Abstract
Background
Increasing evidence points to a direct role for altered microRNA (miRNA or miR) expression levels in cardiovascular remodeling and disease progression. While alterations in miR expression levels have been directly linked to cardiac hypertrophy, fibrosis, and remodeling, their role in regulating gene expression during thoracic aortic aneurysm (TAA) development has yet to be explored.
Methods and Results
The present study examined miR expression levels in aortic tissue specimens collected from patients with ascending TAAs by quantitative real-time PCR, and observed decreased miR expression (miRs -1, -21, -29a, -133a, and -486) as compared to normal aortic specimens. A significant relationship between miR expression levels (miRs -1, -21, -29a, and -133a) and aortic diameter was identified; as aortic diameter increased, miR expression decreased. Using a bioinformatics approach, members of the matrix metalloproteinase (MMP) family, proteins involved in TAA development, were examined for putative miR binding sites. MMP-2 and MMP-9 were identified as potential targets for miR-29a and miR-133a respectively, and MMP-2 was subsequently verified as a miR-29a target in vitro. A significant inverse relationship between miR-29a and total MMP-2 was then identified in the clinical TAA specimens.
Conclusions
These findings demonstrate altered miR expression patterns in clinical TAA specimens, suggesting that the loss of specific miR expression may allow for the elaboration of specific MMPs capable of driving aortic remodeling during TAA development. Importantly, these data suggest that these miRs have biological and clinical relevance to the behavior of TAAs, and may provide significant targets for therapeutic and diagnostic applications.
Keywords: aneurysm, thoracic aorta, microRNA, MMP, remodeling
Introduction
Within the spectrum of cardiovascular diseases, thoracic aortic aneurysms (TAAs) continue to be one of the most dangerous and difficult to treat problems in cardiothoracic surgery. While it is clear that aortic dysfunction and dilatation are a direct result of pathological remodeling of the aortic extracellular matrix (ECM), and that this process is mediated in part by the family of matrix metalloproteinases (MMPs), there remains a paucity of information regarding the upstream mechanisms that regulate these enzymes during TAA development. Recently, a novel class of small non-coding RNA molecules (microRNAs, miRs), 20–25 nucleotides in length, were shown to have important post-transcriptional gene regulatory functions.1 MicroRNAs target short nucleotide sequences within the 3’ untranslated region (UTR) of specific messenger RNAs (mRNAs), and function to induce message degradation, or more typically, translational repression. To date, more than 1000 unique miRs have been identified within the human genome (miRBase statistics),2 and based on computational methodology current predictions suggest that approximately one third of expressed human genes contain miR regulatory target sites.3 Moreover, a single miR is capable of targeting multiple mRNAs, and a single mRNA may contain multiple miR binding sites.4–6 Together, this suggests that a common set of miRs can fine-tune the protein abundance of a cassette of specific genes that together influence specific cellular functions.
As it is becoming increasingly apparent, miRs are important determinants of disease within the cardiovascular system. Clear roles for altered miR expression7 have been implicated in the regulation of smooth muscle cell phenotype, angiogenesis, atherosclerosis, restenosis, and other vascular injury responses.7–10 Moreover, recent work has identified that changes in miR expression may contribute to the pathogenesis of aortic aneurysm or dissection. Liu and colleagues profiled miR expression in a rat model of abdominal aortic aneurysm using a miR microarray, and identified 15 dysregulated miRs with putative targets in multiple signaling pathways, including the mitogen activated protein kinase (MAPK) pathway, which may be important for AAA development.11 Elia and coworkers, examined the miR-143/miR-145 cluster in mouse model of atherosclerosis (ApoE knockout mouse), and reported markedly decreased expression levels in the knockout mice, that were further reduced when the animals were fed a high fat diet.12 They then examined human biopsies from ascending aortic aneurysms, and again observed a loss of expression for miR-143/miR-145, and suggested that the loss of these miRs induced structural modifications in the aorta, mediated in part through the incomplete differentiation of vascular smooth muscle cells. Lastly, Liao and coworkers recently examined miR expression in aortic tissue from a small cohort of patients with thoracic aortic dissections using a miR microarray.13 They likewise reported dysregulated miR expression (18 miRs upregulated, 56 miRs downregulated) with putative targets in the focal adhesion and MAPK pathways. Together, these reports suggest that dysregulated miR expression is a common feature of vascular disease and dysfunction, and that targeting the regulation of miR expression/function may provide significant therapeutic advantages.
Accordingly, it is not unreasonable to postulate that alterations in miR expression may exert an important role in the aortic medial degeneration and dilatation associated with TAA development. Therefore, the present study measured miR expression levels in clinical TAA specimens and examined the coordinate relationships with indices of aneurysm progression.
Methods
Patient Demographics
The study population consisted of aortic tissue specimens obtained from ascending TAA patients with tricuspid aortic valves (n=30) at time of surgical resection. The study excluded specimens from patients with Marfan syndrome, bicuspid aortic valves, or documented aortic dissection. Results were compared to a reference control group consisting of non-aneurysmal aortic specimens (n=10) collected from the ascending aorta of heart donors, and coronary artery bypass graft patients. There were no gender (Control = 80% male, TAA = 83% male; p=0.81 χ2) or age differences (control = 57±4 years, TAA = 62±2 years; p=0.31) between control and ascending TAA patients. All aortic specimens were maintained in frozen storage (−80°C) until time of experimentation. These specimens are a part of a multi-institutional aortic tissue bank located at the Medical University of South Carolina. This study was approved by the Institutional Review Boards of the Medical University of South Carolina, the University of Pennsylvania, and Yale University, and informed consent was obtained from all patients.
Sample Preparation and miR Expression Analysis
Resected aortic tissue specimens were homogenized in cell disruption buffer (mirVana PARIS miRNA Isolation Kit; Applied Biosystems/Ambion, Austin, TX) using a Qiagen Tissuelyser (Qiagen, Valencia, CA) bead-mill homogenizer. Total RNA was isolated (mirVana PARIS miRNA Isolation Kit), and reverse transcribed (TaqMan MicroRNA Reverse Transcription Kit; Applied Biosystems). For quantitative PCR studies, the cDNA was pre-amplified according to the manufacturer recommendations (TaqMan PreAmp Master Mix; Applied Biosystems) and the products used with specific TaqMan MicroRNA Assays (hsa-miR-1, hsa-miR-21, hsa-miR-29a, hsa-miR-133a-1, hsa-miR-208, hsa-miR-486-5p, hsa-miR-760; and snRNA U6 (internal control); Applied Biosystems) to analyze specific miR expression on a CFX96 real-time PCR detection system (Bio-Rad, Hercules ,CA) using the following 2-step procedure: initial denaturation for 10 min at 95°C, was followed by 40 cycles of 15 sec at 95°C, and 60 sec at 60°C.
Microarray analysis
Total RNA isolated from 4 normal aortic specimens and 4 large TAAs was used for miR expression analysis by microarray. The total RNA, containing low molecular weight (LMW) RNA was examined on a Bioanalyzer 2100 (Agilent Technologies) with the Small RNA Chip to ensure LMW RNA content and quality. Total RNA samples (100 ng) were labeled with the FlashTagTM Biotin RNA Labeling Kit (Genisphere LLC) following manufacturer recommendations. Biotin-labeled samples were hybridized to Affymetrix GeneChip® miRNA Arrays that were then washed, fluorescently labeled, and scanned using Affymetrix instrumentation in accordance with Affymetrix and Genisphere protocols. The resulting hybridization data were processed with Affymetrix miRNA QC Tool software (Version 1.0.33.0). Processing settings were as follows: 1) detection scoring was applied; 2) background adjustment used the BC-CG Adjust algorithm; 3) normalization was done using the quantile method; 4) the optional processes ‘added small constant’ and ‘threshold’ were used at default settings; 5) summarization was done using median polish. Resulting summarized hybridization data for the human miR content was then imported into dChip for comparative analysis.14 Differential expression was assessed as either a change in detection or a statistical change in magnitude between aneurysmal and control samples. Differential detection was assigned to miRs scoring entirely undetected in one group but detected in one of more samples of the other group; differential magnitude was assigned to miRs detected in one or more samples of both groups and differing by fold change >1.5, p<0.05 (Student’s t-test, unpaired). False discovery for differential magnitude was estimated as the median number of human miRs discovered by 100 iterations of comparison with randomized sample assignments.
miR Target Prediction
As a first approach to identifying potential miR binding sites in MMP-2 and MMP-9, the full-length transcripts for MMP-2 and MMP-9 were used to search the TargetScanHuman database (version 5.1, http://www.targetscan.org/). The results identified target binding sites for miR-29a (conserved, 7mer-8; context score=−0.01, PCT= 0.82) and miR- 133a (poorly conserved, 7mer-8; context score=−0.04, PCT<0.01) in MMP-2 and MMP-9, respectively.3, 15 To confirm the initial TargetScan results for MMP-2, a series of other bioinformatics databases were also consulted. Of the 8 additional sites that were examined, a target binding site for miR-29a was identified in the 3’UTR of MMP-2 by 6 of the 8 sites including: RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/), NBmiRTar (http://wotan.wistar.upenn.edu/NBmiRTar/), PicTar (http://pictar.bio.nyu.edu/), RNA22 (http://cbcsrv.watson.ibm.com/rna22.html), miRanda (http://www.microrna.org/microrna/home.do), and PITA (http://genie.weizmann.ac.il/). Two sites, DIANA-microT (http://www.diana.pcbi.upenn.edu/cgi-bin/micro_t.cgi), and mirTarget2 (http://mirdb.org/miRDB/) failed to identify a potential target binding domain.
Viral transduction of vascular smooth muscle cells
Human primary aortic vascular smooth muscle cells (PromoCell, Heidelberg, Germany; Cat# C-12533) were maintained in Smooth Muscle Cell Growth Medium 2 (PromoCell, Cat# C-22062) with 10% heat-inactivated Fetal Bovine Serum (Cat#16000-044, Invitrogen, Carlsbad, CA), 5μg/ml Amphotericin B (PromoCell, Cat# C-42040) and 0.5 mg/ml Gentamicin (PromoCell, C-42060) at 37°C in a humidified 5% CO2 incubator. The cells (2×105cells, passage #<8) were seeded into T-25 culture flasks, grown to 70% confluence, and exposed to lentivirus (6.6×106 PFU/ml) encoding either hsa-miR-29a (precursor form; pMIRNA1-hsa-mir-29a, Cat# CS970MR-1, System Biosciences, Mountain View, CA), anti-miR-29a (pmiRZip-29a, Cat# CS970MZ-1, System Biosciences) , or a non-targeting mismatch control (pGreenPuro Scramble Hairpin Control, Cat# MZIP000-VA-1, System Biosciences) using Transdux reagent (LV850A-1, System Biosciences) to improve transduction efficiency, or exposed to Transdux reagent alone (vehicle control). Five days post-transduction the cells were harvested using enzyme-free Cell Dissociation Buffer (Cat# 13151-014, Invitrogen). The cells were centrifuged and resuspended in cold acidic extraction buffer containing protease inhibitor cocktail (Cat# 20-201, Millipore, Billerica, MA), prior to analysis by gelatin zymography.
Gelatin Zymography
The relative abundance of MMP-2 and MMP-9 from the aortic tissue specimens or vascular smooth muscle cells was determined by gelatin zymography.16 The aortic tissue specimens were homogenized in cold acidic extraction buffer using the Qiagen Tissuelyser. Tissue and cell homogenates were centrifuged (4°C, 10 min, 1200xg) and protein concentrations were determined (BCA Protein Assay, Pierce, Rockford, IL). Aortic or cellular extracts (10 μg total protein) were fractionated on non-denaturing 10% polyacrylamide gels containing 0.1% (w/v) gelatin (Invitrogen Corporation, Carlsbad, CA). The gels were then equilibrated and incubated in Zymogram Developing Buffer (Invitrogen) for 18 hours at 37°C. After staining with 0.5% Coomassie Brilliant Blue (2 hours, room temperature), the gels were destained to reveal regions of gelatin clearance. The relative abundance of the active and latent forms of MMP-2 and MMP-9 (as verified by recombinant MMP-2 and MMP-9 standards) were then determined by densitometry using the Gel-Pro Analyzer software package (v3.1.14, Media Cybernetics Inc., Silver Spring, MD).
Confocal Microscopy
Human primary aortic vascular smooth muscle cells were seeded in 35 mm glass-bottom dishes (Cat# P35GC-1.5-20-C; MatTek Corporation, Ashland, MA). When 70% confluency was reached, the cells were transduced with lentiviral constructs, as described above, encoding the hsa-miR-29a precursor or the hsa-anti-miR-29a, or treated with Transdux reagent (vehicle control) alone. Five-days post-transduction the cells were fixed for 20 min with fresh 3.7% paraformaldehyde. The cells were then washed with PBS (6 × 5 min), and then permeabilized with 0.1% Triton X-100 for 10 min. Following a second PBS wash (3 × 5 min), the cells were blocked in 10% normal goat serum for 1 hr. Human MMP-2 (active and latent forms) was then identified by incubating the cells with a rabbit polyclonal antibody (1:200, Cat# ab37150, Abcam, Cambridge, MA) for 2 hr in 3% normal goat serum at room temperature. The cells were again washed with PBS (5 × 3 min), and then incubated in secondary antibody (1:500, AlexaFluor647 goat-anti-rabbit IgG; Cat# A-21244, Invitrogen Corporation) for 30 min at room temperature in the dark. Each viral vector carries a bicistronic copy of green fluorescent protein (GFP) under control of an independent promoter. Cells were then examined by confocal microscopy using a Zeiss LSM 510 Meta Confocal Microscope with a Plan-Neofluar 43X/1.3 oil objective. Dual fluorescence (GFP, ex=488nm, em=band-pass 505–530nm; AlexFluor647, ex=633, em= long pass 650) was recorded and the images exported as .tif files.
Data Analysis
To determine miR expression, the relative change in cycle threshold value (ΔCt) from the internal control snRNAU6, was computed. Expression was then calculated for each normal and TAA specimen using the equation Expression = 2−(ΔCt) which is premised on the fact that each CT value is in direct proportion to the amount of microRNA present at the beginning of the reaction. The results were articulated as a percent change from normal aorta to avoid any bias in total miR concentrations introduced by the pre-amplification step. The mean percent change in miR expression from normal aorta was then calculated. Specimens which did not cycle, or had Ct values higher than 35, were removed from analysis. Additionally, any expression values that fell more than 2 standard deviations away from the mean were considered outliers and removed from analysis so as not to bias the expression results. Results were expressed as mean±SEM.
Relative protein abundance of MMP-2 and MMP-9 were determined from densitometry of zymographic gels. Integrated optical densities were determined for each specimen and the mean percent change of the ratio of active:total MMP from normal aorta was calculated (outliers were identified as values falling more than 2 standard deviations away from the mean and removed from the data set), and the results were expressed as mean±SEM.
All statistical procedures were carried out using the Stata statistical package (Intercooled Stata v8.2; StataCorp LP, College Station, TX). Patient demographics were compared by χ2 analysis (gender) or one-way ANOVA (prcompw module) with Tukey’s post-hoc analysis (age). Changes in miR expression and relative protein abundance were determined using two-tailed one-sample mean comparisons versus normal aorta set at 100%, and differences between groups were determined using one-way ANOVA (prcompw module) with Tukey’s post-hoc analysis; in both cases values of p<0.05 were considered significantly different. Differences in miR expression between the normal and aneurysm groups as determined by microarray, were calculated using a Student’s t-test. Values of p<0.05 were considered to have a significant change in expression. Additionally, differences in miR detectability between the normal and aneurysm groups were calculated using chi-square analysis (tabi command, Stata v8.2). Accordingly, if a given microRNA was detectable in 3 or more specimens of a single group when compared to no detection in the other group, the miR was considered to have significant differential detectability with p<0.05. Relationships between miR expression or MMP protein abundance and aortic size, as well as the relationship between miR expression and total MMP-2 protein abundance, were determined using linear least-squares regression analysis. Results were reported graphically and a correlation constant (r-value) and p-value were determined (pwcorr and regress modules); relationships with p<0.05 were considered significant.
Results
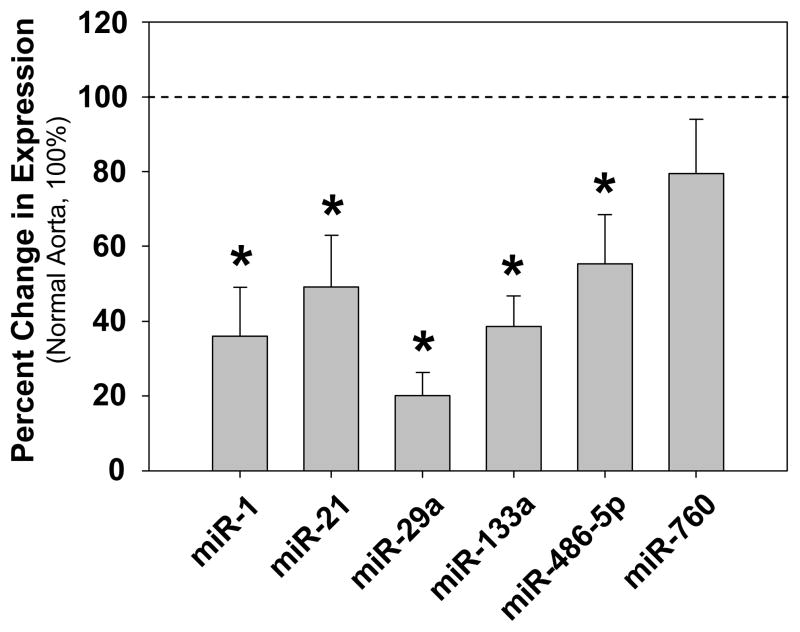
Quantitative real-time polymerase chain reaction (QPCR) was used to determine the relative expression levels of miRs -1, -21, -29a, -133a, -208, -486-5p, and -760 as compared to snRNAU6, a ubiquitous small nuclear RNA component of the spliceosome, used as an endogenous control. The QPCR results revealed significantly reduced expression of five of the seven miRs examined; miRs -1, -21, -29a, -133a, and -486-5p (Fig. 1, and S1). Expression of miR-760 was unchanged, and the α-myosin heavy chain-encoded (myocardial-restricted) miR-208, was not detected in our aortic specimens. To further verify that the loss of miR expression was due specifically to reduced miR levels, and not dramatic changes in snRNAU6, a table of Ct values (mean±SEM) for each miR examined is provided as a part of the Online Supplementary Materials (Table S1).
Figure 1.
Alterations in miR expression in clinical TAA specimens as compared to normal aorta. Mean percent change (±SEM) of miR expression in clinical TAA specimens versus normal aorta. All specimens were analyzed by quantitative PCR. The results for all cycling specimens are shown in the graph. The dashed line represents the results for normal aorta set at 100% for each miR examined (n=minimum of 8). The bars represent results for miR expression in cycling TAA specimens; miR-1 (n=21), miR-21 (n=26), miR-29a (n-27), miR-133a (n=25), miR-486-5p (n=26), and miR-760 (n=26); * indicates p<0.05 versus 100%. Note: expression of the myocardial-specific miR-208 was not detected in our aortic specimens.
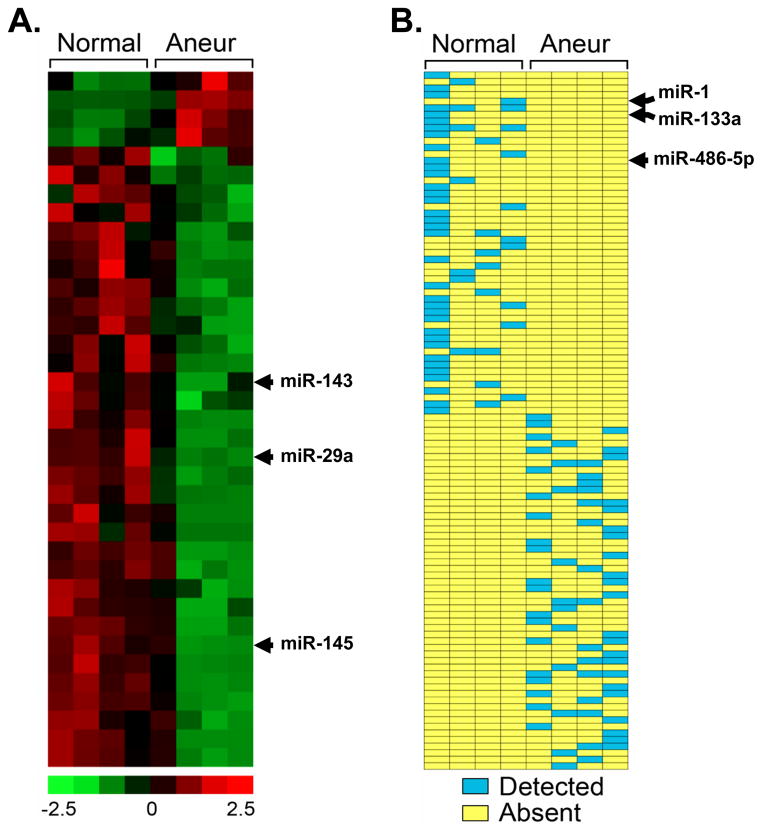
To further examine miR expression in normal and TAA tissues, microarray analysis was performed. The results yielded 37 differentially expressed miRs between normal aorta and aneurysmal aorta, consisting of 4 miRs with increased expression in the TAA group and 33 miRs with decreased expression in the TAA group (Fig 2A). In addition, there were 106 miRs that were differentially detected either in normal aorta or aneurysm, but not both. These results yielded 54 miRs that had increased expression in the aneurysm group, and 52 miRs that had decreased expression (Fig 2B). If a given microRNA was detected in three or more specimens from either group (normal or aneurysm) it was considered to have a significant change in expression (chi-square analysis, p<0.05). These data support the changes in miR expression determined by quantitative PCR for miR-1, miR-29a, miR-133a, and miR-486-5p. Functions for the 37 differentially expressed miRs were estimated based on published manuscripts reported in PubMed (Fig S2). Accordingly, 27% target proliferation pathways, 16% target growth arrest pathways, 8% target ECM structure/function, 5% (each) target apoptosis, migration, and angiogenesis, 3% (each) target drug resistance, immune response, autophagy, and cell adhesion pathways, and 22% to date have no reported function.
Figure 2.
Differentially expressed miRs in clinical TAA specimens as compared to normal aorta. miRs differentially expressed between aneurysmal (Aneur; n=4) and normal (Normal, n=4) aorta were detected by Affymetrix GeneChip analysis. A, Heatmap depicting 37 differentially expresses miRs between Aneur and Normal aorta. Colorimetric scaling for expression values (z-standardized) is shown at bottom. Patterns for miR-29a, and previously describe miR-143/miR-145, are indicated. B, Schematic diagram depicting detection scoring for 106 miRs that changed from detected to undetected or undetected to detected between Aneur and Normal aorta. Positive statistical detection calls for miRs are indicated by blue; absent (i.e., undetected) calls are indicated by yellow. Patterns for miRs-1, -133a, and -486-5p are indicated.
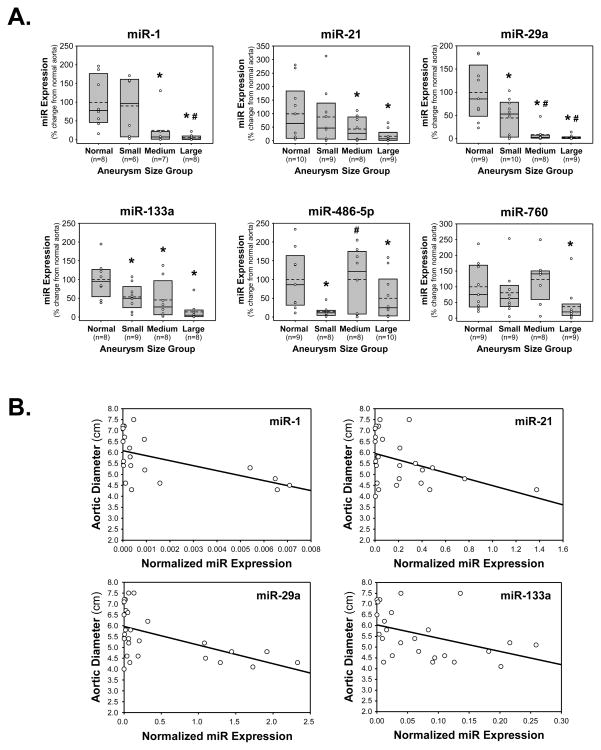
The quantitative PCR results were then stratified based on aortic diameter of the TAAs at time of surgical resection; small (4.0–5.0 cm), medium (5.1–6.0 cm), or large (6.1–7.5 cm). The miR expression levels were determined in each size group and articulated as a percent change from miR expression in normal aorta. Interestingly, each of the miRs detected demonstrated size-dependent changes in expression (Fig. 3A).
Figure 3.
miR expression and relationship to aortic size. A. miR expression was stratified into groups based on aortic diameter (TAA size) defined as: small TAAs (4.0–5.0 cm), medium TAAs (5.1–6.0 cm), or large TAAs (6.1–7.5 cm), and results were compared to normal aorta (Normal). Expression levels were calculated as a percent change form normal aorta (set at 100%) and displayed as box plots showing the median (solid line), interquartile range (25th to 75th percentile; gray box), and the mean (dashed line), overlaid with a scatter plot of each value; * p<0.05 versus 100%, # p<0.05 versus small TAAs. B. Linear least-squares regression analysis demonstrating several significant inverse relationships between miR expression in clinical TAA specimens and aortic diameter; miR-1 (r= −0.5433, p=0.0109, n=21), miR-21 (r=−0.4132, p=0.0359, n=26), miR-29a (r=−0.5364, p=0.0039, n=27), and miR-133a (r=−0.4247, p=0.0344, n=25).
Using linear least-squares regression modeling, miR expression was compared to aortic size for each of the TAA specimens. Several significant inverse relationships were identified; miR-1 (r= −0.5433, p=0.0109, n=21), miR-21 (r=−0.4132, p=0.0359, n=26), miR-29a (r=−0.5364, p=0.0039, n=27), and miR-133a (r=−0.4247, p=0.0344, n=25) (Fig. 3B).
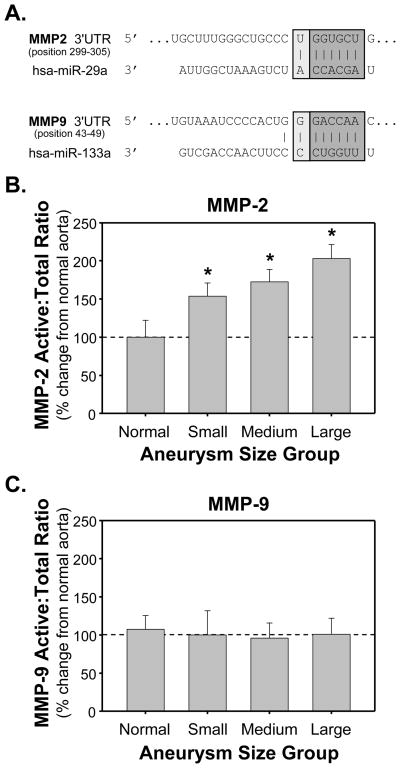
Potential biological targets were searched using the TargetScanHuman database for the presence of conserved nucleotide sequences (7 to 8 nucleotides in length), that significantly matched with the seed regions (nucleotides 2–7) of miRs -1, -21, -29a, and -133a.3 Potential binding sites for miR-29a (conserved, 7mer-8; context score=-0.01, PCT= 0.82) and miR-133a (low species conservation, 7mer-8; context score=−0.04, PCT<0.01) were identified in MMP-2 and MMP-9, respectively (Fig. 4A).3, 5, 15
Figure 4.
Identification and relative abundance of potential biological targets for altered miRs in clinical TAA specimens. A. Significant miR binding sites identified using the TargetScanHuman database (v5.1 released April 2009). B. Relative protein abundance of MMP-2 in clinical TAA specimens as compared to normal aorta as determined by zymographic analysis. C. Relative protein abundance of MMP-9 in clinical TAA specimens as compared to normal aorta as determined by zymographic analysis. Changes in the ratio of active:total forms were determined by densitometry and expressed as mean percent change (±SEM) from normal aorta (set at 100%); * p<0.05 versus 100%.
In order to investigate the relationship between MMP abundance and miR expression, the relative abundance of MMP-2 and MMP-9 protein levels was examined in the same clinical TAA specimens using zymography. Both active and latent bands were identified by recombinant standards and total MMP was calculated (sum of active + latent), and the percent change in ratio of active:total forms as compared to normal aorta were stratified by aneurysm size; providing both a measure of MMP abundance and activational state. The data revealed an increased MMP-2 active:total ratio in each of the aneurysm size groups, while no difference in the MMP-9 ratio was observed (Fig. 4B,C). Using linear-least squares regression modeling, the increase in active:total MMP-2 ratio displayed a significant positive relationship to aneurysm size (r=0.4056, p=0.0262, n=30) suggesting that the abundance and activation of MMP-2 increase with increasing aortic diameter.
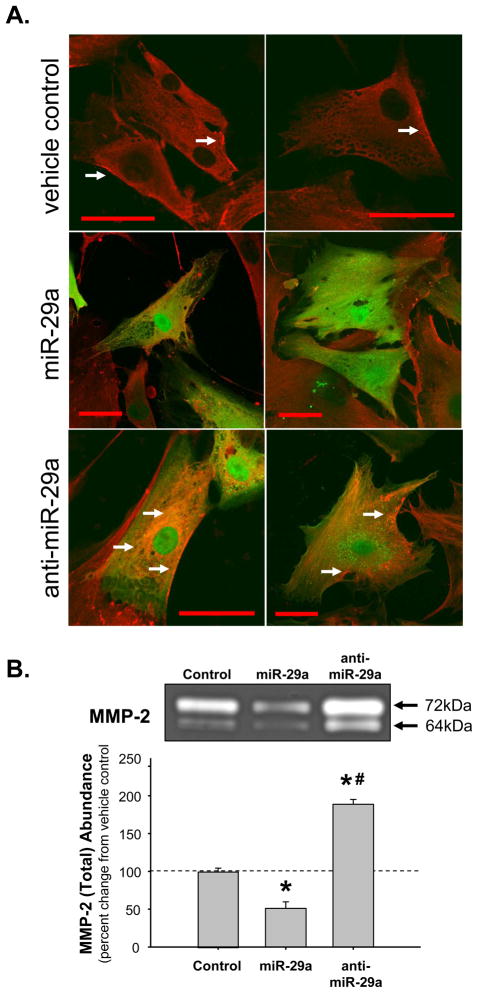
Accordingly, to examine the relationship between MMP-2 abundance and miR expression, human primary aortic vascular smooth muscle cells were transduced with lentiviral constructs encoding the miR-29a precursor or the miR antagonist, anti-miR-29a. Transduced cells were examined by confocal microscopy and gelatin zymography for changes in the abundance of latent and active forms of the MMP-2 protein. Green fluorescent protein (GFP), driven from an independent promoter on each viral construct was used to identify transduced cells. The cells were stained with an antibody for total MMP-2, and dual fluorescence was recorded. As shown in the transduction vehicle controls, MMP-2 is localized in proximity to the plasma membrane surface as identified by the white arrows (Figure 5A, top panels). In miR-29a transduced cells (Fig 5A, middle panels), overexpression of miR-29a resulted in the attenuation of total MMP-2 staining. Conversely, in cells transduced with anti-miR-29a (Fig 5A, bottom panels), overexpression of the miR antagonist resulted in enhanced MMP-2 staining in the peri-nuclear region, as well as at the periphery of the cell (identified by white arrows). In order to quantitate the effects of modulating miR-29a on the abundance of total MMP-2 protein levels, cells were transduced with miR-29a precursor or anti-miR-29a and harvested for gelatin zymography (Fig 5B). The data demonstrated that overexpression of the miR-29a precursor resulted in decreased total MMP-2 abundance. Conversely, overexpression of anti-miR-29a resulted in enhanced total MMP-2 protein abundance. Additionally, when vascular smooth muscle cells were transduced with a non-targeting mismatch control lentivirus, no change in total MMP-2 protein abundance was observed (Fig S3).
Figure 5.
Modulation of miR-29a expression levels in human primary aortic vascular smooth muscle cells. A. Cells were exposed to lentiviral constructs, containing a bicistronic copy of green fluorescent protein (GFP), designed to overexpress miR29a, anti-miR-29a, or to the transduction reagent alone. Five days post-transduction the cells were stained with MMP-2 specific antisera using an AlexFluor647 secondary antibody. Green fluorescence (GFP; ex 488nm/em 509nm) was used to identify transduced cells, while red fluorescence (MMP-2; ex 633nm/em 670) showed the localization and abundance of the active and latent forms of MMP-2. MMP-2 was localized to the cell periphery in vehicle treated cells (transduction vehicle control, top), while in the miR-29a transduced cells (middle panels), MMP-2 abundance was attenuated. In the anti-miR-29a transduced cells (bottom panels), MMP-2 protein levels were enhanced. White arrows show regions of MMP-2 accumulation at the peri-nuclear region, and cell periphery. Red bar = 50μm. B. Cells were exposed to the transduction vehicle alone, miR-29a lentivirus, or anti-miR-29a lentivirus. Five days post-transduction cells were harvested and examined by gelatin zymography. The results demonstrated that overexpression of miR-29a attenuated total MMP-2 protein abundance, while overexpression of anti-miR-29a enhanced total MMP-2 protein abundance (n=3; * p<0.05 versus control, # p<0.05 versus miR-29a transduced cells).
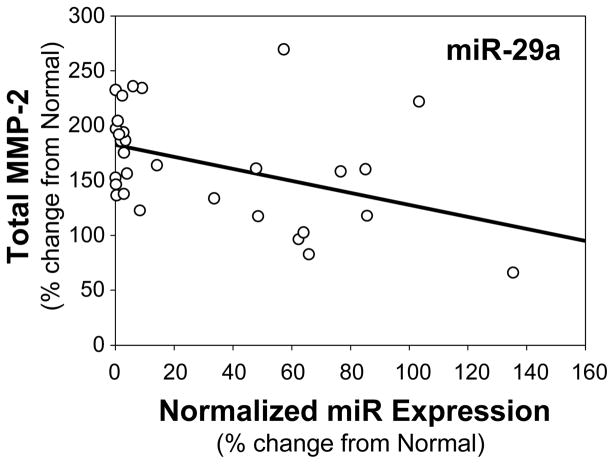
Lastly, to examine the relationship between total MMP-2 protein abundance and miR-29a expression in the aortic specimens, linear least-squares regression modeling was again performed, and a significant inverse relationship between miR-29a expression and total MMP-2 abundance was identified (r=−0.4198, p=0.0209, n=30); indicating that as miR-29a decreased, the relative abundance of MMP-2 increased (Fig. 6).
Figure 6.
Relationship between total MMP-2 abundance and miR-29a expression in clinical TAA specimens. Linear least-squares regression analysis demonstrated a significant inverse relationship between miR-29a expression and total MMP-2 abundance (r= −0.4198, p=0.0209, n=30).
Discussion
Thoracic aortic aneurysm disease results as a consequence of pathological remodeling within the aortic vascular ECM. This remodeling process induces a progressive weakening of the ECM through a variety of mechanisms including altered collagen deposition/processing, and the elevation of proteinase activity.17–19 These changes within the aortic wall result in decreased compliance and competence, culminating in aortic dilatation and eventual rupture. In an effort to further understand the molecular pathogenesis of TAA development, this study examined the expression of a focused set of miRs in clinical TAA specimens compared to aortic expression levels in patients without aneurysm disease. It was hypothesized that TAA development would coincide with alterations in specific miR expression that could affect the induction of target proteins which contribute to aortic vascular remodeling. This study focused on the expression of seven miRs that were selected because of their reported involvement in the regulation of specific protein targets within the cardiovascular system; including several MMPs and ECM components (collagens, elastin, and microfibrillar proteins).7 The unique findings of this study were four-fold.
First, within the focused set of miRs studied, five were found to have decreased expression in clinical ascending TAA specimens as compared to normal aorta (miRs -1, -21, -29a, -133a, and -486) as established by quantitative PCR. MicroRNA expression values were calculated from normalized CT values (ΔCT) using the equation (expression = 2−ΔCT) for each specimen, premised on the fact that each CT value is in direct proportion to the amount of microRNA present at the beginning of the reaction. While factors such as PCR amplification efficiencies can influence the amount of product generated, and thereby influence the sensitivity of an individual reaction, the distribution of measured miR values within the sampled cohort for this study (n=10 normal, and n=30 TAA specimens) provided sufficient power to discriminate a 50%–80% reduction in miR concentration between the normal and TAA groups (corresponding to a CT value increase of 1 to 1.2 cycles). The statistical power for comparing miR values between the two groups was greater than 0.92 for all miRs measured except miR-760, where the power was 0.29. Based on expectations of structural remodeling within the aortic wall in the aneurysm specimens, we anticipated that the protein levels of multiple MMPs and ECM proteins would be induced in response to TAA development.20 Because miRs function to degrade mRNA or repress message translation, we were not surprised to observe a significant loss of miR expression, knowing that many of the putative targets would increase in response to the disease state. This loss of miR expression in vascular disease is consistent with other reports examining miR expression by microarray. For example, Liao and coworkers profiled miR expression in a cohort of ascending aortic specimens by microarray, comparing normal patients to those with aortic dissection. They observed 74 miRs that were differentially expressed with a predominance of miRs (n=56) showing reduced expression in the dissected tissue.13
To further examine miR expression differences between the aneurysmal and normal aorta, microarray analysis was performed, and similar to Liao and coworkers, of the 37 differentially expressed miRs identified, 33 were found to have decreased expression. Included in the group of 33 miRs with decreased expression, were the miR-143/miR-145 cluster, previously reported by Elia and colleagues to be decreased in human ascending aortic aneurysm biopsies,12 and miR-29a, identified in this study as having significantly decreased expression in TAA specimens. Based on previous literature, the majority of these differentially expressed miRs target pathways involved in proliferation, growth arrest, and ECM structure/function. Taken together, this study suggests that the loss of specific miR expression may allow for the elaboration of specific protein targets that contribute to the aortic remodeling process during TAA development.
Second, several miRs displayed a significant relationship between the loss miR expression and the enhancement of aortic diameter (miRs -1, -21, -29a, and -133a). These results suggested that dynamic changes in miR expression levels likely occur during aneurysm progression, and emphasize that the loss of ongoing translational repression may play a key role in modulating the determinants of vascular remodeling.
Third, because alterations in miR expression have been shown to modulate target protein abundance, a bioinformatics approach was used to identify putative miR target sequences in genes known to be involved in TAA formation and progression. As a first approach, the TargetScanHuman database was queried with the sequence of full-length transcripts for several MMPs and ECM structural proteins. Potential biological targets were screened for the presence of conserved nucleotide sequences that significantly matched the seed regions of miRs -1, -21, -29a, and -133a.3 Results identified a highly conserved miR-29a target sequence (7mer-8) in the 3’ UTR of MMP-2, and a less well conserved miR-133a target sequence (7mer-8) in the 3’ UTR of MMP-9. Work by Bartel and colleagues, has suggested that conserved 7mer-8 target sequences are significant predictors for high efficacy miR-mediated message destabilization.4 The MMPs are a diverse family of proteases capable of degrading all components of the vascular ECM. In numerous studies using human specimens and animal models, increased protein levels of MMP-2 and MMP-9 have been directly implicated in aneurysm development in both the abdominal and thoracic aorta.16, 21–29 Therefore, the presence of these predicted target sequences suggests that these proteases may be subject to post-transcriptional or translational regulation by miR expression. 4, 30, 31 The protein levels of MMP-2 and MMP-9 were therefore examined in the clinical TAA specimens, and the indices of MMP-2 abundance were found to be elevated compared to normal aortic specimens. Moreover, a positive relationship was identified between the active:total MMP-2 ratio and aortic diameter, suggesting MMP-2 as an important mediator of aneurysm formation, consistent with previous findings from this laboratory and others.20, 24, 26, 32–35
In order to demonstrate a direct relationship between miR-29a and MMP-2, lentiviral vectors were used to transduce human primary aortic vascular smooth muscle cells with either the miR-29a precursor, or an anti-miR-29a, designed to knockdown cellular miR-29a levels. The transduced cells were then examined for changes in MMP-2 protein levels by confocal microscopy and gelatin zymography. The data demonstrated overexpression of miR-29a resulted in the attenuation of MMP-2 protein levels, while the overexpression of anti-miR-29a resulted in enhanced MMP-2 protein abundance. These results identify MMP-2 as one of the target proteins for miR-29a, and support previous reports that have identified miR-29 as a transcriptional regulator of MMP-2.30, 31
Last, in order to further implicate a role for miR-29a in aneurysm development, regression modeling was performed and a significant inverse relationship between miR-29a expression and total MMP-2 protein abundance was demonstrated. This unique observation suggested that the loss of aortic miR-29a levels may permit the elaboration of MMP-2 translation within the developing aneurysm, and may identify a potential mechanism by which MMP-2 protein induction occurs during TAA development.
While the present study has identified miR-29a as a potential regulator of MMP-2 in the aorta, it is important to note that each miR can target upwards of 200 different mRNAs, many of which may impact TAA development. For example, in silico mapping also identified miR-29a target binding sites in type-I and type-III collagens, elastin, and fibrillin-1. Dysregulation of any of these putative targets maybe deleterious to the structure and function of the thoracic aorta. Thus, altered miR expression and function within clinical TAAs may play a principal role in aneurysm progression by fine-tuning the protein abundance of a cassette of specific genes that together can influence a tissue-specific response.
The unique results of this study carry several significant clinical implications. First, miR expression profiles may provide significant insight into the identification of potential upstream mediators of aortic ECM remodeling and may reveal therapeutic strategies for the treatment of TAA disease. Second, the altered miR expression profiles identified in this study may be expanded to lay the foundation for the development of diagnostic or prognostic bioassays with the potential to define phases of disease progression, informing of the best time to intervene surgically or even indicating the potential for aortic rupture. Lastly, understanding the regulation of specific miR expression in relation to the pathophysiology behind TAA development, may also provide novel therapeutic strategies aimed at modulating miR expression in order to arrest aneurysm development or even reverse dilatation.
Taken together, this study identified that the loss of miR expression during clinical TAA development may play a key role in exacerbating pathological remodeling by removing an inhibitory signal that normally attenuates MMP production. Importantly, these data suggest that these miRs have biological and clinical relevance to the behavior of TAAs, and may provide significant targets for diagnostic and therapeutic applications.
Supplementary Material
Acknowledgments
Funding Sources: This work was supported by the Department of Veterans Affairs: [Career Development Award (CDA-2) and a Merit award to J.A.J] as well as a [Merit Award to F.G.S.], and the National Institutes of Health: [NIH/NCRR RR16434, RR16461 to J.L.B.], [NHLBI/R01s HL057952-08, and HL059165-09 to F.G.S.], and [NHLBI/R21 HL089170-01A1, and R01 HL102121-01A1 to J.S.I.]
Footnotes
Conflict of Interest Disclosures: Drs. Jones, Elefteriades, Bavaria, Joseph Gorman, Robert Gorman, Spinale, and Ikonomidis are all grant recipients. Dr. Spinale consults for Boston Scientific, Acorn Cardiovascular, and Roche Pharmaceuticals. Dr. Ikonomidis consults for W.L. Gore and Associates, and On-X Life Technologies, Inc..
References
- 1.Bartel DP. Micrornas: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Kozomara A, Griffiths-Jones S. Mirbase: Integrating microrna annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microrna targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. Micrornas: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mrnas are conserved targets of micrornas. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microrna target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 7.Small EM, Frost RJ, Olson EN. Micrornas add a new dimension to cardiovascular disease. Circulation. 2010;121:1022–1032. doi: 10.1161/CIRCULATIONAHA.109.889048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albinsson S, Suarez Y, Skoura A, Offermanns S, Miano JM, Sessa WC. Micrornas are necessary for vascular smooth muscle growth, differentiation, and function. Arterioscler Thromb Vasc Biol. 2010;30:1118–1126. doi: 10.1161/ATVBAHA.109.200873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin S, Zhang C. Micrornas in vascular disease. J Cardiovasc Pharmacol. 2011;57:8–12. doi: 10.1097/FJC.0b013e318203759b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sen CK, Gordillo GM, Khanna S, Roy S. Micromanaging vascular biology: Tiny micrornas play big band. J Vasc Res. 2009;46:527–540. doi: 10.1159/000226221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu G, Huang Y, Lu X, Lu M, Huang X, Li W, Jiang M. Identification and characteristics of micrornas with altered expression patterns in a rat model of abdominal aortic aneurysms. Tohoku J Exp Med. 2010;222:187–193. doi: 10.1620/tjem.222.187. [DOI] [PubMed] [Google Scholar]
- 12.Elia L, Quintavalle M, Zhang J, Contu R, Cossu L, Latronico MV, Peterson KL, Indolfi C, Catalucci D, Chen J, Courtneidge SA, Condorelli G. The knockout of mir-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: Correlates with human disease. Cell Death Differ. 2009;16:1590–1598. doi: 10.1038/cdd.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao M, Zou S, Weng J, Hou L, Yang L, Zhao Z, Bao J, Jing Z. A microrna profile comparison between thoracic aortic dissection and normal thoracic aorta indicates the potential role of micrornas in contributing to thoracic aortic dissection pathogenesis. J Vasc Surg. 2011 doi: 10.1016/j.jvs.2010.11.113. [DOI] [PubMed]
- 14.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. Microrna targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikonomidis JS, Jones JA, Barbour JR, Stroud RE, Clark LL, Kaplan BS, Zeeshan A, Bavaria JE, Gorman JH, 3rd, Spinale FG, Gorman RC. Expression of matrix metalloproteinases and endogenous inhibitors within ascending aortic aneurysms of patients with marfan syndrome. Circulation. 2006;114:I365–370. doi: 10.1161/CIRCULATIONAHA.105.000810. [DOI] [PubMed] [Google Scholar]
- 17.Damodarasamy M, Vernon RB, Karres N, Chang CH, Bianchi-Frias D, Nelson PS, Reed MJ. Collagen extracts derived from young and aged mice demonstrate different structural properties and cellular effects in three-dimensional gels. J Gerontol A Biol Sci Med Sci. 2010;65:209–218. doi: 10.1093/gerona/glp202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lakatta EG, Levy D. Arterial and cardiac aging: Major shareholders in cardiovascular disease enterprises: Part ii: The aging heart in health: Links to heart disease. Circulation. 2003;107:346–354. doi: 10.1161/01.cir.0000048893.62841.f7. [DOI] [PubMed] [Google Scholar]
- 19.Lakatta EG, Levy D. Arterial and cardiac aging: Major shareholders in cardiovascular disease enterprises: Part i: Aging arteries: A “set up” for vascular disease. Circulation. 2003;107:139–146. doi: 10.1161/01.cir.0000048892.83521.58. [DOI] [PubMed] [Google Scholar]
- 20.Jones JA, Ruddy JM, Bouges S, Zavadzkas JA, Brinsa TA, Stroud RE, Mukherjee R, Spinale FG, Ikonomidis JS. Alterations in membrane type-1 matrix metalloproteinase abundance after the induction of thoracic aortic aneurysm in a murine model. Am J Physiol Heart Circ Physiol. 2010;299:H114–124. doi: 10.1152/ajpheart.00028.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Absi TS, Sundt TM, 3rd, Tung WS, Moon M, Lee JK, Damiano RR, Jr, Thompson RW. Altered patterns of gene expression distinguishing ascending aortic aneurysms from abdominal aortic aneurysms: Complementary DNA expression profiling in the molecular characterization of aortic disease. J Thorac Cardiovasc Surg. 2003;126:344–357. doi: 10.1016/s0022-5223(02)73576-9. discission 357. [DOI] [PubMed] [Google Scholar]
- 22.Boyum J, Fellinger EK, Schmoker JD, Trombley L, McPartland K, Ittleman FP, Howard AB. Matrix metalloproteinase activity in thoracic aortic aneurysms associated with bicuspid and tricuspid aortic valves. J Thorac Cardiovasc Surg. 2004;127:686–691. doi: 10.1016/j.jtcvs.2003.11.049. [DOI] [PubMed] [Google Scholar]
- 23.Ikonomidis JS, Barbour JR, Amani Z, Stroud RE, Herron AR, McClister DM, Jr, Camens SE, Lindsey ML, Mukherjee R, Spinale FG. Effects of deletion of the matrix metalloproteinase 9 gene on development of murine thoracic aortic aneurysms. Circulation. 2005;112:I242–248. doi: 10.1161/CIRCULATIONAHA.104.526152. [DOI] [PubMed] [Google Scholar]
- 24.Ikonomidis JS, Jones JA, Barbour JR, Stroud RE, Clark LL, Kaplan BS, Zeeshan A, Bavaria JE, Gorman JH, 3rd, Spinale FG, Gorman RC. Expression of matrix metalloproteinases and endogenous inhibitors within ascending aortic aneurysms of patients with bicuspid or tricuspid aortic valves. J Thorac Cardiovasc Surg. 2007;133:1028–1036. doi: 10.1016/j.jtcvs.2006.10.083. [DOI] [PubMed] [Google Scholar]
- 25.Jones JA, Barbour JR, Lowry AS, Bouges S, Beck C, McClister DM, Jr, Mukherjee R, Ikonomidis JS. Spatiotemporal expression and localization of matrix metalloproteinase-9 in a murine model of thoracic aortic aneurysm. J Vasc Surg. 2006;44:1314–1321. doi: 10.1016/j.jvs.2006.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Longo GM, Xiong W, Greiner TC, Zhao Y, Fiotti N, Baxter BT. Matrix metalloproteinases 2 and 9 work in concert to produce aortic aneurysms. J Clin Invest. 2002;110:625–632. doi: 10.1172/JCI15334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McMillan WD, Patterson BK, Keen RR, Pearce WH. In situ localization and quantification of seventy-two-kilodalton type iv collagenase in aneurysmal, occlusive, and normal aorta. J Vasc Surg. 1995;22:295–305. doi: 10.1016/s0741-5214(95)70144-3. [DOI] [PubMed] [Google Scholar]
- 28.McMillan WD, Patterson BK, Keen RR, Shively VP, Cipollone M, Pearce WH. In situ localization and quantification of mrna for 92-kd type iv collagenase and its inhibitor in aneurysmal, occlusive, and normal aorta. Arterioscler Thromb Vasc Biol. 1995;15:1139–1144. doi: 10.1161/01.atv.15.8.1139. [DOI] [PubMed] [Google Scholar]
- 29.Tamarina NA, McMillan WD, Shively VP, Pearce WH. Expression of matrix metalloproteinases and their inhibitors in aneurysms and normal aorta. Surgery. 1997;122:264–271. doi: 10.1016/s0039-6060(97)90017-9. discussion 271–262. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Taylor NE, Lu L, Usa K, Cowley AW, Jr, Ferreri NR, Yeo NC, Liang M. Renal medullary micrornas in dahl salt-sensitive rats: Mir-29b regulates several collagens and related genes. Hypertension. 2010;55:974–982. doi: 10.1161/HYPERTENSIONAHA.109.144428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steele R, Mott JL, Ray RB. Mbp-1 upregulates mir-29b that represses mcl-1, collagens, and matrix-metalloproteinase-2 in prostate cancer cells. Genes Cancer. 2010;1:381–387. doi: 10.1177/1947601910371978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borges LF, Touat Z, Leclercq A, Zen AA, Jondeau G, Franc B, Philippe M, Meilhac O, Gutierrez PS, Michel JB. Tissue diffusion and retention of metalloproteinases in ascending aortic aneurysms and dissections. Human pathology. 2009;40:306–313. doi: 10.1016/j.humpath.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 33.Chung AW, Au Yeung K, Sandor GG, Judge DP, Dietz HC, van Breemen C. Loss of elastic fiber integrity and reduction of vascular smooth muscle contraction resulting from the upregulated activities of matrix metalloproteinase-2 and -9 in the thoracic aortic aneurysm in marfan syndrome. Circ Res. 2007;101:512–522. doi: 10.1161/CIRCRESAHA.107.157776. [DOI] [PubMed] [Google Scholar]
- 34.LeMaire SA, Wang X, Wilks JA, Carter SA, Wen S, Won T, Leonardelli D, Anand G, Conklin LD, Wang XL, Thompson RW, Coselli JS. Matrix metalloproteinases in ascending aortic aneurysms: Bicuspid versus trileaflet aortic valves. J Surg Res. 2005;123:40–48. doi: 10.1016/j.jss.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 35.Sinha I, Bethi S, Cronin P, Williams DM, Roelofs K, Ailawadi G, Henke PK, Eagleton MJ, Deeb GM, Patel HJ, Berguer R, Stanley JC, Upchurch GR., Jr A biologic basis for asymmetric growth in descending thoracic aortic aneurysms: A role for matrix metalloproteinase 9 and 2. J Vasc Surg. 2006;43:342–348. doi: 10.1016/j.jvs.2005.10.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.