP. This work provides a method for the efficient purification of a multi-subunit Pol I transcription initiation factor that may be applicable to other factors for Pol I or other transcription systems.
2. Materials and Methods
2.1 Plasmids and growth conditions
The genes for the three core factor subunits (RRN6, RRN7 and RRN11) were cloned into E. coli overexpression vectors. RRN7 was PCR amplified and subcloned using standard methods into pRSF-Duet (Merck; Darmstadt, Germany). RRN6 and RRN11 were PCR amplified and subcloned into pET-Duet (Merck; Darmstadt, Germany). Sequences encoding a 6-his epitope were fused in frame to the 5′ ends of both RRN7 and RRN11. These dual tags were included to enhance binding to Ni Sepharose, but did not alter the 1:1:1 stoichiometry of the purified complex (Figure 2A). No tag was added to RRN6.
Figure 2.
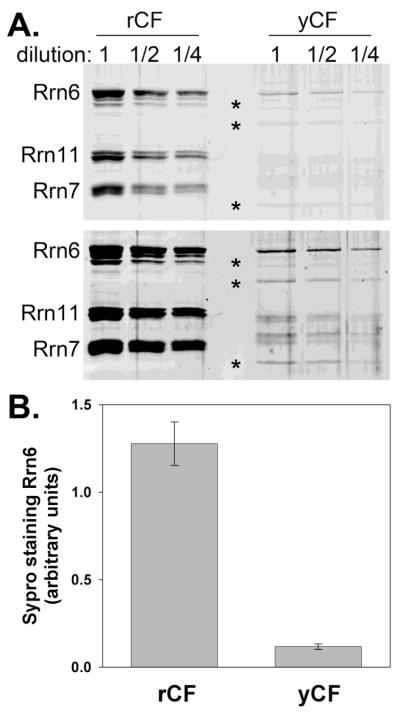
Recombinant core factor is more concentrated than that purified from yeast. (A) Three dilutions of rCF and yCF were run on a 10% SDS PAGE gel and stained with Sypro Ruby Red. The gel was then scanned using a Typhoon fluorescent scanner (excitation, 488 nm; 610 nm emission filter). Two versions of the same scanned gel are shown. Lower image is a higher contrast version with contrast enhancement performed using ImageQuant software (GE, Uppsala, Sweden). Degradation products or impurities are indicated by asterisks. Rrn11 is slightly larger due to an N-terminal his6 tag in rCF, and Rrn7 is larger in yCF due to an N-terminal triple hemagluttinin (HA) tag. In both preparations, Rrn11 and Rrn7 ran as doublets. Every preparation of core factor contains visible Rrn6 degradation products. (B) Rrn6 protein abundance was measured using ImageQuant software. Resulting values were multiplied by their dilution factor, averaged and plotted. Error indicated is one standard deviation + and −.
The plasmids were transformed and co-expressed in BL21(DE3)-pRosetta-2 cells. Induction of RRN6, RRN7, and RRN11 gene expression was accomplished in TB medium (12 g tryptone, 24 g yeast extract and 10 mL of 40% glycerol were added to 870 mL Milli-Q H2O). Just prior to inoculation of the 1 L cultures with overnight cultures, 100 mL of a sterile solution containing 0.17 M KH2PO4 and 0.72 M K2HPO4 and 20 mL of a sterile solution containing 25% (v/v) glycerol, 2.5% (w/v) glucose, and 10% (w/v) lactose was added to the medium.
Following inoculation, the culture was grown at 37°C until A600 = ~0.6. At this point, the cultures were cooled on ice until the media was approximately room temperature. Incubation was continued at 24°C overnight with agitation. The cultures were harvested by centrifugation at 15,000 × g for 20 minutes. The cell pellets were resuspended in breakage buffer (500 mM KCl, 50 mM Tris-HCl pH 7.6, 5 mM MgCl2, 0.1 mM EDTA, 10 mM imidazole, 0.1% (v/v) Tween20, and 20% (v/v) glycerol), and centrifuged again at 30,000 × g for 20 minutes. Cell pellets could be frozen at −80°C or immediately processed.
2.2 Protein purification
The cell pellet was resuspended in breakage buffer [with 2 mM PMSF, 1 mM DTT and “complete” protease inhibitors lacking EDTA; Roche Diagnostics, Indianapolis, IN] and broken with a French press using 3 passes at ~8000 psi. The cell extract was then cleared by centrifugation at 30,000 × g for 20 minutes. The supernatant was added to 5 ml Ni Sepharose 6 Fast Flow resin (GE; Uppsala, Sweden) and incubated at 4°C for 2 hours with gentle agitation. The nickel resin was poured into polypropylene columns (Pierce; Rockford, Illinois) containing 0.75 mL of additional nickel resin and washed with CF wash buffer (200 mM KCl, 50 mM Tris-HCl pH 7.6, 5 mM MgCl2 , 0.1 mM EDTA, 10 mM imidazole, 0.1% (v/v) Tween20, and 20% (v/v) glycerol) and eluted in 7.5 mL of CF elution buffer (200 mM KCl, 50 mM Tris-HCl pH 7.6, 5 mM MgCl2 , 0.1 mM EDTA, 250 mM imidazole, 0.1% (v/v) Tween20, and 20% (v/v) glycerol, 2 mM PMSF).
The nickel eluate was loaded onto a 1 mL HiTrap Heparin HP column (GE; Uppsala, Sweden). A 50 minute gradient was run from 0% Buffer B to 100% Buffer B at 0.5 mL/min, collecting 1 ml fractions. [Buffer A: 200 mM KCl, 50 mM Tris-HCl pH 7.6, 5 mM MgCl2 , 0.1 mM EDTA, 0.1% (v/v) Tween20, and 10% (v/v) glycerol. Buffer B: same as A but contained 1 M KCl.] Peak fractions containing core factor were identified by western blot analysis with a polyclonal anti-Rrn6p antibody or coomassie staining of the gel.
The peak fractions were combined and diluted 1:1 in No-KCl Buffer A and loaded onto a 1 mL Mono-Q 5/50 GL column (GE; Uppsala, Sweden). Using the same Buffer A and Buffer B, a gradient was run from 0 to 80% Buffer B for 50 minutes at 0.5 mL/min, collecting 1 ml fractions. The mono-Q fractions were run on an 8% SDS-PAGE gel, and peak fractions containing core factor were identified by coomassie staining. For comparison of core factor abundance, polyacrylamide gels were stained with Sypro Ruby Red (BioRad Laboratories; Hercules, CA) and scanned on a Typhoon scanner (GE; Uppsala Sweden), according to manufacturers’ instructions.
2.3 In vitro transcription
The Pol I transcription assay was carried out exactly as described (Keener et al., 1998). These assays contain all pure components (linear template DNA, UAF, TBP, Pol I and Rrn3). No semi-pure components or extracts are employed in any transcription analyses described herein. Rrn3 and TBP were recombinantly expressed and purified from E. coli cells, as described (Keener et al., 1998). UAF, Pol I and, where indicated, core factor were purified from yeast cells, also as described previously (Keener et al., 1998). Transcription is initiated by addition of NTPs [200 μM ATP, CTP and UTP; 20 μM GTP (GE, Uppsala, Sweden), and 10 μCi α32P GTP (Perkin Elmer, Waltham, MA)]. No competitor is included, thus these reactions are multi-round assays. After incubation at 23° C for 15 min, assays are stopped by addition of excess phenol. RNA is then harvested by centrifugation and precipitation with ethanol. For the comparison between yeast derived- (yCF) and recombinant core factor (rCF), each master mix was prepared without core factor and the indicated amount of material was added. The +/− UAF and TBP studies were carried out in a similar fashion. Each reaction was run on a 5% TBE-PAGE gel with 7 M urea at 700 V. The gel was dried and exposed to a phosphorimager screen overnight and scanned on a Storm 820 (GE; Uppsala, Sweden).
3. Results
3.1 Purification of recombinant core factor
To facilitate functional characterization of core factor, we developed a recombinant expression and purification strategy for the complex. The three subunits of core factor were co-expressed in E. coli cells carrying the pRosetta-2 plasmid. This plasmid provides tRNAs required for A/U-rich codon usage. Inclusion of this plasmid improved the yield of our proteins considerably.
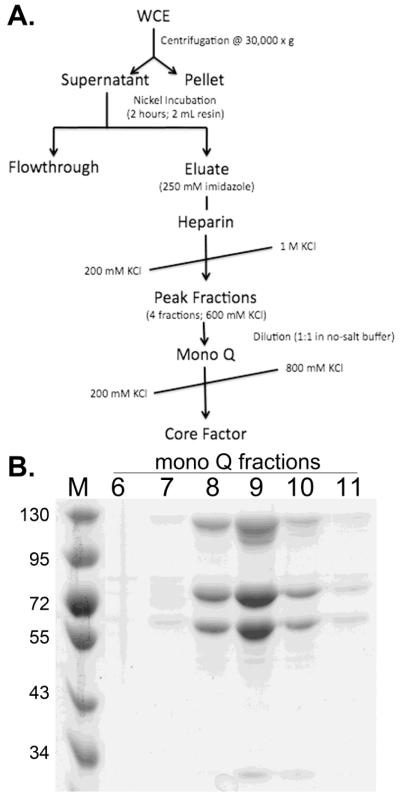
Recombinant core factor used in these studies was purified from 1 L of E. coli using a three-step purification strategy (Figure 1A). To determine the purity of the complex, peak fractions were analyzed by coomassie stain. From the stained gel (Figure 1B, fractions 8-10), three distinct bands are observed. These bands correspond to the Rrn6p, Rrn11p, and Rrn7p subunits of core factor, respectively. The positions of these bands in the gel correlate to the positions of the three core factor subunits previously purified from yeast [(Keener et al., 1998) and Figure 2]. Co-purification of all three subunits of core factor (including untagged Rrn6p) demonstrates that the proteins are assembled into a complex.
Figure 1.
Core factor assembles in E. coli and is purified as a complex. (A) Purification strategy used to obtain pure rCF. (B) Fractions from monoQ gradient were electrophoresed on an 8% SDS-PAGE gel and visualized with Coomasie Blue stain. Fraction numbers are indicated, and the protein molecular weight marker is labeled on the left.
3.2 Relative core factor concentration
Having purified core factor from E. coli, we began characterization of the recombinant material (indicated as rCF) compared to core factor purified from yeast (yCF). For subsequent experiments, we used fraction 9 from the monoQ purification described above (Figure 1B). To measure the relative concentrations of rCF and yCF we ran serial dilutions of rCF and yCF on a 10% SDS PAGE gel and stained the gel with Sypro Ruby Red stain. We used a Typhoon Trio fluorescent scanner for quantitative analysis of protein purity and abundance. We observed substantially higher yield of rCF per volume of sample than yCF (Figure 2A). Using the sensitive Sypro stain, we could readily detect all three core factor subunits in both preparations of core factor (especially with enhanced image contrast, Figure 2A lower panel). Quantification of Sypro signal for each subunit, normalized to the molecular weight of the protein, confirmed 1:1:1 stoichiometry of the 3 subunits in the recombinant complex (Figure 2A, calculation not shown). Furthermore, quantification and comparison of Rrn6 in the rCF and yCF dilutions demonstrated that the rCF sample was approximately 11-fold more concentrated that the yeast-derived core factor (Figure 2B). We note that we used over 200-fold less cell mass to produce rCF than was used for yCF. Thus, expression of the three core factor subunits in E. coli is effective in producing comparatively large amounts of the core factor complex.
3.3 Recombinant core factor activity in vitro
We used our fully reconstituted, promoter-dependent in vitro transcription assay for Pol I (Keener et al., 1998) to compare the activities of rCF and yCF. Recombinant and yeast-derived core factor supported robust transcription, whereas heat inactivation of rCF eliminated transcription entirely (Figure 3A, “H.I.”). Quantification of transcription from a range of yCF and rCF dilutions demonstrated that rCF is ~27-fold more active than yCF per volume (Figure 3B). Based on relative concentrations and relative activities in vitro, we calculated that the specific activity of rCF is ~2.5x higher than yCF (Figure 3B, error propagation is indicated).
Figure 3.
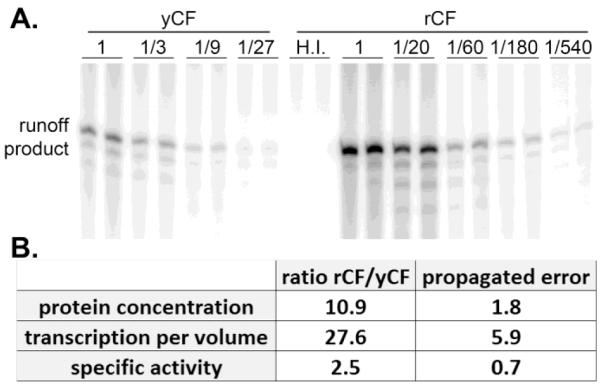
Recombinant core factor has a higher specific activity than yeast-derived core factor. (A) Serial dilutions of yCF and rCF were added to transcription reactions that contained template DNA, UAF, TBP, Pol I and Rrn3. After addition of core factor to the reactions, transcription was initiated by addition of nucleoside triphosphates, including α32P-GTP. Multiple rounds of transcription were permitted and reactions were halted after 15 min by addition of excess phenol. Transcripts were analyzed on denaturing polyacrylamide gels and quantified using a Storm 820 phosphorimager and ImageQuant software (GE, Uppsala, Sweden). Addition of heat-inactivated rCF to the reactions (H.I.) resulted in no transcript accumulation. (B) Product accumulation was measured for all lanes. The resulting values were multiplied by their dilution factor and averaged. Standard deviation of averaged values was calculated. Relative core factor concentration (from Figure 2) and relative transcription per volume of core factor preparation were calculated and are shown. Propagation of error of these values is indicated. Specific activity was calculated as a ratio of these values, again with propagated error indicated.
It is unexpected that a eukaryotic protein complex purified from E. coli should be more active than native product. The yeast-derived product was subject to an additional immunoaffinity purification step in order to achieve the required level of purity. Perhaps this extra handling led to non-specific inactivation of the complex. Additionally, the Rrn7 subunit of yCF carries a relatively bulky triple hemagluttinin (3-HA) tag. Although the addition of this epitope tag did not affect the growth rate of the yeast cells used for purification (unpublished results), it may influence core factor function in vitro, in the absence of potential compensatory effects. Alternatively, there may be covalent modifications of core factor in yeast that reduce activity of the complex. If these modifications were maintained throughout purification, one would expect lower specific activity from yCF. Given the stringency of purification and the resulting stoichiometry of the recombinant core factor complex (Figures 1&2); it is unlikely that we co-purified any specific stimulatory factor or excess core factor subunits from E. coli. Irrespective of the cause for this difference in specific activity, we can conclude that eukaryotic-specific covalent modification of core factor is not required for activity, since we can purify the active complex from E. coli.
3.4 Activation of basal Pol I transcription by TBP
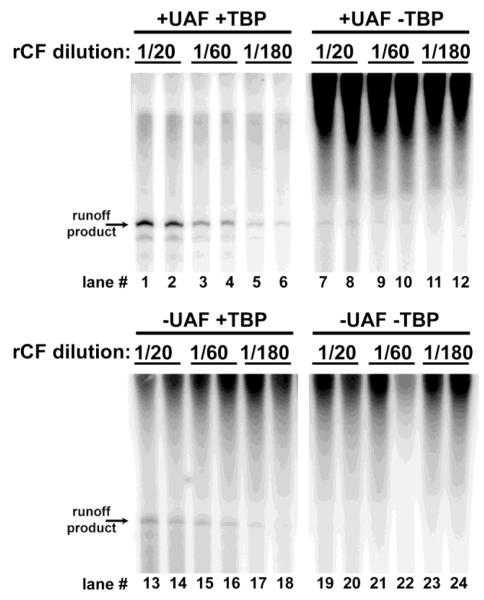
Access to abundant, pure core factor permitted us to reexamine roles for individual factors involved in transcription initiation. In vitro transcription assays performed in the absence of both UAF and TBP revealed very low basal levels of activity, confirming the previous observation that only core factor and Rrn3p are essential for promoter-dependent Pol I transcription in vitro [Figure 4, lanes 19-24; and (Keener et al., 1998)]. Upon the addition of UAF, transcription levels do not change significantly relative to basal levels (Figure 4, lanes 7-12 versus lanes 19-24), indicating that UAF cannot activate transcription without TBP despite a documented interaction between the Rrn9p subunit of UAF and Rrn7p subunit of core factor (Steffan et al., 1996). Addition of TBP to the transcription reactions without UAF, however, does result in activation of promoter-specific transcription (Figure 4, lanes 13-18 versus lanes 19-24). Previous in vivo studies and in vitro studies using whole cell extracts reported TBP-dependent activation of Pol I transcription following overexpression of TBP in yeast (Aprikian et al., 2000). Our results are consistent with these findings. Previous analysis of the role of TBP in Pol I initiation using a reconstituted system did not detect any increase in transcription levels upon the addition of TBP without UAF. The reason for this discrepancy is ultimately unknown. Perhaps the inclusion of the 3-HA tag on Rrn7 influences its association with TBP. Alternatively, there may have been low level contamination of yCF with TBP. In any case, our observations are consistent with those published previously by Reeder and colleagues (Aprikian et al., 2000).
Figure 4.
Core factor is required for transcription in vitro; TBP and UAF enhance activity. Indicated dilutions of rCF were added to in vitro transcription assays with and without UAF and TBP. The gel running conditions were the same as for Figure 3. The −UAF − TBP sample exhibited basal levels of Pol I transcription, whereas the presence of both UAF and TBP induced maximal promoter-specific transcription. The −UAF +TBP sample yielded more product than − UAF −TBP, demonstrating that TBP can activate Pol I transcription without UAF. Lane numbers are indicated for ease of discussion.
Finally, addition of both UAF and TBP yields the highest observed amounts of runoff product (Figure 4, lanes 1-6 versus lanes 7-12). This observation is expected since it is known that UAF, TBP, core factor and Rrn3 are all essential for yeast cell survival.
4. Discussion
4.1 Technical value of rCF purification
Previous methods used to purify core factor for in vitro studies were laborious and expensive. Likewise, previous attempts to express and purify transcriptionally active CF from recombinant systems were unsuccessful (unpublished data). Attempts to purify recombinant SL1 also resulted in very low amounts of pure product due to solubility issues (Comai et al., 1994). We present a successful method for efficient expression and purification of a transcriptionally active multi-subunit Pol I transcription initiation factor. This method is beneficial since amounts of pure CF are no longer limiting for in vitro characterization of Pol I transcription. High yield of the pure, active complex also enables detailed biochemical or biophysical assays of core factor function. Similar procedures can potentially be applied to other transcription factors from yeast and other eukaryotes.
4.2 Biological insights from rCF purification
Comparison of the relative concentration and activity of rCF to yCF revealed that the recombinant product has approximately 2.5-fold higher specific activity than yCF. These data clearly demonstrate that eukaryotic-specific post-translational modifications are not necessary for core factor function in vitro. However, we cannot rule out regulatory modifications in vivo. Previous studies on SL1 have demonstrated that covalent modifications influence its function in vivo (Comai et al., 2000; Banerjee et al., 2005). Given the functional similarity of SL1 to core factor, it is possible core factor subunits are the targets for activating post-translational modifications, and we have failed to identify these sites due to loss during purification or insensitivity of the transcription assay. Alternatively, core factor may be fully functional without covalent modification, and perhaps only down-regulation of activity is controlled by modification.
Our transcription analyses in the presence and absence of various transcription factors (Figure 4) suggests that TBP can activate Pol I transcription in the absence of UAF. We do not know the mechanism by which TBP affects UAF-independent Pol I transcription; however we propose two related but mechanistically distinct models. Since core factor is the functional analogue of SL1, and TBP was identified as a subunit of SL1, perhaps core factor and TBP interact “in solution” prior to interaction of either protein with the rDNA promoter. Alternatively, TBP has known DNA binding affinity, thus it may bind to the promoter and subsequently recruit core factor to the promoter. Reeder and colleagues previously proposed the latter model (Aprikian et al., 2000). However, further studies will be required to determine exactly how core factor is recruited to the rDNA promoter.
4.3 The roles for core factor in Pol I transcription initiation and its relation to other nuclear polymerases
Recent work from the Hahn lab has identified functional and predicted structural similarities between the Rrn7p subunit of core factor and TFIIB (Pol II) and Brf1p (Pol III) (Knutson and Hahn, 2011). This finding is of particular importance, as it demonstrates that similar initiation pathways can be used by each of the three eukaryotic nuclear polymerases. Knutson and Hahn found that recombinantly expressed GST-Rrn7p (and chimeric derivatives) could associate with RNA polymerase I in vitro. These and other data support the conclusion that core factor serves a role in Pol I transcription initiation that is functionally similar to that of TFIIB for Pol II. Future studies using recombinant, intact core factor will lead to a more detailed understanding of the molecular mechanism(s) by which core factor promotes transcription of the rDNA.
If Rrn7p is the functional analogue of TFIIB, what other analogies can be potentially described between the initiation factors for RNA polymerases I and II? The observation that TBP can activate Pol I transcription without UAF suggests that intact core factor may also fulfill roles of the Pol II factor TFIID. Core factor recruits and positions the polymerase on the rDNA promoter by direct interaction with the initiation-competent form of the polymerase (Pol I bound to Rrn3p). We propose that TBP further activates Pol I recruitment by potentially recruiting or stabilizing core factor association with the rDNA promoter. The lack of a true TATA box sequence at the rDNA promoter is also consistent with the previously published observation that TFIID is preferentially associated with TATA-less Pol II promoters (Basehoar et al., 2004; Huisinga and Pugh, 2004). Perhaps, the three subunit core factor complex fills roles of both TFIIB and TFIID in Pol I transcription initiation. This comparison is strictly functional, given the diversity of subunit composition between TFIID and core factor.
As detailed biochemical and genetic analyses accumulate, functional similarities between the initiation pathways for the three nuclear polymerases may continue to emerge (Knutson and Hahn, 2011). However, aggressive, continued investigation is required to characterize the molecular mechanisms by which RNA polymerase I transcription initiation employs comparatively few, simple factors to achieve efficient and robust transcription of the rDNA. Detailed understanding of this initiation pathway and its regulation is critical to understanding or potentially controlling rRNA synthesis during rapid cell proliferation.
5. Conclusions
In this study, we have expressed and purified transcriptionally active core factor from a recombinant system. This result provides technical advantages for future studies. We also performed in vitro assays for Pol I transcription that supported a previously proposed role for TBP in core factor-dependent activation of Pol I transcription. These data demonstrate that this TBP-dependent activation is direct. Since detailed biochemical and biophysical analyses require substantial quantities of pure protein, the purification strategy described here will enable us and others to pursue such studies for Pol I transcription as well as other biological systems.
Highlights.
Co-expression of Rrn6p, Rrn7p and Rrn11p in E. coli produces an active complex.
Recombinant core factor is more active that core factor purified from yeast.
Eukaryotic-specific covalent modifications are not required for core factor activity.
Tata-binding protein can activate Pol I transcription, without UAF, in vitro.
Acknowledgements
We thank Dr. Kirill Popov and his laboratory for use of the french pressure cell and advice. We thank Claudia Blattner (Dr. Patrick Cramer lab) for advice on protein expression conditions. We also thank Dr. Yinfeng Zhang, Krysta Engel and Olga Viktorovskaya for critical analysis of the manuscript and data therein. Funding for this study was generously provided to D.A.S. by the National Institutes of Health, grant #GM84946.
Abbreviations
- RNA polymerase
Pol
- ribosomal RNA
rRNA
- ribosomal DNA
rDNA
- core factor
CF
- upstream activating factor
UAF
- TATA-binding protein
TBP
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aprikian P, Moorefield B, Reeder RH. TATA binding protein can stimulate core-directed transcription by yeast RNA polymerase I. Mol Cell Biol. 2000;20:5269–75. doi: 10.1128/mcb.20.14.5269-5275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee R, Weidman MK, Navarro S, Comai L, Dasgupta A. Modifications of both selectivity factor and upstream binding factor contribute to poliovirus-mediated inhibition of RNA polymerase I transcription. J Gen Virol. 2005;86:2315–22. doi: 10.1099/vir.0.80817-0. [DOI] [PubMed] [Google Scholar]
- Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- Bodem J, Dobreva G, Hoffmann-Rohrer U, Iben S, Zentgraf H, Delius H, Vingron M, Grummt I. TIF-IA, the factor mediating growth-dependent control of ribosomal RNA synthesis, is the mammalian homolog of yeast Rrn3p. EMBO Rep. 2000;1:171–5. doi: 10.1093/embo-reports/kvd032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe SY, Schultz MC, Reeder RH. In vitro definition of the yeast RNA polymerase I promoter. Nucleic Acids Res. 1992;20:279–85. doi: 10.1093/nar/20.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L, Song Y, Tan C, Bui T. Inhibition of RNA polymerase I transcription in differentiated myeloid leukemia cells by inactivation of selectivity factor 1. Cell Growth Differ. 2000;11:63–70. [PubMed] [Google Scholar]
- Comai L, Zomerdijk JC, Beckmann H, Zhou S, Admon A, Tjian R. Reconstitution of transcription factor SL1: exclusive binding of TBP by SL1 or TFIID subunits. Science. 1994;266:1966–72. doi: 10.1126/science.7801123. [DOI] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 2004;13:573–85. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- Keener J, Dodd JA, Lalo D, Nomura M. Histones H3 and H4 are components of upstream activation factor required for the high-level transcription of yeast rDNA by RNA polymerase I. Proc Natl Acad Sci U S A. 1997;94:13458–62. doi: 10.1073/pnas.94.25.13458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener J, Josaitis CA, Dodd JA, Nomura M. Reconstitution of yeast RNA polymerase I transcription in vitro from purified components. TATA-binding protein is not required for basal transcription. J Biol Chem. 1998;273:33795–802. doi: 10.1074/jbc.273.50.33795. [DOI] [PubMed] [Google Scholar]
- Keys DA, Lee BS, Dodd JA, Nguyen TT, Vu L, Fantino E, Burson LM, Nogi Y, Nomura M. Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev. 1996;10:887–903. doi: 10.1101/gad.10.7.887. [DOI] [PubMed] [Google Scholar]
- Keys DA, Vu L, Steffan JS, Dodd JA, Yamamoto RT, Nogi Y, Nomura M. RRN6 and RRN7 encode subunits of a multiprotein complex essential for the initiation of rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae. Genes Dev. 1994;8:2349–62. doi: 10.1101/gad.8.19.2349. [DOI] [PubMed] [Google Scholar]
- Knutson BA, Hahn S. Yeast Rrn7 and human TAF1B are TFIIB-related RNA polymerase I general transcription factors. Science. 2011;333:1637–40. doi: 10.1126/science.1207699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos M, Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol Cell. 2010;37:809–20. doi: 10.1016/j.molcel.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalo D, Steffan JS, Dodd JA, Nomura M. RRN11 encodes the third subunit of the complex containing Rrn6p and Rrn7p that is essential for the initiation of rDNA transcription by yeast RNA polymerase I. J Biol Chem. 1996;271:21062–7. doi: 10.1074/jbc.271.35.21062. [DOI] [PubMed] [Google Scholar]
- Lin CW, Moorefield B, Payne J, Aprikian P, Mitomo K, Reeder RH. A novel 66-kilodalton protein complexes with Rrn6, Rrn7, and TATA-binding protein to promote polymerase I transcription initiation in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:6436–43. doi: 10.1128/mcb.16.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorefield B, Greene EA, Reeder RH. RNA polymerase I transcription factor Rrn3 is functionally conserved between yeast and human. Proc Natl Acad Sci U S A. 2000;97:4724–9. doi: 10.1073/pnas.080063997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M, Nogi Y, Oakes M. Transcription of rDNA in the Yeast Saccharomyces cerevisiae. In: Olson MOJ, editor. The Nucleolus. Kluwer Academic / Plenum Publishers; London: 2004. pp. 128–153. [Google Scholar]
- Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol Cell. 2004;16:943–54. doi: 10.1016/j.molcel.2004.11.031. [DOI] [PubMed] [Google Scholar]
- Peyroche G, Milkereit P, Bischler N, Tschochner H, Schultz P, Sentenac A, Carles C, Riva M. The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. Embo J. 2000;19:5473–82. doi: 10.1093/emboj/19.20.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D. RNA polymerase I activity is regulated at multiple steps in the transcription cycle: recent insights into factors that influence transcription elongation. Gene. 2011 doi: 10.1016/j.gene.2011.08.006. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan JS, Keys DA, Dodd JA, Nomura M. The role of TBP in rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae: TBP is required for upstream activation factor-dependent recruitment of core factor. Genes Dev. 1996;10:2551–63. doi: 10.1101/gad.10.20.2551. [DOI] [PubMed] [Google Scholar]
- Steffan JS, Keys DA, Vu L, Nomura M. Interaction of TATA-binding protein with upstream activation factor is required for activated transcription of ribosomal DNA by RNA polymerase I in Saccharomyces cerevisiae in vivo. Mol Cell Biol. 1998;18:3752–61. doi: 10.1128/mcb.18.7.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–40. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]