Abstract
Background
Chronic rhinosinusitis with nasal polyps (CRSwNP) is associated with Th2-dominant inflammation including eosinophilia, in contrast to non-polypoid CRS (CRSsNP). Chemokine CCL18/PARC (pulmonary and activation regulated chemokine) is known to recruit naïve T cells, B cells, and immature dendritic cells, as well as activate fibroblasts. CCL18is thought to be involved in Th2-related inflammatory diseases including asthma and atopic dermatitis.
Objectives
The objective of this study was to investigate the expression of CCL18 in patients with CRS.
Methods
Using nasal polyp tissue (NP) and uncinate tissue (UT) from controls and patients with CRS, we examined the expression of CCL18 mRNA by real-time PCR and measured CCL18 protein by ELISA, western blot and immunofluorescence.
Results
Compared to UT tissue in control subjects, CCL18 mRNA was significantly increased in NP (p<0.001) and UT (p<0.05) from patients with CRSwNP but not in UT from patients with CRSsNP. Similarly, CCL18 protein was elevated in NP and UT from CRSwNP and levels were even higher in Samter’s triad patients. Immunohistochemical analysis revealed CCL18 expression in inflammatory cells and CCL18+ cells were significantly increased in NP. Immunofluorescence data showed co-localization of CCL18 in CD68+/CD163+/macrophage mannose receptor+ M2 macrophages and tryptase+ mast cells in NP. Levels of CCL18 correlated with markers of M2 macrophages but not with tryptase, suggesting that M2 macrophages are a major CCL18-producing cells in NP.
Conclusion
Overproduction of CCL18 might contribute to the pathogenesis of CRSwNP through its known activities, which include recruitment of lymphocytes and dendritic cells, activation of fibroblasts, and initiation of local inflammation.
Keywords: Chronic rhinosinusitis, Nasal polyps, Samter’s triad, CCL18, PARC, M2 macrophages, Mast cells
INTRODUCTION
Chronic rhinosinusitis (CRS) is a chronic disease that afflicts roughly 10% of the U.S population.1–3 CRS is characterized by inflammation of the sinonasal mucosa. Symptoms of CRS include anterior and/or posterior rhinorrhea, nasal obstruction, decreased sense of smell and nasal pressure, at least two of which persist for 12 weeks despite medical management.1–3 The pathogenesis of CRS is not fully understood at this time, however allergy, bacterial and fungal infections as well as structural anomalies have all been theorized to play a role.2–5 CRS is often divided into two groups based on histology and physical examination: CRS with nasal polyps (CRSwNP) and CRS without nasal polyps (CRSsNP). In general, CRSwNP is associated more closely with clinical complaints of nasal obstruction and olfactory loss and is characterized by eosinophilia and Th2-related inflammation especially in western countries.5, 6 It is also known that B cells, plasma cells, macrophages, neutrophils and dendritic cells (DCs) are present in high concentrations in patients with CRSwNP, making inflammatory cell recruitment a contributor to the pathogenesis of this disease.5–13
Chemokines are a large group of proteins that participate in recruitment of inflammatory cells into tissue sites by binding G protein coupled receptors on their target cells.14, 15 They can be classified into several groups based on their molecular structure. While some chemokines are involved in housekeeping functions of the adaptive immune system, others are produced under pathological conditions in order to mount immune and inflammatory responses and initiate wound healing by selectively recruiting immune cells to the inflamed site.14–16 Exaggerated expression of chemokines can result in tissue damage and inflammatory diseases via excessive recruitment of leukocytes. Therefore, chemokine receptors serve as potential targets for the treatment of many inflammatory diseases.15–18
CCL18, also known as pulmonary and activation regulated chemokine (PARC), MIP-4, AMAC-1 and DC-CK1, is a 7.8 kDa protein composed of 69 amino acids.19–22 CCL18 is thought to have been generated by the fusion of two CCL3-like genes likely occurring after the diversification of rodents and primates.22–24 Therefore the ortholog of CCL18 in rodents has not been discovered. Although its receptor is also unknown at this time, CCL18 is known to be chemotactic for many cell types, including naïve T cells, skin homing memory T cells, Th2 cells and immature DCs.19, 23, 25 CCL18 is known to be expressed in lung, thymus, and lymph nodes and is secreted by monocytes, macrophages, DCs and mast cells.22, 26 Its expression is regulated by the Th2 cytokines IL-4 and IL-13, as well as by IL-10.19, 21, 22 CCL18 is highly expressed in a number of inflammatory diseases including atopic dermatitis, asthma, rheumatoid arthritis and pulmonary fibrosis.22, 25, 27–29 While several chemokines are known to be elevated in CRS, particularly in CRSwNP, expression of CCL18 has been minimally studied in this disease process. In this study, we found that CCL18 was elevated in patients with CRSwNP and examined the cell types that produce it.
METHODS
Patients and biopsies
CRS patients were recruited from the Allergy-Immunology clinic and the Otolaryngology clinic of the Northwestern Medical Faculty Foundation (NMFF) and the Northwestern Sinus Center at NMFF. Sinonasal and NP tissues were obtained from routine Functional Endoscopic Sinus Surgery in patients with CRS. All subjects met the criteria for CRS as defined by the American Academy of Otolaryngology-Head and Neck Surgery Chronic Rhinosinusitis Task Force.1 Patients with an established immunodeficiency, pregnancy, coagulation disorder, Churg-Strauss syndrome, diagnosis of classic allergic fungal sinusitis or cystic fibrosis were excluded from the study. Details of subjects’ characteristics are included in Table I, Table E1–3 and in the Methods section in the Online Repository. All subjects signed informed consent forms and the protocol governing procedures for this study has been approved by the Institutional Review Board of Northwestern University Feinberg School of Medicine.
Table 1.
Subject characteristics
| Control | CRSsNP | CRSwNP | Samter’s | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total no. of subjects | n=28 (14M) | n=55 (26M) | n=92 (65M) | n=12 (6M) | ||||||||
| Age (y), median (range) | 48* (16–63)# | 38 (18–64) | 43 (22–74) | 47 (26–61) | ||||||||
| Y | N | U | Y | N | U | Y | N | U | Y | N | U | |
| Atopy | 0 | 24 | 4 | 24 | 25 | 6 | 42 | 40 | 10 | 7 | 2 | 3 |
| Asthma | 0 | 27 | 2 | 12 | 33 | 10 | 40 | 51 | 1 | 12 | 0 | 0 |
| Methodologies used | UT | UT | UT | NP | NP | |||||||
| Tissue RNA | n=9 (3M) | n=18 (8M) | n=17 (14M) | n=24 (18M) | - | |||||||
| Age | 41 (16–59) | 36 (23–64) | 43 (26–61) | 44 (26–74) | - | |||||||
| Tissue Extract | n=15 (9M) | n=32 (15M) | n=29 (20M) | n=39 (28M) | n=12 (6M) | |||||||
| Age | 49 (19–63) | 37 (18–64) | 41 (18–67) | 41 (22–72) | 47 (26–61) | |||||||
| Immunofluorescence | n=11 (8M) | n=11 (6M) | n=11 (9M) | n=17 (11M) | - | |||||||
| Age | 53 (19–62) | 38 (21–61) | 48 (34–72) | 52 (22–70) | - | |||||||
median,
(range). M; male, Y; yes, N; no, U; unknown.
Real-time PCR
Real-time RT-PCR was performed using the TaqMan method, as described previously.30 Primer and probe sets for CCL18 (sense, 5′-CCAGCTCACTCTGACCACTTCTC-3′; antisense, 5′-GGTGCAGACGAGGACAAGGA-3′; probe, 5′-CTGCCCAGCATCATGAAGGGCCTT-3′) were purchased from Integrated DNA Technologies (Coralville, IA). The mRNA expression levels were normalized to the median expression of the housekeeping gene, β-glucuronidase. Details can be found in the Methods section in the Online Repository.
ELISA, Western blot and cell culture
The concentration of CCL18 in cell free supernatants was determined by specific ELISA kits. The minimal detection limit for these kits is 3.9 pg/ml (R&D systems, Minneapolis, MN). The concentration of CCL18 in tissue homogenate was normalized to the concentration of total protein. Western blot analysis was performed as described previously.31 Human mast cells were obtained as described previously.26 Further details can be found in the Methods section in the Online Repository.
Immunofluorescence assay
Immunofluorescent localization of CCL18 was performed as described previously.13 Briefly, blocked sections were incubated with anti-CCL18 antibody (LifeSpan Biosciences, Seattle, WA) and either anti-CD68 mAb (Thermoscientific), anti-ECP mAb (Diagnostics Development, Sweden) or anti-tryptase mAb (Thermoscientific) at 4°C overnight. After washing, sections were incubated with Alexa Fluor-conjugated secondary antibodies for 1 hour in the dark. After a final washing, coverslips were mounted onto slides using SlowFade Gold with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen). Details can be found in the Methods section in the Online Repository.
Statistics
All data are reported as the mean ± SEM unless otherwise noted. Differences between groups were analyzed using the Mann-Whitney U-test and the one-way ANOVA Kruskal-Wallis test. Correlations were assessed using the Spearman’s rank correlation. A p value of less than 0.05 was considered significant.
RESULTS
CCL18 expression in CRS
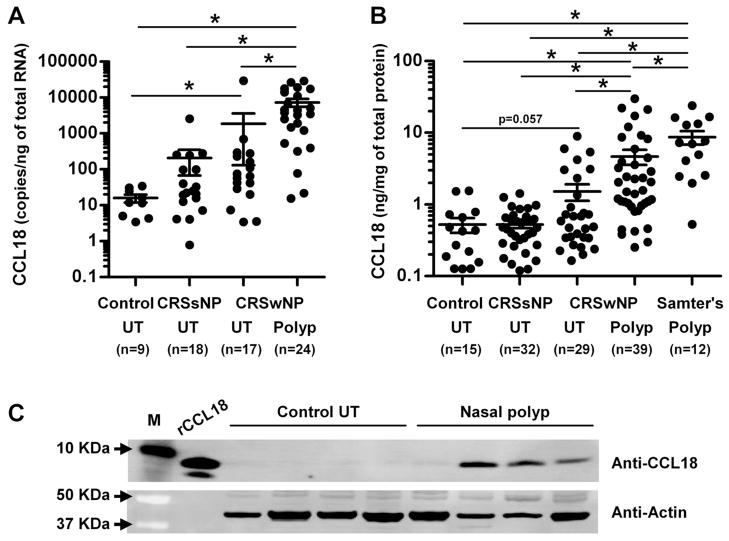
We assessed the expression of CCL18 in UT from patients with CRSsNP, CRSwNP and controls, as well as in NPs from patients with CRSwNP. CCL18 mRNA was significantly increased in NPs from patients with CRSwNP (p<0.001) in comparison to UT from patients with either form of CRS or control subjects (p<0.001, Kruskal-Wallis test; Fig 1, A). CCL18 mRNA was also significantly increased in UT from patients with CRSwNP (p<0.001) in comparison to UT from control subjects (Fig 1, A). To confirm this observation at the protein level, we made detergent extracts from homogenates of UT and NPs and then measured the concentration of CCL18 using ELISA. CCL18 protein was significantly elevated in NPs from CRSwNP (4.75 ± 1.13 ng/mg (median; 1.58 ng/mg); n=39) compared to UT from CRSsNP (0.52 ± 0.06 ng/mg (median; 0.46 ng/mg); n=32), CRSwNP (1.52 ± 0.40 ng/mg (median; 0.59 ng/mg); n=29) and control subjects (0.52 ± 0.12 ng/mg (median; 0.42 ng/ml); n=15) (p<0.001, Kruskal-Wallis test; Fig 1, B). Although not statistically significant, CCL18 protein was also elevated in UT from CRSwNP (p=0.057) compared to UT from controls (Fig 1, B). We further confirmed this finding by western blot. The CCL18 protein was detected as a 7.8 KDa protein and found only in NPs but not in UT from patients with CRSsNP or control subjects (Fig 1, C and data not shown).
Figure 1.
Increased expression of CCL18 in nasal polyps. Total RNA was extracted from uncinate tissue (UT) and nasal polyps (Polyp) and expression of CCL18 RNA was analyzed using real-time PCR (A). Expression of CCL18 protein in tissue homogenates of UT and polyp from patients with aspirin-tolerant CRSwNP and Samter’s triad was measured using ELISA (B) and western blot (C). CCL18 concentration was normalized to the concentration of total protein (B). The results are representative of two separate experiments (C).* p < 0.05.
Nasal polyps in aspirin sensitive patients have generally more inflammation than in aspirin tolerant patients.32 Patients with clinical features including bronchial asthma, aspirin sensitivity and NPs are referred to as having Samter’s triad.32–34 Interestingly, we found that the concentration of CCL18 in NPs was significantly higher in Samter’s triad patients (9.92 ± 1.93 ng/mg (median; 7.13 ng/mg); n=12, p=0.0015) compared to NPs from patients with aspirin tolerant CRSwNP (Fig 1, B).
Identification of CCL18 producing cells in nasal polyps
To examine the distribution of CCL18 in nasal mucosa, we used immunohistochemistry to detect CCL18+ cells in UT and NPs. As shown in Fig 2, CCL18 staining was observed mainly in submucosal inflammatory cells. Although CCL18 staining was also observed in some mucosal and glandular epithelium, levels of mRNA for CCL18 in epithelial cell scrapings from NPs were more than 500-fold lower than levels in whole NP tissue and we could not find elevation of CCL18 protein in nasal lavage from patients with CRSwNP (Fig 1, 2 and E1, and data not shown). We therefore excluded epithelial cells as major CCL18 producing cells and focused on inflammatory cells in this study. We found that CCL18+ inflammatory cells were highly elevated in NPs (Fig 2, D). We counted the number of CCL18+ inflammatory cells using a semi- quantitative method and found that CCL18+ inflammatory cells were significantly elevated in the NPs from CRSwNP (2.37 ± 0.43 CCL18+ cells/high power field (HPF); n=17) compared to UT from CRSsNP (1.16 ± 0.40 CCL18+ cells/HPF, p<0.05; n=11), CRSwNP (0.75 ± 0.18 CCL18+ cells/HPF, p<0.05; n=11) and controls (0.86 ± 0.19 CCL18+ cells/HPF, p<0.05; n=11) (p<0.01, Kruskal-Wallis test; Fig 2, F).
Figure 2.

Immunofluorescence of CCL18 was performed with an anti-human CCL18 antibody. Representative immunostaining (green) for CCL18 in uncinate tissue (UT) from a control subject (A), a patient with CRSsNP (B), a patient with CRSwNP (C), and in nasal polyp tissue (D). Negative control antibody staining in nasal polyp tissue from a patient with CRSwNP (E) is shown. Nuclei were counterstained with DAPI (blue). The number of CCL18 positive cells in UT from control (n=11), CRSsNP (n=11) and CRSwNP (n=11) and in nasal polyps (n=17) was counted using the NIH-issued Image J software (F). * p < 0.05.
We next attempted to identify the CCL18 producing cells in the nasal mucosa of NPs. We first assessed whether the expression of CCL18 correlated with the levels of mRNA expression for markers of inflammatory cells in NPs by real-time PCR. We found that the levels of expression of CCL18 mRNA in NPs were significantly and positively correlated with the expression of macrophage mannose receptor (MMR, r=0.796, p<0.0001) and Charcot-Leyden crystal protein (CLC, r=0.695, p=0.0002) but not with CD3, CD20, CD56, CD138, CD1c, tryptase or CXCR1 (Fig 3, A and data not shown), indicating that CCL18 might be expressed in macrophages and eosinophils.
Figure 3.
Detection of CCL18-producing cells in nasal polyps. Total RNA was extracted from nasal polyp tissue and the expression of CCL18 and cell specific markers was analyzed by real-time PCR (n=24) (A). The correlations were assessed using the Spearman rank correlation. An immunofluorescence assay was performed using anti-CCL18 (green), anti-CD68 mAb (red) for macrophages (B), anti-ECP mAb (red) for eosinophils (C), anti-tryptase mAb (red) for mast cells (D) and control IgG (E). Nuclei were counterstained with DAPI (blue). The results are representative of 5 to 8 separate patients.
We next performed dual immunofluorescence analysis using anti-CCL18 and antibodies against markers of macrophages (CD68) and eosinophils (ECP). We found CCL18 co- localization with CD68+ macrophages (Fig 3, B) but not with ECP+ eosinophils (Fig 3, C) in NPs. In addition to macrophages, we also found co-localization of CCL18 with tryptase+ mast cells (Fig 3, D). Although there was variability from patient to patient, 20–50% of CCL18+ cells were macrophages, 10–60% of CCL18+ cells were mast cells and another 10–50% of CCL18+ cells were unidentified inflammatory cells in NPs (n= 8; data not shown).
We have previously shown that mast cells are able to produce CCL18 during IgE- mediated reactions.26 However, the effect of cytokines on the production of CCL18 is still unknown. Since we observed CCL18+ mast cells in NPs (above), we stimulated IgE-sensitized human peripheral blood-derived mast cells with cytokines TNF, IL-1β, IL-4, IL-13, IFN-γ and IL-17A alone or in combination with anti-IgE antibody in the presence of protease inhibitor cocktail (PIC) or DMSO (vehicle control) for 48 hours to assess production of CCL18. We found that the cytokines alone did not induce CCL18 production but IL-4, IL-13 and IFN-γ significantly enhanced IgE-mediated CCL18 production only in the presence of PIC (Fig E2 and data not shown). However, levels of CCL18 in mast cell culture supernatants were very low. In addition, levels of CCL18 mRNA did not correlate with levels of tryptase in NPs (Fig 3). This suggested to us that mast cells may not be a major CCL18 producing cell type in NPs and we therefore focused on macrophages.
Detection of CCL18 in M2 macrophages
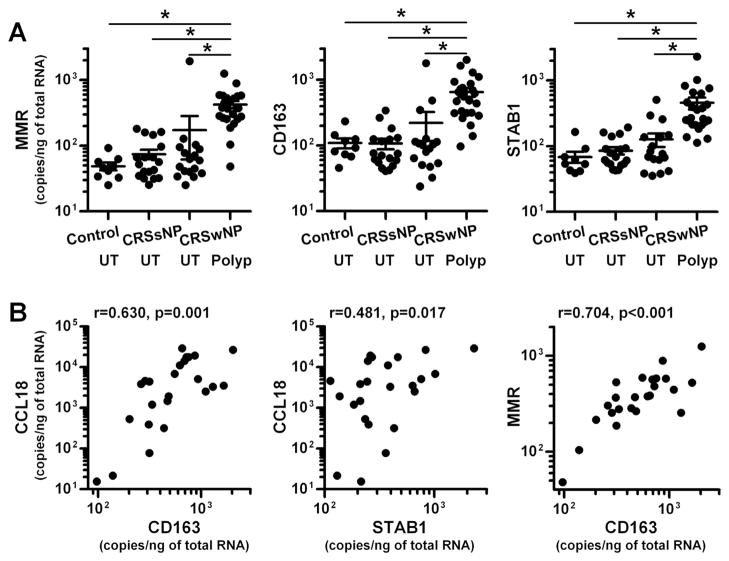
Macrophages are now widely recognized to be polarized by their microenvironment, especially by Th cytokines and pathogens.35–39 Classically activated macrophages (also known as M1 macrophages) are developed by the stimulation with Th1 cytokine IFN-γ and microbial products such as LPS. In contrast, alternatively activated macrophages are primed by Th2 cytokines IL-4 and IL-13, and therefore are called M2 macrophages. Importantly, CCL18 is known as a useful marker of M2 macrophages.36–39 In vitro generated M2 macrophages or IL-4- stimulated monocytes/macrophages are known to release large amounts of CCL18. In addition, we found that expression of CCL18 was positively correlated in NPs with MMR, which is another well-known marker of M2 macrophages (Fig. 3, A). Therefore we next examined whether M2 macrophages were major CCL18 producing cells in NPs. We first determined levels of MMR and other M2 macrophages markers, CD163 and stabilin 1 (STAB1), in NPs by real- time PCR. Levels of mRNA for MMR, CD163 and STAB1 were significantly upregulated in NPs (p<0.05) in comparison with those seen in UT from either patients with CRS or control subjects (Fig 4, A). We also found that expression of CCL18 significantly and positively correlated with CD163 (r=0.630, p=0.001) and STAB1 (r=0.481, p=0.017) (Fig 4, B). Importantly, levels of MMR also significantly correlated with CD163 (r=0.704, p<0.001) and STAB1 (r=0.593, p=0.002) (n=24, Fig 4, B and data not shown).
Figure 4.
Correlation of CCL18 with markers of M2 macrophages in nasal polyps. Total RNA was extracted from UT from controls (n=9), CRSsNP (n=18) and CRSwNP (n=17) and nasal polyp tissue (n=24) (A). The expression of CCL18 and M2 macrophage markers, MMR, CD163 and STAB1 was analyzed by real-time PCR. The correlations in nasal polyps were assessed by using the Spearman rank correlation (B). * p < 0.05.
To further investigate whether M2 macrophages were CCL18 producing cells in NPs, we performed triple immunofluorescence analysis using anti-CCL18 and antibodies against markers of M2 macrophages including MMR and CD163. We detected CCL18 in CD68+, MMR+ cells and CD163+, MMR+ cells in NPs (Fig 5). These data suggest that M2 macrophages are major CCL18 producing cells in NPs.
Figure 5.
Detection of CCL18 in M2 macrophages in nasal polyps. An immunofluorescence assay was performed using anti-CCL18 (A and B; green), anti-CD68 mAb (A; orange), anti-CD163 mAb (B; orange), anti-MMR mAb (A and B; red), and control IgG (C). Nuclei were counterstained with DAPI (blue). The results are representative of four separate patients.
DISCUSSION
It has been reported that CCL18 is elevated in affected tissues in patients with Th2-related inflammatory diseases and that expression of CCL18 is regulated by Th2 cytokines.22, 28, 29 CRSwNP is well known to be characterized by Th2-mediated inflammation and eosinophilia.5, 6 In the current study we demonstrated that CCL18 was significantly increased in NPs (Fig 1). There are very few studies comparing identical tissue taken from patients with CRSsNP and CRSwNP and from controls, and most of those studies show elevation or reduction in NPs compared to non-polypoid tissue. In this study, we present rare evidence that up-regulation of mRNA and protein for CCL18 was also found in UT, which is one of the surrounding mucosal tissues in close proximity to NPs, from patients with CRSwNP compared to UT from control subjects (Fig 1). This suggests that CCL18 may be involved in the initiation of the inflammatory process in patients with CRSwNP. Interestingly CRSwNP in China has been reported to have less eosinophilia and Th2 inflammation than in Western countries.40 It would be of interest to determine whether CCL18 elevation exists in this form of polypoid CRS.
Very recently Plager et al published a microarray study of asthmatic CRSwNP and showed that CCL18 mRNA was upregulated in NPs compared to control tissue.41 Our study unequivocally demonstrates elevation of CCL18 in CRSwNP. Plager et al focused on the expression of CCL18 only in asthmatic CRSwNP.41 When we evaluated whether overexpression of CCL18 in patients with CRSwNP was associated with asthmatic status, atopic status or glucocorticoid treatment, we found no significant difference in CCL18 levels between atopic and non atopic patients, asthmatic and non asthmatic patients or status of nasal steroid treatment (Fig E4). Since we might not have large enough subgroups of patients to make firm conclusions about the effect of oral steroid treatment, future studies will be required to test in larger cohort. In contrast, we found that levels of CCL18 in NPs were highest in patients with Samter’s triad compared to patients with aspirin tolerant CRSwNP (Fig 1). In general, tissues from aspirin sensitive patients are more inflamed than in aspirin tolerant patients.32 This suggests that levels of CCL18 in patients with CRSwNP may be related to severity of inflammation rather than asthmatic or atopic status.
It is well established that CCL18 is produced by monocytes and macrophages. Our immunofluorescence data clearly show that CCL18 was detected in CD68+ macrophages as well as tryptase+ mast cells. This suggests that macrophages and mast cells may be major CCL18 producing cells in NPs. However, the regulation of CCL18 in mast cells is poorly understood. We showed previously that mast cells could release CCL18 upon FcεRI cross-linking.26 In our present study, we investigated the effect of cytokines on the production of CCL18 in mast cells. We found that the Th1 cytokine IFN-γ and Th2 cytokines IL-4 and IL-13 significantly enhanced IgE-mediated CCL18 production in mast cells. This suggests that the effect of IL-4 and IL-13 on the induction of CCL18 is well conserved across cell types. In contrast, IFN-γ is known to inhibit CCL18 expression in monocytes, macrophages and DCs, enhance it in keratinocytes and has no effect in PBMC.21, 25, 29 These data suggest that regulation of CCL18 by IFN-γ may be cell type specific. Although we detected CCL18 protein in mast cells (20–30 pg/106 cells, Fig E2), these levels were more than 50-fold lower than those found to be produced by monocytes/macrophages (1–15 ng/106 cells).25, 37, 42–44 These results suggest that mast cells may be a less important source of CCL18 in NPs.
CCL18 was detected in stimulated mast cells only in the presence of protease inhibitors. This demonstrates that CCL18 is highly sensitive to mast cell proteases. In a separate clinical study, we have found that mast cells are increased in NPs and some of them are degranulated (Takabayashi T, Kato A and Schleimer RP, manuscript in preparation). Although CCL18 mRNA was 500-fold higher in NP than control UT, up-regulation of CCL18 protein was only nine-fold. This suggests that local CCL18 production in NPs may be much more abundant but that a portion may be cleaved immediately by proteases from degranulated mast cells or other tissue cells. This may explain the variability of CCL18 protein levels in NPs. Future studies will be required to identify mast cells proteases that can cleave CCL18 and to investigate the effect of those proteases on the stability of CCL18 in NPs in vitro and in vivo.
Although CRSwNP is well known to be characterized by eosinophilia and Th2-related inflammation, the role of macrophages in the pathogenesis of CRS is poorly understood. Recently, Krysko et al showed that MMR+ M2 macrophages but not M1 macrophages were increased in NPs.12 In our present study, we also found that M2 macrophage markers, MMR, CD163 and STAB1, were significantly upregulated in NPs (Fig 4). This suggests that upregulation of M2 macrophages in NPs should be considered as a feature of this disease. CCL18 is well known to be released from M2 macrophages.37–39 We found that CCL18 was detected in MMR+ and CD163+ macrophages but not in MMR− macrophages in NPs (Fig 5 and data not shown). In addition, we found that levels of M2 macrophage markers correlated with levels of CCL18 (Fig 4). As discussed above, mast cells were able to produce CCL18 but the levels were much weaker than what has been reported in macrophages. These results suggest that M2 macrophages are the major CCL18 producing cell type in NPs. We also found that CCL18 was increased in UT from patients with CRSwNP compared to UT in healthy subjects (Fig 1). In contrast to NPs, we could not find significant elevation of M2 macrophages or mast cells in UT from CRSwNP (Fig 4 and data not shown). Although CCL18 producing cells were not elevated in UT by immunofluorescence, the elevated protein level suggests that local cells produce it. Future studies will be required to identify the CCL18 producing cells in UT from patients with CRSwNP and whether they are distinct from the CCL18 producing cells in NPs.
Although we found that M2 macrophages were major CCL18 producing cells, little is known about the regulation of macrophage recruitment in NPs. Recently we showed that CCL23 was highly upregulated in eosinophilic CRSwNP and eosinophils were a major source of CCL23 in NPs.13 CCL23 is a known ligand of CCR1 and is able to recruit monocytes and macrophages.45 Importantly, we showed that levels of CCL23 were significantly correlated with levels of CCR1 and MMR in NPs.13 Since NPs are usually characterized by eosinophilia and Th2-related inflammation, eosinophil-derived CCL23 might be involved in the recruitment of monocytes and macrophages to NPs, followed by polarization of the recruited cells to the M2 phenotype by Th2 cytokines. M2 macrophages are now known to be a rich source of eotaxins.37–39 We found here that CCL18 was mainly produced by M2 macrophages in NPs. These findings suggest that accumulation and activation of M2 macrophages in NPs may further enhance tissue eosinophilia by the production of eotaxins and may induce local adaptive immunity by the production of CCL18 and its subsequent recruitment of naïve T cells and immature DCs. Importantly, both T cells and DCs were elevated in NPs.5, 6, 13 Future studies will be required to determine the relationship between these chemokines and inflammation in NPs.
We found that CLC, which is a marker of eosinophils, significantly and positively correlated with CCL18 in NPs. Since CCL18 has been described as an antagonist of CCR3, which is predominantly expressed on eosinophils,46 we thought eosinophils may contribute to the production of CCL18. In this regard, Schraufstatter et al showed that peripheral blood eosinophils were able to produce CCL18.42 We also found that peripheral blood eosinophils were able to release small amounts of CCL18 (40–50 pg/106 cells) but it was not enhanced by eosinophil activators or Th2 cytokines (Fig E3). Furthermore, our immunofluorescence data demonstrated that CCL18 was not detected in eosinophils in NPs (Fig 3). Another possibility is that CCL18 may be involved in the recruitment of eosinophils via a CCR3-independent manner. However, Nibbs et al showed that CCL18 inhibited CCL11-dependent peripheral blood eosinophil migration.46 It is known that there are phenotypic differences between peripheral blood eosinophils and tissue eosinophils.47 Therefore it is still possible that CCL18 is involved in the recruitment of NP eosinophils. Future studies will be required to investigate whether CCL18 contributes to the recruitment of eosinophils and whether eosinophils are a source of CCL18 in NPs.
In addition to its chemotactic activity, CCL18 is known to activate fibroblasts. CCL18 activated Sp1 and PKCα, and induced collagen production in lung fibroblasts.48, 49 Proliferation and activation of fibroblasts are considered to be important events leading to NP formation.32 Therefore the role of CCL18 in these processes is worthy of further investigation.
In summary, we report here that M2 macrophages and mast cells produce the chemokine CCL18 and that patients with CRSwNP have increased levels of CCL18 in polypoid tissue as well as in surrounding tissues. Our findings indicate that CCL18 is a novel marker in patients with CRSwNP and that the overproduction of CCL18 in NPs might contribute to the pathogenesis of CRSwNP.
Acknowledgments
Funding: This research was supported in part by NIH grants, R01 HL078860, R01 AI072570 and R37 HL068546 and by a grant from the Ernest S. Bazley Trust.
Abbreviations
- CRS
Chronic rhinosinusitis
- NP
nasal polyp
- CRSwNP
CRS with nasal polyps
- CRSsNP
CRS without nasal polyps
- CCL18
CC Chemokine Ligand 18
- ECP
eosinophil cationic protein
- UT
uncinate tissue
- NP
nasal polyp
- MMR
macrophage mannose receptor
- CLC
Charcot-Leyden crystal protein
- DAPI
4′,6-diamidino-2-phenylindole
Footnotes
Clinical implications: Overexpression of CCL18 in NPs may have a pathogenic role in CRSwNP.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Meltzer EO, Hamilos DL, Hadley JA, Lanza DC, Marple BF, Nicklas RA, et al. Rhinosinusitis: establishing definitions for clinical research and patient care. J Allergy Clin Immunol. 2004;114:155–212. doi: 10.1016/j.jaci.2004.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kern RC, Conley DB, Walsh W, Chandra R, Kato A, Tripathi-Peters A, et al. Perspectives on the etiology of chronic rhinosinusitis: an immune barrier hypothesis. Am J Rhinol. 2008;22:549–59. doi: 10.2500/ajr.2008.22.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dykewicz MS, Hamilos DL. Rhinitis and sinusitis. J Allergy Clin Immunol. 2010;125:S103–15. doi: 10.1016/j.jaci.2009.12.989. [DOI] [PubMed] [Google Scholar]
- 4.Wood AJ, Douglas RG. Pathogenesis and treatment of chronic rhinosinusitis. Postgrad Med J. 2010;86:359–64. doi: 10.1136/pgmj.2009.094813. [DOI] [PubMed] [Google Scholar]
- 5.Bachert C, Van Bruaene N, Toskala E, Zhang N, Olze H, Scadding G, et al. Important research questions in allergy and related diseases: 3-chronic rhinosinusitis and nasal polyposis - a GALEN study. Allergy. 2009;64:520–33. doi: 10.1111/j.1398-9995.2009.01964.x. [DOI] [PubMed] [Google Scholar]
- 6.Van Zele T, Claeys S, Gevaert P, Van Maele G, Holtappels G, Van Cauwenberge P, et al. Differentiation of chronic sinus diseases by measurement of inflammatory mediators. Allergy. 2006;61:1280–9. doi: 10.1111/j.1398-9995.2006.01225.x. [DOI] [PubMed] [Google Scholar]
- 7.Claeys S, De Belder T, Holtappels G, Gevaert P, Verhasselt B, Van Cauwenberge P, et al. Macrophage mannose receptor in chronic sinus disease. Allergy. 2004;59:606–12. doi: 10.1111/j.1398-9995.2004.00471.x. [DOI] [PubMed] [Google Scholar]
- 8.Kato A, Peters A, Suh L, Carter R, Harris KE, Chandra R, et al. Evidence of a role for B cell-activating factor of the TNF family in the pathogenesis of chronic rhinosinusitis with nasal polyps. J Allergy Clin Immunol. 2008;121:1385–92. doi: 10.1016/j.jaci.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schleimer RP, Kato A, Peters A, Conley D, Kim J, Liu MC, et al. Epithelium, inflammation, and immunity in the upper airways of humans: studies in chronic rhinosinusitis. Proc Am Thorac Soc. 2009;6:288–94. doi: 10.1513/pats.200808-088RM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patadia M, Dixon J, Conley D, Chandra R, Peters A, Suh LA, et al. Evaluation of the presence of B-cell attractant chemokines in chronic rhinosinusitis. Am J Rhinol Allergy. 2010;24:11–6. doi: 10.2500/ajra.2010.24.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tieu DD, Peters AT, Carter RT, Suh L, Conley DB, Chandra R, et al. Evidence for diminished levels of epithelial psoriasin and calprotectin in chronic rhinosinusitis. J Allergy Clin Immunol. 2010;125:667–75. doi: 10.1016/j.jaci.2009.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krysko O, Holtappels G, Zhang N, Kubica M, Deswarte K, Derycke L, et al. Alternatively activated macrophages and impaired phagocytosis of S. aureus in chronic rhinosinusitis. Allergy. 2011;66:396–403. doi: 10.1111/j.1398-9995.2010.02498.x. [DOI] [PubMed] [Google Scholar]
- 13.Poposki JA, Uzzaman A, Nagarkar DR, Chustz RT, Peters AT, Suh LA, et al. Increased expression of the chemokine CCL23 in eosinophilic chronic rhinosinusitis with nasal polyps. J Allergy Clin Immunol. 2011;128:73–81. e4. doi: 10.1016/j.jaci.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charo IF, Ransohoff RM. The many roles of chemokines and chemokine receptors in inflammation. N Engl J Med. 2006;354:610–21. doi: 10.1056/NEJMra052723. [DOI] [PubMed] [Google Scholar]
- 15.Viola A, Luster AD. Chemokines and their receptors: drug targets in immunity and inflammation. Annu Rev Pharmacol Toxicol. 2008;48:171–97. doi: 10.1146/annurev.pharmtox.48.121806.154841. [DOI] [PubMed] [Google Scholar]
- 16.Horuk R. Chemokine receptor antagonists: overcoming developmental hurdles. Nat Rev Drug Discov. 2009;8:23–33. doi: 10.1038/nrd2734. [DOI] [PubMed] [Google Scholar]
- 17.Schall TJ, Proudfoot AE. Overcoming hurdles in developing successful drugs targeting chemokine receptors. Nat Rev Immunol. 2011;11:355–63. doi: 10.1038/nri2972. [DOI] [PubMed] [Google Scholar]
- 18.Walters MJ, Wang Y, Lai N, Baumgart T, Zhao BN, Dairaghi DJ, et al. Characterization of CCX282-B, an orally bioavailable antagonist of the CCR9 chemokine receptor, for treatment of inflammatory bowel disease. J Pharmacol Exp Ther. 2010;335:61–9. doi: 10.1124/jpet.110.169714. [DOI] [PubMed] [Google Scholar]
- 19.Adema GJ, Hartgers F, Verstraten R, de Vries E, Marland G, Menon S, et al. A dendritic-cell-derived C-C chemokine that preferentially attracts naive T cells. Nature. 1997;387:713–7. doi: 10.1038/42716. [DOI] [PubMed] [Google Scholar]
- 20.Hieshima K, Imai T, Baba M, Shoudai K, Ishizuka K, Nakagawa T, et al. A novel human CC chemokine PARC that is most homologous to macrophage-inflammatory protein-1 alpha/LD78 alpha and chemotactic for T lymphocytes, but not for monocytes. J Immunol. 1997;159:1140–9. [PubMed] [Google Scholar]
- 21.Kodelja V, Muller C, Politz O, Hakij N, Orfanos CE, Goerdt S. Alternative macrophage activation-associated CC-chemokine-1, a novel structural homologue of macrophage inflammatory protein-1 alpha with a Th2-associated expression pattern. J Immunol. 1998;160:1411–8. [PubMed] [Google Scholar]
- 22.Schutyser E, Richmond A, Van Damme J. Involvement of CC chemokine ligand 18 (CCL18) in normal and pathological processes. J Leukoc Biol. 2005;78:14–26. doi: 10.1189/jlb.1204712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guan P, Burghes AH, Cunningham A, Lira P, Brissette WH, Neote K, et al. Genomic organization and biological characterization of the novel human CC chemokine DC-CK-1/PARC/MIP-4/SCYA18. Genomics. 1999;56:296–302. doi: 10.1006/geno.1998.5635. [DOI] [PubMed] [Google Scholar]
- 24.Tasaki Y, Fukuda S, Iio M, Miura R, Imai T, Sugano S, et al. Chemokine PARC gene (SCYA18) generated by fusion of two MIP-1alpha/LD78alpha-like genes. Genomics. 1999;55:353–7. doi: 10.1006/geno.1998.5670. [DOI] [PubMed] [Google Scholar]
- 25.Schutyser E, Struyf S, Wuyts A, Put W, Geboes K, Grillet B, et al. Selective induction of CCL18/PARC by staphylococcal enterotoxins in mononuclear cells and enhanced levels in septic and rheumatoid arthritis. Eur J Immunol. 2001;31:3755–62. doi: 10.1002/1521-4141(200112)31:12<3755::aid-immu3755>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 26.Kato A, Chustz RT, Ogasawara T, Kulka M, Saito H, Schleimer RP, et al. Dexamethasone and FK506 inhibit expression of distinct subsets of chemokines in human mast cells. J Immunol. 2009;182:7233–43. doi: 10.4049/jimmunol.0801375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nomura I, Gao B, Boguniewicz M, Darst MA, Travers JB, Leung DY. Distinct patterns of gene expression in the skin lesions of atopic dermatitis and psoriasis: a gene microarray analysis. J Allergy Clin Immunol. 2003;112:1195–202. doi: 10.1016/j.jaci.2003.08.049. [DOI] [PubMed] [Google Scholar]
- 28.de Nadai P, Charbonnier AS, Chenivesse C, Senechal S, Fournier C, Gilet J, et al. Involvement of CCL18 in allergic asthma. J Immunol. 2006;176:6286–93. doi: 10.4049/jimmunol.176.10.6286. [DOI] [PubMed] [Google Scholar]
- 29.Pivarcsi A, Gombert M, Dieu-Nosjean MC, Lauerma A, Kubitza R, Meller S, et al. CC chemokine ligand 18, an atopic dermatitis-associated and dendritic cell-derived chemokine, is regulated by staphylococcal products and allergen exposure. J Immunol. 2004;173:5810–7. doi: 10.4049/jimmunol.173.9.5810. [DOI] [PubMed] [Google Scholar]
- 30.Kato A, Truong-Tran AQ, Scott AL, Matsumoto K, Schleimer RP. Airway epithelial cells produce B cell-activating factor of TNF family by an IFN-beta-dependent mechanism. J Immunol. 2006;177:7164–72. doi: 10.4049/jimmunol.177.10.7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chustz RT, Nagarkar DR, Poposki JA, Favoreto S, Jr, Avila PC, Schleimer RP, et al. Regulation and Function of the IL-1 Family Cytokine IL-1F9 in Human Bronchial Epithelial Cells. Am J Respir Cell Mol Biol. 2011;45:145–53. doi: 10.1165/rcmb.2010-0075OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pawliczak R, Lewandowska-Polak A, Kowalski ML. Pathogenesis of nasal polyps: an update. Curr Allergy Asthma Rep. 2005;5:463–71. doi: 10.1007/s11882-005-0027-7. [DOI] [PubMed] [Google Scholar]
- 33.Samter M, Beers RF., Jr Intolerance to aspirin. Clinical studies and consideration of its pathogenesis. Ann Intern Med. 1968;68:975–83. doi: 10.7326/0003-4819-68-5-975. [DOI] [PubMed] [Google Scholar]
- 34.Stevenson DD. Aspirin sensitivity and desensitization for asthma and sinusitis. Curr Allergy Asthma Rep. 2009;9:155–63. doi: 10.1007/s11882-009-0023-4. [DOI] [PubMed] [Google Scholar]
- 35.Kreider T, Anthony RM, Urban JF, Jr, Gause WC. Alternatively activated macrophages in helminth infections. Curr Opin Immunol. 2007;19:448–53. doi: 10.1016/j.coi.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noel W, Raes G, Hassanzadeh Ghassabeh G, De Baetselier P, Beschin A. Alternatively activated macrophages during parasite infections. Trends Parasitol. 2004;20:126–33. doi: 10.1016/j.pt.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 37.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177:7303–11. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 38.Martinez FO, Sica A, Mantovani A, Locati M. Macrophage activation and polarization. Front Biosci. 2008;13:453–61. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- 39.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 40.Zhang N, Van Zele T, Perez-Novo C, Van Bruaene N, Holtappels G, DeRuyck N, et al. Different types of T-effector cells orchestrate mucosal inflammation in chronic sinus disease. J Allergy Clin Immunol. 2008;122:961–8. doi: 10.1016/j.jaci.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 41.Plager DA, Kahl JC, Asmann YW, Nilson AE, Pallanch JF, Friedman O, et al. Gene transcription changes in asthmatic chronic rhinosinusitis with nasal polyps and comparison to those in atopic dermatitis. PLoS One. 2010;5:e11450. doi: 10.1371/journal.pone.0011450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schraufstatter I, Takamori H, Sikora L, Sriramarao P, DiScipio RG. Eosinophils and monocytes produce pulmonary and activation-regulated chemokine, which activates cultured monocytes/macrophages. Am J Physiol Lung Cell Mol Physiol. 2004;286:L494–501. doi: 10.1152/ajplung.00323.2002. [DOI] [PubMed] [Google Scholar]
- 43.van Lieshout AW, van der Voort R, le Blanc LM, Roelofs MF, Schreurs BW, van Riel PL, et al. Novel insights in the regulation of CCL18 secretion by monocytes and dendritic cells via cytokines, toll-like receptors and rheumatoid synovial fluid. BMC Immunol. 2006;7:23. doi: 10.1186/1471-2172-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prasse A, Pechkovsky DV, Toews GB, Jungraithmayr W, Kollert F, Goldmann T, et al. A vicious circle of alveolar macrophages and fibroblasts perpetuates pulmonary fibrosis via CCL18. Am J Respir Crit Care Med. 2006;173:781–92. doi: 10.1164/rccm.200509-1518OC. [DOI] [PubMed] [Google Scholar]
- 45.Nardelli B, Tiffany HL, Bong GW, Yourey PA, Morahan DK, Li Y, et al. Characterization of the signal transduction pathway activated in human monocytes and dendritic cells by MPIF-1, a specific ligand for CC chemokine receptor 1. J Immunol. 1999;162:435–44. [PubMed] [Google Scholar]
- 46.Nibbs RJ, Salcedo TW, Campbell JD, Yao XT, Li Y, Nardelli B, et al. C-C chemokine receptor 3 antagonism by the beta-chemokine macrophage inflammatory protein 4, a property strongly enhanced by an amino-terminal alanine-methionine swap. J Immunol. 2000;164:1488–97. doi: 10.4049/jimmunol.164.3.1488. [DOI] [PubMed] [Google Scholar]
- 47.Ebisawa M, Liu MC, Yamada T, Kato M, Lichtenstein LM, Bochner BS, et al. Eosinophil transendothelial migration induced by cytokines. II. Potentiation of eosinophil transendothelial migration by eosinophil-active cytokines. J Immunol. 1994;152:4590–6. [PubMed] [Google Scholar]
- 48.Atamas SP, Luzina IG, Choi J, Tsymbalyuk N, Carbonetti NH, Singh IS, et al. Pulmonary and activation-regulated chemokine stimulates collagen production in lung fibroblasts. Am J Respir Cell Mol Biol. 2003;29:743–9. doi: 10.1165/rcmb.2003-0078OC. [DOI] [PubMed] [Google Scholar]
- 49.Luzina IG, Tsymbalyuk N, Choi J, Hasday JD, Atamas SP. CCL18-stimulated upregulation of collagen production in lung fibroblasts requires Sp1 signaling and basal Smad3 activity. J Cell Physiol. 2006;206:221–8. doi: 10.1002/jcp.20452. [DOI] [PubMed] [Google Scholar]