Abstract
Experimental evolution is a powerful approach to unravel how selective forces shape microbial genotypes and phenotypes. To this date, the available examples focus on the adaptation to conditions specific to the laboratory. The lactic acid bacterium Lactococcus lactis naturally occurs on plants and in dairy environments, and it is proposed that dairy strains originate from the plant niche. Here we investigate the adaptation of a L. lactis strain isolated from a plant to a dairy niche by propagating it for 1000 generations in milk. Two out of three independently evolved strains displayed significantly increased acidification rates and biomass yields in milk. Genome resequencing, revealed six, seven, and 28 mutations in the three strains, including point mutations in loci related to amino acid biosynthesis and transport and in the gene encoding MutL, which is involved in DNA mismatch repair. Two strains lost a conjugative transposon containing genes important in the plant niche but dispensable in milk. A plasmid carrying an extracellular protease was introduced by transformation. Although improving growth rate and growth yield significantly, the plasmid was rapidly lost. Comparative transcriptome and phenotypic analyses confirmed that major physiological changes associated with improved growth in milk relate to nitrogen metabolism and the loss or down-regulation of several pathways involved in the utilization of complex plant polymers. Reproducing the transition from the plant to the dairy niche through experimental evolution revealed several genome, transcriptome, and phenotype signatures that resemble those seen in strains isolated from either niche.
Due to their short generation times, bacteria are well suited for the investigation of evolutionary strategies in a laboratory setting. The experimental evolution of Escherichia coli for 40,000 generations in a glucose-limited medium is one of the best-known examples in the field (Barrick et al. 2009). In several other studies covering significantly fewer generations, similar approaches were used (Herring et al. 2006; Beaumont et al. 2009; Conrad et al. 2009). These experimental evolution studies typically investigate the adaptation of microbes to specific laboratory conditions (Buckling et al. 2009). It remains to be established whether adaptive events that have occurred naturally can be reproduced in the laboratory.
The lactic acid bacterium Lactococcus lactis grows in a variety of niches. It is frequently encountered on vegetable substrates as well as in the dairy environment (Rademaker et al. 2007; Kelly et al. 2010). Nowadays the organism is widely applied in a broad range of dairy fermentations, and it is a predominant model organism for food fermentations. Most food fermentations originate from ancient processes dating back to the introduction of agriculture and animal husbandry ∼10,000 years ago (Campbell-Platt 1994). Dairy lactococci have been proposed to have evolved from typical plant isolates (Godon et al. 1993; Siezen et al. 2005; van Hylckama Vlieg et al. 2006; Siezen et al. 2008; Kelly et al. 2010; Passerini et al. 2010). The genome sequence of a L. lactis plant isolate revealed several genes that relate to the utilization of plant polymers and are absent in dairy isolates (Siezen et al. 2008). Strains isolated from dairy environments are characterized by a high number of amino acid auxotrophies (Delorme et al. 1993; Godon et al. 1993) and the presence of properties that allow the utilization of milk proteins as a source of amino acids (Siezen et al. 2005; Makarova et al. 2006). These properties are common to dairy strains originating from distant geographic locations in Asia, Europe, North America, and New Zealand (Rademaker et al. 2007; Kelly et al. 2010). In dairy lactococci the utilization of extracellular proteins, like the milk-protein casein, is facilitated by a cell-wall-bound extracellular protease that degrades casein to peptides. These peptides are subsequently imported by dedicated transport systems and subjected to further degradation by intracellular peptidases (Kunji et al. 1996). The extracellular protease of lactococci is often plasmid-encoded. Despite its important role in the dairy environment, the protease is readily lost during propagations in milk (McKay and Baldwin 1974; Hugenholtz et al. 1987; Bachmann et al. 2011).
Here we aim to reproduce the proposed transition of L. lactis from the plant to the dairy niche by propagating three independent cultures of a plant isolate for 1000 generations in milk. The adapted strains grew significantly better in milk and had several characteristics in common with a dairy strain.
Results
Experimental evolution of a plant isolate in milk
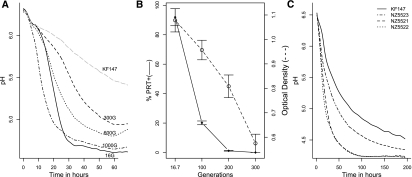
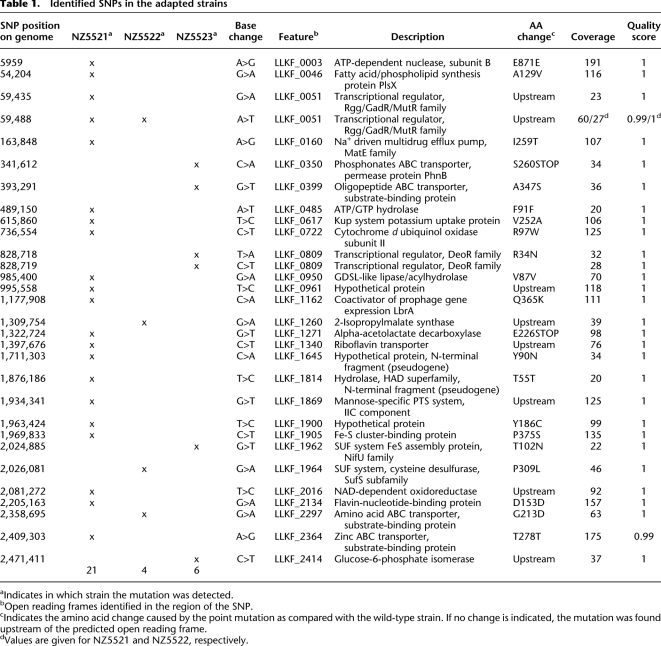
L. lactis KF147 was isolated from mung-bean sprouts (Kelly et al. 1998; Siezen et al. 2008). It grows poorly in milk as compared to typical dairy L. lactis strains. Notably, the introduction of a plasmid, pNZ521 (de Vos et al. 1989), encoding the lactococcal extracellular protease (PrtP) in strain KF147 significantly enhanced growth in milk (Fig. 1A). Strain KF147 harboring pNZ521 (KF147_PRT) was introduced in three parallel adaptive evolution experiments in milk (designated Serial Propagation SP1, SP2, and SP3). The plasmid pNZ521 was introduced for three reasons: (1) extracellular proteases are described to be key enzymes for rapid growth in milk in dairy strains (see the Supplemental Material); (2) to investigate whether coevolution of the usually unstable protease plasmid allows plasmid stabilization; and (3) to guarantee sufficient growth during the initiation of the experiment. Growth is accompanied by acidification and can be monitored by measuring acidification rates. Acidification rates were rapid during the initial cultivations, whereas they decreased after 300 generations (Fig. 1A). Simultaneously, the fraction of protease-positive cells in the culture decreased drastically in all three experiments (Fig. 1B). Recently, we demonstrated that the persistence or loss of the proteolytic trait is the result of a balance between the burden of maintaining and expressing the protease plasmid and the benefit of proteolysis (Bachmann et al. 2011). Since the burden of plasmid maintenance has been shown to decrease during host–plasmid coevolution (Dahlberg and Chao 2003), we reasoned that extended periods of coevolution might increase protease stability in lactococci. For this reason, after 300 generations of evolution, protease-positive cells of each culture were isolated using selective media. Subsequently, they were used for serial propagations in milk. Because of the low frequency of protease-positive cells after 300 generations of propagation, only ∼100 colonies were isolated and propagated from each culture. This introduced a population bottleneck that may have resulted in the loss of beneficial mutations that arose during the first part of the experiment. Further propagation of protease-positive cells revealed that the proteolytic trait could not be stabilized in any of these cultures, and that it was lost at a similar rate as observed during the first 300 generations. After 1000 generations of serial propagation, no protease-positive strains could be detected in any of the parallel cultures. Despite the loss of the proteolytic trait, adapted cultures displayed an increased acidification rate as compared with the parental KF147 strain. From each of the cultures SP1, SP2, and SP3, one colony was isolated, and these strains were designated NZ5521, NZ5522, and NZ5523. These strains displayed acidification characteristics in milk similar to the cultures they were derived from. Further experimentation focused on detailed phenotypic and genetic characterization of these adapted isolates, as well as on the molecular basis of their adaptation to growth in milk.
Figure 1.
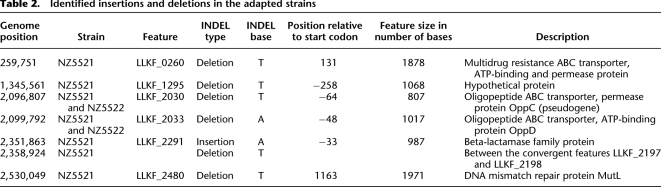
(A) Acidification of milk by the wild-type strain L. lactis KF 147 (long-dashed gray line) and serial propagation of culture SP1 adapted to milk for 16 (solid line), 300 (dashed line), 600 (dotted line), and 1000 (dash-dot line) generations. Acidification profiles are the average of three biological replicates. Standard deviations of the maximum slopes of each curve (calculated over the consecutive measurements of 3 h) were 3.3%, 1.5%, 2.1%, 8.1%, and 6.6% for cultures KF147, 16, 300, 600, and 1000 generations, respectively. (B) Invasion of protease-negative variants during the first 300 generations in milk. The fraction of protease-positive cells (♦, left y-axis), and the final cell density reached (○, right y-axis), during growth in milk decreased throughout the serial propagation of the cultures (x-axis). The averages of three independently propagated cultures (SP1, SP2, and SP3) and standard deviations are shown. (C) Acidification profiles of the wild-type strain KF147 (solid line) and milk adapted single colony isolates NZ5521 (dashed line), NZ5522 (dotted line), and NZ5523 (dash-dot line) grown in milk.
Increased growth rate, yield, and fitness of mutants in milk
First, the growth kinetics and yield of the adapted strains were investigated. The acidification rates of NZ5521 and NZ5522 were increased as compared with KF147, whereas the acidification rate of NZ5523 was only slightly increased (Fig. 1C). Compared with KF147, the maximum cell density reached by the adapted strains was 2.0-fold (p = 0.006), 2.8-fold (p < 0.001), and 1.41-fold (p = 0.31) higher for NZ5521, NZ5522, and NZ5523, respectively. Notably, the strains NZ5521 and NZ5522 reached their maximum cell density within 24 h of cultivation, while with NZ5523 and the parental KF147, this took more than 2 d (Supplemental Fig. 1). The final cell densities in milk cultures of the strains NZ5521 and NZ5522 (∼2 × 109 CFU/mL) were similar to those typically reached by dairy strains grown in milk (Juillard et al. 1995).
Subsequently, relative fitness of the adapted strains was measured by competing for 3 d in a culture with the parental strain KF147. The relative fitness was 1.48 (p > 0.000), 1.1 (p = 0.037), and 1.21 (p = 0.009) for strains NZ5521, NZ5522, and NZ5523, respectively (Supplemental Fig. 2). Remarkably, the relative-fitness increase of NZ5522 was much less pronounced than that of strain NZ5521, while these strains display similar acidification profiles and final cell densities. It should be noted that the competition experiments using the strains KF147, NZ5521, NZ5522, and NZ5523 in coculture with the protease-positive dairy isolate IL594 (Chopin et al. 1984) did not reveal a fitness difference for any of the tested combinations (data not shown). This is probably due to the generation of high amounts of peptides by the protease-positive IL594 strain, which allow high growth rates of both populations. This explanation was corroborated by the observation that the supplementation of milk with a casein-hydrolysate overcomes the growth limitations of KF147 (Supplemental Fig. 3). This emphasizes the importance of nitrogen metabolism as a key performance determinant for the dairy environment.
Adaptation resulted in the loss of mobile elements and generation of a mutator strain
Whole genome resequencing of the adapted strains NZ5521, NZ5522, and NZ5523 revealed 21, four, and six single-nucleotide polymorphisms (SNPs), respectively (Table 1), and seven and two single-base INsertions/DELetions (INDEL) in strains NZ5521 and NZ5522, respectively (Table 2). With the exception of one deletion in a stretch of three adenine residues, all INDELs identified were located in mononucleotide stretches of at least 8 nt, which is consistent with the majority of mutations found in dairy-adapted streptococci (Table 2; Bolotin et al. 2004). Furthermore, strains NZ5522 and NZ5523 had lost an identical 51.3-kb genomic fragment that is similar to a lactococcal sucrose transposon, designated Tn6098 (Kelly et al. 2000; Machielsen et al. 2011). Since Tn6098 contains the only predicted α-galactosidase of the parental KF147 strain, the relative fraction of α-galactosidase-negative strains throughout the evolution experiment could be determined by selective plating. This analysis revealed that between generations 400 and 650, the fraction of α-galactosidase-positive cells declined drastically in cultures SP2 and SP3. Clonal interference most likely explains the observations for culture SP1, where 8% α-galactosidase-negative cells were detected after 650 generations, which then vanished during subsequent propagations (Supplemental Fig. 4; Hegreness et al. 2006). Moreover, the rapid loss of Tn6098 in cultures SP2 and SP3 indicates that maintenance of this transposon poses a burden on fitness under the propagation conditions used in this study.
Table 1.
Identified SNPs in the adapted strains
Table 2.
Identified insertions and deletions in the adapted strains
The higher number of SNPs identified in the genome of strain NZ5521 suggest an increased mutation rate as compared with the other adapted strains. Fluctuation analysis (Luria and Delbruck 1943) revealed that the mutation frequency of NZ5521 is ∼10-fold increased as compared with strains KF147, NZ5522, and NZ5523 (Table 3). Interestingly, the genome of NZ5521 harbors a single-base deletion in a gene involved in DNA mismatch repair, mutL (Table 2). This mutation leads to a 200-amino-acid truncation of the encoded protein that most likely renders it non-functional, potentially explaining the increased mutation rate. As expected for a strain with defects in DNA mismatch repair, the majority of mutations (15 out of 21) in strain NZ5521 were A:T ↔ G:C transitions (Miller 1996).
Table 3.
Fluctuation analysis
Transcriptome of two adapted strains converges toward that of a dairy strain
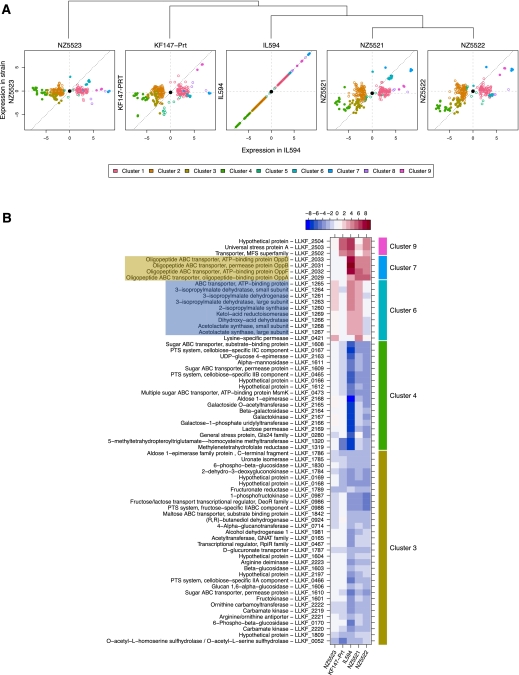
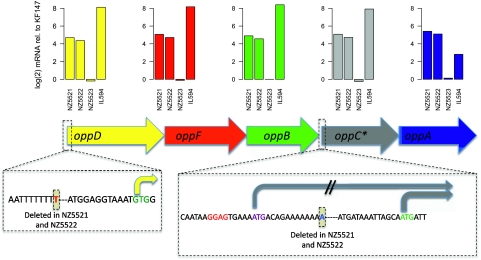
Subsequently we investigated the physiological basis of the milk-adapted phenotype. The transcriptome profiles of exponentially growing cultures of the adapted strains, the parental strain KF147, and KF147_PRT were compared. As a control for growth rate-related changes of transcript levels, KF147_PRT was taken along in the analysis, since this strain does grow well in milk but does not contain mutations. In addition, the dairy strain IL594 was included as a reference. Compared with KF147, the transcription level of 291 genes (out of 1850 genes measured) appeared to be significantly (p < 0.001) differentially expressed in at least one of the adapted strains, or in KF147_PRT or IL594. Nine clusters of differentially expressed genes were defined. An operon encoding an ABC-type oligopeptide transport system displayed the highest differential expression level (∼350-fold higher) in strain IL594 as compared with strain KF147. It was also expressed at a significantly increased level in strains NZ5521 and NZ5522 (Fig. 2, cluster 7). Remarkably, in strains NZ5521 and NZ5522, an identical single base-pair deletion of an adenine residue was identified 48 bases upstream of the first gene in this operon (oppD) (Fig. 3; Table 2). This mutation could be responsible for the increased expression level. Additionally, identical single-base deletions were identified in strains NZ5521 and NZ5522 upstream of the fourth gene in this operon (oppC). In the ancestral strain KF147, oppC is annotated as a pseudogene, which due to a frameshift encodes an N-terminally truncated version of the conserved OppC protein, the permease subunit of the oligopeptide ABC transporter. The single base-pair deletion identified in NZ5521 and NZ5522 restores the normal full-length oppC gene and thereby probably restores its functionality (Fig. 3; Supplemental Fig. 5). The fact that two identical mutations were found in two independently evolved strains indicates that they confer an important selective advantage during growth in milk.
Figure 2.
Genome-wide gene expression profiles during exponential growth in milk. All strains are compared to the wild-type strain KF147, and log2-transformed expression ratios are displayed. (A) Expression levels of all differentially regulated genes compared to KF147 (p < 0.001 in at least one of the strains) in relation to that of the dairy isolate IL594. Genes that are not differentially expressed with respect to strain KF147 map on the center of the plot (•). Expression levels of genes falling either in the bottom left or the top right quadrant show transcript changes toward the levels found in IL594. Genes falling in the top left or bottom right quadrant show changes in the opposite direction compared with IL594. Transcript levels similar to those found in the dairy isolate are identified as dots close to the diagonal of the plot (dotted line). The tree at the top of panel A summarizes the similarity of gene expression between the strains. (B) The differential gene expression in detail for the five most discriminating clusters. Only genes of clusters with an average differential expression of at least fourfold in at least one of the three evolved strains are plotted (corresponding to solid symbols in panel A). Gene functional annotations and identifiers are listed. Brown and blue shading indicates genes/putative operons containing point mutations in the upstream promoter region in at least one of the adapted strains. The heat map on the top of panel B displays the log2-transformed differential expression levels.
Figure 3.
Expression profiles of the opp-operon encoding an oligopeptide ABC-transport system. The bar-plots at the top of the figure display transcript levels of the indicated strains as compared with strain KF147. The deletion of a thymidine residue 64 bases upstream of the GTG-start codon of oppD in strains NZ5521 and NZ5522 is likely to explain the higher expression levels in these strains. In the same two strains, the deletion of an adenine residue (blue) in the upstream region of the truncated oppC gene generates an (alternative) start codon (purple), which is located 78 bases upstream of the native oppC start codon (green) and is preceded by a typical lactococcal ribosome binding site (red) that appears to be absent upstream of the native start codon. (*) Microarray probes designed to detect oppC are not 100% matching to the sequence of strain IL594—the actual expression level of oppC in strain IL594 might therefore be higher than indicated.
The transcription level of a locus encoding genes involved in Branched Chain Amino Acid (BCAA) (leucine, valine, isoleucine) biosynthesis was significantly increased in strains NZ5521 and NZ5523. This locus is also highly expressed in the dairy isolate IL594 (Fig. 2, cluster 6). In contrast, the same locus was transcribed at slightly lower levels in strains NZ5522 and in KF147_PRT (Supplemental Fig. 6). The leucine operon in L. lactis is regulated by transcription attenuation (Bartkus et al. 1991; Godon et al. 1992). A point mutation that is located within the regulatory terminator/antiterminator region of this operon and that may explain the observed decreased expression level in strain NZ5522 was identified in strain NZ5522 (Supplemental Fig. 6; Table 1). The reduced expression level of this locus in strain KF147_PRT is most probably a consequence of the protease-mediated supply of peptides and free amino acids in that culture (Guedon et al. 2005).
Cluster 3 and cluster 4 (Fig. 2) represent transcript levels of ABC-type sugar transporters, cellobiose phospho-transferase systems (PTS), and galactose metabolism, which are expressed at a lower level in IL594 as well as in the adapted strains NZ5521 and NZ5522. The reduced expression of sugar transport systems like the cellobiose-PTS may result from the absence of cellobiose in milk, while this disaccharide can be found in plant-derived materials as a degradation product of cellulose. The decreased transcript levels of the genes involved in galactose utilization (Leloir Pathway) that was observed in strains NZ5521, NZ5521, and the dairy isolate IL594 appear counterintuitive, since the typical milk-disaccharide lactose also contains a galactose moiety. These findings imply that the Leloir Pathway is expressed at a high level during growth on plant material, which may relate to the presence of galactose in typical plant oligosaccharides like stachyose, raffinose, and melibiose. Furthermore, genes involved in the utilization of plant polymers like α- and β-glucosides as well as genes involved in uronic acid metabolism display decreased transcription levels in NZ5521, NZ5522, and IL594, which is in agreement with their redundancy in a dairy environment. Additionally, a transcriptional regulator of the RpiR family, which has been described to regulate phospho-sugar metabolism in E. coli (Sorensen and Hove-Jensen 1996), as well as a transcriptional regulator involved in fructose/lactose transport were down-regulated in the adapted strains and in IL594.
Two point mutations were identified in the NZ5521 genome within the promoter region of a putative transcriptional repressor of the Rgg/GadR/MutR family, one of which was also found in strain NZ5522. This gene shares 41% protein sequence identity with GadR in L. lactis MG1363, which has been shown to regulate the expression of the gadCB operon, involved in glutamate-dependent acid resistance (Sanders et al. 1998). However, the transcriptome data revealed no significant difference in the expression of the gadCB genes in either NZ5521 or NZ5522, but we cannot exclude that the GadR homolog regulates alternative and/or additional genes in KF147 and its derivatives.
Other SNPs and INDELs were found within and around genes coding for transport functions (Tables 1, 2). Several of these, such as ABC-type amino acid and oligopeptide transporters, are likely to influence growth in milk. Many of the mutations identified in the adapted strains are located within the coding regions of genes and therefore may affect the functional properties of the encoded proteins rather than their expression levels. Overall, transcriptome changes occurring in NZ5521 and NZ5522 reflect a convergence toward the transcriptome of IL594 (Fig. 2).
Mutations relating to nitrogen and carbohydrate metabolism are common to all adapted strains
Strain NZ5523 shows fewer transcriptional changes than the other two adapted isolates. NZ5523 shares loss of Tn6098 with NZ5522, but none of the other mutations occurred in both strains. In addition to truncation of a phosphonate ABC transporter, the other two genes affected in NZ5523 are an oligopeptide transporter and a transcriptional regulator of the DeoR family, which is described to regulate sugar catabolism. Therefore, despite the smaller phenotypic changes detected in NZ5523, the mutations identified in this adapted strain are also related to nitrogen and carbohydrate metabolism, which is consistent with multiple mutations selected in strains NZ5521 and NZ5522.
Discussion
The transition from plant to dairy substrates that is proposed to have occurred in the evolution of present dairy lactoccocci (Siezen et al. 2008; Kelly et al. 2010) was experimentally addressed in a controlled laboratory environment. The identification of SNPs and INDELs, as well as transcriptome analysis in milk, established the importance of changes in nitrogen metabolism and the utilization of typical plant polymers.
Mutators can play a role in bacterial domestication
Among the mutations identified in NZ5521 was a single base deletion in the open reading frame of mutL, which is likely to have hitchhiked along with beneficial mutations (Shaver et al. 2002). Analogous to the previous observation of emerging mutator phenotypes during experimental evolution (Sniegowski et al. 1997; Shaver and Sniegowski 2003; Barrick et al. 2009), strain NZ5521 displayed a 10-fold increase in mutation frequency. The selection of mutator phenotypes is consistent with theoretical predictions (Taddei et al. 1997) that suggest a benefit for increased mutation frequencies during adaptation to a new niche.
Mobile elements facilitate genome reduction
The almost simultaneous loss of transposon Tn6098 in cultures SP2 and SP3 indicates that the genes associated with it are dispensable in milk and that its loss confers a fitness advantage. Consequently, we hypothesize that this transposon would eventually also be lost in serial propagation culture SP1 if propagation in milk would be continued.
Genes associated with plant-polymer utilization are down-regulated in milk-adapted strains
In addition to the Tn6098-mediated loss of genes associated with the utilization of α-galactosides, the transcriptome analysis revealed the down-regulation of genes involved in cellobiose, galactose, and uronic-acid metabolism. These carbon sources are particularly abundant in plant-associated niches, and their loss or down-regulation is expected and consistent with earlier descriptions of genomic and phenotypic traits of dairy lactococci (Siezen et al. 2008). Notably, also dairy adapted strains of Streptococcus thermophilus and Lactobacillus bulgaricus show high proportions of pseudogenes related to carbohydrate metabolism (Bolotin et al. 2004; van de Guchte et al. 2006).
Extracellular protease could not be stabilized by host–plasmid coevolution
Despite the positive effects on growth rate and growth yield, the rapid loss of the protease-encoding plasmid suggests that it imposes a high burden on the cell. Since the protease is located extracellularly, peptides produced by it will also be consumed by protease-negative cheater variants, allowing these to invade the culture. Recently, we performed an extensive study on this phenomenon, providing an explanation using a game-theoretical approach. We demonstrated that population composition and cell density play a crucial role in the stabilization of the proteolytic trait (Bachmann et al. 2011). Analogous to the instability of the extracellular protease in L. lactis, yeast cells expressing the extracellular sucrose-degrading enzyme invertase and cheating invertase-negative mutants show comparable population dynamics (Gore et al. 2009). The observation that protease stability could not be increased through host–plasmid coevolution demonstrates the complex dynamics associated with the expression of extracellular substrate-degrading enzymes. Nevertheless, we were able to select mutants that thrive in milk even without an extracellular protease. The inability to degrade extracellular proteins into utilizable peptides is most likely overcome by a more efficient uptake of free peptides in milk combined with amino acid biosynthesis. Interestingly, most of the literature dealing with the loss of protease-encoding plasmids describes protease-negative variants as strains with slower growth and acidification rates in milk. Our results indicate that protease-negative derivatives should be able to enhance their growth rate and overall performance almost to the level of protease-positive strains by adaptive evolution. Alternatively, such an increase in fitness might be limited to lactococci isolated from the plant environment, which have been described to harbor additional genes that allow them to grow in nutritionally poorer and more dynamic environments than milk (van Hylckama Vlieg et al. 2006; Siezen et al. 2008).
Down-regulation of BCAA biosynthesis is consistent with genome decay of dairy isolates
A point mutation upstream of a genomic locus involved in the biosynthesis of branched-chain amino acids coincides with a reduced expression level in NZ5522. The BCAA genomic locus was shown to be non-functional in 17 tested dairy isolates (Godon et al. 1993), and pseudogenes were identified in the leucine operon of the three sequenced dairy strains IL1403, SK11, and MG1363 (Bolotin et al. 2001; Makarova et al. 2006; Wegmann et al. 2007). However, compared with KF147, this operon is expressed at a higher level in strains NZ5521, NZ5523, and IL594. The IL1403 BCAA biosynthesis genes were described to be expressed in a regulated manner, despite BCAA auxotrophy (Godon et al. 1993). The investigators provided two possible explanations for this observation. The first explanation proposes that the operon also encodes genes with essential functions that are not related to the BCAA biosynthesis, for which the ABC-transporter ATP-binding protein (Ymeb) could be a candidate (Supplemental Fig. 6). The second explanation postulates that the observations may reflect the ongoing process of genome decay during adaptation to the dairy environment. Our finding that strain NZ5522 performs well in milk despite the low expression of the BCAA biosynthesis operon appears to falsify the first and strongly favors the second explanation.
Oligo-peptide transport plays a key role during the adaptation to growth in milk
The identical mutations in the promoter region of a highly up-regulated oligopeptide transport system in two adapted strains represent an astonishing parallel to dairy L. lactis isolates. This oligopeptide transport system has been shown to be indispensable for the transport of casein-derived peptides (Kunji et al. 1995). Comparative genomics has revealed that the acquisition of genes related to amino acid degradation and transport, as well as the loss of genes involved in the utilization of carbohydrates that do not occur in milk, are prominent dairy-specific adaptations (Siezen et al. 2005; Makarova et al. 2006; Makarova and Koonin 2007). Moreover, several genes involved in nitrogen metabolism and peptide transport were induced after transition to a dairy environment (Gitton et al. 2005; Bachmann et al. 2010). These observations, in combination with the stimulation of growth in milk of the plant isolate KF147 by the addition of amino acids (Supplemental Fig. 3), are consistent with the notion that the elevated expression of the oligopeptide transport system is an important adaptation to the dairy niche.
Conclusion
Overall, numerous aspects of our findings are in agreement with an evolutionary process that is proposed to have occurred naturally. The observation that niche-specific adaptations occurring in environmental isolates can be reproduced by experimental evolution opens exciting possibilities for the investigation of evolutionary strategies as they occur in complex environmental samples.
Methods
Bacterial strains and DNA techniques
The strain used for experimental evolution was L. lactis KF147 (Kelly et al. 1998; Siezen et al. 2008). Laboratory propagation of KF147 was kept to a minimum to ensure a minimal laboratory history for the strain. Strains were grown either in M17 medium (Merck) supplemented either with 0.5% lactose (LM17) or 0.5% glucose (GM17), or in reconstituted skimmed milk (RSM) (Promex Spray 1% skimmed milk powder; Friesland Foods Butter). The plasmid pNZ521 (de Vos et al. 1989) was introduced into strain KF147 by electroporation (Wells et al. 1993), and transformants were selected by plating on medium supplemented with 5 μg/mL chloramphenicol. KF147 and all evolved variants were grown at 30°C. Rifampicin was used at a concentration of 50 μg/mL. DNA techniques were carried out as described elsewhere (Sambrook et al. 1989).
Experimental evolution
KF147 harboring pNZ521 (KF147_PRT) was used to inoculate 10 mL of skimmed milk and grown until the milk coagulated (∼pH 5.1). The coagulation of milk was considered an indicator of fully grown cultures since lactococcal growth in milk is tightly coupled to lactic acid production and ceases after milk coagulation (Supplemental Fig. 7). After coagulation, a 105-fold dilution was inoculated into 10 mL of milk again. Calculating with an initial population size (N0) after inoculation of 105 cells and a final population size (Nf) in a fully grown culture of 1010 cells, the number of generations (g) per propagation step is log2(Nf/N0), which equals 16.6 generations. The effective population size (Ne) is N0 × g and equals 1.66 × 106 (Lenski et al. 1991). After 300 generations, a sample of the culture was plated on medium containing 5 μg of chloramphenicol/mL, to select for strains harboring plasmid pNZ521. For each parallel culture, about 100 colonies were washed off the plates, and the suspension was used for further propagation. The propagation process was continued as described above until the cultures had grown for 1000 generations in milk (∼5 mo). Throughout the experiment, stock solutions were prepared and frozen at regular intervals.
Phenotypic characterization
The number of colony forming units (CFU) was determined by plating dilutions on LM17 or indirectly by measuring the optical density at 600 nm after clearing milk. Clearing of milk was done by briefly mixing milk cultures with a solution of 0.2% sodium hydroxide and 0.2% EDTA using a ratio of 1:9, respectively, and incubating for 5 min.
Acidification profiles were determined in milk by using an automated pH electrode system to follow the pH (CINAC; Ysebaert) or by using microplates containing optical pH indicators (Precision Sensing GmbH).
After serial propagation cultures SP1, SP2, and SP3 had been propagated for 1000 generations in milk, three randomly picked single-colony isolates were obtained from each individually adapted culture. Each isolate was tested for CFUs, acidification rates, and fitness in milk. No significant differences between isolates from each culture were found, and subsequently one isolate of each independently evolved culture was chosen for further evaluation. These single-colony isolates were assigned the names NZ5521, NZ5522, and NZ5523, and they were isolated from cultures SP1, SP2, and SP3, respectively. α-Galactosidase-negative mutants of KF147 were identified by their white phenotype on LM17 agar plates containing 40 μg/mL 5-bromo-4-chloro-3-indolyl-α-D-galactopyranoside (X-α-Gal).
Fluctuation analysis
Fluctuation analysis (Luria and Delbruck 1943) was performed by inoculating 24 individual 200-μL cultures of each strain with about 200 cells. The cultures were grown under non-selective conditions (GM17 medium) until they reached stationary phase (∼2 × 109 cells/mL). A fraction of each culture was plated on GM17 supplemented with rifampicin (50 μg/mL), and after 1 d of incubation at 30°C the CFUs were determined. The total number of cells plated on medium supplemented with rifampicin was determined by plating dilutions of the cultures on GM17. Mutation frequencies were calculated with the Fluctuation Analysis Calculator (FALCOR) web tool using the maximum likelihood estimator (Hall et al. 2009). The results were corrected for plating efficiency/sampling as described in the FALCOR instructions.
Determination of relative fitness
A spontaneous rifampicin-resistant mutant of strain KF147 was isolated by plating 108 cells on LM17 medium, supplemented with 50 μg of rifampicin per milliliter. After 3 d of incubation, single colonies were isolated. A rifampicin-resistant strain that showed no growth difference compared with the wild-type strain throughout the growth curve was designated KF147RIF and used for further experiments. The relative fitness of the evolved strains was determined in competition assays of the adapted strains and the rifampicin-resistant mutant KF147RIF. The two competitors were inoculated in a 1:1 ratio. The number of CFUs was determined immediately after inoculation, and 1 and 3 d after inoculation by plating on either LM17 or LM17 supplemented with rifampicin. The number of CFUs for each adapted strain in the competition experiment was determined by subtracting the number of rifampicin-resistant colonies from the total number of colonies found. Relative fitness (W) was calculated according to the following formula (de Visser and Lenski 2002), where Nj and Ni represent the abundance of the two competing strains directly after inoculation (0) and after competition (1):
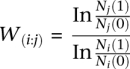 |
Genotypic characterization
Full-genome resequencing using the Illumina platform was performed by GATC-Biotech. The sequencing data are deposited in the NCBI Sequence Read Archive (accession number SRP004629). Sequence alignment against the reference genome of KF147 was performed with standard settings using the ELAND software package of the Illumina analysis pipeline. To increase the accuracy of Single Nucleotide Polymorphism (SNP) and INsertion/DELetion (INDEL) identification, the reference genome was also resequenced with the same technology and confirmed the sequence previously published for this strain (Siezen et al. 2010). SNPs and INDELs were identified with an in-house Perl script by GATC-Biotech. Quality scores indicating the fraction of reads consistent with the new consensus sequence were assigned to identified SNPs. Only unambiguous SNPs with a sequence coverage of more than 20-fold and a quality score >0.99 were used for further analysis. To validate the identified mutations, 24 SNPs and INDELs were amplified by PCR amplification of the region of interest in the wild-type and adapted strains followed by amplicon sequence determination. The amplification of genomic DNA was performed using Taq polymerase according to the manufacturer's instructions (Promega). Amplicon sequencing was carried out by BaseClear. This strategy allowed the confirmation of all except one incorrectly predicted INDEL mutation. The deletion of the transposon Tn6098 was confirmed by the amplification across the deletion junction using genomic DNA as a template in combination with primers HB03delFW (5′-gtcatttaagaagcctttcgcatagag-3′) and HB03delREV (5′-gaacatagaactccctgctcttggag-3′), and the resulting amplicon was subjected to sequence determination, confirming the anticipated transposon excision site.
Design of microarrays
Microarrays containing in situ–synthesized 60-mer oligomers were produced by Agilent Technologies, according to a custom probe design based on the complete genome sequence of L. lactis subsp. lactis IL1403 (NCBI Reference Sequence: NC_002662.1) (Bolotin et al. 2001) and the incomplete genome sequence of L. lactis subsp. lactis KF147 (Siezen et al. 2008). A total of 13,392 unique 60-mers with a theoretical melting temperature of ∼82°C were selected. The melting temperature was calculated using nearest-neighbor calculations (Peyret et al. 1999), assuming an Na+ concentration of 1 M and an oligonucleotide concentration of 10−12 M. The sequencing and annotation of strain KF147 was an ongoing process during this project, but probes were eventually remapped to the full genome sequence and annotation of KF147 (NCBI Reference Sequence: NC_013656). The 2721 putative genes were represented by one (5.4%), two (6.7%), three to six (68%), or seven or more probes (11.7%). A total of 218 putative genes were not represented on the array because no unique probe satisfying the selection criteria could be selected. Many of the putative genes not represented on the array were relatively short: 55% had less than 50 amino acids, and 80% had less than 100 amino acids, indicating that not all of them might be coding genes. The array design has been deposited in GEO (accession number GPL7410).
RNA isolation, labeling, and hybridization
RNA isolation from milk was performed either during logarithmic growth or from stationary-phase cells as described earlier (Sieuwerts et al. 2010) with the deviation that the first step (resuspension in 60% glycerol) was omitted. In short, the milk was cleared at −20°C using sodium citrate. Cells were pelleted, and after a washing step, RNA was isolated using a phenol–chloroform extraction protocol followed by a column purification step. Synthesis of cDNA, labeling, hybridization, and scanning of the microarrays was performed as described before (Sieuwerts et al. 2010). The microarray hybridization scheme for the transcriptome analyses after growth in milk consisted of a compound loop design with 16 arrays (Supplemental Table S1).
Array data analysis
After blank spots had been removed, array measurements were normalized by local fitting of an M-A plot using the Loess algorithm and the Limma package (Smyth et al. 2003) in R (http://www.r-project.org). Normalized intensities were used for further analysis. The regulation ratios between the different samples were calculated per probe from the hybridizations using linear modeling functions from the Limma package. The statistical significance of regulation ratios was calculated from variation in biological duplicates, using the eBayes function in Limma (cross-probe variance estimation) and false discovery rate (FDR) adjustment of the P-values (Smyth et al. 2003). Subsequently, the probe regulation ratios and the P-values were averaged per gene over all probes targeting the gene. Although clearly the averaged P-values do not represent an FDR-corrected P-value anymore, low average P-values for a gene are still indicative of highly significant regulation ratios, and, in fact, they are likely to underestimate the significance of the observed regulation.
If not indicated differently, the comparison between strain KF147 including its derivatives and IL594 (the ancestor of IL1403), the analysis was performed using only those probes that display 100% sequence identity in both strains (7725 distinct probes covering 1850 genes). This avoided scoring of expression differences confounded by sequence mismatches.
Genes differentially regulated (p < 0.001) in at least one of the assayed strains were clustered hierarchically using Euclidian distance measure and complete linkage as the agglomeration method. Clustering of strains was performed using the Euclidian distance and Ward's minimum variance agglomeration method. Both analyses were preformed using the “hclust” function in R (http://www.r-project.org).
Data access
Microarray data have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GPL7410. The Illumina sequencing data have been submitted to the NCBI Sequence Read Archive (SRA) (http://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession number SRP004629.
Acknowledgments
We thank Mark van der Garde for technical assistance and Jumamurat Bayjanov and Roland Siezen for access to the assembled and annotated genome of L. lactis KF147 prior to publication. Furthermore, we thank Christof Francke for useful discussion on the functional annotation of adapted strains, and Rebecca Painter for proofreading the manuscript. This project was carried out within the research program of the Kluyver Centre for Genomics of Industrial Fermentation, which is part of The Netherlands Genomics Initiative/Netherlands Organization for Scientific Research.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.121285.111.
References
- Bachmann H, de Wilt L, Kleerebezem M, van Hylckama Vlieg JE 2010. Time-resolved genetic responses of Lactococcus lactis to a dairy environment. Environ Microbiol 12: 1260–1270 [DOI] [PubMed] [Google Scholar]
- Bachmann H, Molenaar D, Kleerebezem M, van Hylckama Vlieg JE 2011. High local substrate availability stabilizes a cooperative trait. ISME J 5: 929–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461: 1243–1247 [DOI] [PubMed] [Google Scholar]
- Bartkus JM, Tyler B, Calvo JM 1991. Transcription attenuation-mediated control of leu operon expression: Influence of the number of Leu control codons. J Bacteriol 173: 1634–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont HJ, Gallie J, Kost C, Ferguson GC, Rainey PB 2009. Experimental evolution of bet hedging. Nature 462: 90–93 [DOI] [PubMed] [Google Scholar]
- Bolotin A, Wincker P, Mauger S, Jaillon O, Malarme K, Weissenbach J, Ehrlich SD, Sorokin A 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res 11: 731–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Renault P, Sorokin A, Ehrlich SD, Kulakauskas S, Lapidus A, Goltsman E, Mazur M, Pusch GD, et al. 2004. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat Biotechnol 22: 1554–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckling A, Craig Maclean R, Brockhurst MA, Colegrave N 2009. The Beagle in a bottle. Nature 457: 824–829 [DOI] [PubMed] [Google Scholar]
- Campbell-Platt G 1994. Fermented foods—a world perspective. Food Res Int 27: 253–257 [Google Scholar]
- Chopin A, Chopin MC, Moillo-Batt A, Langella P 1984. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid 11: 260–263 [DOI] [PubMed] [Google Scholar]
- Conrad TM, Joyce AR, Applebee MK, Barrett CL, Xie B, Gao Y, Palsson BO 2009. Whole-genome resequencing of Escherichia coli K-12 MG1655 undergoing short-term laboratory evolution in lactate minimal media reveals flexible selection of adaptive mutations. Genome Biol 10: R118 doi: 10.1186/gb-2009-10-10-r118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg C, Chao L 2003. Amelioration of the cost of conjugative plasmid carriage in Eschericha coli K12. Genetics 165: 1641–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme C, Godon JJ, Ehrlich SD, Renault P 1993. Gene inactivation in Lactococcus lactis: Histidine biosynthesis. J Bacteriol 175: 4391–4399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Visser JA, Lenski RE 2002. Long-term experimental evolution in Escherichia coli. XI. Rejection of non-transitive interactions as cause of declining rate of adaptation. BMC Evol Biol 2: 19 doi: 10.1186/1471-2148-2-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos WM, Vos P, de Haard H, Boerrigter I 1989. Cloning and expression of the Lactococcus lactis subsp. cremoris SK11 gene encoding an extracellular serine proteinase. Gene 85: 169–176 [DOI] [PubMed] [Google Scholar]
- Gitton C, Meyrand M, Wang J, Caron C, Trubuil A, Guillot A, Mistou MY 2005. Proteomic signature of Lactococcus lactis NCDO763 cultivated in milk. Appl Environ Microbiol 71: 7152–7163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godon JJ, Chopin MC, Ehrlich SD 1992. Branched-chain amino acid biosynthesis genes in Lactococcus lactis subsp. lactis. J Bacteriol 174: 6580–6589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godon JJ, Delorme C, Bardowski J, Chopin MC, Ehrlich SD, Renault P 1993. Gene inactivation in Lactococcus lactis: Branched-chain amino acid biosynthesis. J Bacteriol 175: 4383–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore J, Youk H, van Oudenaarden A 2009. Snowdrift game dynamics and facultative cheating in yeast. Nature 459: 253–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedon E, Sperandio B, Pons N, Ehrlich SD, Renault P 2005. Overall control of nitrogen metabolism in Lactococcus lactis by CodY, and possible models for CodY regulation in Firmicutes. Microbiology 151: 3895–3909 [DOI] [PubMed] [Google Scholar]
- Hall BM, Ma CX, Liang P, Singh KK 2009. Fluctuation analysis CalculatOR: a web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics 25: 1564–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegreness M, Shoresh N, Hartl D, Kishony R 2006. An equivalence principle for the incorporation of favorable mutations in asexual populations. Science 311: 1615–1617 [DOI] [PubMed] [Google Scholar]
- Herring CD, Raghunathan A, Honisch C, Patel T, Applebee MK, Joyce AR, Albert TJ, Blattner FR, van den Boom D, Cantor CR, et al. 2006. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet 38: 1406–1412 [DOI] [PubMed] [Google Scholar]
- Hugenholtz J, Splint R, Konings WN, Veldkamp H 1987. Selection of protease-positive and protease-negative variants of Streptococcus cremoris. Appl Environ Microbiol 53: 309–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juillard V, Le Bars D, Kunji ER, Konings WN, Gripon JC, Richard J 1995. Oligopeptides are the main source of nitrogen for Lactococcus lactis during growth in milk. Appl Environ Microbiol 61: 3024–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WJ, Davey GP, Ward LJ 1998. Characterization of lactococci isolated from minimally processed fresh fruit and vegetables. Int J Food Microbiol 45: 85–92 [DOI] [PubMed] [Google Scholar]
- Kelly WJ, Davey GP, Ward LJ 2000. Novel sucrose transposons from plant strains of Lactococcus lactis. FEMS Microbiol Lett 190: 237–240 [DOI] [PubMed] [Google Scholar]
- Kelly WJ, Ward LJ, Leahy SC 2010. Chromosomal diversity in Lactococcus lactis and the origin of dairy starter cultures. Genome Biol Evol 2: 729–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunji ER, Hagting A, De Vries CJ, Juillard V, Haandrikman AJ, Poolman B, Konings WN 1995. Transport of β-casein-derived peptides by the oligopeptide transport system is a crucial step in the proteolytic pathway of Lactococcus lactis. J Biol Chem 270: 1569–1574 [DOI] [PubMed] [Google Scholar]
- Kunji ER, Mierau I, Hagting A, Poolman B, Konings WN 1996. The proteolytic systems of lactic acid bacteria. Antonie van Leeuwenhoek 70: 187–221 [DOI] [PubMed] [Google Scholar]
- Lenski RE, Rose MR, Simpson SC, Tadler SC 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat 138: 1315–1341 [Google Scholar]
- Luria SE, Delbruck M 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28: 491–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machielsen R, Siezen RJ, van Hijum SA, van Hylckama Vlieg JE 2011. Molecular description and industrial potential of Tn6098 conjugative transfer conferring α-galactoside metabolism in Lactococcus lactis. Appl Environ Microbiol 77: 555–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Koonin EV 2007. Evolutionary genomics of lactic acid bacteria. J Bacteriol 189: 1199–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine N, et al. 2006. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci 103: 15611–15616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay LL, Baldwin KA 1974. Simultaneous loss of proteinase- and lactose-utilizing enzyme activities in Streptococcus lactis and reversal of loss by transduction. Appl Microbiol 28: 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH 1996. Spontaneous mutators in bacteria: Insights into pathways of mutagenesis and repair. Annu Rev Microbiol 50: 625–643 [DOI] [PubMed] [Google Scholar]
- Passerini D, Beltramo C, Coddeville M, Quentin Y, Ritzenthaler P, Daveran-Mingot ML, Le Bourgeois P 2010. Genes but not genomes reveal bacterial domestication of Lactococcus lactis. PLoS ONE 5: e15306 doi: 10.1371/journal.pone.0015306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyret N, Seneviratne PA, Allawi HT, SantaLucia J Jr 1999. Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A·A, C·C, G·G, and T·T mismatches. Biochemistry 38: 3468–3477 [DOI] [PubMed] [Google Scholar]
- Rademaker JL, Herbet H, Starrenburg MJ, Naser SM, Gevers D, Kelly WJ, Hugenholtz J, Swings J, van Hylckama Vlieg JE 2007. Diversity analysis of dairy and nondairy Lactococcus lactis isolates, using a novel multilocus sequence analysis scheme and (GTG)5-PCR fingerprinting. Appl Environ Microbiol 73: 7128–7137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T 1989. Molecular cloning: A laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Sanders JW, Leenhouts K, Burghoorn J, Brands JR, Venema G, Kok J 1998. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol Microbiol 27: 299–310 [DOI] [PubMed] [Google Scholar]
- Shaver AC, Sniegowski PD 2003. Spontaneously arising mutL mutators in evolving Escherichia coli populations are the result of changes in repeat length. J Bacteriol 185: 6076–6082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaver AC, Dombrowski PG, Sweeney JY, Treis T, Zappala RM, Sniegowski PD 2002. Fitness evolution and the rise of mutator alleles in experimental Escherichia coli populations. Genetics 162: 557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieuwerts S, Molenaar D, van Hijum SA, Beerthuyzen M, Stevens MJ, Janssen PW, Ingham CJ, de Bok FA, de Vos WM, van Hylckama Vlieg JE 2010. Mixed-culture transcriptome analysis reveals the molecular basis of mixed-culture growth in Streptococcus thermophilus and Lactobacillus bulgaricus. Appl Environ Microbiol 76: 7775–7784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siezen RJ, Renckens B, van Swam I, Peters S, van Kranenburg R, Kleerebezem M, de Vos WM 2005. Complete sequences of four plasmids of Lactococcus lactis subsp. cremoris SK11 reveal extensive adaptation to the dairy environment. Appl Environ Microbiol 71: 8371–8382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siezen RJ, Starrenburg MJ, Boekhorst J, Renckens B, Molenaar D, van Hylckama Vlieg JE 2008. Genome-scale genotype–phenotype matching of two Lactococcus lactis isolates from plants identifies mechanisms of adaptation to the plant niche. Appl Environ Microbiol 74: 424–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siezen RJ, Bayjanov J, Renckens B, Wels M, van Hijum SA, Molenaar D, van Hylckama Vlieg JE 2010. Complete genome sequence of Lactococcus lactis subsp. lactis KF147, a plant-associated lactic acid bacterium. J Bacteriol 192: 2649–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK, Yang YH, Speed T 2003. Statistical issues in cDNA microarray data analysis. Methods Mol Biol 224: 111–136 [DOI] [PubMed] [Google Scholar]
- Sniegowski PD, Gerrish PJ, Lenski RE 1997. Evolution of high mutation rates in experimental populations of E. coli. Nature 387: 703–705 [DOI] [PubMed] [Google Scholar]
- Sorensen KI, Hove-Jensen B 1996. Ribose catabolism of Escherichia coli: Characterization of the rpiB gene encoding ribose phosphate isomerase B and of the rpiR gene, which is involved in regulation of rpiB expression. J Bacteriol 178: 1003–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei F, Radman M, Maynard-Smith J, Toupance B, Gouyon PH, Godelle B 1997. Role of mutator alleles in adaptive evolution. Nature 387: 700–702 [DOI] [PubMed] [Google Scholar]
- van de Guchte M, Penaud S, Grimaldi C, Barbe V, Bryson K, Nicolas P, Robert C, Oztas S, Mangenot S, Couloux A, et al. 2006. The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc Natl Acad Sci 103: 9274–9279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hylckama Vlieg JE, Rademaker JL, Bachmann H, Molenaar D, Kelly WJ, Siezen RJ 2006. Natural diversity and adaptive responses of Lactococcus lactis. Curr Opin Biotechnol 17: 183–190 [DOI] [PubMed] [Google Scholar]
- Wegmann U, O'Connell-Motherway M, Zomer A, Buist G, Shearman C, Canchaya C, Ventura M, Goesmann A, Gasson MJ, Kuipers OP, et al. 2007. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J Bacteriol 189: 3256–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JM, Wilson PW, Le Page RW 1993. Improved cloning vectors and transformation procedure for Lactococcus lactis. J Appl Bacteriol 74: 629–636 [DOI] [PubMed] [Google Scholar]