Abstract
A recent study of CDK4/6-inhibitors in glioblastoma (GBM) xenografts identified retinoblastoma tumor suppressor protein RB1 status as a determinant of tumor therapeutic efficacy. Because of the need for clinically applicable RB1 testing, we assessed the utility of 2 complementary methods for determining RB1 status in GBM. Using fluorescence in situ hybridization (FISH) and immunohistochemistry (IHC), we analyzed 34 GBMs that had also undergone molecular characterization as part of The Cancer Genome Atlas (TCGA). By IHC, 4 tumors (11.8%) had complete loss of RB protein expression, including 2 with homozygous deletion of RB1 by FISH and 1 with hemizygous deletion of RB1 by FISH combined with a novel nonsense mutation in RB1. Consistent with these results, in an independent set of 51 GBMs tested by IHC we demonstrated loss of RB1 protein in 5 (9.8%). In GBM molecular subtype analysis of TCGA data, complete loss of RB1 transcript expression was seen in 18 of 170 tumors (10.6%) and these were highly enriched for, but not exclusive to, the proneural subtype (p < 0.01). These data support the use of IHC for determining RB1 status in clinical GBM specimens and suggest that RB1 alterations may be more common in certain GBM subgroups.
Keywords: Fluorescence in situ hybridization, Glioblastoma, Immunohistochemistry, Patient stratification, RB1, The Cancer Genome Atlas (TCGA)
INTRODUCTION
Glioblastoma (GBM) is the most common primary malignant brain tumor of adults with a median survival of less than 2 years (1-3). Novel therapeutic options are desperately needed. GBMs are characteristically heterogeneous and contain different cell signaling pathway alterations (4, 5). Novel therapeutics are being designed to target some of these pathways but their success likely depends on patient stratification into molecular subgroups so that therapies are to specific subsets of patients with predicted favorable responses.
Aberrations in the CDKN2A/p16-CDK4/6-RB pathway are common in GBM (6-8) and they have been shown to be critical in gliomagenesis or tumor progression from lower-grade astrocytomas (9). CDK4 and CDK6 phosphorylate RB1, which induces the release of the transcription factor E2F, thus facilitating the transition of the cell cycle from the G1 to S phase. In normal cells this cell cycle transition is negatively regulated by the p16 protein, which binds and inhibits CDK4/6 function. Thus, alterations in expression of p16, CDK4, CDK6, or RB1 can result in dysregulation of the cell cycle (10). Indeed, The Cancer Genome Atlas (TCGA) project has shown that this pathway is altered in nearly 80% of primary GBMs with the most frequent genetic alterations being CDKN2A gene deletion or mutation, CDK4 amplification, and RB1 mutation or deletion (11). A recent preclinical study investigated the efficacy of a CDK4/6-specific inhibitor (PD-0332991) in intracranial GBM xenograft tumors and found it to be a potent inhibitor of GBM growth (12). As expected, however, this antitumor effect was not seen in RB1-deficient tumors because the latter cell signaling alteration is downstream of the drug target. Thus, stratification of patient tumors based upon RB1 status may be important.
Multiple mechanisms of bi-allelic RB1 gene inactivation have been identified in tumors, including combinations of deletions and point mutations (13). In astrocytomas, alterations in RB1 expression have been associated with increased tumor cell proliferation and decreased survival (14, 15). Assessment of RB1 status, however, is not currently performed clinically. Here, we used immunohistochemistry (IHC) for determining RB1 status in formalin fixed paraffin-embedded (FFPE) clinical GBM samples and validated this assay using profiling data from TCGA (11), including relative RB1 gene copy numbers and transcript expression levels. Copy number assessment was additionally assessed by FISH and concordance levels between the methodologies were determined.
MATERIALS AND METHODS
Tumor Material and Study Design
FFPE tumor tissue was obtained from the UCSF Brain Tumor SPORE Tissue Bank (CHR #10-01318) in accordance with ethical standards of the UCSF Institutional Review Board. GBM tissue microarrays containing central regions of tumor were constructed from 34 tumors previously profiled by TCGA (11). Each tumor was represented on the array with at least 2 tissue cores. FISH results were compared to corresponding TCGA array comparative genome hybridization (CGH) data (30 cases available). Immunohistochemistry (IHC) data for 33 cases were compared to corresponding TCGA transcript profiling data. A total of 33 tumors were analyzed by both FISH and IHC. A second independent set of 51 GBMs was also analyzed.
FISH Analysis
FISH was performed on FFPE tissue microarray slides as described (16). Hybridization was achieved using Spectrum Orange-labeled RB1 (13q14) probe (Abbott Molecular, Abbott Park, IL). For each sample with adequate signal, a minimum of 100 non-overlapping nuclei was enumerated by 2 investigators (A.P., P.G.). A hemizygous RB1 deletion was defined as >50% of tumor nuclei showing only 1 signal (17). Homozygous deletion was defined by the lack of any RB1 signal in the majority of tumor cells despite the presence of signals within intratumoral non-neoplastic elements such as endothelial cells; this was required to rule out hybridization failure. FISH images were captured using an Olympus BX60 fluorescence microscope with charge coupled device camera, Z-stack motor and a CytoVision basic workstation (Applied Imaging, Santa Clara, CA). FISH signals were scored as not deleted, hemizygous deletion, or homozygous deletion, and scoring was blinded to the results of IHC and profiling.
IHC
IHC was performed for RB1 (BD Biosciences, San Jose, CA, G3-245) and Iba1 (Wako Chemicals USA, Inc., Richmond, VA, 019-19741) following antigen retrieval. The latter antibody identified intratumoral microglia and macrophages. An automated IHC staining process was used for all biopsies (Benchmark XT, Ventana Medical Systems, Inc., Tucson, AZ). Tumor RB1 protein status was determined by counting cells with positive nuclear staining in 3 20x fields. RB1-positive endothelial cells served as an internal positive control. All slides were reviewed by the same neuropathologist (J.J.P.) who was blinded to genomics and FISH data. IHC scores were scored as follows: 0 for expression in <20% of tumor cells; 1+ for expression in 20%-50% of tumor cells; and 2+ for positive expression in >50% of tumor cells. Digital images were captured using a microscope (Olympus, Center Valley, PA, Model BX41TF) and digital camera (Olympus, Model DP70).
Genomic Data Analysis
Copy number data (HG-CGH-244) and gene expression data (AgilentG4502A) log2(Tumor/Normal) were obtained from the TCGA Data Portal on December 20, 2010 and March 23, 2011. Gene deletion of RB1 was defined as a copy number loss resulting in a log2(Tumor/Normal) less than or equal to −0.4 (fold-change of copy number in tumor vs. normal less than 0.76), which included the RB1 gene (chr13:48,877,883-49,056,024). Loss of RB1 transcript expression was defined as a log2 (Tumor/Normal) of less than or equal to zero (fold change of gene expression in tumor vs. normal less than 1).
Statistics
A two-tailed t-test was used to compare mean values. For Kaplan-Meier survival analysis, groups were compared using the Log rank test, and categories were compared using adjusted 2-sided Fisher exact test.
RESULTS
Concordance of RB1 FISH and Array CGH
We examined 34 GBMs by FISH and IHC and compared the results to corresponding array CGH and transcript profiling data (11). Eleven of 33 tumors (34%) exhibited deletion of at least 1 copy of RB1 and 2 tumors had homozygous deletion; 1 tumor did not have adequate tissue for evaluation (Fig. 1A, C, E). The FISH results were 100% concordant with array CGH data (11/11 tumors with homozygous or hemizygous loss of RB1 by FISH had decreased copy number of RB1 by array CGH; Table). Both cases with homozygous deletion of RB1 by FISH showed loss of transcript expression.
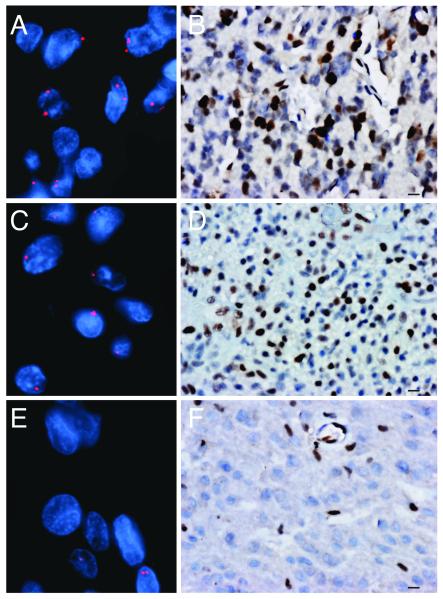
Figure 1.
Determination of RB1 status in glioblastoma (GBM) by fluorescence in situ hybridization (FISH) and immunohistochemistry (IHC). Representative images of tumors analyzed for RB1 copy number by FISH (red) and for RB1 protein expression by IHC (brown). (A-D) Cells of tumors with intact RB1 primarily have 2 signals per nucleus by FISH (A) and are positive for RB1 by IHC (B). A hemizygously deleted tumor primarily has 1 RB1 signal per nucleus by FISH (C) and is positive for RB1 by IHC (D). A homozygously deleted tumor primarily has no signal for RB1 by FISH (E). The presence of 2 signals in nucleus at bottom right likely represents a non-neoplastic cell and no positivity for RB1 protein by IHC (F). The positive nuclei of endothelial and inflammatory cells provide a positive internal control. Bar: A, C, E = 10 μm; B, D, F = 40x.
Table.
Concordance of RB1 Status as Determined by Fluorescence In Situ Hybridization and Immunohistochemistry Compared to The Cancer Genome Atlas
| Tumor ID |
RB1 deletion by FISH |
Copy number (log2 ratio)a |
RB1 protein by IHCb, c |
RB1 expression (log2 ratio)d |
RB1 somatic mutatione |
GBM Subtypef |
|---|---|---|---|---|---|---|
| 08-0520 | Homozygous | −2.406 | 0 | −0.255 | NA | N |
| 08-0350 | Homozygous | −2.146 | 0 | −0.145 | NA | P |
| 08-0386 | Hemizygous | −0.399 | 0 | −0.33 | NA | N |
| 08-0389 | Hemizygous | −0.682 | 0 | 0.093 | Novel nonsense mutation |
NA |
| 08-0349 | Hemizygous | −0.638 | 1 | 0.036 | NA | N |
| 08-0359 | Hemizygous | −0.549 | 1 | 0.584 | No | P |
| 08-0347 | Hemizygous | −0.473 | 1 | 0.544 | No | P |
| 08-0511 | Hemizygous | −0.704 | 2 | 0.474 | NA | C |
| 08-0517 | Hemizygous | −0.828 | 2 | 0.515 | NA | P |
| 08-0516 | Hemizygous | −0.606 | 2 | 0.402 | NA | NA |
| 08-0357 | Hemizygous | −0.571 | 2 | 0.735 | NA | C |
| 08-0246 | Not Del | −0.322 | 1 | 0.835 | No | C |
| 08-0512 | Not Del | −0.302 | 1 | −0.7509 | NA | M |
| 08-0360 | Not Del | −0.270 | 1 | 0.648 | No | M |
| 08-0375 | Not Del | −0.050 | 1 | 0.691 | No | C |
| 08-0380 | Not Del | −0.041 | 1 | 0.959 | No | |
| 08-0518 | Not Del | −0.036 | 1 | 1.006 | NA | C |
| 08-0356 | Not Del | −0.032 | 1 | 1.061 | NA | NA |
| 08-0355 | Not Del | −0.016 | 1 | 1.016 | NA | C |
| 08-0385 | Not Del | NA | 1 | 0.532 | No | P |
| 08-0345 | NA | −0.080 | 2 | 1.333 | NA | NA |
| 08-0510 | Not Del | −0.132 | 2 | 1.108 | NA | M |
| 08-0514 | Not Del | −0.088 | 2 | 1.063 | NA | C |
| 08-0353 | Not Del | −0.083 | 2 | 1.158 | No | NA |
| 08-0390 | Not Del | −0.003 | 2 | 1.148 | No | M |
| 08-0522 | Not Del | 0.048 | 2 | 0.986 | NA | M |
| 08-0392 | Not Del | −0.083 | 2 | 0.625 | NA | M |
| 08-0531 | Not Del | −0.063 | 2 | 0.777 | NA | C |
| 08-0244 | Not Del | −0.056 | 2 | 0.711 | NA | NA |
| 08-0521 | Not Del | −0.004 | 2 | 0.591 | NA | NA |
| 08-0358 | Not Del | −0.003 | 2 | 0.85 | NA | C |
| 08-0344 | Not Del | 0.020 | 2 | 0.615 | No | P |
| 08-0525 | Not Del | 0.041 | 2 | 1.399 | NA | NA |
| 08-0381 | Not Del | NA | 2 | NA | NA | NA |
CGH, comparative genomic hybridization; FISH, fluorescence in situ hybridization; GBM, glioblastoma; IHC, immunohistochemistry; NA, data not available; No Del, no deletion; RB1, Retinoblastoma Tumor Suppressor Protein; TCGA, The Cancer Genome Atlas.
Copy number [Tumor/Normal] data from TCGA, HMS.harvard HG-CGH-244A; gene deletion defined as [log2(Tumor/Normal) less than or equal to −0.4 (fold change of copy number in tumor vs. normal less than or equal to 0.758)] (11).
Mean IHC score for tumors with decreased RB1 copy number by array CGH was significantly different than the mean IHC score for tumors with intact RB1 (0.95 vs. 1.48, p<0.05; n=32).
The mean IHC score for tumors with loss of RB1 transcript expression was significantly different than the mean IHC score for tumors with intact RB1 expression (0 vs. 1.42, p<0.00005; n=33).
Expression [Tumor/Normal] data from TCGA, AgilentG4502A_07; loss of RB1 expression defined as [log2(Tumor/Normal) less than 0 (fold-change of expression in tumor vs. normal less than 1)].
Sequencing data from TCGA, hgsc.bcm.edu (11).
GBM subtype as determined by Verhaak et al (18). P, proneural; N, M, Mesenchymal; C, Classical
Concordance of RB1 Immunohistochemistry and RB1 Expression Data
RB1 protein status was assessed in the same cohort of tumors by IHC (Figs. 1B, D, F, 2B, D, F). RB1-positive microglia/macrophages and endothelial cells served as internal positive controls. Due to tumor heterogeneity, including heterogeneity of tumor cell expression of RB1, known to be influenced by many factors including cell cycle (18, 19), and variation in density of the tumor-associated microglia/macrophages, tumors were determined to be RB1-intact if a minimum threshold of 20% of tumor nuclei were RB1-positive. In addition, immunostaining for microglia/macrophage marker Iba1 was used to highlight non-neoplastic cells that retain RB1 protein (Fig. 2A, C, E, F). Loss of RB1 as determined by IHC (4 of 34 tumors; 11.8%) was very similar to RB1 loss based upon expression profiling with an overall concordance of 94% (31/33; Table). Although other non-neoplastic cells in the tumor microenvironment might express RB1, our threshold of 20% was sufficiently high to distinguish tumors with RB1 loss (mean RB1-positive nuclei 14.1% (range 11%-18%), n = 4) from tumors with intact but heterogeneous RB1-expression (mean RB1-positive nuclei 62% (range 36%-93%), n = 12, p < 0.001). Interestingly, one discrepant tumor lacking protein expression by IHC was hemizygously deleted for RB1 by FISH and had a novel RB1 nonsense point mutation in the remaining allele (TCGA-08-0389). In that case, it seems likely that the TCGA expression data resulted from the detection of mutant RB1 transcript that does not encode detectable protein. In the case of TCGA-08-0512, the basis of undetectable protein expression is less clear. Nevertheless, these data suggest that RB1 IHC is highly effective for detecting alterations in RB1 protein expression.
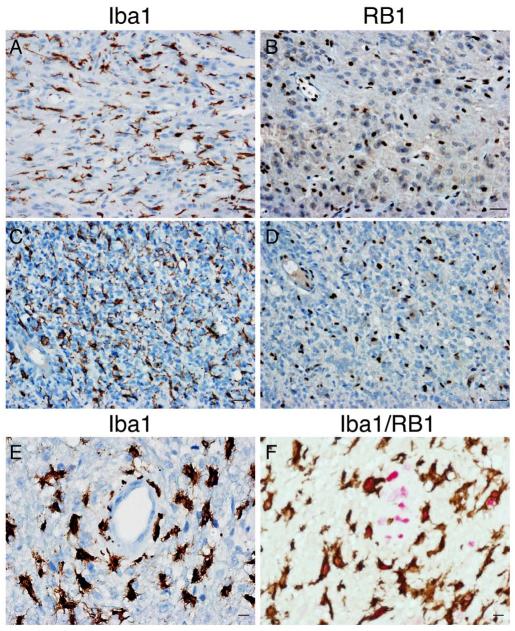
Figure 2.
Assessment of RB1 protein loss in tumor cells. (A-E) Two RB1-immunonegative tumors were either homozygous (A, B) or hemizygous (C, D) deleted for RB1 by fluorescence in situ hybridization. A cut-off value for RB1-positivity of 20% tumor nuclei was used to distinguish RB1-negative tumor cells (B, D) from RB1-positive non-neoplastic elements including endothelial cells and microglia/macrophages (A, C, E; Iba1-positive). Tumor cell nuclei Are generally larger than those of non-neoplastic cells. (F) Double immunostaining for Iba1 (brown) and RB1 (red) highlights RB1-positive microglia/macrophages. Bars: A-D = 30 μm; E, F = 10 μm.
Concordance of RB1 FISH and Immunohistochemistry
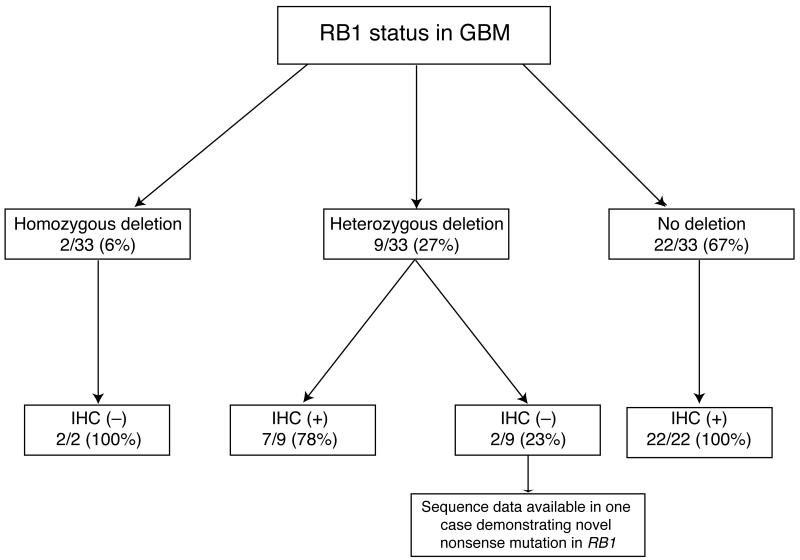
Both tumors with homozygous deletions for RB1 were negative for RB1 by IHC (100%) (Fig. 3). Similarly, the 22/22 (100%) GBMs intact for RB1 by FISH were RB1-immunopositive. Of the 9 GBMs with hemizygous deletions of RB1 by FISH, 7 (78%) were positive by IHC. The remaining 2 hemizygously deleted cases were negative by IHC, suggesting inactivation of the second allele with loss of RB1 protein expression. In fact, 1 of these tumors contained a novel nonsense mutation in RB1 further supporting that notion. The second tumor had no sequencing data available (Table). These data suggest that when there is homozygous deletion of RB1 by FISH, one can reasonably infer that the gene is inactivated whereas the gene is unlikely to be inactivated in cases with retained copy numbers. On the other hand, hemizygous deletion is unreliable for predicting tumor RB1 status because this assay alone cannot predict whether the retained allele is wild type or mutant.
Figure 3.
Agreement between fluorescence in situ hybridization (FISH) and immunohistochemistry (IHC) for detecting RB1 loss in 33 glioblastomas (GBM). IHC and FISHshowed high concordance when FISH showed either retained copy numbers or homozygous deletions. The majority of tumors with hemizygous RB1 deletion by FISH retained RB1 expression by IHC.
Associations of RB1 Status with GBM Subtype and Clinical Parameters
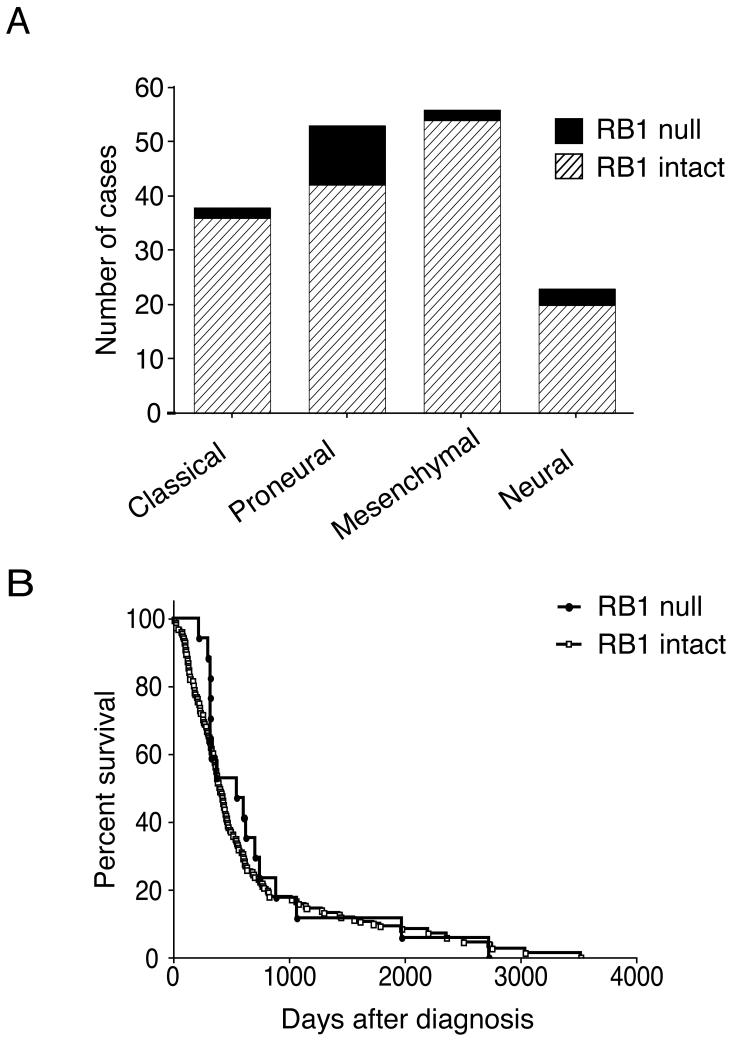
To determine the prevalence of loss of RB1 transcript expression in GBMs, we examined the available data from 170 tumors from TCGA (11). RB1 transcript expression was lost in 18 of 170 tumors (10.6%). To determine the frequency of RB1 loss in an independent set of GBMs, we immunostained an additional 51 FFPE GBMs. Similar to the frequency of RB1 loss noted for the TCGA analyzed tumors, we identified 5 GBMs with loss of RB1 protein expression (9.8%). GBMs have been stratified into different subtypes based upon patterns of genomic alteration and transcript expression level (20-22). To determine whether loss of RB1 expression was over-represented in any one GBM subgroup, we analyzed RB1 status in previously subtyped tumors (18). Loss of RB1 expression was a more frequent event in the proneural subtype (p < 0.01, adjusted 2-sided Fisher exact test; Fig. 4A). Furthermore, gene deletion of RB1was more frequent in the proneural subtype, although this difference did not reach statistical significance (p = 0.08, adjusted 2-sided Fisher exact test). However, RB1 loss was not exclusive to the proneural subtype and occurred in all 4 subtypes. The clinical characteristics of patients with and without loss of RB1 expression were similar with respect to overall survival (Fig. 4B; n = 160), sex (female to male ratio of 64% vs. 58%, respectively; n = 170), and age (56 vs. 50 years, respectively; n = 170).
Figure 4.
Loss of RB1 transcript expression is associated with the proneural glioblastoma (GBM) subtype, but is not associated with altered survival. (A) Loss of RB1 transcript expression was identified in 18 of 168 (10.5%) of The Cancer Genome Atlas (TCGA) tumors; these were over-represented in the proneural subtype as defined by Verhaak et al (18) (p < 0.01, adjusted two-sided Fisher exact test). (B) Loss of RB1 transcript expression was not associated with a difference in overall survival on univariate analysis (11).
DISCUSSION
GBM is a heterogeneous disease and stratification of patients based upon predicted response to specific agents may be critical for the success of targeted therapeutics. Because current diagnostic neuropathology relies on the analysis of FFPE material, broad applicability of a new assay currently requires it to be paraffin-based. Using FFPE material, we used FISH to identify tumors with deletions of RB1 and IHC to identify tumors with loss of RB1 protein expression. In combination, the 2 methods provided information regarding potential mechanisms of RB1 alteration and RB1 status. Our data demonstrate loss of RB1 protein in roughly 10% of GBMs, consistent with previous estimates (11, 19).
Loss of heterozygosity of the RB1 gene on chromosome 13q is a relatively common event in astrocytoma (11, 23, 24). In a subset of tumors, alteration of the remaining RB1 allele and subsequent inactivation of RB1 disrupts the p16-CDK4/6-RB1 pathway (8, 19, 25, 26). The most common alteration of this pathway, homozygous CDKN2A deletion, results in suppression of RB1 protein function via elevated CDK4/6 activity. CDK4/6 is an attractive therapeutic target in tumors lacking CDKN2A/p16 function, but not in tumors lacking RB1 protein. Using IHC on clinical tumor samples we stratified patients into an RB1-intact group that was expected to benefit from CDK4/6 inhibitors and a group with loss of RB1 expression that would not be expected to benefit. IHC was a reliable method to detect RB1 status as predicted by our analysis of RB1 copy number by FISH and large-scale genomic data from TCGA, including expression array, array CGH, and sequencing (11). These data support the use of IHC to identify RB1 status for potential stratification of patients for clinical trials. Furthermore, they confirm the robust nature of the TCGA data and demonstrate its usefulness in the development and validation of clinical tests for patient stratification. In fact, expression array data available on the TCGA Data Portal (downloaded October 24, 2011) predict that 47 of 424 (11.1%) tumors analyzed have loss of RB1 expression. This estimate is consistent with our data showing loss of RB1 protein expression in 9 of 85 (10.6%) GBMs.
Two of 33 tumors were identified with hemizygous gene deletion by FISH but absent protein expression. Typical of tumor suppressor genes, multiple mechanisms may explain complete loss of RB1 protein in the setting of a hemizygous deletion, including point mutation, deletion, or alterations in transcriptional or translational regulation (27, 28). Consistent with this line of reasoning, sequencing of one of these cases demonstrated a novel nonsense point mutation in the remaining intact copy of RB1 predicted to result in a premature stop codon. Sequencing data was not available from the second case. We did not identify any tumors with loss of RB1 protein expression in the absence of gene deletion, but such occurrences have been described in other cancers (29, 30). Our data indicate that determination of RB1 protein status in tumor tissue reliably identifies the majority of cases with homozygous RB1 gene inactivation. It is possible, however, that in some cases epitope expression may persist in the context of a truncated, dysfunctional protein, yielding a false negative result. Such results are anticipated to be very infrequent.
We did not identify an association between RB1 status and survival in either our subset of tumors analyzed by IHC or in the 170 TCGA tumors analyzed by expression array. Although some reports have suggested such an association in GBM (31), RB1 may have a more direct correlation with prognosis in lower-grade astrocytomas including World Health Organization grade II and III tumors (14, 15, 32). Our finding that alterations in RB1 are more common in the proneural subtype of GBM may reflect similarities between genetic alterations in a subset of GBMs and lower-grade astrocytomas. Future studies will address this possibility and determine the utility of RB1 FISH and IHC for patient stratification in both low- and high-grade astrocytomas.
ACKNOWLEDGMENTS
We thank Cynthia Cowdrey and Yunita Lim for assistance with obtaining human tissue and preparing the tissue microarrays. This work was supported by funds from the National Institutes of Health (K08 NS063456 from the National Institute of Neurological Disorders and Stroke to J.J.P. and the UCSF Brain Tumor SPORE P50CA097257 to A.P., M.D.P., and M.S.B).
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Hess KR, Broglio KR, Bondy ML. Adult glioma incidence trends in the United States, 1977-2000. Cancer. 2004;101:2293–9. doi: 10.1002/cncr.20621. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–66. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 4.Stommel JM, Kimmelman AC, Ying H, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–90. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 5.Brennan C, Momota H, Hambardzumyan D, et al. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PloS One. 2009;4:e7752. doi: 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He J, Olson JJ, James CD. Lack of p16INK4 or retinoblastoma protein (pRb), or amplification-associated overexpression of cdk4 is observed in distinct subsets of malignant glial tumors and cell lines. Cancer Res. 1995;55:4833–6. [PubMed] [Google Scholar]
- 7.Jen J, Harper JW, Bigner SH, et al. Deletion of p16 and p15 genes in brain tumors. Cancer Res. 1994;54:6353–8. [PubMed] [Google Scholar]
- 8.Ichimura K, Schmidt EE, Goike HM, et al. Human glioblastomas with no alterations of the CDKN2A (p16INK4A, MTS1) and CDK4 genes have frequent mutations of the retinoblastoma gene. Oncogene. 1996;13:1065–72. [PubMed] [Google Scholar]
- 9.Chow LM, Endersby R, Zhu X, et al. Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain. Cancer Cell. 2011;19:305–16. doi: 10.1016/j.ccr.2011.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biernat W, Tohma Y, Yonekawa Y, et al. Alterations of cell cycle regulatory genes in primary (de novo) and secondary glioblastomas. Acta Neuropathol. 1997;94:303–9. doi: 10.1007/s004010050711. [DOI] [PubMed] [Google Scholar]
- 11.The Cancer Genome Atlas Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michaud K, Solomon DA, Oermann E, et al. Pharmacologic inhibition of cyclindependent kinases 4 and 6 arrests the growth of glioblastoma multiforme intracranial xenografts. Cancer Res. 2010;70:3228–38. doi: 10.1158/0008-5472.CAN-09-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lohmann DR, Brandt B, Hopping W, et al. The spectrum of RB1 germ-line mutations in hereditary retinoblastoma. Am J Hum Genet. 1996;58:940–9. [PMC free article] [PubMed] [Google Scholar]
- 14.Kim YH, Lachuer J, Mittelbronn M, et al. Alterations in the RB1 Pathway in Low-grade Diffuse Gliomas Lacking Common Genetic Alterations. Brain Pathol. 2011;21:645–51. doi: 10.1111/j.1750-3639.2011.00492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura M, Konishi N, Tsunoda S, et al. Retinoblastoma protein expression and MIB-1 correlate with survival of patients with malignant astrocytoma. Cancer. 1997;80:242–9. doi: 10.1002/(sici)1097-0142(19970715)80:2<242::aid-cncr12>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 16.Lusis EA, Chicoine MR, Perry A. High throughput screening of meningioma biomarkers using a tissue microarray. J Neurooncol. 2005;73:219–23. doi: 10.1007/s11060-004-5233-y. [DOI] [PubMed] [Google Scholar]
- 17.Perry A, Nobori T, Ru N, et al. Detection of p16 gene deletions in gliomas: a comparison of fluorescence in situ hybridization (FISH) versus quantitative PCR. J Neuropathol Exp Neurol. 1997;56:999–1008. doi: 10.1097/00005072-199709000-00005. [DOI] [PubMed] [Google Scholar]
- 18.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mancini MA, Shan B, Nickerson JA, et al. The retinoblastoma gene product is a cell cycle-dependent, nuclear matrix-associated protein. Proc Natl Acad Sci USA. 1994;91:418–22. doi: 10.1073/pnas.91.1.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henson JW, Schnitker BL, Correa KM, et al. The retinoblastoma gene is involved in malignant progression of astrocytomas. Ann Neurol. 1994;36:714–21. doi: 10.1002/ana.410360505. [DOI] [PubMed] [Google Scholar]
- 21.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–73. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 22.Mischel PS, Shai R, Shi T, et al. Identification of molecular subtypes of glioblastoma by gene expression profiling. Oncogene. 2003;22:2361–73. doi: 10.1038/sj.onc.1206344. [DOI] [PubMed] [Google Scholar]
- 23.James CD, Carlbom E, Dumanski JP, et al. Clonal genomic alterations in glioma malignancy stages. Cancer Res. 1988;48:5546–51. [PubMed] [Google Scholar]
- 24.Ransom DT, Ritland SR, Moertel CA, et al. Correlation of cytogenetic analysis and loss of heterozygosity studies in human diffuse astrocytomas and mixed oligo-astrocytomas. Genes Chromosomes Cancer. 1992;5:357–74. doi: 10.1002/gcc.2870050412. [DOI] [PubMed] [Google Scholar]
- 25.Ueki K, Ono Y, Henson JW, et al. CDKN2/p16 or RB alterations occur in the majority of glioblastomas and are inversely correlated. Cancer Res. 1996;56:150–3. [PubMed] [Google Scholar]
- 26.Burns KL, Ueki K, Jhung SL, et al. Molecular genetic correlates of p16, cdk4, and pRb immunohistochemistry in glioblastomas. J Neuropathol Exp Neurol. 1998;57:122–30. doi: 10.1097/00005072-199802000-00003. [DOI] [PubMed] [Google Scholar]
- 27.Kim H, Huang W, Jiang X, et al. Integrative genome analysis reveals an oncomir/oncogene cluster regulating glioblastoma survivorship. Proc Natl Acad Sci USA. 2010;107:2183–8. doi: 10.1073/pnas.0909896107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura M, Yonekawa Y, Kleihues P, et al. Promoter hypermethylation of the RB1 gene in glioblastomas. Lab Invest. 2001;81:77–82. doi: 10.1038/labinvest.3780213. [DOI] [PubMed] [Google Scholar]
- 29.Horowitz JM, Park SH, Bogenmann E, et al. Frequent inactivation of the retinoblastoma anti-oncogene is restricted to a subset of human tumor cells. Proc Natl Acad Sci USA. 1990;87:2775–9. doi: 10.1073/pnas.87.7.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knudson AG., Jr. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820–3. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Backlund LM, Nilsson BR, Goike HM, et al. Short postoperative survival for glioblastoma patients with a dysfunctional Rb1 pathway in combination with no wild-type PTEN. Clin Cancer Res. 2003;9:4151–8. [PubMed] [Google Scholar]
- 32.Backlund LM, Nilsson BR, Liu L, et al. Mutations in Rb1 pathway-related genes are associated with poor prognosis in anaplastic astrocytomas. Br J Cancer. 2005;93:124–30. doi: 10.1038/sj.bjc.6602661. [DOI] [PMC free article] [PubMed] [Google Scholar]