Abstract
New sequencing technologies can address diverse biomedical questions but are limited by a minimum required DNA input of typically 1 μg. We describe how sequencing libraries can be reproducibly created from 20 pg of input DNA using a modified transpososome-mediated fragmentation technique. Resulting libraries incorporate in-line bar-coding, which facilitates sample multiplexes that can be sequenced using Illumina platforms with the manufacturer's sequencing primer. We demonstrate this technique by providing deep coverage sequence of the Escherichia coli K-12 genome that shows equivalent target coverage to a 1-μg input library prepared using standard Illumina methods. Reducing template quantity does, however, increase the proportion of duplicate reads and enriches coverage in low-GC regions. This finding was confirmed with exhaustive resequencing of a mouse library constructed from 20 pg of gDNA input (about seven haploid genomes) resulting in ∼0.4-fold statistical coverage of uniquely mapped fragments. This implies that a near-complete coverage of the mouse genome is obtainable with this approach using 20 genomes as input. Application of this new method now allows genomic studies from low mass samples and routine preparation of sequencing libraries from enrichment procedures.
Next-generation sequencing (NGS) methods produce millions of short reads that are either subsequently compared to a reference genome in silico or reassembled to provide de novo target sequence data (Metzker. 2010). In addition to creating large data sets from an individual target sequence, methods exist to pool multiple, uniquely identifiable, sample libraries that can be demultiplexed in silico following sequencing, thereby lowering overall costs of acquiring data sets for low-complexity samples.
A common starting point for template preparation for NGS platforms is random fragmentation of target DNA and addition of platform-specific adapter sequences to flanking ends. Protocols typically use sonication to shear input DNA, coupled with several rounds of enzymatic modification to produce a sequencer-ready product. In addition to being labor-intensive and difficult to automate, manufacturers' protocols commonly need starting DNA quantities in the microgram range, most of which is lost during preparation, with only a small fraction being present in the final library. The requirement for large quantities of DNA to prepare NGS libraries makes the sequencing of many limited-material protocols—such as forensic and ChIP-seq samples, and single-cell studies such as genotyping, sequencing, or RNA-seq—challenging.
Recently, an alternative to the standard methods of fragmentation and adapter ligation has become available (Syed et al. 2009a,b). Recombinant transposon-derived dsDNA integration complexes, transpososomes, have been produced with a pre-adsorbed 19-bp core transpososome recognition motif (TRM) containing oligonucleotides. Upon transpososome integration, target DNA is simultaneously fragmented and TRM oligos ligated to the 5′ end of each double-stranded target (Syed et al. 2009a,b). A series of platform-specific PCR amplifications is then required to produce sequencer-ready libraries. This technique allows NGS library synthesis from 50 ng using the manufacturer's standard protocols. Further titration of this method has been reported to produce unvalidated libraries from as little as 10 pg of template (Adey et al. 2010). Although a major advance, an important limitation of this technique is the incompatibility of tagmented libraries with standard platform-specific sequencing primers.
Here we describe a modified tagmentation technique that permits picogram quantities of target DNA to be sequenced reproducibly on Illumina platforms with the standard Illumina paired-end (PE) sequencing primer. Our modified adapter sequences also incorporate an “in-line” bar-coding system that allows sample multiplexing without the need for additional index-specific reads. To ensure accurate input of picogram levels of DNA, we have developed a high-sensitivity fluorescence-based quantitation system that reproducibly reports sample DNA concentrations in the femtogram range. To validate our approach, we used this technique to deeply sequence an Escherichia coli K-12 genome from as little as 20 pg of input genomic DNA. Parallel data sets obtained using sonication-based Illumina library preparation methods produced from 1 μg of genomic DNA provide comparative information on target GC bias, levels of library diversity, and coverage. Furthermore, we show that reducing input quantities from 1 μg to 2 ng, 200 pg, or even 20 pg results in libraries with comprehensive sequence coverage and high degrees of fragment diversity. These findings were confirmed with low-coverage resequencing of 20 pg of mouse genomic DNA.
Results
Quantitation of target input
The manufacturer's standard tagmentation procedure requires the use of a predetermined transpososome-to-DNA ratio. To maintain this relationship when titrating down levels of target, it becomes necessary to measure very low concentrations of DNA accurately. We have developed a highly reproducible DNA sample quantitation method using a fluorescent DNA reporter dye, background signal quencher, and highly sensitive optical plate reader (see Methods). We are able to reproducibly measure 500 fg/μL in the final 20-μL reaction, equivalent to 10 pg/μL in the initial sample when using our standard 1:20 dilution (Methods) (Supplemental Fig. 1).
In-line bar-coding and read quality
The alteration to the manufacturer's tagmentation protocol described here uses oligos complementary to the 19-bp TRM sequence with an additional recognition site for a remote cutting type IIG restriction endonuclease. These enzymes are subsequently used to remove the majority of the 29-bp flanking sequence, including the TRM, to leave a mandatory 2-bp 3′-TG overhang at both ends of all library fragments that is subsequently used to ligate adapters in a highly efficient reaction (Fig. 1). The chosen restriction endonucleases have short recognition sequences that also occur in the input DNA. Using such enzymes in the preparation of NGS library fragments will, in addition to trimming away transposon core sequences, also lead to cleavage of a number of fragments that will be lost from the library, leading to reduced coverage of target DNA around endogenous enzyme-recognition sites. To overcome this, we used different type IIG restriction enzyme and tailed primer combinations (AcuI, BpmI, BsgI, and BpuEI) to produce library fragments with identical 2-bp 3′-GT overhangs in the flanking core oligo sequence. Endogenous cleavage sites for each enzyme are largely non-overlapping. Hence, sequencing libraries created by pooling sublibraries from separate enzyme/primer combinations should minimize coverage reduction at endogenous recognition motifs.
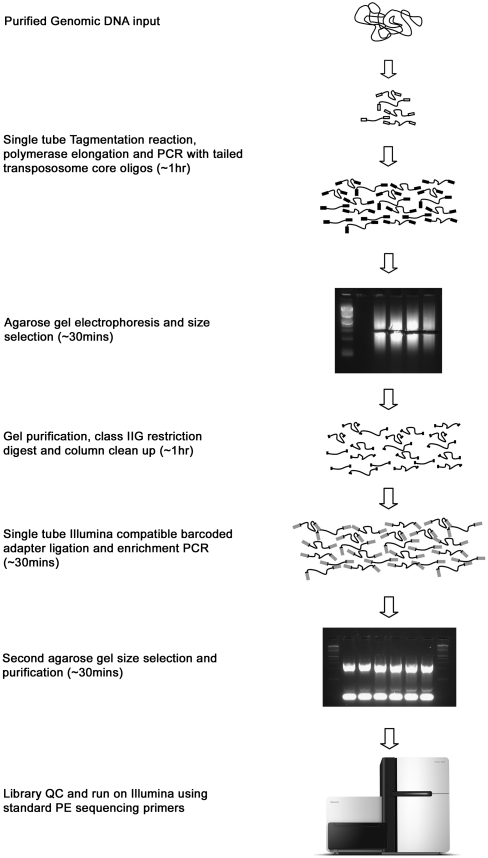
Figure 1.
Schematic overview of modified Tagmentation procedure. Purified genomic DNA is tagmented using a specific ratio of enzyme and target (see Methods). Following tagmentation (addition of standard TRM oligo shown as white box), the reaction is quenched through the addition of premixed PCR reagents, subject to a brief extension step, and amplified with limited PCR cycles using a single tailed-oligo (black boxes) resulting in a library of fragments flanked by identical 29-bp sequences that can be size-selected by standard gel electrophoresis techniques. Tailed oligos contain a recognition site for a remote cutting type IIG restriction endonuclease that is used to remove the majority of the 29-bp flanking sequence, including the core transposon motif, and leaves a mandatory 2-bp 3′-TG overhang at both ends of all amplified library fragments. Modified Illumina sequencing adapters (gray boxes), incorporating variable length in-line bar-coding sequences, are then ligated to this 2-bp overhang in a highly efficient reaction. A second limited cycle PCR is performed directly on the ligation reaction and products run on an agarose gel and subject to a second round of size selection. The purified product is then sequenced on an Illumina GAII sequencer using the manufacturer's standard methods.
Separate E. coli K12 genomic DNA libraries were prepared in parallel using either sonication-based Illumina-compatible techniques (1 μg of input DNA) (see Methods) or our altered tagmentation method (with 1-ng, 100-pg, and 10-pg levels of input DNA) (see Methods). For the tagmentation method, libraries were created with separate restriction enzyme/primer combinations, and additional libraries were made by pooling equimolar amounts of either two (BpmI and BsgI) or four (AcuI, BpmI, BsgI, and BpuEI) independent sublibraries prior to adapter ligation. Each library used specific staggered length in-line bar-coded adapters that allow indexing of both reads of a read pair while simultaneously offsetting a common three-base sequence found at the start of all tagmentation reads (Supplemental Table 2). Libraries were multiplexed, sequenced on an Illumina GAII 51bp paired-end (PE) flowcell, and data retrieved. Resultant data files were converted to fastq format, demultiplexed into constituent libraries, and filtered for reads where all constituent base calls exceeded a phred quality of 20 (Methods).
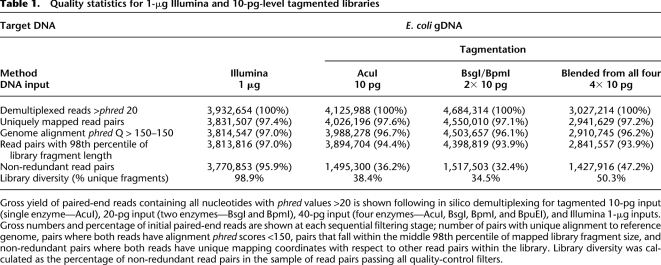
At the 10-pg level, multiplexed libraries for one enzyme (10-pg input), two enzymes (20-pg total input), and four enzymes (40-pg total input) yielded 3.1 × 107 paired 51-bp reads from the flowcell lane. 1.3 × 107 (42%) of these pairs contained constituent nucleotides that fell below the arbitrary phred 20 threshold (Methods) (Table 1) and were discarded. Of the remaining data, 3.7 × 105 pairs (1.2% of the total) had chimeric or unrecognized bar codes. Thus our in-line bar-coding method successfully demultiplexed 98.9% of all high-quality paired reads resulting in sublibraries with an average of 4.0 × 106 paired reads (∼3.5 × 108 bp of sequence data) per library, equal to ∼75-fold statistical coverage of the E. coli 4.6-Mb genome (Table 1). To prevent alignment biases due to unequal read lengths, we removed 7 bp, corresponding to the longest bar code, from the 5′ end of all reads for all libraries.
Table 1.
Quality statistics for 1-μg Illumina and 10-pg-level tagmented libraries
The yields and quality of tagmented libraries outlined here are equivalent to data obtained from non-bar-coded “standard” Illumina GAII 51-bp PE libraries run by our laboratory (Supplemental Table 1).
Read alignment quality and library diversity
Each 44-bp PE data set was aligned separately to an E. coli K-12 reference genome (Methods). Resultant data sets were quality-filtered for uniquely mapping read pairs where both reads exceeded a mapping phred of 150 (Table 1).
PCR is used to increase the available material for sequencing in both standard Illumina and our modified tagmentation library preparation methods. This leads to the amplification of library fragments and a consequent increase in DNA mass with a reduction in fragment diversity due to amplification bias. For this reason, we sought to compare the relative diversity, and hence information content, of the aligned data sets. PCR duplicates within each filtered data set were identified and excluded (Methods) (Table 1). The 1-μg Illumina libraries were found to contain ∼1% library fragment redundancy, whereas duplication levels for the 10-pg tagmentation library set varied between 49% and 64%. Repeating the tagmentation process with higher DNA input amounts (100-pg and 1-ng levels) produced libraries with lower degrees of redundancy, 16%–23% and 12%–18%, respecxtively (Table 1; Supplemental Table 3).
To allow direct comparison between preparation methods, we randomly selected subsets of 1 × 106 (1 million) non-redundant, uniquely mapping 44-bp paired reads for all further analyses (see Methods).
Relative coverage
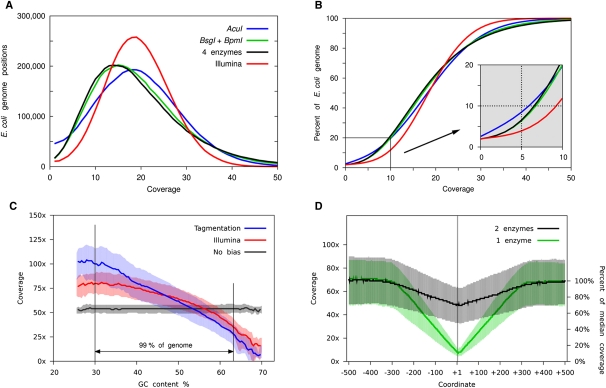
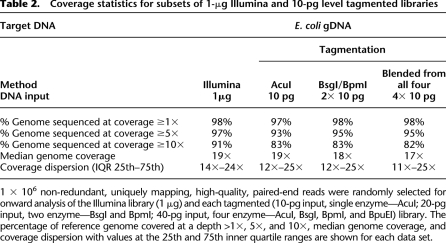
We first considered whether specific biases exist in statistical coverage between the two library preparation methods. Comparison of each library shows that the tagmentation method generates larger variation in coverage across the target genome when compared to the standard Illumina method (Fig. 2A). As expected, tagmentation libraries produced using a single restriction endonuclease cover large numbers of genomic regions at zero (Fig. 2A; Table 2) or low (Fig. 2B; Table 2) statistical coverage. However, also as predicted, blended libraries from two or four independent tagmented reactions digested with separate enzymes substantially reduce the frequency of low-coverage regions to levels similar to those observed for the 1-μg Illumina library (Fig. 2A,B; Table 2). Equivalent results were observed with 1-ng- and 100-pg-level tagmentation library data sets (Supplemental Table 4).
Figure 2.
Analysis of bias in 10-pg level libraries. (A,B) 1 × 106 non-redundant, uniquely mapping, high-quality, paired-end reads were randomly selected for further analysis of the Illumina library (red; 1 μg input) and each tagmented library. Tagmented libraries used 10 pg of input (blue; single enzyme—AcuI), 20 pg of input (black; two enzymes—BsgI and BpmI), and 40 pg of input (green; four enzymes—AcuI, BsgI, BpmI, and BpuEI). Coverage depth across the genome and percentage of genome covered at increasing cumulative coverage depths for each library were compared. (C) Median coverage depth for genomic regions defined by GC content was also analyzed for a single enzyme tagmentation library AcuI (blue), an Illumina library (red), and 1 × 106 size-matched fragments randomly selected in silico from the reference genome to model coverage in a non-biased manner (black). Shaded regions represent 25th and 75th inner quartile regions for each data set. The region between vertical black lines represents 99% of the total reference genome. (D) Fold coverage across 1-kb genomic regions containing endogenous cleavage sites for single enzyme (green; AcuI, AcuI sites shown) and two enzyme (black; BsgI and BpmI; BpmI sites shown) tagmented libraries. Approximately 3000 genomic regions are represented in each analysis. Absolute coverage and percentage median coverage by base position are shown for each region.
Table 2.
Coverage statistics for subsets of 1-μg Illumina and 10-pg level tagmented libraries
Relative GC content bias
We next considered whether coverage from the amplified libraries exhibited a bias in GC content. To compare relative and absolute sequence biases between Illumina and tagmented libraries, we compared data sets to an unbiased in silico library of 1 million randomly sampled uniquely mappable, non-redundant, E. coli K-12 genome fragments of equivalent fragment insert lengths to the test libraries (Fig. 2C; Supplemental Fig. 2). Statistical coverage levels between the two experimental data sets were most similar in genomic regions exceeding 50% GC content. Here both libraries showed under-representations of expected coverage levels compared to the simulated unbiased set. Overall, coverage for both libraries was biased toward AT-rich sequences with the tagmentation data set showing greatest deviation.
Effect of enzymatic digest on local coverage
To quantitate the effect of using single enzymatic digests to produce tagmented libraries, we analyzed sequence coverage in the vicinity of endogenous recognition sites for the endonuclease used in the library preparation. Our data show that, as predicted, library fragments were reduced to 7% of normal coverage levels, spanning a few base pairs across the enzyme binding site (Fig. 2D). Full coverage depth was restored within one insert length immediately flanking the recognition motif. Analysis of libraries made from two independent enzymatic digests showed that minimum coverage levels are restored to 70% of normal levels at any individual enzyme site (Fig. 2D). Blending four separately digested libraries increases the coverage at endogenous sites still further to 83% of median levels.
Sequence preference for transpososome insertion
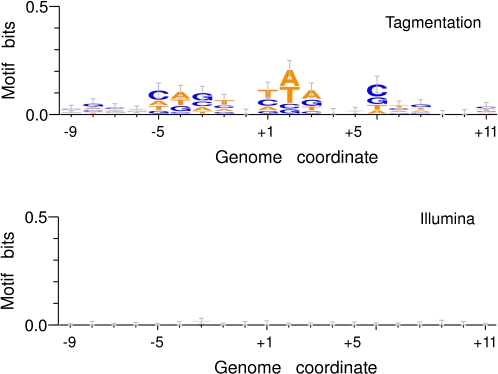
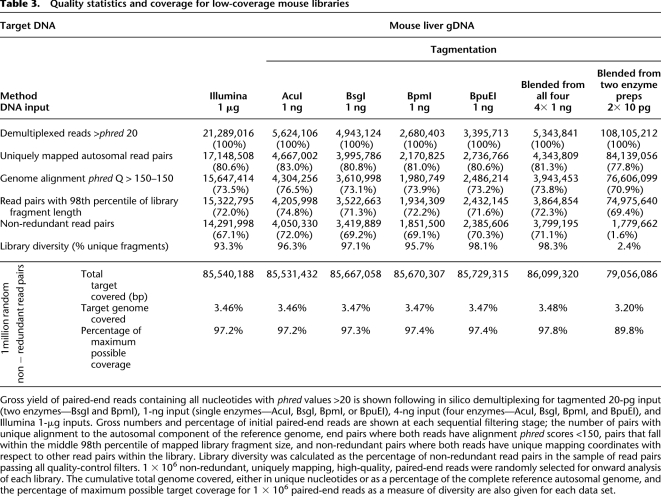
The use of an enzymatic reaction to fragment target DNA as an alternative to sonication immediately raises the question of whether preferred transpososome sequence binding motifs exist and, if so, how this may introduce further bias in local sequence coverage. Consequently, we next sought enriched sequence motifs at transpososome integration sites. Analysis of transpososome integration sites in our 10-pg-, 100-pg-, and 1-ng-level tagmented library sets provided evidence for a weak ∼13-bp motif centered at the point of fragmentation (Fig. 3), consistent with a preference reported independently (Adey et al. 2010). Analysis of the Illumina library yielded no evidence of sequence enrichments at sites of template fragmentation.
Figure 3.
Sequence motif analyses at tagmentation and Illumina fragmentation sites. Genomic sequences at 1000 randomly chosen sites of transpososome integration (top; single enzyme, AcuI 10-pg input tagmentation library) or physical sonication induced shearing positions (bottom; Illumina library) were analyzed for over-represented sequence motifs and plotted by nucleotide prevalence at each base position. The fragmentation site is shown at base +1. A weak sequence preference spanning an ∼13-bp region was found in the tagmented library. No sequence preference was found in the Illumina library.
Tagmentation with 20 pg of mouse genomic input
To investigate the utility of this technique with complex animal genomes, seven separate C57BL/6J mouse liver genomic DNA libraries were prepared using either sonicated (1-μg input) or our modified tagmentation method (20-pg, 1-ng, or 4-ng input). Tagmented libraries with inputs at 1 ng and 4 ng were multiplexed and run on a single Illumina GAII lane. The library produced using the sonication-based Illumina protocol was run on a separate lane. Both lanes were run as 51-bp PE sequences on the same GAII flowcell. The 20-pg input tagmented library was run using two lanes of an Illumina HiSeq 2000 platform at 100 bp PE. As before, resultant data files were converted to fastq format, demultiplexed into constituent libraries, trimmed to 44 bp residual length, and filtered for reads where all constituent nucleotides exceeded a base call phred score of 20 (Methods). Approximately 3.2 × 107 51-bp paired reads were recovered from the tagmentation multiplex lane. 8.5 × 106 (26.5%) of these fell below the quality threshold and were discarded. Of the remaining data set, 6.5 × 105 paired reads (2.0% of the total) had chimeric or unrecognized barcodes. 2.2 × 107 paired reads (68.8% of the lane) contained identifiable bar codes, passed the phred 20 filter, were successfully demultiplexed into constituent libraries (average 4.4 × 106 ± 1.3 × 106 paired reads per library), and were trimmed to remove the bar code sequence and produce 44-bp residual length reads. Each individual library therefore represents 3.9 × 108 bp of sequence data. 2.9 × 107 51-bp paired reads were recovered from the mouse genomic DNA library lane prepared using the sonication-based Illumina method. 2.1 × 107 PE reads (72.4% of lane) contained all constituent base calls exceeding the phred 20 threshold, representing 1.8 × 109 bp of sequence data. A total of 2.3 × 108 100-bp PE reads were produced from the 20-pg input mouse genomic DNA tagmented library. 1.1 × 108 PE reads (47.5%) contained identifiable bar codes, passed the phred 20 filter, and were trimmed to 44 bp representing an equivalent of 9.7 × 109 bp of sequence data.
As with the E. coli analyses, each 44-bp PE data set was aligned separately to the C57BL/6J reference assembly (Methods). Resultant data sets were quality-filtered for uniquely mapping paired ends with alignment scores above our arbitrary threshold (Table 3). PCR duplicates within each filtered data set were identified based on mapping coordinates and 1 × 106 (1 million) non-redundant, uniquely mapping PE reads from each library were selected for further analyses. Finally, gross target genome coverage and constituent fragment diversity were derived by computing unique nucleotide coverage and comparing with the maximum predicted unique nucleotide coverage for 1 million 44-bp PE reads (Table 3).
Table 3.
Quality statistics and coverage for low-coverage mouse libraries
No gross differences were observed in the levels of library quality, fragment redundancy, target coverage, or fragment diversity between the 1-μg input sonication-based Illumina and 1-ng input tagmentation libraries (Table 3). PCR duplication rates of up to 39-fold those of the other libraries were, however, seen in the 20-pg library and strongly suggest that at these input levels, diversity of the starting material becomes a limiting factor.
Discussion
Standard protocols for next-generation library synthesis typically require ∼1 μg of input DNA. Some sequencing centers have successfully generated libraries from ∼2 ng (P Piazza, pers. comm.). Tagmentation methods allow reduction to 50 ng but require sample-specific sequencing primers (Syed et al. 2009a,b). Tagmentation has been reported to recover sequence from 10 pg of genomic DNA; however, methods for accurately quantitating the DNA input and the quality of the resultant libraries are not reported (Adey et al. 2010). We have modified tagmentation technology to allow routine preparation of fully platform-compatible NGS libraries from picogram quantities of DNA. Our study carried out detailed analysis of low-input sequencing data sets and showed that biases are equivalent to standard Illumina 1-μg input libraries.
The production of picogram-level libraries requires the accurate quantification of target DNA. We have therefore developed a method for reliably quantifying sample concentrations down to the femtogram/microliter range.
Our protocol uses more PCR than standard sonication-based Illumina methods, resulting in increased duplicate sequences and an AT-rich sequence bias. Bacterial or mouse libraries produced from 1 ng of material displayed a maximum of 1.2× more duplicates when compared to the 1-μg sonication-based Illumina preparation. Interestingly, at the 10-pg level, there is a greater discord between the relative duplicate levels observed between the E. coli 2.9×, and mouse 38.9×, libraries compared to the 1-μg sonication-based Illumina preparation. This apparent reduction in the final information content of the libraries can largely be attributed to the limiting input material. Twenty picograms of gDNA represents a theoretical maximum of ∼6.7 × 107 300-bp average insert fragments. Diversity of both mouse and E. coli genomes are sufficient that false duplicates due to fragmentation at identical genomic coordinates should not be a significant issue at this level of input material. In our E. coli libraries we recovered 1.5 × 106 of the available input fragments, representing a capture of 2.2% of the theoretical maximum available starting material. Our mouse library recovered 1.8 × 106 of such fragments, ∼2.7% of the maximum available starting material. Approximately 17-fold greater high-quality uniquely mapped reads were sequenced for the mouse 20-pg input library compared to the E. coli counterpart. Thus, the 13-fold increase in duplicate rates observed between these libraries suggests that the majority of available fragment diversity in the mouse library as fixed before PCR amplification has been sequenced to exhaustion with the large number of additional sequencing reads increasing the apparent duplicate rate without adding to overall library information.
As input DNA quantity in NGS library synthesis is reduced to low levels, two process-related factors may reduce diversity in the resultant data set: first, only a fraction of the initial sample contributes directly to the final library; and, second, increased loss of effective template necessitates the use of more PCR, ultimately raising the duplication frequency in the final data set. The input template quantity contributing to the final library is set by both the DNA fraction fragmented within a selected size range and by the amount of template that becomes successfully ligated and amplified. Our tagmentation protocol uses a single tube reaction to fragment input DNA, ligate TRM-containing oligos, and amplify fragments. Restriction endonucleases are then used to create a ligatable 2-bp overhang as a more efficient alternative to the analogous “A-tailing” Illumina stages. Hence, in our method, process-induced reduction in diversity mainly arises from the efficiency of enzymatic fragmentation and controlled rejection of material at size selection. In current Illumina protocols, substantial additional losses are incurred during the sonication process, through inefficiencies incurred at several enzymatic manipulations and through multiple sample purifications. Our 20-pg input library data suggest that 2%–3% of the starting material is captured with our method. An analogous figure has not been reported for the Illumina method. Thus, when our modified tagmentation method is used to synthesize libraries from 1 ng of input material, acceptable levels of diversity are observed in the final data set when sequenced at this depth. However, our data suggest that further reducing input material to the 100-pg and 10-pg levels has a limiting effect on final library diversity. Despite the loss of ∼95% of the initial material in our library preparation, it is worthy of note that our 20-pg mouse libraries recovered an ∼0.4× coverage of unique mapped fragments of the non-repetitive mouse genome from only 7× genome equivalents of starting material. This is a significant recovery of mouse genomic data from genome-level inputs and has not currently been achieved by any other technique. Extrapolation from this result suggests that near-complete coverage of mapped fragments for the mouse genome should be possible with ∼50 pg of input. This figure would be closer to 60 pg if coverage were measured as sequenced bases using 100-bp PE reads. Further improvements in diversity should be achievable, relative to those reported here, by increasing the size range of library fragments selected and by titrating down the use of PCR. These alterations should decrease the amount of starting material required to achieve a particular coverage. However, it should be noted that it will never be possible to recover a full genome coverage for samples where the quantity of starting material is close to or less than a single genome equivalent. As a complement to whole genome studies, this technique is also likely to have widespread utility in the analysis of samples derived from enrichment procedures that typically result in low DNA yields but where coverage over target regions is very high.
Techniques are currently available for amplifying low-input samples prior to NGS library preparation (Tang et al. 2009). However, these approaches are likely to introduce sample bias and amplification artifacts that are impossible to distinguish in the final library. Our technique avoids amplification prior to fragmentation and uses an in silico paired-end-read duplicate filter to exclude gross artifacts, providing a more accurate representation of the relative relationship between input molecules. This is an important consideration when attempting to capture relative data from very-low-quantity starting procedures such as single-cell transcription profiling, which is currently only possible with target pre-amplification (Tang et al. 2009).
The use of type IIG endonucleases in our library synthesis was shown to cause predictable, highly localized coverage loss at endogenous recognition sites. Blending sublibraries created with separate restriction enzymes resolves this issue but requires parallel production of multiple samples. We have shown that blending two libraries is sufficient to increase target coverage to levels similar to the Illumina preparation. Hence, our laboratory uses a two-restriction-enzyme method as standard. The use of alternative type IIG restriction enzymes with less frequent endogenous sequence motifs may be explored so that a single preparation technique giving acceptable coverage might be used.
Our modification to the standard Illumina PE adapter sequence results in the repeated sequencing of a mandatory CAG motif at the start of all reads. This may result in a failure of some Illumina base-calling software. To avoid this, we use a variable length in-line bar-coding region in our adapter to offset these constant sequences and simultaneously allow library multiplexing. Our in-line bar-coding system indexes both reads of a paired-end fragment, allowing important quality checks such as the identification of interlibrary fragment chimeras, which are not possible using standard indexing systems.
This research has focused on the application of this technology to Illumina library synthesis. However, the basic fragmentation, ligation, and digestion protocol described here provides a universal entry point to the creation of libraries for any current NGS or third-generation platform that requires the ligation of an adapter to the ends of fragmented target DNAs during library preparation.
Overall, we believe that the modified tagmentation method presented here realizes the true potential of transpososome-mediated NGS library preparation technology. It allows reproducible sequencing of picogram quantities of target DNA in a fully platform-compatible method. Application of this technique will finally make practicable many studies in which amplification prior to fragmentation is undesirable or in which limited genetic materials are available such as genomic analysis of unculturable bacteria, low mass forensic or museum samples, low input ChIP-seq and genotyping, and sequencing or RNA-seq transcription analysis of single cells.
Methods
Input sample quantitation
A 2× serial dilution of lambda genomic DNA (Invitrogen) in 1× TE (Promega) was produced to give final concentrations from 100 pg/μL to 390 fg/μL in 1× TE. Ten-microliter aliquots of each standard were added to a 384-well Optiplate-F (Perkin-Elmer) in triplicate. Ten-microliter aliquots of 1:10 sample dilutions were added to separate plate wells. Ten microliters of PicoGreen working solution, 1:100 Quant-iT PicoGreen dsDNA reagent (Invitrogen), and 1:10 AccuBlue High Sensitivity Enhancer 1000× (Biotium) in 1× TE (Promega) were added to each sample well. The plate was centrifuged briefly at 1000g and placed into a BMG PHERAstar. Optimal focal height and gain were set, samples were shaken for 30 sec, and then fluorescence values were read with 485-nm excitation and 520-nm emission filters. Standard dilutions (final plate standard dilutions 50 pg/μL to 195 fg/μL) were plotted against blank normalized relative fluorescence using MARS Data Analysis Software (BMG LABTECH), and concentrations of unknown samples were extrapolated.
Illumina library preparation
One microgram of non-methylated E. coli K-12 MG1651 gDNA (Zymo #D5016) was placed in a DNA LoBind microcentrifuge tube (Eppendorf) and diluted to a total volume of 40 μL in 1× TE. Sonication was performed in a Misonix 4000 sonicator (Misonix) using a cup horn with circulating ice-cold water (Amplitude 100, 4× cycles of 60 sec of sonication with samples left to cool on wet ice for 60 sec between cycles). Following sonication, 1 μL of neat sample was run on a Bioanalyser 2100 (Agilent) using a HS DNA chip (Agilent) to monitor for optimal sample fragmentation. Ten microliters of 6× gel loading buffer (Maniatis et al. 1982) was added to the remaining sample. This was then mixed and split into two equal aliquots, which were loaded on adjacent wells of a 3% Nusieve agarose gel (Lonza) pre-stained with 0.5× GelRed (Biotium) and subjected to 35 min of electrophoresis at 100 V in 1× TAE. One hundred nanograms of 1-kb ladder (Promega) was run in a parallel lane. Following electrophoresis, the gel was imaged in a UVP bioimaging system adapted for use with hyper-bright green 528-nm LEDs (RS) and visualized using a 617 ± 73-nm filter (Semrock). A 250–350-bp region of fragmented target sample was excised, and DNA was recovered using the Zymoclean Gel DNA recovery kit (Zymo Research). Target DNA was then end-repaired in a 100-μL total volume reaction (1× NEB phosphorylation buffer, 0.5 mM NEB dNTPs, 5 μL of NEB T4 DNA polymerase E6003S, 1 μL of NEB Klenow fragment E6004S, 5 μL of NEB T4 PNK E6005S) for 60 min at 20°C. The reaction mix was then cleaned using a DNA Clean and Concentrator Kit (Zymo Research) and eluted twice with 10 μL of 1× TE. Pooled eluates were then subject to a 50-μL total volume A-Tailing reaction (1× NEB NEBuffer 2, 0.2 mM NEB dATP, 3 μL of NEB Klenow 3′–5′ Exo− E6006S) for 30 min at 37°C. The completed reaction mix was then cleaned using a DNA Clean and Concentrator Kit (Zymo Research) and eluted twice with 10 μL of 1× TE and pooled, and 1 μL of eluate was subjected to high-sensitivity DNA quantitation in a final volume of 20 μL. Samples were ligated to standard Illumina Paired End adapters (Illumina) in a 15-μL reaction (1× NEB T4 DNA ligase buffer with 1 mM final concentration ATP, 10× molar excess Illumina PE adapter, 2000 units of NEB T4 DNA ligase) for 30 min at 20°C. Following incubation, 2 μL of neat ligation reaction was used as a template in a 25-μL PCR enrichment reaction (1× Finnzymes Phusion HF master mix, 1 μM Illumina PE PCR primer 1, 1 μM Illumina PE PCR primer 2). PCR was performed in a Piko Thermal cycler (Finnzymes) with the following cycling parameters: once for 30 sec at 98°C, 14 times for 10 sec at 98°C, 30 sec at 65°C, and 30 sec at 72°C; once for 5 min at 72°C. Following amplification, 1 μL of a 1:100 dilution of the neat PCR sample was run on a Bioanalyser 2100 (Agilent) using a HS DNA chip (Agilent) to check for appropriate amplification. Following confirmation of enrichment, the remaining sample was mixed with 5 μL of 6× gel loading buffer (Maniatis et al. 1982), loaded on a 3% Nusieve agarose gel (Lonza) pre-stained with 0.5× GelRed (Biotium), and subjected to 35 min of electrophoresis at 100 V in 1× TAE. Fifty nanograms of 1-kb ladder (Promega) was run in a parallel lane. Following electrophoresis, the gel was imaged in a UVP bioimaging system (UVP) adapted for use with hyper-bright green 528-nm LEDs (RS) and visualized using a 617 ± 73 nm filter (Semrock). Amplified product, corresponding to the target library, was excised, and DNA was recovered in 10 μL of 1× TE using the Zymoclean Gel DNA recovery kit (Zymo Research). A 1-μL aliquot of the final library was subjected to high-sensitivity DNA quantitation in a final volume of 20 μL. Library dilutions were adjusted to 10 nmol and used for cluster generation and sequence analysis on Illumina GAII Genome Analyser or HiSeq 2000 platforms.
Tagmentation library preparation
Dilutions (250 pg/μL, 25 pg/μL, or 2.5 pg/μL) of non-methylated E. coli K-12 gDNA (NEB) were produced by serial dilution in 1× TE, and concentrations were confirmed with high-sensitivity DNA quantitation in a final volume of 20 μL. Four-microliter aliquots of input samples were added to the appropriate wells of a 24-well PCR plate (Finnzymes). One-microliter aliquots of a 5× reaction master mix (5× Epibio Nextera LMW Reaction buffer, 0.2 μL of EpiBio 454 Life Sciences (Roche) FLX–compatible Nextera Enzyme mix at dilutions 1/10, 1/100, and 1/1000 for 10-ng, 100-pg, and 10-pg input dilutions, respectively) were added, and samples were sealed with cap strips, combined by brief centrifugation, and incubated for 5 min at 55°C in a Piko thermal cycler (Finnzymes) followed by an immediate hold step at 4°C. Twenty-microliter aliquots of a PCR master mix were added to give a final concentration of 1× Phusion HF reaction master mix (Finnzymes) and 2 μM tailed primer in a 25-μL final reaction volume. Tailed primers are specific for the endonuclease to be used in the preparation (AcuI tailed oligo, GCGCGCCTGAAGATGTGTATAAGAGACAG; BsgI tailed oligo, GCGCGCGTGCAGATGTGTATAAGAGACAG; BpmI tailed oligo, GCGCGCCTGGAGATGTGTATAAGAGACAG; BpuEI tailed oligo, GCGCGCCTTGAGATGTGTATAAGAGACAG) and were synthesized and PAGE-purified (IDT). Reactions were immediately cycled at one time for 5 min at 72°C, one time for 30 sec at 98°C; 10 times for 10 sec at 98°C, 30 sec at 65°C, and 45 sec at 72°C; and one time for 5 min at 72°C in a Piko thermal cycler (Finnzymes). The number of cycles of PCR used was varied dependent on library input quantity such that 1 ng = 10× cycles, 100 pg = 12× cycles, and 10 pg = 14× cycles. Following amplification, 5 μL of 6× gel loading buffer (Maniatis et al. 1982) was added to the sample, mixed, and loaded on a 3% Nusieve agarose gel (Lonza) pre-stained with 0.5× GelRed (Biotium) and subjected to 35 min of electrophoresis at 100 V in 1× TAE. Fifty nanograms of 1-kb ladder (Promega) was run in a parallel lane. Following electrophoresis, the gel was imaged in a UVP bioimaging system (UVP) adapted for use with hyper-bright green 528-nm LEDs (RS) and visualized using a 617 ± 73-nm filter (Semrock). A 250–350-bp region of tagmented target sample was excised using 4-mm Genecatcher disposable gel excision pipette tips (Gel Company) and DNA recovered using the Zymoclean Gel DNA recovery kit (Zymo Research) with a final elution of 2 aliquots of 10 μL of 1× TE. Recovered DNA was then digested with 5 units of the appropriate class III endonuclease (AcuI, BpmI, BsgI, BpuEI, NEB) using the manufacturer's recommended conditions in a 30-μL final volume for 30 min at 37°C. Digested samples were cleaned using a DNA Clean and Concentrator Kit (Zymo Research) and eluted with 10 μL of 1× TE. One microliter of eluate was subjected to high-sensitivity DNA quantitation in a final volume of 20 μL. Twenty-microliter ligation reactions were set up using all remaining target template (final concentration 1× NEB T4 DNA ligase buffer, 2000 units of NEB T4 DNA ligase, 10× molar excess of Illumina compatible adapter) for 30 min at 20°C. Following incubation, 4× 5-μL aliquots of neat ligation reaction were used as a template in replicate 25-μL final volume PCR enrichment reactions (1× Finnzymes Phusion HF master mix, 1 μM Illumina PE PCR primer 1, 1 μM Illumina PE PCR primer 2). PCR was performed in a Piko Thermal cycler (Finnzymes) with the following cycling parameters: one time for 30 sec at 98°C, eight times for 10 sec at 98°C, 30 sec at 65°C, and 30 sec at 72°C; and one time for 5 min at 72°C. Following amplification, 1 μL of a 1:100 dilution of the neat PCR sample was run on a Bioanalyser 2100 (Agilent) using a HS DNA chip (Agilent) to check for appropriate amplification. Following confirmation of enrichment, the remaining samples were mixed with 5 μL of 6× gel loading buffer (Maniatis et al. 1982), loaded on a 3% Nusieve agarose gel (Lonza) pre-stained with 0.5× GelRed (Biotium), and subjected to 35 min of electrophoresis at 100 V in 1× TAE. Fifty nanograms of a 1-kb ladder (Promega) was run in a parallel lane. Following electrophoresis, the gel was imaged in a UVP bioimaging system (UVP) adapted for use with hyper-bright green 528-nm LEDs (RS) and visualized using a 617 ± 73-nm filter (Semrock). Amplified sample, corresponding to the target library, was excised using 4-mm Genecatcher disposable gel excision pipette tips (Gel Company), gel slices from amplifications of the same target library were pooled, and DNA was recovered into 10 μL of 1× TE using a Zymoclean Gel DNA recovery kit (Zymo Research). One microliters of eluate was subjected to high-sensitivity DNA quantitation in a final volume of 20 μL. Library dilutions were adjusted to 10 nmol and used for cluster generation and sequence analysis on an Illumina GAII Genome Analyser and delivered to our local NGS service provider, who sequenced the library using standard manufacturer's procedures.
Sequencing adapters
Modified Illumina adapters were synthesized to allow ligation of the GT sticky end left by the class III endonuclease digests. Five pairs of adapters (consisting of partially complementary oligos 1 and 2) were produced with staggered bar-code sequences (Supplemental Table 2). Adapter primers were synthesized using PAGE purification (IDT technologies) and diluted to 100 μM in 1× TE. A 50-μL adapter annealing reaction was carried out (final concentration 20 μM adapter 1, 20 μM adapter 2, 1× T4 DNA Ligase buffer) on a Pico thermocycler (Finnzymes) using the following cycling parameters: once for 2 min at 95°C, 130 times at 85°C–0.5°C, followed by a 4°C hold. Final annealed adapter concentrations were 20 μM per adapter.
Sample demultiplexing, trimming, and quality control
Raw Illumina format PE data sets were simultaneously filtered for reads containing base calls with phred scores <20, demultiplexed into constituent sublibraries based on in-line bar codes, trimmed to remove bar-coding nucleotides, and converted into fastq format using our own software. Overall library quality before and after demultiplexing, filtering, and trimming were monitored using the FastQC software package (Andrews 2010).
Genome alignment
Individual sequenced data sets were aligned to the reference genomes of the bacterium E. coli K-12 MG1655 (http://www.ncbi.nlm.nih.gov/sites/entrez?db=genome&cmd=Retrieve&dopt=Overview&list_uids=115) and repeat-masked mouse Mus musculus C57BL/6J assembly m37 (http://www.ncbi.nlm.nih.gov/mapview/stats/BuildStats.cgi?taxid=10090&build=36&ver=1 Build 36.1). Genome alignment was carried out with Novoalign v.2.05.33 (Novocraft; http://novocraft.com/main/page.php?s=novoalign), currently the most accurate algorithm for short-read alignment to reference genomes (Li and Homer 2010; Lunter and Goodson 2010). Novoalign was run in the paired-end mode and with default program settings.
Duplicate filtering
Duplicate paired-end reads within the same library were identified based on alignment coordinates of both reads within a pair using our SAM file duplicate filtering utility (Maslau 2011) to leave a subset of non-degenerate read pairs. This utility has been made available to the community under the GNU General Public License.
Data analysis
Successfully demultiplexed, trimmed, and quality thresholded data sets were further filtered. Read pairs were kept only when both reads aligned to unique locations in the reference genome, had mapping phred scores >150, and possessed a fragment size within the central 98% distribution of apparent mapped library fragment insert size.
Genome coverage statistics were calculated using the sequenced bases of the forward and reverse reads mapped to the reference genome. The GC content-dependent bias in the genome coverage was calculated using the complete inferred genomic fragments. GC content was assigned to the middle base of each 250-bp window of the genome, with 1 bp relative offset. The statistical dispersion in genome coverage at each position was described using median values and inner-quartile ranges (25th and 75th percentiles).
For the analysis of coverage at class IIG endonuclease sites, the full complement of enzyme recognition sites for the reference genome were initially identified in silico. Genome coverage at each nucleotide across a 1-kb region centered at each enzyme cut site (coordinate +1) was calculated, and data for all cut sites were overlaid. The statistical dispersion in genome coverage at each position was described with median values and inner-quartile ranges (25th and 75th percentiles).
Putative consensus sequence motifs at fragmentation sites were investigated within 50-bp regions centered at each of 1000 randomly selected fragmentation sites. Sequence conservation was displayed using the Weblogo program (Crooks et al. 2004) and annotated with the Gnuplot program (Janert. 2010). Three replications resulted in the identification of essentially identical sequence motifs.
Data access
Sequencing data sets used in this study are available through the EMBL-EBI European Nucleotide Archive (http://www.ebi.ac.uk/ena/). Accession numbers of alignment files for each data set are as follows; E. coli genomic DNA 1-μg Illumina library (accession number ERS066939), E. coli genomic DNA 1-ng AcuI Tagmentation library (accession number ERS066940), E. coli genomic DNA 1-ng BsgI Tagmentation library (accession number ERS066941), E. coli genomic DNA 1-ng BpuEI Tagmentation library (accession number ERS066943), E. coli genomic DNA 1-ng BpmI Tagmentation library (accession number ERS066942), E. coli genomic DNA 1-ng all enzymes Tagmentation library (accession number ERS066944), E. coli genomic DNA 100-pg AcuI Tagmentation library (accession number ERS066945), E. coli genomic DNA 100-pg BsgI/BpmI Tagmentation library (accession number ERS066946), E. coli genomic DNA 100-pg all enzymes Tagmentation library (accession number ERS066947), E. coli genomic DNA 10-pg AcuI Tagmentation library (accession number ERS066948), E. coli genomic DNA 10-pg BsgI/BpmI Tagmentation library (accession number ERS066949), E. coli genomic DNA 10-pg all enzymes Tagmentation library (accession number ERS066950), Mouse genomic DNA 1-μg Illumina library (accession number ERS066931), Mouse genomic DNA 1-ng AcuI Tagmentation library (accession number ERS066932), Mouse genomic DNA 1-ng BsgI Tagmentation library (accession number ERS066933), Mouse genomic DNA 1-ng BpuEI Tagmentation library (accession number ERS066935), Mouse genomic DNA 1-ng BpmI Tagmentation library (accession number ERS066934), Mouse genomic DNA 1-ng all enzymes Tagmentation library (accession number ERS066936), Mouse genomic DNA 10-pg BsgI/BpmI Tagmentation library lane 1 (accession number ERS066937), Mouse genomic DNA 10-pg BsgI/BpmI Tagmentation library lane 2 (accession number ERS066938).
Acknowledgments
This work was made possible with Alamy grants administered by the Fischer Family Trust (M.D.F, N.J.P., G.Z, B.F., and S.M.) and Medical Research Council, UK funding (C.P.P.). We acknowledge Christoffer Nellåker and Andreas Heger at the MRC Functional Genomics Unit, University of Oxford and Nick Carruccio at EpiBio for useful discussions and technical information.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.124016.111.
References
- Adey A, Morrison HG, Asan, Xun X, Kitzman JO, Turner EH, Stackhouse B, Mackenzie AP, Caruccio NC, Zhang X, et al. 2010. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol 11: R119 doi: 10.1186/gb-2010-11-12-r119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S 2010. FastQC High Throughput Sequence QC Report. 0.7.0. 00. http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/
- Crooks GE, Hon G, Chandonia JM, Brenner SE 2004. WebLogo: A sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janert PK 2010. Gnuplot in action: Understanding data with graphs. Manning Publications, Shelter Island, NY [Google Scholar]
- Li H, Homer N 2010. A survey of sequence alignment algorithms for next-generation sequencing. Brief Bioinform 11: 473–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunter G, Goodson M 2010. Stampy: A statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res 21: 936–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J 1982. Molecular cloning: A laboratory manual, 2nd ed Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- Maslau S 2011. NGS PCR duplicate removal utility. http://genserv.anat.ox.ac.uk/software/ngs_duplicate_filter
- Metzker ML 2010. Sequencing technologies—the next generation. Nat Rev Genet 11: 31–46 [DOI] [PubMed] [Google Scholar]
- Syed F, Grunenwald H, Caruccio N 2009a. Next-generation sequencing library preparation: Simultaneous fragmentation and tagging using in vitro transposition. Nat Methods Application Note 6 http://www.nature.com/nmeth/journal/v6/n11/full/nmeth.f.272.html [Google Scholar]
- Syed F, Grunenwald H, Caruccio N 2009b. Optimized library preparation method for next-generation sequencing. Nat Methods Application Note 6 http://www.nature.com/nmeth/journal/v6/n10/abs/nmeth.f.269.html [Google Scholar]
- Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, et al. 2009. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 6: 377–382 [DOI] [PubMed] [Google Scholar]