Abstract
Methotrexate is used to treat autoimmune diseases and malignancies, including acute lymphoblastic leukemia (ALL). Inter-individual variation in clearance of methotrexate results in heterogeneous systemic exposure, clinical efficacy, and toxicity. In a genome-wide association study of children with ALL, we identified SLCO1B1 as harboring multiple common polymorphisms associated with methotrexate clearance. The extent of influence of rare versus common variants on pharmacogenomic phenotypes remains largely unexplored. We tested the hypothesis that rare variants in SLCO1B1 could affect methotrexate clearance and compared the influence of common versus rare variants in addition to clinical covariates on clearance. From deep resequencing of SLCO1B1 exons in 699 children, we identified 93 SNPs, 15 of which were non-synonymous (NS). Three of these NS SNPs were common, with a minor allele frequency (MAF) >5%, one had low frequency (MAF 1%–5%), and 11 were rare (MAF <1%). NS SNPs (common or rare) predicted to be functionally damaging were more likely to be found among patients with the lowest methotrexate clearance than patients with high clearance. We verified lower function in vitro of four SLCO1B1 haplotypes that were associated with reduced methotrexate clearance. In a multivariate stepwise regression analysis adjusting for other genetic and non-genetic covariates, SLCO1B1 variants accounted for 10.7% of the population variability in clearance. Of that variability, common NS variants accounted for the majority, but rare damaging NS variants constituted 17.8% of SLCO1B1's effects (1.9% of total variation) and had larger effect sizes than common NS variants. Our results show that rare variants are likely to have an important effect on pharmacogenetic phenotypes.
Methotrexate is an antifolate agent used to treat malignancies, including acute lymphoblastic leukemia (ALL), as well as for autoimmune diseases, such as rheumatoid arthritis. Methotrexate inhibits folate synthesis and hence DNA synthesis, causing proliferating cells to undergo apoptosis. Inter-individual variation in clearance of methotrexate results in vastly different exposure to the drug, which affects its clinical effectiveness and toxicity (Evans et al. 1984, 1986, 1998). In a genome-wide association study (GWAS) of children with ALL, we identified SLCO1B1 as the only locus harboring single nucleotide polymorphisms (SNPs) associated with methotrexate clearance at a genome-wide significance level (Supplemental Fig. 1; Trevino et al. 2009). Preclinical (van de Steeg et al. 2009, 2010) and clinical (Lopez-Lopez et al. 2011) in vivo studies have confirmed the association and the importance of this gene to methotrexate disposition and effect.
SLCO1B1 encodes SLCO1B1 (also known as OATP1B1), a hepatic organic anion transporter that also mediates disposition of many medications (including statins and irinotecan) and biochemicals (Konig et al. 2000; Abe et al. 2001; Hirano et al. 2004; Nozawa et al. 2005; Weaver and Hagenbuch 2010). Although many SNPs have been identified in SLCO1B1, only a few are known to have functional effects (Franke et al. 2010). The common T521C variant (rs4149056), which produces a V174A substitution, is present at a minor allele frequency (MAF) of ∼13% across most populations (Pasanen et al. 2008). This substitution results in decreased transport function in vitro (Tirona et al. 2001; Iwai et al. 2004; Kameyama et al. 2005) and lower drug clearance in vivo (Niemi et al. 2004, 2005a,b; Pasanen et al. 2006, 2007). We confirmed the reduced clearance of methotrexate in those carrying the C allele of rs4149056 (Trevino et al. 2009), and it has since been validated (Lopez-Lopez et al. 2011). However, we found multiple linkage disequilibrium (LD) blocks in SLCO1B1 harboring SNPs independently associated with methotrexate clearance (Supplemental Fig. 2), suggesting that variation throughout the SLCO1B1 gene may be important for methotrexate disposition.
Although many of the SNPs previously identified in SLCO1B1 were common (MAF > 5%), our goal was to identify all variants, both common and rare, and determine whether rare variants also have a role in methotrexate disposition. There has been little phenotypic analysis of rare variants in pharmacogenomics to date, particularly in the setting of follow-up of a pharmacogenetics GWAS signal (Asimit and Zeggini 2011). As genetic testing moves from research to clinical implementation, it is important to define the relative contribution of common versus rare genetic variants in influencing phenotypic variation (Bodmer and Bonilla 2008; Bansal et al. 2010; Liu and Leal 2010; Zhu et al. 2011). Several current clinically available pharmacogenetic tests focus only on common variants (Altman et al. 2011); if rare variants must be interrogated, clinical genetic testing will likely need to become 100% sequence-based to be adequate, which will involve technical and informatic challenges. Therefore, we comprehensively interrogated SLCO1B1 genetic variations in DNA from patients who had been carefully phenotyped for methotrexate clearance.
Results
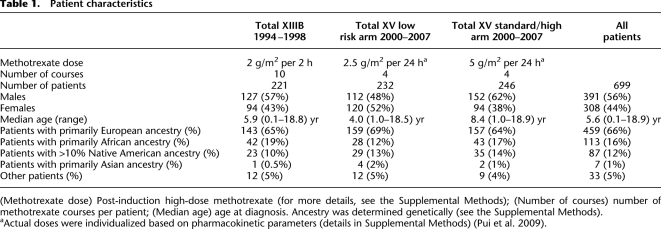
After our GWAS identified SLCO1B1 as the only locus harboring SNPs associated with methotrexate clearance at the genome-wide significance level (Supplemental Fig. 1; Trevino et al. 2009), we sequenced the gene in a large group of well-phenotyped patients (Table 1). We used a combination of sequencing exons in the entire patient group and a strategy of focusing on the phenotypic outliers (Cohen et al. 2006) (highest vs. lowest methotrexate clearance patients) to detect important variants; we verified that a strategy of focusing on the outliers identified the important variants (Supplemental Fig. 3). We found 39 SNPs in exons 10–15 among all patients, and 21 SNPs in exons 1–9 among the phenotypic outliers; we genotyped all of these, in addition to all other published non-synonymous (NS) SNPs, in all patients (Supplemental Fig. 4). In total, we genotyped 104 SNPs, 93 of which were polymorphic in our 699 patients (Supplemental Table 1).
Table 1.
Patient characteristics
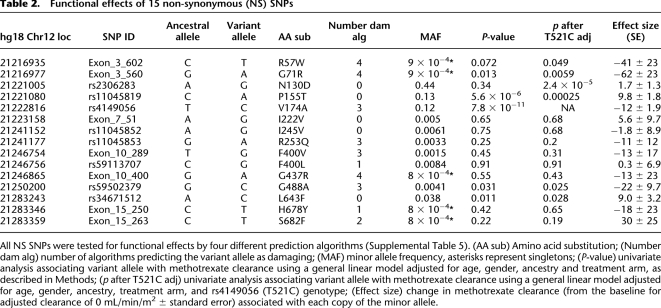
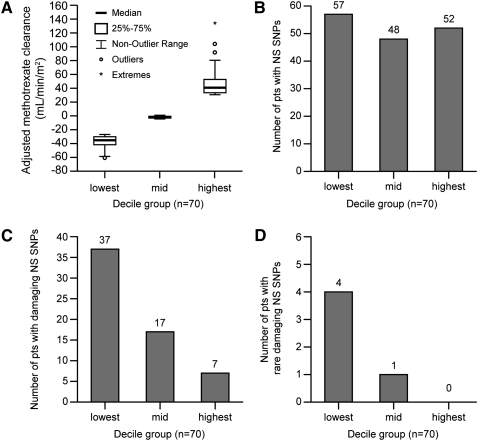
Of the 93 SNPs, 15 were NS, and seven of these were categorized as functionally damaging by at least three of four computational prediction algorithms (Ng and Henikoff 2001; Ferrer-Costa et al. 2004; Yue et al. 2006; Adzhubei et al. 2010), whereas the other eight NS SNPs were categorized as tolerated (Table 2). Three of these NS SNPs were common, with a minor allele frequency (MAF) >5%, one had low frequency (MAF 1%–5%), and 11 were rare (MAF < 1%). Among patients with extreme phenotypes (Fig. 1A), we found an overrepresentation of damaging NS SNPs in those with the lowest decile for clearance compared with those with the average (45th–55th percentile) or highest deciles (p = 2.3 × 10−8) (Fig. 1C). Even after excluding patients with the damaging C alleles at the common rs4149056 polymorphism, the same trend holds for rare damaging NS SNPs (p = 0.026) (Fig. 1D). When all NS SNPs (irrespective of functional status) were considered, there was no evidence of overrepresentation of variant alleles in patients with extremely high or low clearances (p = 0.3) (Fig. 1B).
Table 2.
Functional effects of 15 non-synonymous (NS) SNPs
Figure 1.
Methotrexate clearance and presence of non-synonymous (NS) SNPs among outliers. (A) The average methotrexate clearance (adjusted for age, gender, ancestry, and treatment arm) is depicted for the lowest, middle, and highest tenth percentile groups (n = 70 patients per decile). The number of patients harboring NS SNPs did not differ among clearance decile groups (p = 0.3, Cochran-Armitage Trend Test) (B), whereas the number of patients harboring damaging NS SNPs differed by clearance decile group (p = 2.3 × 10−8, Cochran-Armitage Trend Test) (C), as did the number of patients harboring rare (MAF < 1%) damaging NS SNPs (p = 0.026, Cochran-Armitage Trend Test) (D).
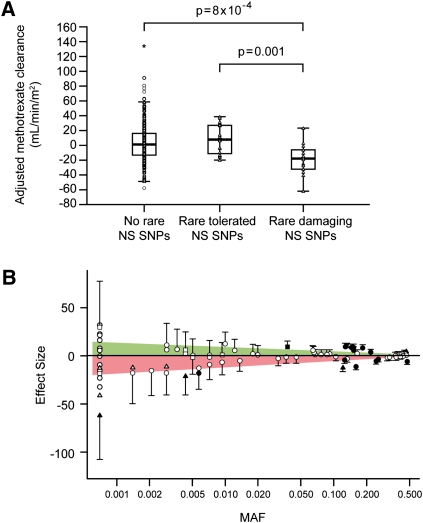
Because it has already been established that the rs4149056 C allele confers reduced transporter function, we examined which other SLCO1B1 polymorphisms were associated with methotrexate clearance. Among the 542 patients who were homozygous for the rs4149056 T allele, the 14 patients with rare damaging NS SNPs had lower methotrexate clearance than the 20 patients with rare tolerated NS SNPs (p = 0.001) (Fig. 2A) and the other 508 patients without any rare NS SNPs (p = 8 × 10−4). Effect size tended to be inversely related to MAF (Fig. 2B), although small sample sizes (inherent in rare variant analysis) precluded statistical significance in many cases. These results are consistent with the hypothesis that rare damaging SNPs may have more profound effects on phenotype than common SNPs (Zhu et al. 2011).
Figure 2.
Patients with rare damaging NS SNPs have reduced methotrexate clearance. (A) Among the 542 patients who did not carry the variant C allele at rs4149056, median adjusted methotrexate clearance is lower in patients with rare damaging NS SNPs (n = 14) than those having no rare NS SNPs (n = 508, p = 8 × 10−4) or having rare tolerated NS SNPs (n = 20, p = 0.001, Wilcoxon rank-sum test). (B) SNPs with smaller MAFs have larger effect sizes. (Triangles) NS damaging SNPs; (squares) NS tolerated SNPs; (circles) other SNP types. Shapes filled in black indicate p < 0.05; others do not reach statistical significance. Effect size indicates the change in adjusted methotrexate clearance (in milliliters per minute per meter squared) attributable to each copy of the minor allele based on the general linear model, and error bars represent standard error within the model. Shading indicates area under a linear regression fit line for negative effect sizes (pink, lower clearance) or positive effect sizes (green, higher clearance) versus MAF.
Considering all 93 SNPs (adjusting for treatment arm, ancestry, gender, and age), 27 SNP genotypes (Supplemental Table 2) were associated with methotrexate clearance (p < 0.05). To identify which SNP genotypes were associated with clearance independent of their LD with the common rs4149056 NS SNP, we performed multivariate linear regression analysis with rs4149056 as a covariate: 23 of the 27 SNPs retained their association (p < 0.05) (Supplemental Table 2). Interestingly, an additional 12 SNPs were associated (p < 0.05) with clearance only after adjustment for rs4149056, indicating that the effect of these variants may be masked by the variation due to this common functional variant. Within the entire cohort of 699 patients, common low-function rs4149056 C alleles were present in 157 patients (22.4% of all children, MAF 12%), and an additional 15 (2.1% of children) had one rare allele associated with reduced methotrexate clearance (Exon3_560: one patient; Exon3_602: one patient; rs59502379: six patients; rs79661732: seven patients). Taken together, our results indicate that rare NS SNPs account for substantial variability in a small but non-negligible percentage of patients.
Of the seven damaging NS SNPs, the minor allele for three was related to lower methotrexate clearance in the entire group of 699 patients (p < 0.05) (Table 2). Conversely, the minor allele for three of the eight tolerated NS SNPs was associated with increased methotrexate clearance (p < 0.05; rs2306283 only after adjusting for rs4149056). The three damaging and three tolerated SNPs associated with clearance remain significant after adjusting for rs4149056. The rs2306283 SNP encodes an N130D substitution and becomes significant only after adjusting for rs4149056 (p = 0.34 before and p = 2.4 × 10−5 after adjustment) by conferring higher methotrexate clearance in the patients heterozygous for rs4149056 (Supplemental Fig. 5), which was confirmed in a classification and regression tree analysis (Supplemental Fig. 6).
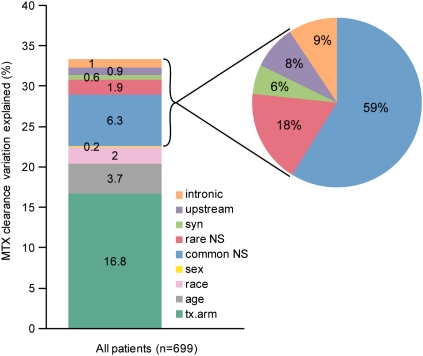
To estimate the percentages of variability in methotrexate clearance accounted for by non-genetic and genetic variation, a multivariate analysis with forward variable selection revealed that the clinical covariates of treatment arm, age, ancestry, and gender accounted for 16.8%, 3.7%, 2%, and 0.2% of inter-patient variability in clearance, respectively. Variants in SLCO1B1 accounted for 10.7% of inter-patient variability in clearance, with rare damaging and common coding SNPs accounting for 1.9% and 6.3%, respectively (Fig. 3). Of the clearance variation accounted for by SLCO1B1 (10.7% of the total), rare damaging NS variants constituted 17.8% of SLCO1B1's effect on clearance (Fig. 3). The rare damaging SNPs explained a significant portion of the variability in MTX clearance (p = 0.00014). Overall, clinical and genetic covariates accounted for one-third of the population variation in clearance.
Figure 3.
One-third of inter-patient variability in methotrexate clearance can be explained by clinical covariates and SLCO1B1 SNP genotypes. Forward variable selection was used to select covariates, including SNPs in each category (common NS, rare NS, synonymous [syn], 5′ upstream, or intronic) as input based on each covariate's increase in R2 for explaining variability in clearance (see Methods). SNPs included in this model are listed in Supplemental Table 2. Of the variation in clearance explained by SLCO1B1 polymorphisms, the percent apportioned to each category of SNP (common NS, rare NS, synonymous, 5′ upstream, or intronic) is illustrated in the pie chart at right.
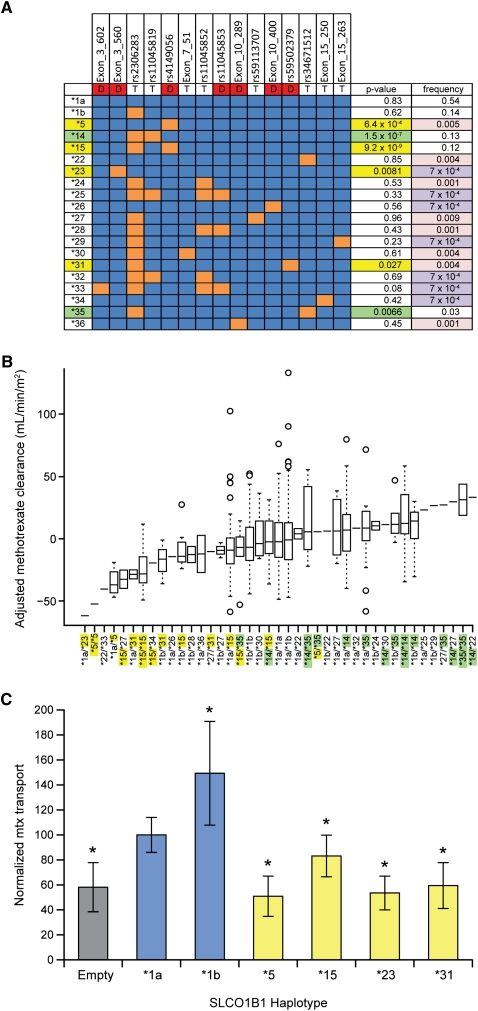
Based on 15 NS SNPs, we observed 20 SLCO1B1 haplotypes (Fig. 4A), five of which were previously reported (Tirona et al. 2001; Pasanen et al. 2008). In a univariate linear regression model, the *5 (p = 6.4 × 10−4) and *15 (p = 9.2 × 10−9) haplotypes and two novel haplotypes (*23, p < 0.009 and *31, p < 0.03) were associated with low methotrexate clearance. The *14 haplotype (p = 1.5 × 10−7) and one novel haplotype (*35, p < 0.007) were associated with high clearance. All four haplotypes that were associated with low clearance contain an NS SNP predicted to be damaging, whereas the two haplotypes associated with high clearance only harbor NS SNPs predicted to be tolerated by all four prediction algorithms. The haplotypes harboring damaging NS SNPs are clustered with low-clearance patients (Fig. 4B), haplotypes harboring tolerated NS SNPs are clustered with high-clearance patients, and those heterozygous for high-clearance and low-clearance haplotypes are clustered with patients with no variant alleles (and intermediate clearance), suggesting that the low- and high-activity alleles offset each other's effects.
Figure 4.
SLCO1B1 haplotypes affect average adjusted methotrexate clearance. (A) Haplotypes are based on 15 NS SNPs. (Red boxes with D) Damaging SNPs; (T) tolerated SNPs; (blue boxes) the ancestral allele; (orange boxes) the variant allele. The univariate P-value for the association with methotrexate clearance is indicated after adjustment for age, gender, ancestry, and treatment arm. (Yellow P-values) The haplotype confers significantly lower methotrexate clearance; (green P-values) significantly higher methotrexate clearance; (purple frequencies) singletons; (pink) those that are rare (frequency < 1%). (B) Diplotypes (x-axis) are sorted by median adjusted methotrexate clearance (y-axis). (Yellow haplotypes) Associated with lower methotrexate clearance (p < 0.05); (green haplotypes) associated with higher methotrexate clearance (p < 0.05). (Horizontal line) Median; (boxes) 25th to 75th percentiles; (whiskers) non-outlier range; (circles) outliers. (C) Methotrexate (mtx) uptake by HEK293 cells stably transfected with SLCO1B1 coding variants, plotted as percentage of SLCO1B1*1a-expressing cells, which averaged 30.8 pmol/mg. (Empty) HEK293 cells expressing the empty vector; (asterisks) transport that differs significantly (at p < 0.05, Student's t-test) compared with the SLCO1B1*1a haplotype.
To test for the functional consequences of novel variants discovered in our patients, we stably expressed SLCO1B1 variants in mammalian HEK293 cells and measured the rate of methotrexate transport. The in vivo pharmacokinetic findings were supported by these in vitro functional studies: Cellular uptake of methotrexate in vitro was reduced in cells stably transfected with the SLCO1B1 haplotypes associated with reduced methotrexate clearance in patients (*5, *15, *23, and *31) (Fig. 4C).
Discussion
Our study demonstrates that both rare and common genomic variations contribute to meaningful phenotypic variation in drug disposition. Using computational tools for estimating functional effects of NS substitutions (Carr et al. 2009; Gonzalez-Perez and Lopez-Bigas 2011), we found that damaging SNP genotypes concentrated among those with the lowest methotrexate clearance, with similar results when only rare damaging SNPs were considered (Fig. 1). Also important is that patients with intermediate clearance carried a burden of damaging SNPs in between the two outlying clearance groups, and in patients without the common damaging rs4149056 C allele, clearance was lower in those who harbored rare damaging SNP genotypes compared with those who harbored rare tolerated NS SNPs or no rare NS SNPs (Fig. 2). Despite some concerns about performance of computational tools to estimate functional effects of NS SNPs, the approach worked well in this study: Even combining the SNPs predicted to be damaging and tolerated into diplotypes yielded the expected intermediate level of methotrexate clearances (Fig. 4B). Moreover, our in vitro functional experiments validated the algorithms used to predict damaging SNPs and mimicked our in vivo assessments of clearance for each of the haplotypes with SNPs predicted to be damaging (Fig. 4C).
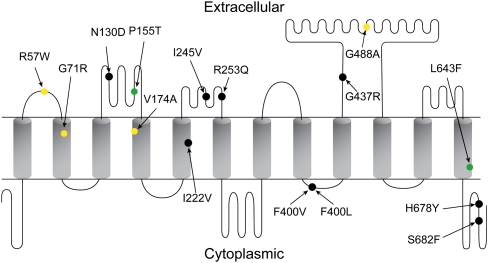
For example, one novel SNP (Exon_3_602) was observed only once among nearly 1400 chromosomes in our patients and resulted in the substitution of arginine at position 57 in the first extracellular loop (Fig. 5). In vitro mutagenesis had suggested that this site might be important for substrate binding, because it is a highly conserved, positively charged amino acid facing the putative binding pocket (Weaver and Hagenbuch 2010). We found that the *23 haplotype, which harbors a variant predicted to be damaging at position 71, has severely reduced methotrexate uptake in vitro (Fig. 4C), consistent with our in vivo assessments of clearance (Fig. 4B). Additionally, the *5 haplotype displayed a greater magnitude of a reduction in methotrexate transport in vitro than did the *15 haplotype, mimicking the patient data (i.e., *1a/*5 patients had lower clearance than *1a/*15 patients).
Figure 5.
Extracellular, transmembrane, and intracellular cytoplasmic domains predicted for SLCO1B1, indicating all 15 non-synonymous SNPs. Shown are SNPs associated with lower methotrexate clearance (p < 0.05) (yellow), higher clearance (green), or no association with clearance (black).
Epistasis is illustrated by the rs2306283 N130D SNP genotype in the second extracellular loop (Fig. 5), which was related to higher methotrexate clearance in patients carrying the lower-function rs4149056 T521C allele (which resides in the fourth transmembrane domain) (Supplemental Fig. 5; Pasanen et al. 2008). The G allele of rs2306283 is predicted by all four algorithms to be tolerated and is associated with increased clearance of methotrexate only after considering the rs4149056 genotype. Even though there was no statistically significant interaction between rs4149056 and rs2306283 (p = 0.08), the effect size of the G allele of rs2306283 was much greater in the rs4149056 TT/CC patients (36 ± 9.8 mL/min per meter squared) than in the patients with the rs4149056 TT genotype (8.3 ± 2.1 mL/min per meter squared) (Supplemental Table 4).
It is important that pharmacogenetic studies include all possible non-genetic covariates as part of the process for evaluating the quantitative contribution of genetic variation to the phenotype. Because all 699 patients were participating in clinical trials, we had outstanding ascertainment of all covariates. We could account for one-third of population variability in clearance, with 10.7% of all variation due to SLCO1B1 variants. Of that 10.7%, rare variants accounted for 17.8% of the variation attributed to SLCO1B1 SNPs. Thus, 15 of 699 unselected patients (2.1%) carried a rare NS allele in SLCO1B1, indicating that for the phenotype of methotrexate clearance, rare variation has an effect for a non-negligible proportion of patients.
The percentage of variation in methotrexate clearance accounted for by clinical covariates (22.7%) is comparable to the 19.7% described for a complex phenotype such has hypertriglyceridemia (Johansen et al. 2010), although for our pharmacogenetic phenotype, rare variants constituted a greater proportion of genetic variation (17.8%) than was true for rare variants associated with hypertriglyceridemia. Rare NS SNPs conferring either lower (if damaging) or higher (if tolerated) function for non-pharmacogenetic phenotypes, such as blood pressure (Tomaszewski et al. 2010) and other phenotypes, has been demonstrated (Cohen et al. 2006; Momozawa et al. 2010). Thus, evidence is mounting that rare variants may be important to capture for the level of comprehensive interrogation of genetic variation that will be needed in clinical genetic testing.
Although we could explain a third of the inter-individual variation in methotrexate clearance based on SLCO1B1 variation and non-genetic covariates, the other two-thirds of inter-patient variability remains unexplained. This variation could be due to epigenetic variation, genomic variation not assessed (in non-coding or coding regions), differences in regulatory miRNA, post-translational modification or gene expression, or other clinical or environmental variables not assessed. Moreover, additional studies of methotrexate disposition with low-dose methotrexate and with other regimens of high-dose methotrexate will be needed to assess the role of SLCO1B1 variation in other clinical contexts.
Our study serves as a proof-of-principle for follow-up of a gene hit resulting from a GWAS in pharmacogenomics; here, a single gene (SLCO1B1) had common variants associated with drug clearance, but deep sequencing yielded additional rare variants that are likely to be functional and have penetrant effects. As is true for some disease phenotypes, our findings support the hypothesis that a combination of common and rare variants is likely to be important for pharmacogenetic phenotypes. There will be challenges when genotyping tests are deployed in the clinic, because it is likely that sequencing will be required for detection of rare variants for that small but non-negligible group of patients who harbor them.
Methods
Methotrexate clearance was assessed for a total of 4184 courses administered at 2–5 g/m2 in 699 patients; doses for patients enrolled on the Total XV trial were adjusted based on pharmacokinetic parameters to achieve a desired plasma exposure, as described (Pui et al. 2009). Parents and patients gave informed consent or assent where appropriate, and the study was approved by the Institutional Review Board. Details of the two clinical trials and pharmacokinetic analysis have been previously published (Pui et al. 2004, 2009; Trevino et al. 2009). The genome-wide array data were previously published on 640 of these patients (Trevino et al. 2009); the current cohort includes an additional 59 patients not reported in the original GWAS (Supplemental Fig. 7).
Genotyping
In addition to all common variants (n = 24) interrogated on a genome-wide array (Trevino et al. 2009), we extensively sequenced and genotyped all SLCO1B1 exons and surrounding introns and all reported non-synonymous variants in a group of 699 children with acute lymphoblastic leukemia (ALL) treated on St. Jude Children's Research Hospital Total XIIIB and Total XV protocols. Details of patients included in the current analysis and previous analysis (Trevino et al. 2009) are included in Supplemental Figure 7. Germline DNA was extracted from blood after remission was achieved. Genotypes were generated and calls made as described (Trevino et al. 2009), using GeneChip Human Mapping 500K Array sets or the Genome-Wide Human SNP Array 6.0 (Affymetrix). The SLCO1B1 T521C (rs4149056) SNP was genotyped as described (Trevino et al. 2009). Genotyping and sequencing strategies are detailed in the Supplemental Material.
The methods used to generate each SNP genotype are included in Supplemental Table 1. The P-values (before and after adjustment for the T521C SNP, rs4149056), effect size, and SNPs included in the multivariate model are included in Supplemental Table 2. The Hardy–Weinberg equilibrium P-value and MAF in each ancestry group are included in Supplemental Table 3.
Bioinformatics
We used four prediction algorithms—SIFT (Ng and Henikoff 2001), PMUT (Ferrer-Costa et al. 2004), SNPS3D (Yue et al. 2006), and Polyphen2 (Adzhubei et al. 2010)—to categorize NS SNPs into damaging or tolerated groups. The SNPs in the damaging group (n = 7) were predicted to be damaging by three or four of the algorithms, while the other NS SNPs were included in the tolerated group (Table 2). Supplemental Table 5 contains the individual SNP scores for each algorithm.
Cellular uptake assays
The plasmid pCMV6-XL4 vector, encoding SLCO1B1*1b, was purchased from Origene, and SLCO1B1 was subsequently subcloned into pcDNA5/FRT/TO (Invitrogen). Mutations were introduced by the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies) and confirmed by Sanger sequencing. Stable, isogenic HEK293 cell lines expressing inducible SLCO1B1 variants were constructed using the Flp-In T-REx System according to the manufacturer's instructions (Invitrogen). Equal expression among variants was confirmed by RT-PCR (data not shown). Methotrexate uptake was performed using [3H]methotrexate as described in detail in the Supplemental Methods. The uptake of SLCO1B1 variant–expressing cells was normalized to the uptake of SLCO1B1*1a (WT)–expressing cells and expressed as the average uptake from two or three independent assays done in triplicate. Results were compared using a two-tailed t-test.
Statistical analysis
Missing SNP genotypes among the 93 SNPs analyzed in 699 patients were imputed using the fastPHASE software (Scheet and Stephens 2006). Haplotypes were inferred using PHASE (Stephens et al. 2001). Genetic ancestries of patients were estimated using STRUCTURE (Pritchard et al. 2000) based on genotypes from the SNP array (Yang et al. 2010). Imputation of variants from the 1000 Genomes Project was performed using MACH1.0 (Li et al. 2010) based on HapMap CEU, YRI, CHB, and JPT reference populations (additional details in the Supplemental Methods).
The Cochran-Armitage Trend Test (prop.trend.test in R) was used to compare the proportion of patients in each clearance decile group harboring at least one NS, damaging NS, or rare damaging NS SNP (Fig. 1). The Wilcoxon rank-sum test was used to compare adjusted methotrexate clearance in patients (excluding carriers of the rs4149056 C allele) with rare damaging NS SNPs, rare tolerated NS SNPs, and those with no rare NS SNPs (Fig. 2A). This non-parametric test was used because the sample size was small (n = 20 patients with rare damaging NS SNPs, n = 14 patients with rare tolerated NS SNPs).
Average methotrexate clearance was used as the phenotype because linear mixed effect models considering the clearance of each course of methotrexate individually did not alter the association between genotypes and methotrexate clearance when compared with using the average methotrexate clearance across all courses (Trevino et al. 2009). Associations between germline SNP genotypes and methotrexate clearance were evaluated using a general linear model assuming an additive effect of allelic dosage, with all models including age, ancestry, gender, and treatment regimen as “clinical” covariates (Supplemental Table 2). Effect size refers to the beta from the linear regression (Table 2; Supplemental Tables 2, 4). “Adjusted methotrexate clearance” refers to the residual from the linear regression including the clinical covariates. Age was coded in years and treated as a continuous variable. The treatment regimen was coded as a categorical variable. Clinical covariates were always included in any analyses of genetic (single SNP or multiple SNPs) association with clearance.
Multiple linear regression analysis was used to estimate percent variation explained by multiple genotypes and other covariates. All SNPs were grouped into the following categories: common NS, rare NS, synonymous, 5′ upstream, UTR, or intronic. Linear regression models were fit by including each category of SNPs sequentially (in the order above). We performed a forward selection strategy, adding SNPs that were significant at the 0.05 level in the presence of the SNPs and additional covariates already in the model. The significance of the SNP was determined using ANOVA comparing the models with the SNP versus without the SNP. The difference in R2 values between the models was used to calculate the percentage of variation attributable to each variable. Clinical covariates were included in all models. Statistical analyses were conducted using R (http://www.r-project.org/) and Statistica 10 (StatSoft, Inc). All P-values presented are two-sided and were not adjusted for multiple testing.
Data access
The data have been submitted to dbGaP under accession number phs000426.v1.p1 (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000426.v1.p1).
Acknowledgments
We thank our protocol co-investigators, clinical and research staff, particularly Pamela McGill, Jeremy Hunt, Nancy Kornegay, and Xiangjun Cai, as well as patients and families for their participation. We thank the staff from the St. Jude Hartwell Center for Bioinformatics and Biotechnology. We thank Jun Yang for a critical review of the manuscript. This work is supported by NCI grants CA 36401 and CA 21765 and the NIH/NIGMS Pharmacogenomics Research Network (U01 GM92666 and U01 HL65899), and by the American Lebanese Syrian Associated Charities (ALSAC).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.129668.111.
References
- Abe T, Unno M, Onogawa T, Tokui T, Kondo TN, Nakagomi R, Adachi H, Fujiwara K, Okabe M, Suzuki T, et al. 2001. LST-2, a human liver-specific organic anion transporter, determines methotrexate sensitivity in gastrointestinal cancers. Gastroenterology 120: 1689–1699 [DOI] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR 2010. A method and server for predicting damaging missense mutations. Nat Methods 7: 248–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman RB, Kroemer HK, McCarty CA, Ratain MJ, Roden D 2011. Pharmacogenomics: Will the promise be fulfilled? Nat Rev Genet 12: 69–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asimit J, Zeggini E 2011. Rare variant association analysis methods for complex traits. Annu Rev Genet 44: 293–308 [DOI] [PubMed] [Google Scholar]
- Bansal V, Libiger O, Torkamani A, Schork NJ 2010. Statistical analysis strategies for association studies involving rare variants. Nat Rev Genet 11: 773–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer W, Bonilla C 2008. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet 40: 695–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr DF, Whiteley G, Alfirevic A, Pirmohamed M 2009. Investigation of inter-individual variability of the one-carbon folate pathway: A bioinformatic and genetic review. Pharmacogenomics J 9: 291–305 [DOI] [PubMed] [Google Scholar]
- Cohen JC, Pertsemlidis A, Fahmi S, Esmail S, Vega GL, Grundy SM, Hobbs HH 2006. Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels. Proc Natl Acad Sci 103: 1810–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans WE, Crom WR, Stewart CF, Bowman WP, Chen CH, Abromowitch M, Simone JV 1984. Methotrexate systemic clearance influences probability of relapse in children with standard-risk acute lymphocytic leukaemia. Lancet 1: 359–362 [DOI] [PubMed] [Google Scholar]
- Evans WE, Crom WR, Abromowitch M, Dodge R, Look AT, Bowman WP, George SL, Pui CH 1986. Clinical pharmacodynamics of high-dose methotrexate in acute lymphocytic leukemia. Identification of a relation between concentration and effect. N Engl J Med 314: 471–477 [DOI] [PubMed] [Google Scholar]
- Evans WE, Relling MV, Rodman JH, Crom WR, Boyett JM, Pui CH 1998. Conventional compared with individualized chemotherapy for childhood acute lymphoblastic leukemia. N Engl J Med 338: 499–505 [DOI] [PubMed] [Google Scholar]
- Ferrer-Costa C, Orozco M, de la Cruz X 2004. Sequence-based prediction of pathological mutations. Proteins 57: 811–819 [DOI] [PubMed] [Google Scholar]
- Franke RM, Gardner ER, Sparreboom A 2010. Pharmacogenetics of drug transporters. Curr Pharm Des 16: 220–230 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Perez A, Lopez-Bigas N 2011. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet 88: 440–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Maeda K, Shitara Y, Sugiyama Y 2004. Contribution of OATP2 (OATP1B1) and OATP8 (OATP1B3) to the hepatic uptake of pitavastatin in humans. J Pharmacol Exp Ther 311: 139–146 [DOI] [PubMed] [Google Scholar]
- Iwai M, Suzuki H, Ieiri I, Otsubo K, Sugiyama Y 2004. Functional analysis of single nucleotide polymorphisms of hepatic organic anion transporter OATP1B1 (OATP-C). Pharmacogenetics 14: 749–757 [DOI] [PubMed] [Google Scholar]
- Johansen CT, Wang J, Lanktree MB, Cao H, McIntyre AD, Ban MR, Martins RA, Kennedy BA, Hassell RG, Visser ME, et al. 2010. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet 42: 684–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameyama Y, Yamashita K, Kobayashi K, Hosokawa M, Chiba K 2005. Functional characterization of SLCO1B1 (OATP-C) variants, SLCO1B1*5, SLCO1B1*15 and SLCO1B1*15+C1007G, by using transient expression systems of HeLa and HEK293 cells. Pharmacogenet Genomics 15: 513–522 [DOI] [PubMed] [Google Scholar]
- Konig J, Cui Y, Nies AT, Keppler D 2000. A novel human organic anion transporting polypeptide localized to the basolateral hepatocyte membrane. Am J Physiol Gastrointest Liver Physiol 278: G156–G164 [DOI] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR 2010. MaCH: Using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 34: 816–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DJ, Leal SM 2010. Replication strategies for rare variant complex trait association studies via next-generation sequencing. Am J Hum Genet 87: 790–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Lopez E, Martin-Guerrero I, Ballesteros J, Pinan MA, Garcia-Miguel P, Navajas A, Garcia-Orad A 2011. Polymorphisms of the SLCO1B1 gene predict methotrexate-related toxicity in childhood acute lymphoblastic leukemia. Pediatr Blood Cancer 57: 612–619 [DOI] [PubMed] [Google Scholar]
- Momozawa Y, Mni M, Nakamura K, Coppieters W, Almer S, Amininejad L, Cleynen I, Colombel JF, de Rijk P, Dewit O, et al. 2010. Resequencing of positional candidates identifies low frequency IL23R coding variants protecting against inflammatory bowel disease. Nat Genet 43: 43–47 [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S 2001. Predicting deleterious amino acid substitutions. Genome Res 11: 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemi M, Schaeffeler E, Lang T, Fromm MF, Neuvonen M, Kyrklund C, Backman JT, Kerb R, Schwab M, Neuvonen PJ, et al. 2004. High plasma pravastatin concentrations are associated with single nucleotide polymorphisms and haplotypes of organic anion transporting polypeptide-C (OATP-C, SLCO1B1). Pharmacogenetics 14: 429–440 [DOI] [PubMed] [Google Scholar]
- Niemi M, Backman JT, Kajosaari LI, Leathart JB, Neuvonen M, Daly AK, Eichelbaum M, Kivisto KT, Neuvonen PJ 2005a. Polymorphic organic anion transporting polypeptide 1B1 is a major determinant of repaglinide pharmacokinetics. Clin Pharmacol Ther 77: 468–478 [DOI] [PubMed] [Google Scholar]
- Niemi M, Kivisto KT, Hofmann U, Schwab M, Eichelbaum M, Fromm MF 2005b. Fexofenadine pharmacokinetics are associated with a polymorphism of the SLCO1B1 gene (encoding OATP1B1). Br J Clin Pharmacol 59: 602–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa T, Minami H, Sugiura S, Tsuji A, Tamai I 2005. Role of organic anion transporter OATP1B1 (OATP-C) in hepatic uptake of irinotecan and its active metabolite, 7-ethyl-10-hydroxycamptothecin: In vitro evidence and effect of single nucleotide polymorphisms. Drug Metab Dispos 33: 434–439 [DOI] [PubMed] [Google Scholar]
- Pasanen MK, Neuvonen M, Neuvonen PJ, Niemi M 2006. SLCO1B1 polymorphism markedly affects the pharmacokinetics of simvastatin acid. Pharmacogenet Genomics 16: 873–879 [DOI] [PubMed] [Google Scholar]
- Pasanen MK, Fredrikson H, Neuvonen PJ, Niemi M 2007. Different effects of SLCO1B1 polymorphism on the pharmacokinetics of atorvastatin and rosuvastatin. Clin Pharmacol Ther 82: 726–733 [DOI] [PubMed] [Google Scholar]
- Pasanen MK, Neuvonen PJ, Niemi M 2008. Global analysis of genetic variation in SLCO1B1. Pharmacogenomics 9: 19–33 [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui CH, Sandlund JT, Pei D, Campana D, Rivera GK, Ribeiro RC, Rubnitz JE, Razzouk BI, Howard SC, Hudson MM, et al. 2004. Improved outcome for children with acute lymphoblastic leukemia: Results of Total Therapy Study XIIIB at St Jude Children's Research Hospital. Blood 104: 2690–2696 [DOI] [PubMed] [Google Scholar]
- Pui CH, Campana D, Pei D, Bowman WP, Sandlund JT, Kaste SC, Ribeiro RC, Rubnitz JE, Raimondi SC, Onciu M, et al. 2009. Treating childhood acute lymphoblastic leukemia without cranial irradiation. N Engl J Med 360: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheet P, Stephens M 2006. A fast and flexible statistical model for large-scale population genotype data: Applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet 78: 629–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P 2001. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68: 978–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirona RG, Leake BF, Merino G, Kim RB 2001. Polymorphisms in OATP-C: Identification of multiple allelic variants associated with altered transport activity among European- and African-Americans. J Biol Chem 276: 35669–35675 [DOI] [PubMed] [Google Scholar]
- Tomaszewski M, Debiec R, Braund PS, Nelson CP, Hardwick R, Christofidou P, Denniff M, Codd V, Rafelt S, van der Harst P, et al. 2010. Genetic architecture of ambulatory blood pressure in the general population: Insights from cardiovascular gene-centric array. Hypertension 56: 1069–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevino LR, Shimasaki N, Yang W, Panetta JC, Cheng C, Pei D, Chan D, Sparreboom A, Giacomini KM, Pui CH, et al. 2009. Germline genetic variation in an organic anion transporter polypeptide associated with methotrexate pharmacokinetics and clinical effects. J Clin Oncol 27: 5972–5978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Steeg E, van der Kruijssen CM, Wagenaar E, Burggraaff JE, Mesman E, Kenworthy KE, Schinkel AH 2009. Methotrexate pharmacokinetics in transgenic mice with liver-specific expression of human organic anion-transporting polypeptide 1B1 (SLCO1B1). Drug Metab Dispos 37: 277–281 [DOI] [PubMed] [Google Scholar]
- van de Steeg E, Wagenaar E, van der Kruijssen CM, Burggraaff JE, de Waart DR, Elferink RP, Kenworthy KE, Schinkel AH 2010. Organic anion transporting polypeptide 1a/1b-knockout mice provide insights into hepatic handling of bilirubin, bile acids, and drugs. J Clin Invest 120: 2942–2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver YM, Hagenbuch B 2010. Several conserved positively charged amino acids in OATP1B1 are involved in binding or translocation of different substrates. J Membr Biol 236: 279–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Trevino LR, Yang JJ, Scheet P, Pui CH, Evans WE, Relling MV 2010. ARID5B SNP rs10821936 is associated with risk of childhood acute lymphoblastic leukemia in blacks and contributes to racial differences in leukemia incidence. Leukemia 24: 894–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue P, Melamud E, Moult J 2006. SNPs3D: Candidate gene and SNP selection for association studies. BMC Bioinformatics 7: 166 doi: 10.1186/1471-2105-7-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Ge D, Maia JM, Zhu M, Petrovski S, Dickson SP, Heinzen EL, Shianna KV, Goldstein DB 2011. A genome-wide comparison of the functional properties of rare and common genetic variants in humans. Am J Hum Genet 88: 458–468 [DOI] [PMC free article] [PubMed] [Google Scholar]