Abstract
Small non-coding RNAs (ncRNAs) are key regulators of plant development through modulation of the processing, stability, and translation of larger RNAs. We present small RNA data sets comprising more than 200 million aligned Illumina sequence reads covering all major cell types of the root as well as four distinct developmental zones. MicroRNAs (miRNAs) constitute a class of small ncRNAs that are particularly important for development. Of the 243 known miRNAs, 133 were found to be expressed in the root, and most showed tissue- or zone-specific expression patterns. We identified 66 new high-confidence miRNAs using a computational pipeline, PIPmiR, specifically developed for the identification of plant miRNAs. PIPmiR uses a probabilistic model that combines RNA structure and expression information to identify miRNAs with high precision. Knockdown of three of the newly identified miRNAs results in altered root growth phenotypes, confirming that novel miRNAs predicted by PIPmiR have functional relevance.
Small regulatory non-coding RNAs (ncRNAs) were discovered only 20 years ago (Lee et al. 1993) but have been implicated as key regulators in both development and disease for most eukaryotes (Farazi et al. 2011). Plants have a diverse array of small RNAs, including transposon-derived small interfering RNAs (siRNAs), trans-acting small interfering RNAs (tasiRNAs), natural antisense RNAs (nat-siRNAs), heterochromatin- and repeat-associated siRNAs, and microRNAs (miRNAs) (Chen 2010). In plants, miRNAs can both transcriptionally and post-transcriptionally repress expression of their targets (Cuperus et al. 2010).
In Arabidopsis thaliana, miRNA biogenesis begins with a DNA-dependent RNA polymerase II–produced primary transcript, which folds into a stem–loop structure (Voinnet 2009). The stem portion of the primary transcript is cleaved by a protein complex that contains a DICER-LIKE endonuclease (DCL) to produce the miRNA precursor. The precursor is further cleaved to remove the loop portion of the stem–loop by the same processing enzymes. The remaining duplex contains the mature miRNA and a complementary miRNA* sequence. The mature miRNA sequence is loaded into the RNA induced silencing complex (RISC), which contains an ARGONAUTE (AGO) protein. It is in this complex that the miRNA finds its messenger RNA target by base-pair complementarity and represses target expression by either causing mRNA degradation or by blocking translation (Huntzinger and Izaurralde 2011). Alternatively, activated RISC can cause methylation and transcriptional silencing of target loci (Khraiwesh et al. 2010; Wu et al. 2010). There are four DCL and 10 AGO proteins in Arabidopsis, which can be used to produce a variety of small RNAs with different first nucleotide specificity and varying functionality. For example, canonical miRNA precursors are processed by DCL1, and the resulting 21-nt species with a 5′ uridine is selectively incorporated with AGO1 (Mi et al. 2008).
The variable size and structure of plant miRNA precursors have made the identification of new miRNA genes a challenge both computationally and experimentally. To date, two types of approaches have been used for the high-throughput identification of new plant miRNA genes. The first approach is purely computational and involves the systematic folding of all intergenic regions into miRNA-like hairpin structures while including additional features such as conservation or simultaneous prediction of stem–loops and their targets (Bonnet et al. 2004; Jones-Rhoades and Bartel 2004; Adai et al. 2005). The downside to this approach is that there are a vast number of putative miRNA genes with little experimental validation. The second major approach has been to analyze all sequences from small RNA deep-sequencing data sets to see if they meet a given set of rules. If a sequence meets the necessary rule requirements and the surrounding sequence is able to fold into a stem–loop like structure, then it is automatically classified as a new miRNA (Moxon et al. 2008; Hendrix et al. 2010). Currently there is only a single tool, miRDeep (Friedlander et al. 2008), that uses a probabilistic model based on features from both small RNA deep-sequencing data and genomic data; however, this tool was developed for animal miRNA genes and is not able to identify the types of complex miRNA precursors seen in plants.
Several groups have undertaken studies to identify miRNAs, along with other small ncRNAs, using sequencing technologies in Arabidopsis shoots, flowers, and mature pollen (Llave et al. 2002; Gustafson et al. 2005; Fahlgren et al. 2007; Grant-Downton et al. 2009). These studies identified miRNAs that control leaf, floral, shoot, and vascular development. Additionally, miRNAs have been shown to modulate miRNA and siRNA biogenesis by targeting genes within the small RNA processing pathways. Plant miRNAs have been implicated in hormone signal transduction pathways and in responses to plant pathogens and environmental stresses. Furthermore, miRNA expression patterns were precisely defined in both space and time during the development of floral and seed tissues (Valoczi et al. 2006). Taken together, these studies led us to hypothesize that miRNAs exist that are specific not only to the root as a whole, but also to precise spatial and temporal locations during root development.
The Arabidopsis root offers an exceptional opportunity for the study of development at cellular resolution because of its radially symmetric structure (Fig. 1A). The different cell types of the root are arranged in concentric cylinders around a central core of vasculature. In the stem cell niche located close to the tip of the root, stem cells divide to generate specific cell types that are constrained to longitudinal cell files. Additionally, the developmental age of any given cell can be ascertained from its distance from the stem cell niche along the longitudinal axis, with the youngest cells at the root tip and the oldest cells furthest from the tip. This effectively reduces the four dimensions of development (three spatial dimensions plus time) to two dimensions (the radial axis corresponds to cell type and the longitudinal axis to age), making the root a useful and powerful system in which to identify and study molecules and networks that control development (Benfey and Scheres 2000).
Figure 1.
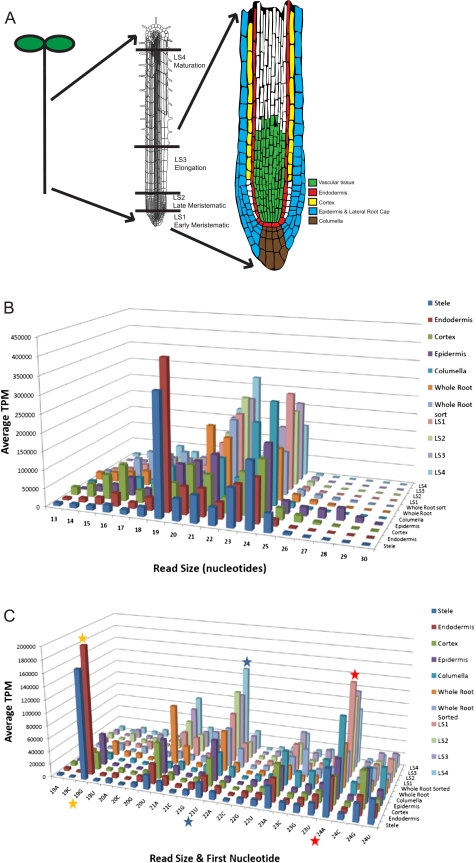
Small RNA characterization in the Arabidopsis thaliana root. (A) Overview of the Arabidopsis root developmental zones and cell types used in this study. (From left to right) Schematic drawing of an Arabidopsis seedling; the root denoting developmental zones isolated by hand sectioning; and the root tip displaying the cell types analyzed in this study. The colors indicate the regions covered by the GFP marker lines used for isolation of cell types by cell sorting. (B) Size distributions display preferences for small RNAs of different types. Size distribution of the reads in the radial and longitudinal data sets is shown. Reads were normalized to transcripts per million (TPM), and the two biological replicates of each radial cell type were averaged. (C) Highly abundant small RNA species are reflected in the read size and the identity of the first nucleotide. Read size by first nucleotide of the reads from the radial and longitudinal data sets is depicted. Canonical miRNAs are 21 nt and begin with a U (blue star), while heterochromatin-associated siRNAs are 24 nt and begin with an A (red star) (Mi et al. 2008). The 19-nt peak corresponds to tRNA fragments, similar to what was reported in roots (orange star) (Hsieh et al. 2009).
Using GFP marker lines, we generated cell type–specific small RNA deep-sequencing profiles of each of the major cell types in the Arabidopsis root (Fig. 1A). In addition, we obtained profiles of the early and late meristematic, elongation, and early maturation developmental zones. This information provides us with an atlas of small RNA expression, most notably miRNA expression, across the entire root organ. Furthermore, we present a probabilistic classifier, PIPmiR (pipeline for the identification of plant miRNAs), which uses both genomic and small RNA deep-sequencing information to identify 66 novel miRNAs with high confidence.
Results
Radial and developmental data sets reveal small ncRNA expression
We determined the cell type specificity of small ncRNAs in the root by using cell-sorting technology followed by deep sequencing. We used five independent lines expressing GFP in the stele, endodermis, cortex, epidermis, and columella, which cover the major cell types of the root tip. Isolation of individual cell types by fluorescence-activated cell sorting was then coupled with Illumina small RNA deep sequencing to generate two independent libraries for each cell type. We also generated a root developmental time course by hand-sectioning 100 Columbia-0 roots into early and late meristematic, elongation, and differentiation zones. In addition, we made four libraries consisting of whole root tissue (WT): two unsorted and two mock sorted. An overview of the data sets is given in Figure 1A.
Approximately 200 million short reads were mapped to the Arabidopsis (TAIR9) genome (Supplemental Table 1). The number of raw reads per sample was normalized to transcripts per million (TPM) to correct for a varying number of reads in the different sequencing lanes. This type of normalization is still open to bias from PCR amplification, RNA ligase preference, and the reverse transcription reaction, but is currently the standard procedure (Linsen et al. 2009; Meyer et al. 2010).
A measure of the high quality of these data can be seen in the size distributions of the sequence reads (Fig. 1B). As expected for known small ncRNAs, 77% of the reads fall within the range of 19–24 nt. Small RNAs of different read lengths show a bias in their most 5′ nucleotide because the 5′ base of small RNAs is a characteristic of the AGO with which it associates (Mi et al. 2008). Of the reads that are 21 nt in length, 62% have a 5′ uridine (Fig. 1C). This is characteristic of the DCL1 cleavage and AGO1 association found in most known miRNAs (Llave et al. 2002; Park et al. 2002; Reinhart et al. 2002; Rajagopalan et al. 2006; Fahlgren et al. 2007; Grant-Downton et al. 2009). In addition, 58% of the 24-nt length reads have a 5′ adenosine, characteristic of AGO4 association (Mi et al. 2008).
Roots have recently been shown to contain an abundant class of 19-nt small RNAs derived from fragmentation of tRNAs (Hsieh et al. 2009). This size class is abundantly represented in our data (15% of all reads 19–24 nt) and is highly enriched in the stele and endodermis. Taken together, our data represent a rich resource of root small RNA expression profiles.
Known miRNAs display cell type and developmental zone enrichment
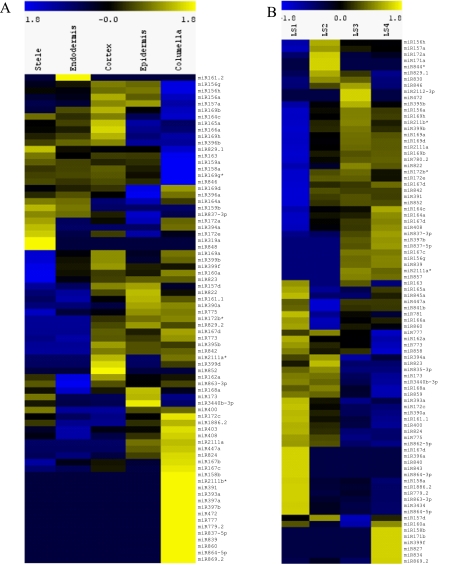
We profiled the expression of the 243 currently annotated mature miRNAs that originate from 213 precursors as defined by MirBase v.16 (Griffiths-Jones et al. 2008) and found 133 mature miRNAs derived from 127 precursors represented in our data sets (Supplemental Data Set 1). The expression of each miRNA gene in a cell type or longitudinal section was normalized by a scaling factor designed for read count data (Anders and Huber 2010). For the cell type and whole root samples with a biological replicate, average expression values were used for further analyses since the biological replicates demonstrated high reproducibility (r = 0.81–0.97) (Supplemental Table 2). We found both broad and very specific expression patterns for known miRNAs (Fig. 2A). The miRNAs with the lowest variance across tissues, i.e., those that would be most useful for qPCR normalization, are the miR169 and miR162 families (Supplemental Table 3), while the miRNAs with the lowest variance across longitudinal sections were the miR156 and miR157 families (Supplemental Table 4). Overall, clustering of miRNA expression profiles resulted in nine distinct radial expression patterns (Supplemental Fig. 1). Intriguingly, many miRNAs displayed cell type specificity as determined by the information content of their expression profiles (Supplemental Fig. 2A).
Figure 2.
Cell type– and developmental zone–specific enrichment of known miRNAs. MiRNA expression profiles from the radial data sets represented as heat maps. (Yellow) Enrichment; (blue) under-representation. (A) Radial expression map, in which columns are cell types and rows are mature miRNAs. Note that some miRNAs are enriched in only one cell type, while others are enriched in multiple cell types. Individual clusters are shown in more detail in Supplemental Figure 1. (B) Developmental zone expression map, in which columns are developmental zones and rows are mature miRNAs. Note that some miRNAs are enriched in a specific developmental zone, while other miRNAs are under-represented in specific developmental zones. Individual clusters are shown in more detail in Supplemental Figure 2. The miRNAs listed are the representative members of the family as listed in Supplemental Figure 10.
The expression values of the longitudinal data sets were similarly analyzed and clustered (Fig. 2B; Supplemental Fig. 2B). Each of the developmental zones had specifically enriched groups of mature miRNAs. We also observed patterns that show fluctuations in miRNA expression across the developmental zones (Supplemental Fig. 3).
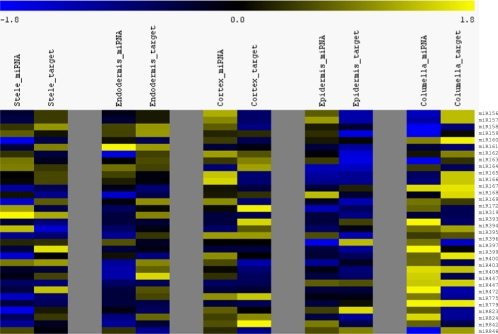
In the simplest case, miRNAs and the mRNA of their targets are expected to have reciprocal expression patterns. To determine how often this occurs, we compared our miRNA expression profiles with target mRNA expression profiles from a previous microarray-based root study (Brady et al. 2007). First, we obtained validated targets for known miRNAs expressed in our data sets from the Arabidopsis thaliana Small RNA Project (ASRP) website (Gustafson et al. 2005; Backman et al. 2008). While using only validated targets lowered our sample size, we wanted to remove the error that would be associated with false-positive target predictions. Expression values of all targets for a particular miRNA were averaged within each cell type and compared with the log2-normalized expression of the miRNA in the same cell types and zones (Fig. 3). This method will not identify miRNAs that show translational repression of their targets. Analysis of the correlations show three miRNA:mRNA sets with significant P-values for a strong negative correlation: miR156 with squamosa promoter binding like proteins (SPLs), miR157 with SPLs, and miR396 with growth-regulating factors (Supplemental Table 5). Additionally, two sets show an unexpected strong positive correlation: miR447 with 2-phosphoglycerate kinase-related proteins and miR779 with a leucine-rich repeat protein kinase. The sets with no correlation included miR397 with laccases, miR172 with APETALA2-domain containing transcription factors, miR159 with MYBs, and two sets that target members of the miRNA biogenesis pathway, miR168 with AGO1 and miR162 with DCL1. Overall, many known miRNAs have enrichment in one or more cell types and/or developmental zones (Supplemental Fig. 2). These spatio-temporal patterns will be very useful in the elucidation of specific functional roles for root miRNAs in biological circuits.
Figure 3.
Expression of miRNAs and their validated targets varies by cell type and developmental zone. Heat map representation of known miRNA expression and average miRNA target expression z-scores side by side for each of the cell types. An inverse relationship between the expression level of miRNAs and their validated targets was found for many known miRNAs. (Yellow) High expression; (blue) low expression.
A new classifier, PIPmiR, is used to predict miRNA foldback structures
Plant miRNA precursor sequences are much more diverse in both length and secondary structure than those in animals, thus making them more challenging to identify computationally (Jones-Rhoades and Bartel 2004). There are two precursor-processing pathways that have been identified for plant miRNA genes. The first, and primary, pathway involves stem-to-loop processing in which the sequence and structure beyond the miRNA–miRNA* site are necessary and used by the cleavage pathway components to excise the mature sequence(s) (Reinhart et al. 2002; Kurihara and Watanabe 2004; Cuperus et al. 2010; Mateos et al. 2010; Song et al. 2010; Werner et al. 2010). The second pathway involves loop-to-stem processing in which only the structure between the miRNA and miRNA* is necessary for the cleavage pathway components to excise the mature sequence(s) (Addo-Quaye et al. 2008; Bologna et al. 2009).
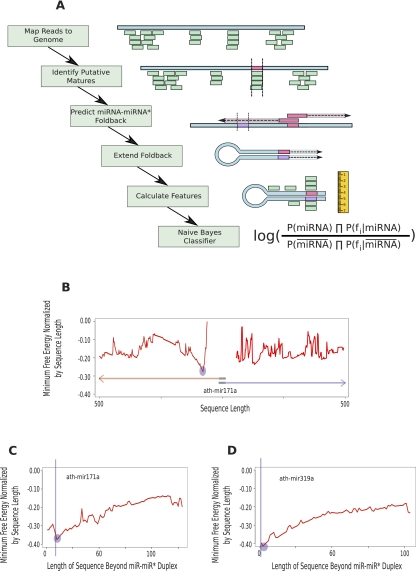
We developed a computational pipeline that, beginning with a mature miRNA candidate, can accurately identify the precursor sequence necessary for proper processing of the miRNA gene (Fig. 4). We applied our pipeline to all mature sequences listed in miRBase v16 (Jones-Rhoades and Bartel 2004). Our pipeline identified valid precursor sequences for all known mature miRNAs, except ath-mir406 and ath-mir824. The precursor sequence defined in miRBase for ath-mir406 places the mature miRNA within a loop such that there is no matching miRNA* sequence. This structure is not consistent with established miRNA processing pathways. The size of the precursor identified for ath-mir824 is >500 nt long, the current limit of our method. We found that the majority of our predicted precursors were very similar to what is currently listed in miRBase, although they were frequently a few nucleotides shorter or longer than the current annotation. There were six exceptions in which we found substantially different precursor sequences than those currently annotated (Supplemental Data Set 1). For these transcripts, our predictor identifies a precursor that places the mature sequence on the arm opposite from the current miRBase annotation (i.e., the stem–loop structure is upstream of the mature miRNA instead of downstream, or vice versa).
Figure 4.
PIPmiR pipeline. (A) A schematic depiction of each step of the PIPmiR pipeline. (B) The minimum normalized free energy by sequence length calculated while determining the miRNA–miRNA* foldback structure for ath-mir171a. The blue circle highlights the sequence with the overall minimum value that was used in the subsequent step. (C,D) The minimum normalized free energy by sequence length calculated while extending the miRNA–miRNA* foldback structure for ath-mir171a and ath-mir319a, respectively. The blue circles highlight the location identified by PIPmiR as the correct foldback structure. The vertical blue lines represent the sequence identified as necessary for the proper processing of the miRNA.
Recent studies have experimentally validated the length of the precursor sequence necessary for proper processing of eight different miRNA genes. Two of these genes, ath-mir159a and ath-mir319a, have been shown to be processed by the loop-to-stem pathway, meaning that the structure and sequence required for processing reside between the miRNA and the miRNA* and that structure and sequence beyond this site are not necessary (Bologna et al. 2009). For these experimentally validated examples, our pipeline predicts a precursor structure that ends within 2 nt of the miRNA–miRNA* duplex. In contrast, there are six miRNAs—ath-mir164c, ath-mir171a, ath-mir172a, ath-mir167a, ath-mir390a, and ath-mir398a—known to require certain nucleotides beyond the miRNA–miRNA* duplex for proper processing (Cuperus et al. 2010; Mateos et al. 2010; Song et al. 2010). We accurately predict the necessary precursor sequence that contains these nucleotides for four of the six. Our predictions for both ath-mir167a and ath-mir390a are that the precursor sequence ends at the miRNA–miRNA* duplex. Overall, we predict that 149 of the 213 precursor sequences extend >2 nt beyond the miRNA–miRNA* duplex, consistent with the majority of precursors being processed by the stem-to-loop pathway (Supplemental Data Set 1).
The processing mechanisms that result in the generation of multiple mature miRNAs from a single precursor are not known. There are currently 29 precursors in miRBase that are annotated to contain more than one mature miRNA sequence. Regardless of the starting mature sequence within these precursors, our pipeline identified a precursor sequence that contains all of the mature miRNAs in 26 of the 29 instances. The three exceptions are ath-mir779, for which folding by starting with miR779.2 does not extend the precursor to include miR779.1; ath-mir774b, for which folding by starting with miR774b does not include the full miR774b*; and ath-mir829, for which folding by starting with miR829.1 stops 1 nt short of including the complete sequence of miR829.2. For subsequent analysis, we used the fold that contained all mature sequences for the precursor or used the longest predicted form of the precursor.
PIPmiR classifier accurately predicts miRNAs
The second phase of PIPmiR uses the short read data in combination with genomic features to specifically identify miRNAs present in the sequence libraries and to distinguish miRNAs from other small RNAs. These features were selected based on known biogenesis, processing, and targeting properties of plant miRNAs. The classifier was trained on small RNA deep-sequencing data from five different experiments, which cover a variety of Arabidopsis tissues (Montgomery et al. 2008; Hsieh et al. 2009; Fahlgren et al. 2010; Kanno et al. 2010). Our positive set consisted of 144 known miRNAs that were expressed with 10 or more reads in at least one of the five training libraries. The negative set consisted of intergenic or intronic genomic locations where at least 10 reads mapped and a potential precursor foldback structure was identified using the pipeline. Overall, we included 42,916 genomic locations in our negative training set. A fourfold cross-validation test of our model revealed that PIPmiR separated the known miRNAs from other ncRNAs with a sensitivity of 91.7% and a specificity of 99.9%. This is the minimum specificity possible since some members of the negative set may correspond to currently unannotated mature miRNAs. For comparison purposes, we used the rule-based plant miRNA identifier miRCat (Moxon et al. 2008) on the same training data. At a specificity of 99.9%, this algorithm correctly identified 113 (78%) of the known miRNAs expressed (Supplemental Table 6).
PIPmiR identifies novel miRNAs expressed in the root
We next applied PIPmiR to each of our root data sets to identify new miRNA genes. Across all libraries, we identified 420,974 unique putative mature miRNA sequences from intergenic or intronic regions that had a valid pre-miRNA fold structure. Putative mature miRNAs that overlapped by at least 90% were grouped into a single mature sequence. Our algorithm identified 183 putative mature sequences from 168 precursors that were classified as an miRNA in at least one of the data sets. Of these potential mature miRNAs, 145 from 135 precursors had a median classification score >0 across all of the data sets in which they were observed (Supplemental Data Set 2).
All of these candidates bear the hallmarks of products of the small RNA processing pathways in plants and exceed the evidence available for many recent miRBase additions based on high-throughput sequencing. However, the presence of a small RNA pathway substrate does not imply its consistent physiological function. Here, we took advantage of our biological replicates and applied a more rigorous filter to identify a more stringent set of novel miRNAs for further analysis. We additionally required that each of the putative miRNAs be expressed in both biological replicates for at least a single tissue and have a positive score in each of those replicates. To account for the single replicates of the developmental zones, we required a minimum raw transcript count of 25 and a positive score. Overall, the pipeline reported 66 novel miRNAs from 64 precursors (Table 1; Supplemental Table 7). Fifty-eight of the 66 previously unknown miRNAs (88%) are reported to have at least one target using the WMD3 target predictor (Ossowski et al. 2008; Supplemental Data Sets 1–3). This is highly similar to the number of known miRNAs with predicted targets (220 of 243, 91%).
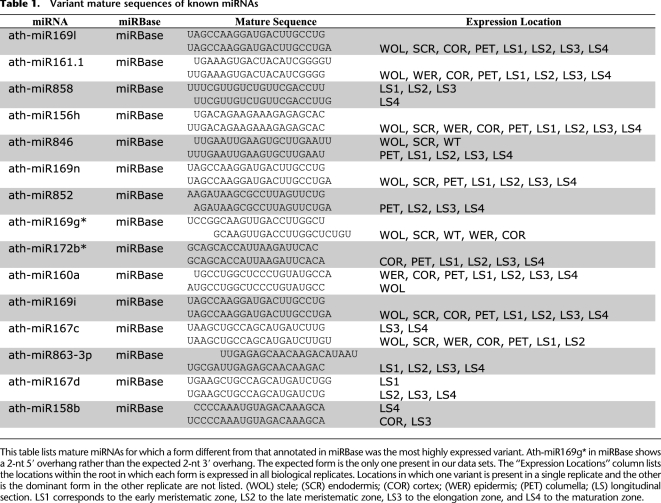
Table 1.
Variant mature sequences of known miRNAs
Additional mature miRNAs emerge from currently known precursors
Of the 133 currently annotated miRNAs found in our expression data, 23 had a dominant form different from miRBase v16 (Supplemental Data Set 3) in at least one of the data sets. Fifteen of those were the more dominant form in both replicates of a single cell type, 13 of which were the dominant form in both replicates of more cell types than the current miRBase annotation (Table 1). Most of these alternate forms differed by only a single nucleotide, with the exception of ath-miR169g*, which differed by 4 nt. Contrary to all known processing pathways, this particular miRNA would produce a 2-nt 5′ overhang when in an miRNA–miRNA* duplex as annotated in miRBase. The miRNA* form corresponding to the expected 2-nt 3′ overhang is the only one present in our data sets.
In addition to the adjusted forms of currently annotated miRNAs, we also observed additional mature miRNA sequences arising from the previously identified precursor sequences. We identified 62 miRNA* sequences from currently known miRNAs that may function as independent mature miRNAs (Supplemental Table 8). These miRNA* sequences met the most stringent thresholds of the PIPmiR classifier in that they were expressed in both replicates of a tissue (or at the higher level in the longitudinal sections), had a score above 0 in both replicates, and had a median score >0 for all of the tissues in which they were found. Further validation is needed to determine how many of these function as independent mature miRNAs.
Similar to what was observed by others (Zhang et al. 2010), we found that mature miRNAs may also arise from regions of the precursor that are not currently annotated as either a mature miRNA or miRNA*. In addition to miRNA precursors known to contain more than one mature miRNA (ath-mir319a, ath-mir319b, ath-mir447a, and ath-mir822), we found 11 additional precursors that give rise to a novel mature miRNA. These were each expressed in both replicates of at least a single tissue and had a median classification score >0 (Supplemental Table 9; Supplemental Data Set 3).
New miRNAs are validated
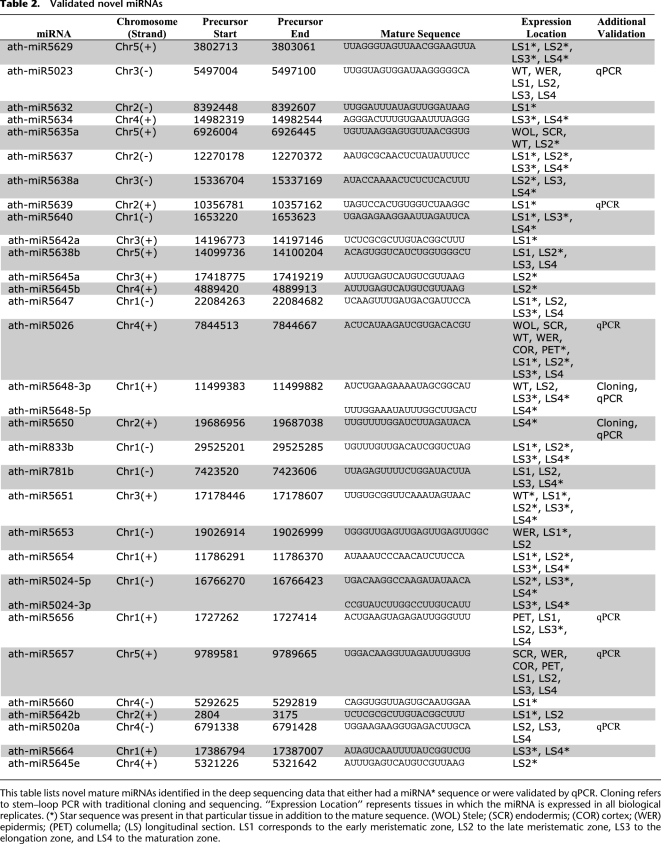
Twenty-nine of the miRNAs we identified could immediately be classified as new miRNAs based on standard annotation criteria (Meyers et al. 2008) that accept as evidence the presence of both miRNA and miRNA* sequences on predicted hairpin structures. We failed to detect miRNA* for the remaining putative new miRNAs, which is not unexpected considering the low abundance of the mature miRNA. We further validated eight new miRNAs using stem–loop PCR (Table 2). Seven of these miRNAs had T-DNA insertion lines readily available (Sessions et al. 2002; Alonso et al. 2003). For the eighth, we generated a target mimicry line (Franco-Zorrilla et al. 2007). To assess whether the disruptions in our new miRNAs resulted in root development phenotypes, we compared growth rates of the primary root of the knockout lines with a wild-type or empty vector control.
Table 2.
Validated novel miRNAs
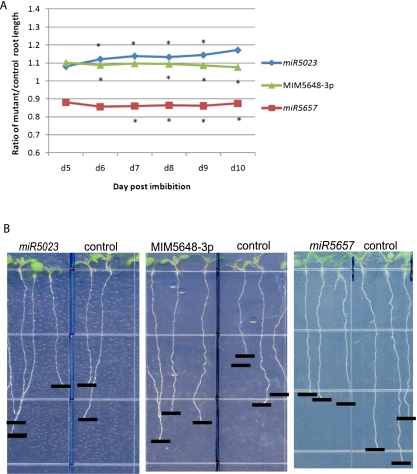
Mutations in three of the new miRNAs—miR5023, miR5648-3p, and miR5657—resulted in a root growth phenotype (Fig. 5). miR5023 was found in the epidermis and all developmental zones, with highest expression in the late meristematic zone. The T-DNA insertion in this line would interrupt the predicted hairpin of the precursor, and the phenotype of the mutant was enhanced root growth. MiR5648-3p was expressed in the late meristematic, elongation, and maturation zones, with the highest expression in the late meristematic zone. No T-DNA insertion lines were available, but a line using suppression by target mimicry (MIM5648-3p) also had an enhanced root growth phenotype. MiR5657 was found in all radial cell types and all developmental zones, with the highest expression in the epidermis, and a knockout line exhibited retarded root growth. The knockdown in mutants miR5023 and miR5657 is shown in Supplemental Figure 4A, while the increased relative expression of targets and the complementary miRNA regions in MIM5648-3p are shown in Supplemental Figure 4, B and C. While further analysis is needed to determine the exact role of these miRNAs in root development, these results suggest that new miRNAs we identified have functional relevance.
Figure 5.
Novel miRNAs display root length phenotypes. (A) Mutants in two novel miRNAs (miR5023 and MIM5648-3p) have longer roots than their Columbia-0 wild-type or empty vector control, respectively, while one novel miRNA mutant (miR5657) has shorter roots than the wild-type control. (*) P-value <0.05 from Student's t-test (see Supplemental Table 10). (B) Mutant and control roots grown vertically on plates display root length phenotypes. Pictures were taken at 8 d post-imbibition.
Discussion
In this study, we present analyses of small RNA deep-sequencing data sets generated from the major cell types and developmental zones of the Arabidopsis root. Using these data, we describe the expression profiles not only of known miRNAs, but also identify a substantial number of new mature miRNAs. We observed that many known and new miRNAs have developmental zone specificity, with a large number of known miRNAs enriched in the early maturation zone. This suggests that the miRNAs function to repress targets after the developmental program is initiated.
High-resolution profiling of individual cell types can provide insights into the role of miRNAs that are masked by profiling entire organs (Fig. 3). For example, miR156/157 was the most highly expressed miRNA family in all of our data sets, and the known targets had very low expression. Looking at the developmental zones, however, miR156/157 targets had higher and lower expression in the elongation and maturation zones, respectively, while miR156/157 had the opposite expression pattern. Since this particular family of miRNAs has a known role in vegetative phase change in leaves (Poethig 2009), it is possible they could be performing a similar role in the root, delineating the region between elongating and mature cells.
Another known miRNA family, miR165/166, was recently shown to play a role in the specification of the xylem in roots (Carlsbecker et al. 2010), leading us to examine the expression pattern of these miRNAs in our data sets. Carlsbecker et al. found that these miRNAs are highly expressed in the endodermis and quiescent center with weaker expression in the cortex and epidermis. We found a similar pattern of miR165/166 expression in our data sets, but our cortex expression was higher than in the endodermis (Fig. 2A). This could be due to the expression of the cortex GFP marker used for sorting, which labels the cortex cells in the elongation to maturation zones. While previous studies focused on the meristematic zone, we have additional information about miR165/166 localization. Our data show that the highest levels of miR165/166 are in the early meristematic zone, and these levels drop in the late meristematic zone (Fig. 2B; Supplemental Data Set 1). This indicates that the action of this miRNA could be to set up a gradient of target expression necessary for proper pattern formation in the meristematic zone.
In addition to describing the root expression profile of currently known miRNA genes, including their variants, we developed a computational pipeline capable of identifying precursor sequences, which are then used in a probabilistic framework to accurately distinguish new miRNAs from other small ncRNAs. We identified 183 new miRNA candidates across all of our data sets. When we required consistency in classification score and expression across biological replicates, we arrived at a subset of 66 new mature miRNAs from 64 precursor sequences for which there is high confidence.
The PIPmiR classifier also identified new mature miRNAs that emerge from the precursor sequences of known miRNA genes. We found that there are 62 mature miRNAs for which their miRNA* sequence could be considered as a separate, functional mature miRNA. In addition, we identify 15 mature sequences that originate from precursor sequences not currently annotated as either a mature miRNA or its miRNA* sequence. Unlike animal miRNAs that can have hundreds of targets (Friedman et al. 2009), plant miRNAs have only a few targets but may diversify their mRNA targets through the production of multiple mature miRNAs from the same precursor.
Reduction in activity for three new miRNA genes resulted in significant differences in root growth rate when compared with wild-type controls. Reduction in activity for two of the miRNAs resulted in increased root growth, whereas reduction in the third miRNA resulted in reduced root growth. As has been previously reported, regulators that affect root growth are likely to have specific expression patterns across developmental zones (Tsukagoshi et al. 2010), and we see this for two new miRNAs with root growth phenotypes. MiR5023 and miR5657 show enrichment in specific developmental zones, namely, the two meristematic sections (Supplemental Fig. 2B; Supplemental Data Set 2). Further analysis is needed to clarify the specific roles of these miRNAs in the regulation of root growth.
Finally, our small RNA deep-sequencing data sets provide detailed expression profiles of classes of ncRNAs in the Arabidopsis root other than miRNAs. An example is the distribution of small ncRNA sizes found in the different developmental zones. A larger number of 21-nt small ncRNAs, the canonical miRNA size, were found in the meristematic zone, while more 24-nt small RNAs, the canonical heterochromatin-associated siRNA size, were found in the early maturation zone. This could indicate a functional role of the maturation zone in silencing that has not previously been recognized, and future work will examine additional classes of small RNAs in these data sets, because they can also be expected to exhibit distinct spatio-temporal expression patterns. In summary, our study demonstrates the power of isolating individual cell types and developmental zones in combination with deep sequencing and computational analyses to obtain detailed profiles of ncRNAs, as well as to significantly extend the compendium of known functional RNAs.
Methods
Cell sorting and Illumina small RNA library preparation
Cell type–specific sorting was performed using GFP-labeled lines (Birnbaum et al. 2003). The stele was marked by pWOODEN LEG::GFP (WOL) (Mahonen et al. 2000), endodermis and quiescent center by pSCARECROW::GFP (SCR) (Birnbaum et al. 2003), the cortex by pCORTEX::GFP (COR) (Lee et al. 2006), the epidermis and lateral root cap by pWEREWOLF::GFP (WER) (Lee and Schiefelbein 1999; Sena et al. 2004), and columella by enhancer trap PET111 (PET) (Nawy et al. 2005).
At least 1 million GFP-positive cells (or mock-sorted cells in the case of whole root sorted samples) were collected directly into miRVana (Ambion) lysis buffer and stored at −80°C until extraction. The total RNA extraction protocol was used. For the longitudinal sections, 100 6-d-old Columbia-0 wild-type roots were hand-dissected into four pieces: two meristematic zone sections, one elongation zone section, and one ∼2-mm maturation zone section. The sections were placed in the miRVana kit lysis buffer immediately after dissection, and then the 100 roots for each section were combined before total RNA extraction.
High-molecular-weight RNA was precipitated using 5% PEG8000 and 0.5 M NaCl, and low-molecular-weight (LMW) RNA was ethanol-precipitated from the supernatant as described in Lu et al. (2007). LMW RNA used in small RNA library construction had an RIN score of 8 or above by Agilent Bioanalyzer. Illumina small RNA sequencing libraries were prepared as described in the v.1 protocol. To validate each library before using for sequencing, <1 ng was traditionally cloned and about 100 colonies were Sanger-sequenced. Libraries that contained <5% adapter:adapter sequences (indicating no small RNA insert) with small RNA sequences that could be aligned to the Arabidopsis genome were used for Illumina sequencing on an Illumina Genome Analyzer II. Two biological replicates of each cell type, two biological replicates of the whole root sorted, two biological replicates of the whole root unsorted, and one of each longitudinal section were constructed and sequenced.
Small RNA data set processing and alignment
All small RNA libraries, including those from previous studies (Montgomery et al. 2008; Hsieh et al. 2009; Fahlgren et al. 2010; Kanno et al. 2010), were stripped of their respective 3′-adapter sequences using the FASTX toolkit (Blankenberg et al. 2010). Reads that were <13 nt in length or contained an ambiguous nucleotide were discarded. The remaining reads were aligned to the Arabidopsis thaliana genome (TAIR9) allowing up to three mismatches using the Bowtie (Langmead et al. 2009) algorithm. We allowed each sequence to map to multiple locations with a maximum of 25 locations per sequence because many families of small ncRNAs share precisely the same sequence. Reads that failed to map to the genome or mapped to the genome in more than 25 locations within their optimal mismatch stratum were discarded. Mapped locations where at least one read mapped with no mismatches were used for further analysis; this removed mapped locations where all of the reads mapping to that particular location contained one or more mismatches. Only intergenic and intronic sequences were considered for further analysis. Of the locations where 10 or more reads mapped and at least one read was a perfect match, <18% of the total reads contained a mismatch.
miRNA expression profiles
All reads that mapped to a known mature miRNA as identified in miRBase v16 (Griffiths-Jones et al. 2008) or overlapped at least 90% with a known miRNA were counted toward the expression value. The count value was normalized by the size factor as defined by Anders and Huber (2010). As to not be biased by miRNA family size, one representative member of each of the families was used in the calculation of this normalizing factor (Supplemental Table 10). The normalizing factors were calculated using the “DESeq” (Anders and Huber 2010) library in the R statistical software package (R Development Team 2009).
Identification of putative miRNA mature sequences
Initially, we constructed a set of putative hairpin sequences based on the aligned short sequence reads. We then used those reads to derive a subset of putative miRNA precursor sequences that are used as input to a probabilistic binary classifier. In the remainder of this section, we describe all the analysis steps in more detail.
Aligned reads that overlapped were grouped together, and the most abundant read within that group was considered the putative mature miRNA; only putative mature miRNAs with more than 10 reads were considered for further analysis. For the purposes of training PIPmiR, exact matches to miRBase v16 (Griffiths-Jones et al. 2008) mature sequences were used as the positive set, regardless of whether that was the most abundant form of reads at that location. In addition, no unannotated mature miRNA sequences that overlapped a known precursor were used in training the model.
PIPmiR precursor prediction
Given the large variation as to the location of a mature miRNA in the precursor sequence, all sequences starting with the potential mature miRNA were extended upstream of and downstream from 50 to 500 nt with a step size of 2 nt. Each sequence was folded with RNAFold (Zuker and Stiegler 1981), and the minimum free energy, normalized by sequence length, was recorded if there was a valid miRNA* sequence. A valid miRNA* sequence was required to be at least half the length of the mature, no more than 1.5× the length of the mature, and to contain no hairpin structures. The precursor sequence that contained the lowest normalized free energy was truncated back to the miRNA–miRNA* sequence containing the stem–loop. This new hairpin sequence was then extended up to a total length of 500 nt stepwise by adding 2 nt to both ends of the sequence. At each step, the sequence was folded and the normalized minimum free energy recorded. The sequence with the overall lowest minimum free energy normalized by length was kept as the miRNA precursor sequence. If the miRNA* sequence in this putative precursor sequence did not overlap with the miRNA* sequence identified at the initial stem–loop identification stage by at least 90%, then the sequence was discarded and this putative miRNA was not considered further.
To combine precursors that contained multiple mature sequences, we first required that such precursors overlapped by at least 90%. Then they were combined to the most upstream and the most downstream location of all of the predictions. This was done to ensure that we included all mature sequences.
PIPmiR classifier
PIPmiR applies a naive Bayes classifier that uses 15 different features (listed below) collected from either genomic information or small RNA deep-sequencing experiments. The resulting score from the classifier is the log-odds of the probability of a sequence being a mature miRNA versus that sequence being another type of ncRNA. Therefore, any sequence with a score above 0 is classified as a mature miRNA. All continuous features were converted to 10 equal-sized bins between the minimum and maximum values seen in the training data. Values observed in the data that were either less than or greater than the range of the training data were set to the minimum or maximum value found in the training data. The classification was implemented using the naive Bayes function in the e1071 library (Dimitriadou et al. 2010) from the R statistical software package (R Development Team 2009).
A total of five different data sets of A. thaliana small RNA sequencing were used in the training and evaluation of PIPmiR. Four data sets were downloaded from the NCBI Sequence Read Archive (Leinonen et al. 2010) and consisted of data from different plant tissues: SRX014817 (shoot) (Hsieh et al. 2009), SRX003110 (inflorescence tissue: flower stages 1–12) (Montgomery et al. 2008), SRX021356 (total aerial, bolting, and flowering) (Fahlgren et al. 2010), and SRX016973 (mixed-stage inflorescence tissue) (Kanno et al. 2010). The fifth data set used in the training and evaluation was from this study (whole root; replicate 2).
All mature miRNAs listed in miRBase v16 (Griffiths-Jones et al. 2008) with 10 or more reads in any individual data set were used in the positive set; this resulted in 144 mature sequences being included. The data set that had the largest number of reads for each mature sequence was used to calculate the features for that particular miRNA.
The negative training set consisted of all putative mature sequences found in each of the five data sets that mapped to intergenic or intronic regions. If a putative mature miRNA was present in multiple data sets, the data set with the largest number of reads at that location was used to calculate the features for that particular negative control. To be included in the negative set, the putative mature miRNA had to have more than 10 reads and be predicted to have a valid precursor sequence. There were a total of 42,916 putative mature miRNAs used in the negative set. If a putative mature miRNA mapped to a location of a known miRNA precursor sequence but was not a known mature miRNA as defined by miRBase v16, then it was discarded from being either in the positive or the negative set.
Feature list
GC% of first half of miRNA: Percent of “G” or “C” nucleotides in the first half (5′ end) of the mature miRNA sequence
GC% of second half of miRNA: Percent of “G” or “C” nucleotides in the second half (3′ end) of the mature miRNA sequence
Nucleotide 10: Tenth nucleotide from the 5′ end of the mature miRNA sequence
Nucleotide 11: Eleventh nucleotide from the 5′ end of the mature miRNA sequence
First nucleotide: 5′-most nucleotide of the mature miRNA sequence
Last nucleotide: 3′-most nucleotide of the mature miRNA sequence
Mature length: Number of nucleotides in the mature miRNA sequence
Star length: Number of nucleotides in the miRNA star sequence
Normalized energy: Minimum free energy of the precursor foldback structure divided by the length of the precursor sequence
Consecutive mismatches: The maximum number of consecutive non-base pairings between the mature miRNA and the miRNA* sequence
miRNA–miRNA* match: The percent of nucleotides in the mature sequence that have a base-pairing to a nucleotide in the miRNA* sequence
miRNA*–miRNA ratio: The ratio of the number of reads that exactly map to the miRNA* sequence divided by the number of reads that map to the mature miRNA sequence. Any value >1 for this feature is set to a value of 1.
Overhang ratio: The number of reads that overlap 85% or less of the mature miRNA divided by the number of reads that map exactly to the mature sequence. Any value >1 for this feature is set to a value of 1.
miRNA and miRNA* compared to entire precursor: The number of reads that map exactly to the mature miRNA or the miRNA* divided by the number of reads that map to the precursor sequence
Opposite strand ratio: The number of reads that map to the precursor sequence, but on the opposite strand, divided by the number of reads that map to the precursor on the transcribed strand. Any value >1 for this feature is set to a value of 1.
miRNA expression analysis
Heat maps were produced using the multiexperiment viewer (MeV) that is part of the TM4 microarray software suite (Saeed et al. 2006). Z-scores for miRNA expression and z-scores for their average target(s) expression value were calculated before entering into MeV for heat map production. The Z-score was defined as [(log2-normalized expression) − (mean of log2-normalized expression of group)]/SD. After loading data, the gene expression profiles were hierarchically clustered using Euclidean distance. A figure of merit was generated with 100 iterations to obtain the approximate number of clusters appropriate for k-means clustering. k-means clustering was performed using Euclidean distance and up to 50 iterations.
For known miRNA target expression analysis, the list of validated targets was downloaded from the ASRP website (Backman et al. 2008), and the average expression value was calculated per family by averaging the expression values of the targets of that family from laboratory microarray data (Brady et al. 2007). Pearson product moment correlation coefficients (r) and P-values were calculated using the “HMisc” library in the R statistical software package (R Development Team 2009), and correlations of the miRNA:mRNAs sets were ordered from lowest to highest P-values.
miRNA validation, target prediction, and phenotyping
Verification of the expression of novel miRNAs was performed using whole root Col-0 total RNA and stem–loop RT-PCR (Varkonyi-Gasic et al. 2007) using miR156 as the normalizing control. Some miRNAs were further validated (designated “cloning” in Table 2) using stem–loop endpoint PCR, and these PCR products were cloned into a pCR-Blunt-TOPO vector (Invitrogen) and their sequences confirmed by Sanger sequencing (Varkonyi-Gasic et al. 2007).
Targets for both known and novel mature miRNAs were predicted using the WMD3 web microRNA designer using default settings (Ossowski et al. 2008).
Homozygous T-DNA insertion mutants (usually producing knockout mutants) were identified for the novel miR candidates (McElver et al. 2001; Sessions et al. 2002; Alonso et al. 2003). The insertion line for mir5023 was CS824777–SAIL_582_B05, and for mir5657 was CS833058–SAIL_739_F11. The target mimicry line for miR5648-3p was produced as described (Franco-Zorrilla et al. 2007). Root lengths were assayed by growing mutant seeds versus control seeds vertically on 1× Murashige and Skoog salt mixture, 1% sucrose, 2.3 mM 2-(N-morpholino)ethanesulfonic acid (pH 5.8), and 1% agar plates. Root lengths were measured using ImageJ (Abramoff et al. 2004), and the Student's t-test was used to determine statistical significance.
Data access
The raw sequencing files have been submitted to the NCBI Sequence Read Archive (SRA) (http://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession number SRA037191 and study number SRP006839. The PIPmiR pipeline can be found at http://www.genome.duke.edu/labs/ohler/research/PIPmiR/.
Acknowledgments
We thank Jaimie Van Norman and Siobhan Brady for help with figure illustrations, and the Benfey laboratory for comments on the manuscript. Thanks to Heather Belcher and Beth Harvat for assistance with the cell sorting, and to Ramiro Rodriguez for help with target validation protocols. Thanks to Kevin Shianna of the Duke University Genomic Analysis Facility for sequencing the first Illumina test run, and Lisa Bukovnik at the Institute for Genome Science and Policy Core Sequencing Facility for sequencing the subsequent runs. This work was funded by a grant to P.N.B. and U.O. from the NIH for the Duke Center for Systems Biology (P50-GM081883). N.W.B. was supported by NIH/NIGMS training grant (5T32 GM007184-32). J.J.P. was supported by a Ruth L. Kirschstein National Research Service Award (F32GM086976). N.W.B., J.S.-S., and J.S. were supported by a Howard Hughes Medical Institute Vertically Integrated Partners grant (52005871). MiRNA studies in the Weigel laboratory are supported by European Community FP6 IP SIROCCO (contract LSHG-CT-2006-037900).
Note added in proof
We note that while our paper was under review, another study (Borges et al. 2011) also identified four of our high-confidence novel miRNAs (miR5014, miR5020a, miR5024, and miR5026). A second study (Yang et al. 2011) reported an additional two of our high-confidence novel miRNAs in their supplemental materials. The miRBase names we obtained for these miRNAs are miR5650 and miR5654.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.123547.111.
References
- Abramoff MD, Magelhaes PJ, Ram SJ 2004. Image processing with ImageJ. Biophotonics Int 11: 36–42 [Google Scholar]
- Adai A, Johnson C, Mlotshwa S, Archer-Evans S, Manocha V, Vance V, Sundaresan V 2005. Computational prediction of miRNAs in Arabidopsis thaliana. Genome Res 15: 78–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addo-Quaye C, Eshoo T, Bartel DP, Axtell MJ 2008. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol 18: 758–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106 doi: 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backman TW, Sullivan CM, Cumbie JS, Miller ZA, Chapman EJ, Fahlgren N, Givan SA, Carrington JC, Kasschau KD 2008. Update of ASRP: The Arabidopsis Small RNA Project database. Nucleic Acids Res 36: D982–D985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Scheres B 2000. Root development. Curr Biol 10: R813–R814 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN 2003. A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Blankenberg D, Gordon A, Von Kuster G, Coraor N, Taylor J, Nekrutenko A 2010. Manipulation of FASTQ data with Galaxy. Bioinformatics 26: 1783–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna NG, Mateos JL, Bresso EG, Palatnik JF 2009. A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. EMBO J 28: 3646–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet E, Wuyts J, Rouze P, Van de Peer Y 2004. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc Natl Acad Sci 101: 11511–11516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F, Pereira PA, Slotkin RK, Martienssen RA, Becker JD 2011. MicroRNA activity in the Arabidopsis male germline. J Exp Bot 62: 1611–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee J-Y, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN 2007. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Lee J-Y, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vate'n A, Thitamadee S, et al. 2010. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X 2010. Small RNAs—secrets and surprises of the genome. Plant J 61: 941–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus JT, Montgomery TA, Fahlgren N, Burke RT, Townsend T, Sullivan CM, Carrington JC 2010. Identification of MIR390a precursor processing-defective mutants in Arabidopsis by direct genome sequencing. Proc Natl Acad Sci 107: 466–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitriadou E, Hornik K, Leisch F, Meyer D, Weingessel A 2010. e1071: Misc functions of the Department of Statistics (e1071), TU Wien. R package version 1.5-24 [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, et al. 2007. High-throughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of MIRNA genes. PLoS ONE 2: e219 doi: 10.1371/journal.pone.0000219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Jogdeo S, Kasschau KD, Sullivan CM, Chapman EJ, Laubinger S, Smith LM, Dasenko M, Givan SA, Weigel D, et al. 2010. MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana. Plant Cell 22: 1074–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farazi TA, Spitzer JI, Morozov P, Tuschl T 2011. miRNAs in human cancer. J Pathol 223: 102–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J 2007. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N 2008. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol 26: 407–415 [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant-Downton R, Trionnaire GL, Schmid R, Rodriguez-Enriquez J, Hafidh S, Mehdi S, Twell D, Dickinson H 2009. MicroRNA and tasiRNA diversity in mature pollen of Arabidopsis thaliana. BMC Genomics 10: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, Dongen Sv, Enright AJ 2008. miRBase: Tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson AM, Allen E, Givan S, Smith D, Carrington JC, Kasschau KD 2005. ASRP: The Arabidopsis Small RNA Project database. Nucleic Acids Res 33: D637–D640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix D, Levine M, Shi W 2010. miRTRAP, a computational method for the systematic identification of miRNAs from high throughput sequencing data. Genome Biol 11: R39 doi: 10.1186/gb-2010-11-4-r39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY, Li WH, Chiou TJ 2009. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol 151: 2120–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E 2011. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat Rev Genet 12: 99–110 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP 2004. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14: 787–799 [DOI] [PubMed] [Google Scholar]
- Kanno T, Bucher E, Daxinger L, Huettel B, Kreil DP, Breinig F, Lind M, Schmitt MJ, Simon SA, Gurazada SG, et al. 2010. RNA-directed DNA methylation and plant development require an IWR1-type transcription factor. EMBO Rep 11: 65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khraiwesh B, Arif MA, Seumel GI, Ossowski S, Weigel D, Reski R, Frank W 2010. Transcriptional control of gene expression by microRNAs. Cell 140: 111–122 [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Watanabe Y 2004. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci 101: 12753–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J 1999. WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99: 473–483 [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854 [DOI] [PubMed] [Google Scholar]
- Lee J-Y, Colinas J, Wang JY, Mace D, Ohler U, Benfey PN 2006. Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc Natl Acad Sci 103: 6055–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen R, Sugawara H, Shumway M 2010. The sequence read archive. Nucleic Acids Res 39: D19–D21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsen SE, de Wit E, Janssens G, Heater S, Chapman L, Parkin RK, Fritz B, Wyman SK, de Bruijn E, Voest EE, et al. 2009. Limitations and possibilities of small RNA digital gene expression profiling. Nat Methods 6: 474–476 [DOI] [PubMed] [Google Scholar]
- Llave C, Kasschau KD, Rector MA, Carrington JC 2002. Endogenous and silencing-associated small RNAs in plants. Plant Cell 14: 1605–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Meyers BC, Green PJ 2007. Construction of small RNA cDNA libraries for deep sequencing. Methods 43: 110–117 [DOI] [PubMed] [Google Scholar]
- Mahonen AP, Bonke M, Kauppinen L, Riikonen M, Benfey PN, Helariutta Y 2000. A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev 14: 2938–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos JL, Bologna NG, Chorostecki U, Palatnik JF 2010. Identification of microRNA processing determinants by random mutagenesis of Arabidopsis MIR172a precursor. Curr Biol 20: 49–54 [DOI] [PubMed] [Google Scholar]
- McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, Smith K, Thomas C, Schetter A, Zhou Q, Cushman MA, et al. 2001. Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159: 1751–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer SU, Pfaffl MW, Ulbrich SE 2010. Normalization strategies for microRNA profiling experiments: A ‘normal’ way to a hidden layer of complexity? Biotechnol Lett 32: 1777–1788 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carrington JC, Chen X, Green PJ, et al. 2008. Criteria for annotation of plant microRNAs. Plant Cell 20: 3186–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C 2008. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery TA, Yoo SJ, Fahlgren N, Gilbert SD, Howell MD, Sullivan CM, Alexander A, Nguyen G, Allen E, Ahn JH, et al. 2008. AGO1–miR173 complex initiates phased siRNA formation in plants. Proc Natl Acad Sci 105: 20055–20062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon S, Schwach F, Dalmay T, Maclean D, Studholme DJ, Moulton V 2008. A toolkit for analysing large-scale plant small RNA datasets. Bioinformatics 24: 2252–2253 [DOI] [PubMed] [Google Scholar]
- Nawy T, Lee JY, Colinas J, Wang JY, Thongrod SC, Malamy JE, Birnbaum K, Benfey PN 2005. Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell 17: 1908–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D 2008. Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X 2002. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12: 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig RS 2009. Small RNAs and developmental timing in plants. Curr Opin Genet Dev 19: 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Team 2009. R: A language and environment for statistical computing. http://www.mendeley.com/research/r-a-language-and-environment-for-statistical-computing-2/#page-1 [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP 2006. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20: 3407–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP 2002. MicroRNAs in plants. Genes Dev 16: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J 2006. TM4 microarray software suite. Methods Enzymol 411: 134–193 [DOI] [PubMed] [Google Scholar]
- Sena G, Jung JW, Benfey PN 2004. A broad competence to respond to SHORT ROOT revealed by tissue-specific ectopic expression. Development 131: 2817–2826 [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. 2002. A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Axtell MJ, Fedoroff NV 2010. RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr Biol 20: 37–41 [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H, Busch W, Benfey PN 2010. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143: 606–616 [DOI] [PubMed] [Google Scholar]
- Valoczi A, Varallyay E, Kauppinen S, Burgyan J, Havelda Z 2006. Spatio-temporal accumulation of microRNAs is highly coordinated in developing plant tissues. Plant J 47: 140–151 [DOI] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP 2007. Protocol: A highly-sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3: 12 doi: 10.1186/1746-4811-3-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O 2009. Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Werner S, Wollmann H, Schneeberger K, Weigel D 2010. Structure determinants for accurate processing of miR172a in Arabidopsis thaliana. Curr Biol 20: 42–48 [DOI] [PubMed] [Google Scholar]
- Wu L, Zhou H, Zhang Q, Zhang J, Ni F, Liu C, Qi Y 2010. DNA methylation mediated by a microRNA pathway. Mol Cell 38: 465–475 [DOI] [PubMed] [Google Scholar]
- Yang X, Zhang H, Li L 2011. Global analysis of gene-level microRNA expression in Arabidopsis using deep sequencing data. Genomics 98: 40–46 [DOI] [PubMed] [Google Scholar]
- Zhang W, Gao S, Zhou X, Xia J, Chellappan P, Zhang X, Jin H 2010. Multiple distinct small RNAs originate from the same microRNA precursors. Genome Biol 11: R81 doi: 10.1186/gb-2010-11-8-r81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M, Stiegler P 1981. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res 9: 133–148 [DOI] [PMC free article] [PubMed] [Google Scholar]